Abstract
Purpose
To assess the possible utility of machine learning for classifying subjects with and subjects without osteoarthritis (OA) using sodium magnetic resonance imaging (MRI) data.
Theory
Support vector machine (SVM), k-nearest neighbors (KNN), naïve Bayes (NAB), discriminant analysis (DIA), linear regression (LNR), logistic regression (LGR), neural networks (NNE), decision tree (DTR) and tree bagging (TBG) were tested.
Methods
Sodium MRI with and without fluid suppression by inversion recovery was acquired on the knee cartilage of 19 controls and 28 OA patients. Sodium concentrations were measured in regions-of-interest (ROIs) in the knee for both acquisitions. Mean (MEAN) and standard deviation (STD) of these concentrations were measured in each ROI, and the minimum, maximum and mean of these two measurements were calculated over all ROIs for each subject. The resulting 12 variables per subject were used as predictors for classification.
Results
Either Min[STD] alone, or in combination with Mean[MEAN] or Min[MEAN], all from fluid suppressed data, were the best predictors with an accuracy >74%, mainly with linear LGR and linear SVM. Other good classifiers include DIA, LNR and NAB.
Conclusion
Machine learning is a promising technique for classifying OA patients and controls from sodium MRI data.
Keywords: sodium MRI, classification, machine leaning, cartilage, osteoarthritis
INTRODUCTION
Osteoarthritis (OA) is a degenerative disease of the articular cartilage that can be associated with a reduction in glycosaminoglycan (GAG) concentration, changes in the size and organization of collagen fibers, and increased water content [1]. Many magnetic resonance imaging (MRI) methods for assessing osteoarthritis in cartilage are under development, such as T2 mapping [2], T1ρ mapping [3], GAG chemical exchange saturation transfer (gagCEST) [4], delayed gadolinium-enhanced MR imaging of cartilage (dGEMRIC) [5], diffusion tensor imaging (DTI) [6], and sodium MRI [7]. All of these methods have their advantages and weaknesses [8], but quantitative sodium MRI [9] has been shown to strongly correlate with the GAG concentration in the cartilage [1,10,11]. Our team recently developed a sodium MRI method in which the synovial fluid signal around the cartilage is suppressed by adiabatic inversion recovery [12, 13]. Fluid suppression reduces significantly partial volume effect and thus increases the sensitivity of the technique to changes in GAG content within the cartilage. This promising technique could be a useful complementary tool to other imaging techniques (standard MRI, radiography) for detecting early signs of OA (loss of GAG), or follow-up of treatment of OA and cartilage repair, through direct assessment of the GAG content in cartilage. In a previous study [13], we found that the sodium concentration measurements (means and standard deviations) from fluid-suppressed sodium MRI of articular cartilage were best predictors of OA when compared with asymptomatic controls. In this latter study, we used logistic regression on full data to find the best individual variables (or predictors) for classifying OA subjects and controls, from the accuracy point-of-view. In the present work, we used the same data as Ref. [13] and applied different methods of machine learning to assess which variables or group of variables from sodium MRI generate the most efficient classification between control and OA. Efficiency of classification was assessed by adjusted accuracy, which is a modification of the accuracy definition that take into account the difference between sensitivity and specificity. Data classification using machine learning is a growing field of interest in medical diagnosis [14], and in multivariate data analysis of medical imaging, particularly in brain [15–17] and cartilage [18,19]. Nowadays, medical data collected is increasing in size and complexity, and machine learning could be of importance for interpreting and classifying these datasets as an aid to clinicians in the decision making process [14]. Medical data classification with machine learning would therefore allow to help develop an automatic and objective way of making decisions based on multiple parameters from one or different modalities (MRI, x-ray, blood test, biopsy etc.). Machine learning is a branch of artificial intelligence that focuses on algorithms capable of learning or adapting their structure (or model parameters) based on a training dataset, through optimization of a cost function [14,20]. A more detailed description of machine learning and of the methods used in the present work can be found in the following Theory section, and in references within. The aim of this exploratory study is to estimate if statistical/machine learning methods have the potential to be of utility for detecting OA in articular cartilage with sodium MRI in a robust and objective way.
THEORY
Machine learning can be defined as “a set of methods that can automatically detect patterns in data, and then use the uncovered patterns to predict future data, or to perform other forms of decision making under uncertainty” [20]. In general, an original data set is divided in 2 types of subsets: train datasets will be used to optimize a model that best represent the patterns in data (learning/training phase), and tests datasets that are used to assess the performance of the model for recognizing these patterns (testing phase). Machine learning can be separated in two types: supervised (or predictive) and unsupervised (or descriptive). In supervised learning, the goal is to learn a model from a vector of input data x to outputs y, given a labeled set of input-output pairs with Strain the training set and N the number of training samples. Classification is a type of supervised machine learning for which the goal is to separate the data in different categories (or classes) y. In the present study, we will only talk about binary classification, where there are only two classes y ∈ {0,1}. For unsupervised learning, there is no known categories y in the data and the goal is to use only the input vectors x to find non-labeled patterns in the data (clustering). In this section we will present a short and very simplified description of the basic principles of the different methods of classification that used in this study (Fig. 1 and 2). More detailed descriptions of these methods may be found in references [20–25]. In general, the values of input vectors x are called variables, or features, or predictors, while the output values y are called categories or classes.
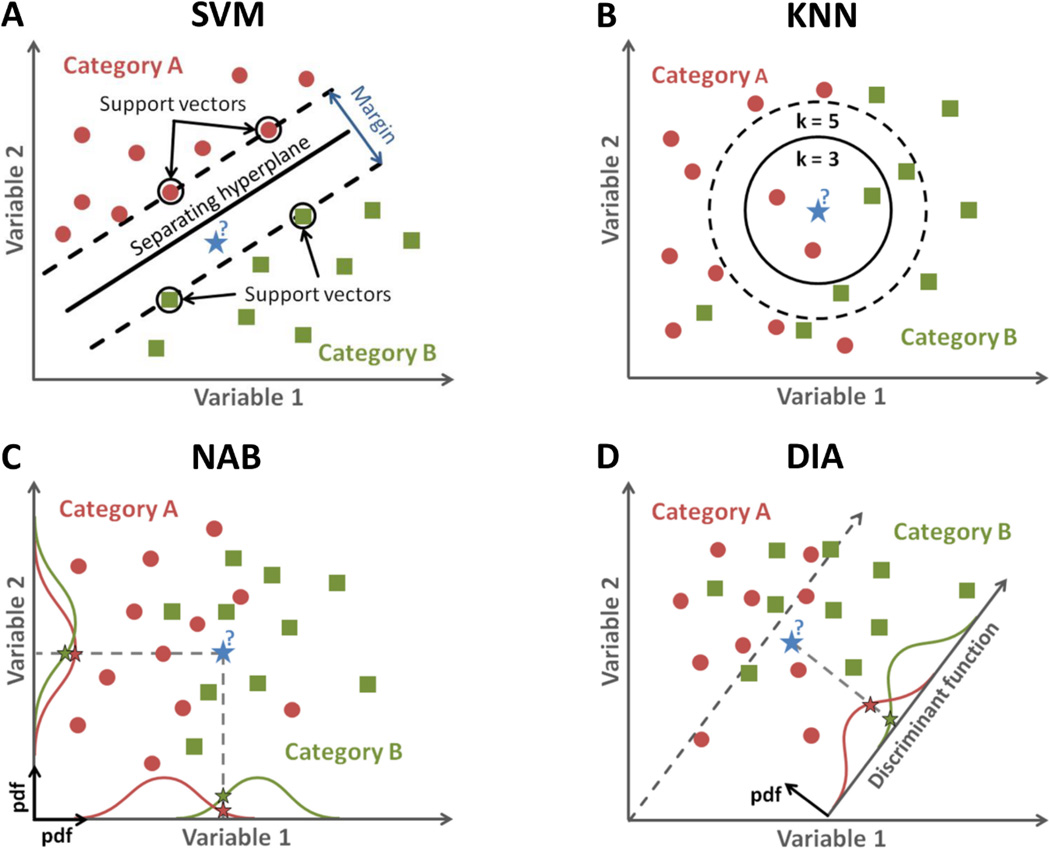
Figure 1.
(A) Example of support vector machine (SVM) classification with 2 variables. In this example, the new data point (blue star) is classified as green category B. (B) Example of k-nearest neighbors (KNN) classification with 2 variables and k=3 or 5. For k=3, new data is classified as red category A (2 red neighbors against 1 green), while for k=5, it is classified as green category B (3 green neighbors against 2 reds) (C) Example of naïve Bayes (NAB) classification for 2 variables. In this example, the numbers of samples in each category is Ngreen = 10 and Nred = 12, and therefore the prior probabilities for each category are Prior(A)=12/22=0.55 and Prior(B)=0.45. The posterior probabilities for a a data point to belong to category X (X=a or B) is Posterior(X)~Prior(X)×pdf(X,Variable 1)×pdf(X,Variable 2). The new data point is then classified in the category with the highest posterior probability. (D) Example of discriminant analysis (DIA) classification for 2 variables. The method is similar to NAB, but with a likelihood function = pdf from the discriminant function for each category.
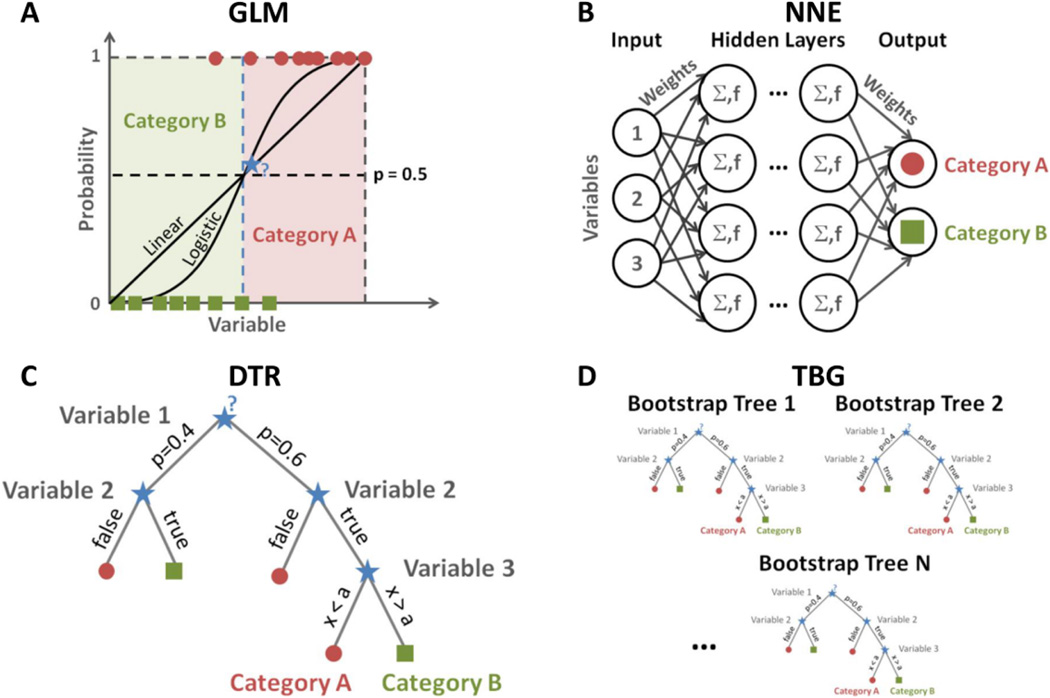
Figure 2.
(A) Examples of linear regression (LNR) and logistic regression (LGR) classifications for 1 variable, which are particular cases of generalized linear model (GLM). In this example, the cutoff probability for making a decision is 0.5, and therefore the new data sample (blue star) is classified as category A with both classifiers. (B) Schematics of feed-forward neural networks (NNE) binary classification for 3 input variables. Σ represents the weighted sum of all inputs arriving to a neuron, and f is the activation function (generally a step or a sigmoid function) that generates the output of each neuron. (C) Example of decision tree (DTR) classification with 3 types of variables (probability, categorical, numerical). (D) Example of tree bagging (TBG) for N trees.
SUPPORT VECTOR MACHINE (SVM)
Support vector machine [26] is a classifier for which the goal is to find the best hyperplane in the variable space that separates two categories of data with the largest margin possible. Margin means the maximal width of the slab parallel to the hyperplane that has no interior data points. The margin is said soft when the algorithm allows a few data to be misclassified and to be located within the margin. The support vectors are the data points of each class that are closest to the separating hyperplane and form the boundaries of the margin. For linear SVM, the separating hyperplane can be described by the dot product equation w·x+b=0, and the limits of the margin are defined by w·x+b=1 and w·x+b=−1, with b a constant and w a vector (normal to the hyperplane). The goal of SVM algorithm is to find the optimum b and w that separate the two classes of data with the largest margin possible using the train dataset. For quadratic SVM, the dot product equation can be replaced by a kernel function which includes w and also quadratic terms of x (nonlinear classification). A 2D linear example of SVM classification is illustrated in Fig. 1A.
K-NEAREST NEIGHBORS (KNN)
K-nearest neighbors [27] is a simple classifier in which a data point xi is classified by a majority vote of its k neighbors (k is generally an odd number to avoid ties). A commonly used distance metric for detecting neighbors in the variable space is the Euclidean distance. The training phase of the method consists simply of storing the variable vectors and class labels (xi, yi) of the train dataset. A 2D example of KNN classification with Euclidean distance, for k=3 and 5, is illustrated in Fig. 1B.
NAIVE BAYES (NAB)
Naïve Bayes [28] is a probabilistic classifier based on the Bayes’ theorem with strong (naïve) independence assumption between variables, which appears to work well in practice even when this assumption is not valid. In the training phase, the method estimates the prior probability and likelihood that the sample belongs to each class using the train dataset. In the test phase, the method computes the posterior probability that a test sample belongs to each class. The method then classifies the test sample according the largest posterior probability. Written in a simple form, the posterior probability that a data point with vector of variables xi belongs to a certain class yi is Posterior = (Prior × Likelihood)/Evidence, with: (1) Prior = prior probability of any data to be in category y. The empirical prior probability of a class is the number of training samples of this class divided by the total number of training samples. (2) Likelihood = probability density functions (pdf) of each variable in the training dataset, which are assumed to be normal (Gaussian) distributions. (3) Evidence = a normalizing constant that scales both posterior probabilities equally. Therefore it does not affect classification and can be ignored. A 2D example of NAB classification with normal pdfs is illustrated in Fig. 1C.
DISCRIMINANT ANALYSIS (DIA)
Discriminant analysis [29] is a probabilistic classifier which assumes that the data has a Gaussian mixture distribution. For linear discriminant analysis, the model has the same covariance matrix for each class; only the means vary. For quadratic discriminant analysis, both means and covariances of each class vary. The method then find a weighted combination of variables (the discriminant function) in order to maximize difference between the classes pdfs. Similarly to NAB, the posterior probability that a data point xi belongs to a certain class is the product of the prior probability by the discriminant function (or likelihood, or multivariate normal density). DIA is closely related to NAB, analysis of variance (ANOVA) and principal component analysis (PCA). A 2D example of linear DIA classification with empirical prior is illustrated in Fig. 1D.
LINEAR AND LOGISTIC REGRESSIONS (LNR, LGR)
Linear and logistic regression classifiers can be described within the framework of generalized linear models (GLM) [30]. A GLM has 3 characteristics: (1) at each set of values for the predictors, the response has a distribution that can be normal, binomial, Poisson, gamma, or inverse Gaussian, with parameters including a mean µ; (2) a coefficient vector b defines a linear combination xb of the predictors x; (3) a link function f defines the model as f(µ) = xb. For LNR, the distribution is normal and the link function is defined as the mean function f(µ)=µ=xb (identity). For LGR, the distribution is binomial and the link function is defined as f(µ) =ln[µ/(1-µ)]=xb (logit), and the mean function is µ =1/[1+exp(−xb)]. An extension to a nonlinear model can be made by including squared terms and products of pairs of distinct predictors in the model included in the link function (quadratic model). The unknown parameters b are typically estimated with maximum likelihood or Bayesian techniques using the training data. The outcome of both LNR or LGR is a probability 0<p<1. A cutoff probability is therefore defined (generally p=0.5) for deciding to which class the new data sample xi belongs to (yi=0 if p<0.5 or yi=1 if p>0.5). An 1D example of LNR and LGR with threshold p=0.5 is illustrated in Fig. 2A.
NEURAL NETWORKS (NNE)
Neural networks [31] classifiers are loosely based on biological neural networks architecture in the brain, where neurons connected to each others by axons are used to process information. In the original version of NNE, an artificial neuron computes a weighted sum of its n input signals xj (j=1,2,…,n), and generates an output response r=1 if this sum is above a threshold u, and r=0 otherwise. The response output of each neuron can be computed as , with f an activation function, which is generally defined as a step function or a sigmoid (logistic) function. NNE can be described as a directed graph in which artificial neurons are nodes and directed edges (with weights) are connections between neuron outputs and inputs. There are two categories of NNE: (1) feed-forward networks, in which there is no loop, and (2) recurrent/feedback, which include loops of feedback connections. The most common NNEs are feed-forward networks, also called multilayer perceptron, where neurons are organized into multiple hidden layers with unidirectional connections between these layers (see Fig. 2B). The training phase of NNE consists of optimizing the weights wi using the train dataset, an optimization algorithm and a performance function (such as cross-entropy [32]). The train dataset is divided into 3 subsets: a training subset to adjust the weights, a validation subset to minimize overfitting and a testing subset to test the final solution.
DECISION TREE (DTR)
A decision tree [33] classifier is a tree-like graph or model in which each internal node represents a test on an attribute (or predictor), each branch represents the outcome of the test and each leaf node represents a class label where a decision is taken after computing all attributes (see Fig. 2C). The 3 types of nodes are: (1) a root node that has no incoming edge (or branch) and one or more outgoing edges; (2) an internal (or test) node that has exactly one incoming edge and two or more outgoing edges; (3) a leaf (or terminal) node that has exactly one incoming edge and no outgoing edges. Each internal node splits the instance space into two or more subspaces according to a certain discrete function of the input attribute values. Each leaf node is assigned a class label representing the most appropriate target value, or a probability (using Bayes’ theorem for example) for the target attribute to have a certain value. Data samples are classified by navigating the tree from the root node to a leaf node according to the outcomes of the tests for each internal node along the path. Decision tree training is performed on a train dataset for optimizing decision (probability) thresholds of the internal nodes, that will then be applied on the test samples. Pruning is a technique that can be implemented in the algorithm in order to reduce the size of the decision tree by removing sections of the tree that provide little power to classify instances (branch trimming). This will reduce the complexity of the final classifier as well as provide a better predictive accuracy by reducing overfitting.
TREE BAGGING (TBG)
Bootstrap aggregating (or bagging) [34] is a method used for enhancing the performance of weak learner classifiers such as decision trees, which are adaptive and robust but do not generalize well in general. Tree bagging consists in fitting many large trees to bootstrap-resampled versions of the training data and classify by majority vote. Bagging helps reducing the variance of the classifications and helps avoiding overfitting. See Fig. 2D.
METHODS
SODIUM MRI DATA
In the present work, we used the same sodium MRI data of articular cartilage in the knee joint that was acquired in a previous study [13]. Sodium data acquisition and processing are summarized below. We refer the reader to this previous study [13] for more details on the data and for examples of distributions of sodium concentration values in different regions in cartilage for controls and OA patients.
Volunteers
19 asymptomatic volunteers (controls) and 28 symptomatic volunteers (OA patients) were recruited from the general public and from the New York University-Hospital of Joint Diseases knee osteoarthritis cohort, respectively. These OA patients fulfilled the criteria for clinical osteoarthritis symptoms defined by the American College of Rheumatology [35] and had radiographic evidence of tibial-femoral knee osteoarthritis, with a Kellgren-Lawrence (KL) grade of 1–4 on standardized weight-bearing fixed flexion posterior-anterior knee radiographs [36]. Among the 28 OA patients, 16 had a diagnosis of OA with KL=1, 7 with KL=2, 4 with KL=3 and 1 with KL=4. Patients with KL=1–4 are referred as ‘all OA’ population, and patient with KL1–2 are referred as ‘early OA’ population (n=23). This study was approved by the institutional review board and performed in compliance with the Health Insurance Portability and Accountability Act. All subjects provided written informed consent.
Hardware
Sodium MRI data was acquired on a 7 T whole-body scanner (Siemens Healthcare, Erlangen, Germany) with either a single-tuned sodium birdcage radiofrequency (RF) knee coil (Rapid MR International, Columbus, Ohio, USA), or a home-made multichannel dual-tuned 1H/23Na coil (NYU Center for Biomedical Imaging, RF Core, New York, NY) [37].
Sodium MRI acquisition and reconstruction
Sodium MRI data were acquired with a radial 3D sequence [38] and images were reconstructed offline in Matlab (Mathworks, Natick, Mass) by using a nonuniform fast Fourier transform algorithm [39]. Fluid suppression was obtained with inversion recovery (IR) by using an adiabatic pulse and appropriate inversion time before the radial 3D acquisition. The adiabatic pulse was the wideband uniform rate and smooth truncation (WURST) pulse [40] with a sweep range of 2 kHz. In the present article, we will refer to the acquisition sequence without fluid suppression as radial 3D (abbreviation: R3D), and the sequence with fluid suppression as IR WURST (IRW). The parameters for R3D acquisition were: 10,000 projections, TR = 100 ms, TE = 0.4 ms, flip angle = 90°, isotropic field-of-view (FOV) = 200 mm, dwell time = 80 µs, nominal (reconstructed) resolution = 2 mm, real (acquired) resolution = 3.3 mm, total acquisition time (TA) = 16:44 min. The parameters for IRW acquisition were the same as for R3D except TR = 140 ms, WURST pulse amplitude/duration = 240 Hz / 10 ms, inversion time (TI) = 24 ms, and TA = 23:25 min.
Image post-processing
All images were acquired in the presence of calibration phantoms placed within the FOV. These phantoms were made of 4% agar gel with different sodium concentrations (100, 150, 200, 250, and 300 mmol/L) and with known relaxation times. Sodium concentration maps (for both R3D and IRW) were calculated by using linear regression of the phantom signals after relaxation correction of the gels. The sodium maps were then corrected for the average T1 and biexponential T2* of cartilage in vivo [41] to achieve a more accurate quantification of the sodium concentration in cartilage. Because, on average, 25% of the volume in cartilage is made up of solids without any sodium, the values of the voxels of the final sodium maps were divided by a factor of 0.75 [42, 43]. Prior to this sodium quantification processing, all sodium images were corrected by the signal-to-noise ratio (SNR) map of each coil acquired on a large solution phantom filling the whole volume inside the coil. This RF correction was necessary mainly for correcting for sensitivity inhomogeneities of the multichannel RF coil.
Sodium data measurements
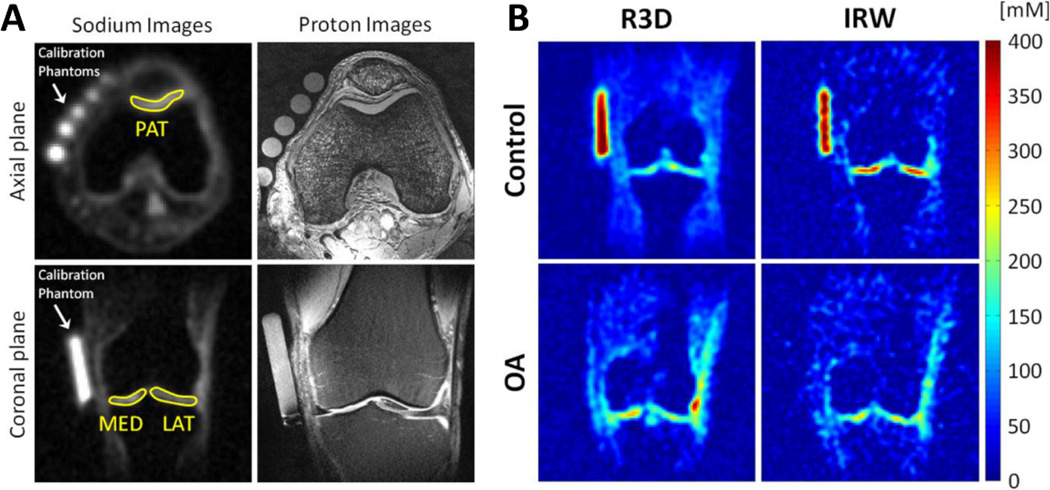
Three regions of interest (ROIs) of 30 pixels were drawn on the patellar (PAT), femorotibial lateral (MED), and femorotibial medial (LAT) cartilage on four consecutive sections of the sodium maps. Sodium images with ROIs are presented in Fig. 3A alongside proton images. Sodium maps from R3D and IRW in OA and control subjects are shown in Fig. 3B. The mean (MEAN) and standard deviation (STD) of sodium concentration were then calculated for each ROI (in mmol/L, or mM). For each subject and each sequence (R3D, IRW), the minimum, maximum, and mean of sodium MEAN and STD were calculated over the 12 ROI measurements (3 compartments: PAT, MED and LAT; and 4 slices). Therefore, each subject will be assigned a set of 12 global statistical measures (or predictors) that will be used for classification. These measures will be abbreviated as following in the rest of this article: Min[MEAN]R3D, Max[MEAN]R3D, Mean[MEAN]R3D, Min[STD]R3D, Max[STD]R3D, Mean[STD]R3D, and Min[MEAN]IRW, Max[MEAN]IRW, Mean[MEAN]IRW, Min[STD]IRW, Max[STD]IRW, Mean[STD]IRW. The standard deviations (std) of MEAN and STD were also calculated over the 12 ROIs, but were found non-significant in the classification analysis, and therefore will be ignored in this study. A z-score transformation was also tested on all data, but with no effect on the final results, and therefore will not be discussed. The coefficient of determination R2 (the square of the coefficient of correlation) was calculated between all predictors for data including all OA, and early OA subjects only. This coefficient measures the strength of the linear association between two variables.
Figure 3.
(A) Examples of sodium images of articular cartilage in the knee at 7 T (1st column) and proton images (2nd column). (B) Examples of sodium concentration maps in a control subject and a patient with OA, with and without fluid suppression (IRW and R3D respectively). R3D = Radial 3D, IRW = IR WURST. Figures from Madelin G. et al., Radiology 268(2), 481–491, 2013. Reproduced with permission from RSNA.
CLASSIFICATION
Classifiers
A total of sixteen classifiers were tested, from nine classification methods with different options (see Table 1):
DIA (2 classifiers): Two types of discriminant analyses were tested: linear and quadratic. The prior probability of each class was chosen as empirical.
LGR (2 classifiers): Logistic regression was tested with both linear and quadratic models. The linear model contains an intercept and linear terms for each predictor. The quadratic model contains the linear model completed with all product pairs of predictors (including squared terms). Cutoff probability was chosen as 0.5.
LNR (2 classifiers): Linear regression was also tested with both linear and quadratic models and cutoff probability = 0.5.
KNN (2 classifiers): k-nearest neighbor was tested with k=3 and k=5, with Euclidean distance between variables.
NAB (1 classifier): Naïve Bayes was tested with normal distribution model and empirical prior probability.
NNE (2 classifiers): Neural networks were tested using feed-forward network architecture, sig-moid output neurons, and with 10 or 20 hidden layers. Pattern recognition was performed with a random selection of values of the training dataset, with local train/valid/test ratios = 0.6/0.2/0.2. The training algorithm was chosen as scaled conjugate gradient and the training performance was calculated using the cross-entropy method.
SVM (2 classifiers): Support vector machine was tested with two kernels: linear and quadratic. Sequential minimal optimization (SMO) algorithm was used for finding the separating hyper-plane.
DTR (1 classifiers): Decision tree was tested with empirical prior probability, and pruning option.
TBG (2 classifiers): Tree bagging was tested with 10 and 20 trees.
Table 1. List of classification methods with options.
NHL = number of hidden layers, NT = number of trees.
| Methods | Option 1 | Option 2 | Option 3 | Option 4 | Option 5 | |
|---|---|---|---|---|---|---|
| 1 | DIA | Type = Linear | Prior = Empirical | |||
| 2 | DIA | Type = Quadratic | Prior = Empirical | |||
| 3 | LGR | Model = Linear | Cutoff probability = 0.5 | |||
| 4 | LGR | Model = Quadratic | Cutoff probability = 0.5 | |||
| 5 | LNR | Model = Linear | Cutoff probability = 0.5 | |||
| 6 | LNR | Model = Quadratic | Cutoff probability = 0.5 | |||
| 7 | KNN | k = 3 | Distance = Euclidean | Rule = Nearest | ||
| 8 | KNN | k = 5 | Distance = Euclidean | Rule = Nearest | ||
| 9 | NAB | Distribution = Normal | Prior = Empirical | |||
| 10 | NNE | NHL = 10 | Divide function = Random | Train function = SCG* | Performance function = Cross-entropy | Train/Valid/Test ratios = 0.6/0.2/0.2 |
| 11 | NNE | NHL = 20 | Divide function = Random | Train function = SCG* | Performance function = Cross-entropy | Train/Valid/Test ratios = 0.6/0.2/0.2 |
| 12 | SVM | Kernel = Linear | Method = SMO” | |||
| 13 | SVM | Kernel = Quadratic | Method = SMO” | |||
| 14 | DTR | Prior = Empirical | Pruning = Yes | |||
| 15 | TBG | NT= 10 | ||||
| 16 | TBG | NT=20 |
SCG = Scaled Conjugate Gradient
SMO = Sequential Minimal Optimization
Cross-validation partition
Two partition methods were used to classify the data: holdout cross-validation and resubstitution [22, 24, 44, 45]. For holdout cross-validation, the algorithm divides randomly the values of the predictors into a test (or holdout) set, with 0<test ratio<1, and a train set, with train ratio = 1-test ratio. This process was performed 100 times for each predictor, or set of predictors. The average and standard deviation of the sensitivity, specificity and (adjusted) accuracy of each classifier was then calculated for comparison of their classification performance. In this study, we tested the classifiers with train/test ratios = 0.5/0.5, 0.6/0.4 and 0.7/0.3. As the results with train/test ratios = 0.6/0.4 and 0.7/0.3 were identical to the ones with 0.5/0.5, we will only show and discuss results of these latter ratios without loss of generality. For resubstitution, both the training and test sets contain all of the original values of the predictors (train ratio = test ratio = 1), and only one iteration of the classification process was therefore needed.
Feature selection and PCA
Feature (or variable) selection (FS) was used to select a subset of relevant predictors for use in the model construction of each classifier. These selected predictors can be different for each classifier, as they all use different methods of classification. It is therefore expected that some groups of features might be good predictors for certain classifiers but not for others. The criterion for selecting features was minimization of the misclassification from the confusion matrix for each classifier (sum of false positive rate + false negative rate). Feature selection was performed sequentially on full data resubstitution, either in forward or backward direction. In forward FS, an initial candidate set includes only one predictor and the algorithm add predictors sequentially until the criterion increases. For backward FS, an initial candidate set includes all the predictors and the algorithm removes predictors sequentially until the criterion increases. Principal component analysis (PCA) was also applied for reducing the dimensionality of the data (see Fig. S1 in Supplementary Material).
Classifications
In this study, we performed first, classifications on all individual predictors separately with resubstitution (as a means to determine the optimum performance for each classifier with this data), and then with holdout cross-validation. The classification methods were then applied on the predictors selected by feature selection (both backward and forward) for each classifier with holdout cross-validation. Finally, classifications were also applied on the main principal components calculated from PCA with holdout cross-validation (train/test ratios = 0.5/0.5).
SENSITIVITY, SPECIFICITY, ACCURACY, ROC
We used the standard definitions for sensitivity, specificity and accuracy of classification, and we defined OA as ‘Positive’ class and control as ‘Negative’ class. Using the standard abbreviations: TP = number of True Positives (OA classified as OA), TN = number of True Negatives (control classified as control), FP = number of False Positives (control classified as OA), FN = number of False Negatives (OA classified as control), we have the following formulas:
For ranking the classifiers, we defined the adjusted accuracy metric, which favors classifiers with higher accuracy which minimize the difference between sensitivity and specificity:
Receiver operating characteristics (ROC) analysis was also performed for all classification methods with the best predictor and holdout cross-validation (train ratio = test ratio = 0.5) and 100 iterations. Mean ROC curves and mean areas under the curve (AUC) were then calculated for each case, to compare the performance of the classifiers between each other, but also to compare this performance with the ranking of classifiers using adjusted accuracy.
All classification processing was performed in Matlab using functions from the Statistics toolbox.
RESULTS
The coefficients of determination (R2) between all predictors, for data including all OA and early OA subjects, are shown in Table 2. For R3D data, a strong correlation (R2≥0.7) is observed between Mean values of MEAN or STD of sodium concentrations and their Min and Max counterparts. For IRW data, a strong correlation (R2≥0.7) is observed between Mean values of MEAN sodium concentrations and their Min and Max counterparts, and between Mean of STD and Max of STD. Tables 3 and 4 present the best results (from the adjusted accuracy point of view) for each classifier with full data resubstitution, when all OA subjects and only early OA subjects are included, respectively. Only individual sodium measures were included in each classification. In both cases (all OA and early OA), we can see that a majority of the best classifications for each classifier were obtained when using data acquired with IRW (16/20 instances for all OA, 20/22 instances for early OA), and particularly Min[STD]IRW, as predictors. However, the very best classifiers (adjusted ac-curacy>80%) were TBG, DTR and KNN and were mostly using the MEAN sodium values (either from R3D or IRW) as best predictors.
Table 2. Coefficients of determination (R2).
between all different predictors for data including all OA and early OA (left value/right value, respectively). Moderate and strong coefficients of determination with mean R2>0.7 are shown in bold font.
| R2 | R3D |
IRW |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Min[MEAN] | Max[MEAN] | Mean[MEAN] | Min[STD] | Max[STD] | Mean[STD] | Min[MEAN] | Max[MEAN] | Mean[MEAN] | Min[STD] | Max[STD] | Mean[STD] | ||
| R3D | Min[MEAN] | 1 | |||||||||||
| Max[MEAN] | 0.53/0.58 | 1 | |||||||||||
| Mean[MEAN] | 0.80/0.82 | 0.84/0.86 | 1 | ||||||||||
| Min[STD] | 0.37/0.31 | 0.25/0.26 | 0.35/0.33 | 1 | |||||||||
| Max[STD] | 0.25/0.19 | 0.21/0.21 | 0.26/0.25 | 0.52/0.46 | 1 | ||||||||
| Mean[STD] | 0.48/0.43 | 0.36/0.41 | 0.48/0.50 | 0.72/0.67 | 0.75/0.71 | 1 | |||||||
| IRW | Min[MEAN] | 0.58/0.61 | 0.24/0.26 | 0.43/0.45 | 0.17/0.14 | 0.10/0.07 | 0.18/0.16 | 1 | |||||
| Max[MEAN] | 0.44/0.57 | 0.37/0.38 | 0.51/0.55 | 0.27/0.32 | 0.11/0.14 | 0.26/0.34 | 0.60/0.66 | 1 | |||||
| Mean[MEAN] | 0.55/0.64 | 0.32/0.33 | 0.51/0.55 | 0.28/0.30 | 0.13/0.13 | 0.26/0.29 | 0.81/0.83 | 0.91/0.93 | 1 | ||||
| Min[STD] | 0.20/0.31 | 0.03/0.05 | 0.11/0.15 | 0.20/0.21 | 0.13/0.14 | 0.17/0.19 | 0.34/0.38 | 0.35/0.37 | 0.43/0.44 | 1 | |||
| Max[STD] | 0.14/0.15 | 0.12/0.12 | 0.16/0.15 | 0.33/0.32 | 0.22/0.22 | 0.29/0.28 | 0.15/0.17 | 0.41/0.42 | 0.36/0.38 | 0.26/0.24 | 1 | ||
| Mean[STD] | 0.24/0.30 | 0.09/0.11 | 0.18/0.20 | 0.41/0.42 | 0.20/0.20 | 0.31/0.32 | 0.27/0.29 | 0.52/0.53 | 0.52/0.53 | 0.57/0.55 | 0.75/0.74 | 1 | |
Table 3. Classification of all OA vs. controls for individual sodium measurements, with full resubmission of the data (train ratio = test ratio = 1).
The best results for each classifier were selected and then ranked by level of adjusted (Adj.) accuracy. NHL = number of hidden layers, NT = number of trees.
| Rank | Methods | Options | Measurements | Adj. Accuracy (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|
| 1 | TBG | NT = 20 | Min[MEAN]IRW | 100 | 100 | 100 | 100 |
| TBG | NT = 20 | Mean[MEAN]R3D | 100 | 100 | 100 | 100 | |
| TBG | NT = 20 | Min[MEAN]R3D | 100 | 100 | 100 | 100 | |
| 2 | TBG | NT= 10 | Max[MEAN]R3D | 96.91 | 97.9 | 96.4 | 100 |
| TBG | NT= 20 | Min[MEAN]R3D | 96.91 | 97.9 | 96.4 | 100 | |
| 3 | DTR | Min[STD]IRW | 86.22 | 87.2 | 85.7 | 89.5 | |
| DTR | Mean[MEAN]IRW | 86.22 | 87.2 | 85.7 | 89.5 | ||
| 4 | KNN | k = 3 | Max[MEAN]IRW | 81.71 | 87.2 | 92.9 | 78.9 |
| 5 | KNN | k = 5 | Min[STD]IRW | 79.58 | 80.9 | 82.1 | 78.9 |
| 6 | NAB | Min[STD]IRW | 78.62 | 78.7 | 78.6 | 78.9 | |
| DIA | Quadratic | Min[STD]IRW | 78.62 | 78.7 | 78.6 | 78.9 | |
| NNE | NHL = 10 | Min[STD]IRW | 78.62 | 78.7 | 78.6 | 78.9 | |
| 7 | NNE | NHL = 20 | Max[MEAN]IRW | 76.07 | 80.9 | 85.7 | 73.7 |
| 8 | LGR | Quadratic | Min[STD]IRW | 75.53 | 76.6 | 75.0 | 78.9 |
| 9 | DIA | Linear | Min[STD]IRW | 75.36 | 78.7 | 82.1 | 73.7 |
| LGR | Linear | Min[STD]IRW | 75.36 | 78.7 | 82.1 | 73.7 | |
| LNR | Linear | Min[STD]IRW | 75.36 | 78.7 | 82.1 | 73.7 | |
| LNR | Quadratic | Min[STD]IRW | 75.36 | 78.7 | 82.1 | 73.7 | |
| 10 | SVM | Linear | Min[STD]IRW | 72.44 | 74.5 | 71.4 | 78.9 |
| 11 | SVM | Quadratic | Min[STD]IRW | 66.97 | 72.3 | 64.3 | 84.2 |
Table 4. Classification of early OA vs. controls for individual sodium measurements, with full resubmission of the data (train ratio = test ratio = 1).
The best results for each classifier were selected and then ranked by level of adjusted (Adj.) accuracy. NHL = number of hidden layers, NT = number of trees.
| Rank | Methods | Options | Measurements | Adj. Accuracy (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|
| 1 | TBG | NT= 10 | Mean[MEAN]IRW | 100 | 100 | 100 | 100 |
| TBG | NT= 10 | Max[MEAN]IRW | 100 | 100 | 100 | 100 | |
| TBG | NT= 10 | Mean[MEAN]R3D | 100 | 100 | 100 | 100 | |
| TBG | NT= 20 | Mean[STD]IRW | 100 | 100 | 100 | 100 | |
| TBG | NT= 20 | Mean[MEAN]IRW | 100 | 100 | 100 | 100 | |
| TBG | NT= 20 | Max[MEAN]IRW | 100 | 100 | 100 | 100 | |
| 2 | DTR | Min[STD]IRW | 85.51 | 88.1 | 91.3 | 84.2 | |
| DTR | Min[MEAN]R3D | 85.51 | 88.1 | 91.3 | 84.2 | ||
| 3 | KNN | k = 3 | Max[MEAN]IRW | 84.71 | 85.7 | 87.0 | 84.2 |
| 4 | NNE | NHL= 20 | Max[MEAN]IRW | 79.62 | 81.0 | 82.6 | 78.9 |
| LGR | Linear | Min[STD]IRW | 79.62 | 81.0 | 82.6 | 78.9 | |
| DIA | Linear | Min[STD]IRW | 79.62 | 81.0 | 82.6 | 78.9 | |
| KNN | k = 5 | Min[STD]IRW | 79.62 | 81.0 | 82.6 | 78.9 | |
| 5 | LNR | Linear | Min[STD]IRW | 78.36 | 78.6 | 78.3 | 78.9 |
| DIA | Quadratic | Min[STD]IRW | 78.36 | 78.6 | 78.3 | 78.9 | |
| NAB | Min[STD]IRW | 78.36 | 78.6 | 78.3 | 78.9 | ||
| 6 | LGR | Quadratic | Mean[STD]IRW | 75.47 | 78.6 | 73.9 | 84.2 |
| 7 | LNR | Quadratic | Mean[STD]IRW | 74.67 | 76.2 | 73.9 | 78.9 |
| SVM | Linear | Mean[STD]IRW | 74.67 | 76.2 | 73.9 | 78.9 | |
| SVM | Linear | Min[STD]IRW | 74.67 | 76.2 | 73.9 | 78.9 | |
| 8 | NNE | NHL= 10 | Mean[STD]IRW | 69.42 | 71.4 | 73.9 | 68.4 |
| 9 | SVM | Quadratic | Min[STD]IRW | 68.08 | 73.8 | 65.2 | 84.2 |
Tables 5 and 6 present the best results (from the adjusted accuracy point of view) for each classifier with holdout cross-validation of the data and 100 iterations, when all OA subjects and only early OA subjects are included, respectively. Only individual sodium measures were included in each classification. In the holdout case, the best mean results for each classifier were obtained only when using data acquired with IRW (16/16 instances for both all and early OA), and particularly Min[STD]IRW, as predictors (14/16 instances for all OA, 12/16 instances for early OA). In this case, the very best classifications (mean adjusted accuracy>70%) were obtained with LGR, DIA and LNR for all OA, and LGR, DIA, LNR, SVM and NAB for early OA, while TBG, DTR and KNN performed poorer. In both cases, linear LGR with Min[STD]IRW was ranked first. Table 7 shows the best classifications (mean adjusted accuracy≥65%) for all OA and for early OA data, after feature selection of the predictors, and with holdout cross-validation and 100 iterations. For both all OA and early OA, the very best classification was obtained when using only a simple predictor, Min[STD]IRW and DIA method (quadratic for all OA, linear for early OA), with a mean adjusted accuracy ≥70% and mean accuracy, sensitivity and specificity all ≥75%. For both cases, we can also notice that linear SVM performed reasonably, with mean adjusted accuracy > 68% and mean accuracy, sensitivity and specificity all ≥72%, when using two IRW predictors for classification: Min[STD]IRW associated with either Min[MEAN]IRW (for early OA) or Mean[MEAN]IRW (for all OA). Linear LGR also performed quite well (mean adjusted accuracy≥66%, with high mean accuracy ≥75%, high mean sensitivity ≥80% but moderate mean specificity ≥65–68%) with the same pair of predictors for both early OA and all OA: Min[STD]IRW and Mean[MEAN]IRW. LNR and NAB are the only other classifiers ranked in this table as all other classifiers generated a mean adjusted accuracy ⩽65%.
Table 5. Classification of all OA vs. controls for individual sodium measurements, with holdout cross-validation of the data (train ratio = test ratio = 0.5) and 100 iterations.
The best results for each classifier were selected and then ranked by level of adjusted (Adj.) accuracy. Results are shown as mean ± standard deviation. Results with Adj. Accuracy ≥ 70% are shown in bold font. NHL = number of hidden layers, NT = number of trees.
| Rank | Methods | Options | Measurements | Adj. Accuracy (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|
| 1 | LGR | Linear | Min[STD]IRW | 71.17±10.39 | 77.9±6.9 | 80.9±11.2 | 73.3±15.3 |
| 2 | DIA | Quadratic | Min[STD]IRW | 70.78±9.36 | 75.7±7.1 | 77.0±9.8 | 73.8±13.5 |
| 3 | DIA | Linear | Min[STD]IRW | 70.08±9.87 | 76.9±6.2 | 80.8±9.8 | 70.9±15.1 |
| 4 | LNR | Linear | Min[STD]IRW | 70.38±8.03 | 76.4±5.8 | 78.9±9.8 | 72.6±12.9 |
| 5 | SVM | Linear | Min[STD]IRW | 69.68±7.44 | 74.2±6.8 | 69.8±9.8 | 81.1±10.8 |
| 6 | NAB | Min[STD]IRW | 68.39±9.63 | 74.1±7.3 | 76.1±10.4 | 71.0±14.3 | |
| 7 | LNR | Quadratic | Min[STD]IRW | 68.15±9.95 | 74.3±7.1 | 73.6±11.8 | 75.4±15.9 |
| 8 | LGR | Quadratic | Min[STD]IRW | 67.98±9.37 | 73.0±7.2 | 71.4±11.7 | 75.6±13.3 |
| 9 | KNN | k = 5 | Min[STD]IRW | 66.95±8.92 | 72.5±6.3 | 74.9±9.2 | 68.7±14.1 |
| 10 | SVM | Quadratic | Min[STD]IRW | 63.99±9.76 | 70.7±7.0 | 64.4±12.2 | 80.4±15.0 |
| 11 | KNN | k = 3 | Min[STD]IRW | 63.56±9.84 | 69.9±7.7 | 71.8±12.2 | 67.0±14.9 |
| 12 | DTR | Min[STD]IRW | 60.89±14.98 | 69.3±10.2 | 72.1±15.2 | 65.0±21.0 | |
| 13 | TBG | NT = 10 | Max[MEAN]IRW | 60.44±12.47 | 68.8±9.2 | 72.3±14.8 | 63.3±19.1 |
| 14 | TBG | NT = 20 | Max[MEAN]IRW | 60.17±12.66 | 69.9±9.2 | 76.3±13.4 | 60.0±19.3 |
| 15 | NNE | NHL = 10 | Min[STD]IRW | 50.72±19.58 | 65.3±11.7 | 76.6±13.8 | 47.7±28.7 |
| 16 | NNE | NHL = 20 | Min[STD]IRW | 49.02±18.25 | 62.7±11.6 | 72.9±16.0 | 46.8±26.9 |
Table 6. Classification of early OA vs. controls for individual sodium measurements, with holdout cross-validation of the data (train ratio = test ratio = 0.5) and 100 iterations.
The best results for each classifier were selected and then ranked by level of adjusted (Adj.) accuracy. Results are shown as mean ± standard deviation. Results with Adj. Accuracy ≥ 70% are shown in bold font. NHL = number of hidden layers, NT = number of trees.
| Rank | Methods | Options | Measurements | Adj. Accuracy (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|
| 1 | LGR | Linear | Min[STD]IRW | 73.63±8.42 | 78.0±6.9 | 77.9±11.7 | 78.2±10.2 |
| 2 | DIA | Linear | Min[STD]IRW | 73.19±8.56 | 78.3±6.9 | 79.4±11.7 | 77.0±11.9 |
| 3 | LNR | Linear | Min[STD]IRW | 72.51±8.93 | 77.4±7.4 | 78.3±11.3 | 76.2±11.9 |
| 4 | SVM | Linear | Mean[STD]IRW | 70.46±9.36 | 74.5±7.9 | 72.1±10.9 | 77.4±11.3 |
| 5 | DIA | Quadratic | Min[STD]IRW | 70.25±8.02 | 75.3±6.7 | 73.9±11.6 | 77.1±12.2 |
| 6 | NAB | Min[STD]IRW | 70.00±9.74 | 75.5±7.9 | 75.8±12.8 | 75.0±12.6 | |
| 7 | KNN | k = 5 | Min[STD]IRW | 68.94±8.76 | 74.1±6.8 | 72.0±12.5 | 76.7±11.3 |
| 8 | LNR | Quadratic | Min[STD]IRW | 68.09±9.81 | 73.7±7.6 | 70.1±13.7 | 78.0±11.9 |
| 9 | LGR | Quadratic | Min[STD]IRW | 67.53±9.99 | 73.1±7.7 | 70.7±13.5 | 76.1±13.1 |
| 10 | KNN | k = 3 | Min[STD]IRW | 65.48±10.38 | 71.3±7.6 | 71.4±13.2 | 71.2±14.4 |
| 11 | SVM | Quadratic | Min[STD]IRW | 64.82±9.42 | 72.2±7.4 | 62.7±11.9 | 83.8±12.8 |
| 12 | TBG | NT= 20 | Max[MEAN]IRW | 63.16±12.33 | 70.1±10.1 | 73.5±13.2 | 65.9±17.7 |
| 13 | DTR | Mean[STD]IRW | 61.72±13.61 | 69.6±9.1 | 66.3±16.1 | 73.7±19.6 | |
| 14 | TBG | NT= 10 | Max[MEAN]IRW | 61.41±12.83 | 68.9±9.9 | 70.5±16.6 | 67.0±18.5 |
| 15 | NNE | NHL= 10 | Min[STD]IRW | 53.07±21.0 | 65.7±13.8 | 74.8±16.6 | 54.6±31.2 |
| 16 | NNE | NHL= 20 | Min[STD]IRW | 52.70±22.24 | 64.5±15.0 | 73.2±16.4 | 53.8±31.1 |
Table 7. Classification of all OA vs. controls, and early OA vs. controls, for multiple sodium measurements chosen from sequential feature selection, with holdout cross-validation of the data (train ratio = test ratio = 0.5) and 100 iterations.
The classifiers are ranked by level of adjusted accuracy (Adj. Acc). Results are shown as mean ± standard deviation, and only methods with Adj. Acc.[02D7E]65% are shown. Abbreviations: FS = feature selection, B = backward, F = forward, Sens. = sensitivity, Spec. = specificity.
| Rank | Methods | Options | FS | Adj. Acc. (%) | Acc. (%) | Sens. (%) | Spec. (%) | Measurements | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All OA vs. controls | |||||||||||
| 1 | DIA | Quadratic | F | 71.17±6.91 | 76.0±5.8 | 76.4±10.3 | 75.2±10.8 | Min[STD]IRW | |||
| 2 | LNR | Quadratic | F | 68.65±10.19 | 75.1 ±7.5 | 76.6±11.3 | 72.7±15.7 | Min[STD]IRW | |||
| 3 | SVM | Linear | F | 68.17±10.32 | 74.2±7.7 | 71.9±12.8 | 77.8±15.1 | Mean[MEAN]IRW | Min[STD]IRW | ||
| 4 | LNR | Linear | B | 67.22±11 | 74.0±8.7 | 76.4±12.2 | 70.3±16.8 | Mean[MEAN]R3D Min[MEAN]IRW |
Min[STD]R3D Max[MEANIRW |
Max[STD]R3D Mean[MEAN]IRW |
Mean[STD]R3D |
| 5 | NAB | B | 66.51 ±9.45 | 71.9±7.7 | 71.6±11.9 | 72.4±13.4 | Mean[STD]R3D | Min[STD]IRW | Mean[STD]IRW | ||
| 6 | LGR | Linear | F | 66.40±11.60 | 75.5±7.0 | 82.0±10.1 | 65.3±18.2 | Mean[MEAN]IRW | Min[STD]IRW | ||
| DIA | Linear | B | 66.40±11.70 | 72.8±9.7 | 73.6±14.3 | 71.4±15.4 | Mean[MEAN]R3D Min[MEAN]IRW |
Min[STD]R3D Max[MEAN]IRW |
Max[STD]R3D Mean[MEAN]IRW |
Mean[STD]R3D | |
| 7 | NAB | F | 66.07±10.51 | 71.9±8.1 | 69.9±12.7 | 75.1±14.9 | Min[STD]IRW | Max[STD]IRW | |||
| 8 | DIA | Linear | F | 65.56±10.79 | 72.9±7.0 | 76.2±17.9 | 67.7±17.3 | Min[MEAN]R3D | Min[STD]R3D | Min[STD]IRW | |
| 9 | SVM | Quadratic | F | 65.24±9.82 | 71.4±8.1 | 70.1±12.5 | 73.4±16.3 | Max[STD]R3D | Min[STD]IRW | Max[STD]IRW | |
| Early OA vs. controls | |||||||||||
| 1 | DIA | Linear | F | 73.38±8.36 | 78.6±6.9 | 80.5±11.4 | 76.3±11.9 | Min[STD]IRW | |||
| 2 | SVM | Linear | F | 68.8±12.18 | 75.6±9.1 | 77.3±14.1 | 73.6±16.9 | Min[MEAN]IRW | Min[STD]IRW | ||
| 3 | LGR | Linear | F | 67.33±15.29 | 75.2±10.0 | 81.3±12.8 | 67.8±20.3 | Mean[MEAN]IRW | Min[STD]IRW | ||
| 4 | NAB | B | 67.16±11.31 | 73.7±8.4 | 76.2±13.1 | 70.7±16.1 | Min[MEAN]IRW | Min[STD]IRW | |||
| 5 | LNR | Linear | B | 65.65±9.42 | 71.7±7.7 | 71.0±13.5 | 72.7±14.3 | Min[STD]R3D | Mean[STD]R3D | Max[MEAN]IRW | Mean[MEAN]IRW |
| 6 | LNR | Linear | F | 65.38±12.18 | 72.8±8.0 | 77.4±11.9 | 67.2±17.8 | Max[MEAN]R3D | Mean[MEAN]IRW | Min[STD]IRW | |
Tables with more detailed results are shown on Supplementary Material (Tables S1 to S6). Results from PCA are shown in Supplementary Material (Fig. S1 and S2, and Tables S7 and S8). We can observe in the 2D space of the two first principal components (biplot Fig. S2) that the variables from IRW are regrouped together (showing therefore moderate to strong correlation between them), and the variables from R3D are also regrouped together (also showing therefore moderate to strong correlation between them). Variables from IRW and R3D show weak correlation (the angles between the lines are close to 90°). We can also detect that Min[STD]IRW and Mean[STD]IRW have the slightly longest lines in this 2D space (each line represents the approximate variance of the variable). Classifications were applied on the 4 first principal components calculated from PCA, which correspond altogether to 91% of the variance of the whole data initially represented by the 12 sodium variables. Results of the classifications are presented in Tables S7 and S8, but did not show any improvements compared to classifications with single variables (such as Min[STD]IRW and Mean[STD]IRW) or feature selection. Adjusted accuracies (and simple accuracies) from classification using principal components were all below the values obtained from data after feature selection, for both all OA and early OA data versus control data: <63% adjusted accuracy (<71% accuracy) for PCA, 65–73% adjusted accuracy (71–79% accuracy) for feature selection.
ROC analysis was also performed for all classifications of all OA vs. controls data with the Min[STD]IRW feature only (as it was found the more relevant feature in this study), with holdout cross-validation and 100 iterations. Mean ROC curves and mean AUCs over these 100 iterations are presented in Fig. S3 and S4 of supplementary material. It was observed that the eight best classifiers, with AUC in the range 0.80–0.85, were the same as the eight best classifiers ranked from the (adjusted) accuracy viewpoint, as shown in Table 5, through not exactly in the same order.
DISCUSSION AND CONCLUSIONS
Results from the classification with data resubstitution and individual predictors confirm the results from our previous analysis [13] where Min[STD]IRW was found the best predictor for differentiating controls from both all OA and early OA patients, using logistic regression as a classifier. In the present analysis, surprisingly, weak learner classifiers such as KNN, DTR, and its extension TBG, generated the highest adjusted accuracies for classification, for both data including all OA and early OA, and with different predictors from IRW or R3D data. Other classifiers also generated reasonable adjusted accuracies that can still put them as good candidates for optimum classification with cross-validation and for multivariate analysis. When using the classifiers in the training/testing framework (cross-validation on individual predictors) however, the tendency was inverted and LGR, DIA, LNR and SVM performed better than DTR, KNN and TBG, all with the same predictor Min[STD]IRW. This shows that these latter classifiers are strong learners and present more robust and consistent results with our data. As previously found in Ref. [13], synovial fluid suppression (by IR in our case) was found primordial for allowing to differentiate healthy cartilage from cartilage of patient with a diagnosis of OA (both early OA KL1–2 or all OA late KL 1–4) when using sodium MRI. Fluid suppression reduces significantly partial volume effect in the images and allows to measure sodium (and GAG) content in cartilage more accurately.
After feature selection, two main classifiers were found to give some of the highest adjusted accuracies, in common for both early OA and all OA data: linear SVM and linear LGR, with only two predictors involved: Min[STD]IRW associated with either Mean[MEAN]IRW or Min[MEAN]IRW. As these two [MEAN]IRW predictors are strongly correlated (R2~0.82), we can consider using either of them for future classifications. Mean[MEAN] (with or without fluid suppression) was usually used in previous sodium MRI studies as the main variable for detecting loss of GAG in cartilage (with generally a threshold of around 220 mM such as Mean[MEAN]<220 mM ≡ OA, Mean[MEAN]>220 mM ≡ control) [1, 12, 13, 43]. Due to their similar standard deviations over 100 iterations (~10%), we can consider that either linear LGR, quadratic DIA, linear DIA, and linear LNR with single predictor Min[STD]IRW, or linear SVM and linear LGR with two predictors Min[STD]IRW and Mean[MEAN]IRW (or Min[MEAN]IRW) give very similar results and can be useful classifiers from machine learning for classifying OA patients and controls with this kind of sodium MRI data. PCA did not improve the classification accuracies compared to feature selection. Moreover, PCA results can generally be difficult to interpret from a biological point-of-view. A biological interpretation would be more practical for understanding the disease and for comparing methods for its detection and/or grading assessment, or for understanding the effects of potential drugs or repair operation on the cartilage. In our study, we found that measuring the minimum variation of sodium concentration in cartilage Min[STD]IRW over different cartilage regions in the knee seems to be a good indicator of OA. One reason advanced in Ref. [13] was that the range of GAG (and sodium) concentrations within healthy cartilage is higher compared to OA cartilage, with high concentrations in the radial zone (next to the bone) and lower concentrations in the tangential zone (near the surface). Combining Min[STD]IRW with Mean[MEAN]IRW or Min[MEAN]IRW could therefore probably improve the performance of sodium MRI for assessing OA, as it will give information on both the mean and the variance of GAG content within articular cartilage. Further investigations are needed to assess this hypothesis, either on animal model of OA, or by comparing sodium MRI data with diffusion MRI [46] and dGEMRIC [5] data acquired on a larger population of controls and OA patients with different KL grades. This will be the next step of our study.
The difference of performance for the different classifiers used in this study can be due to their bias-variance trade-off characteristics, when applied to this kind of small size data (12 variables of 47 data values). Bias can be defined as the tendency of the learning algorithm to “consistently learn the same wrong thing” [47], or the error introduced due to approximation of complex problems by a much simpler model [24]. The variance can be defined as the tendency of the learning algorithm to “consistently learn random things irrespective of the real signal” [47], or the error due to the amount of change of the learning algorithm if it is estimated using a different training dataset [24]. Basically, bias is a measure of how much far off is the model’s prediction from the correct value, while variance is a measure of how much different predictions from the model vary. Flexible (non-parametric) classifiers – such as DTR, KNN, NNE and TBG – have generally low bias/high variance and will have a tendency to overfit the data. These classifiers will therefore perform best when using resubsitution (train data = test data = all data), but will generate a higher error rate when applied with cross-validation, as they overfit the train dataset and thus misclassify the test dataset. This is what we observed with our data: DTR, KNN, NNE and TBG had the highest accuracies with resubstitution. On the other hand, these classifiers performed poorly with cross-validation, contrarily to less flexible parametric classifiers – such as LNR, LGR, DIA, SVM and NAB. These latter learning algorithms have generally high bias/low variance and therefore don’t overfit the training data (they even probably underfit it) and generate lower misclassification of the testing data. As a consequence, when using a small train dataset as in our case, it is recommended to use such low variance methods for classification.
In addition to the bias-variance trade-off and overfitting characterictics of the different methods, failures in the classifications may come from many different factors (maximum adjusted accuracy obtained was~74% with holdout cross-validation): wrong diagnosis and grading of OA from radiographs, wrong classification due to the learning method (small size of training dataset), inaccurate sodium quantification due to noisy MRI data, inaccurate sodium calibration, subject movement, RF coil inefficiency. We expect that errors from sodium data acquisition (sequence, calibration, linear regression, relaxation corrections) are probably the main sources of failure of the classifications, and work to significantly improve the signal-to-noise ratio and the resolution of the sodium images is under progress (new sequences [48,49], new images reconstruction methods [50]).
A significant improvement of the classification would include a multiparametric approach from different MRI acquisitions as suggested in Ref. [18], including sodium MRI along proton MRI data such as such as apparent diffusion coefficient (ADC, linked to GAG content) and fractional anisotropy (FA, linked to collagen matrix) from DTI [46], T2 and T1ρ, dGEMRIC data, or magnetization transfer ratio, in the machine learning algorithm in order to increase the (adjusted) accuracy, sensitivity and specificity, and thus help the diagnosis of early OA or even help understand what is OA from an imaging point-of-view [51]. T1 and T2 (or T2*) sodium relaxation times measurements could also help improve the accuracy of sodium MRI for detecting early signs of OA. These relaxation times are highly correlated with the electric field gradients surrounding the sodium ions, through quadrupolar interaction, and therefore depend on the concentration of GAG molecules and also on collagen architecture of the extracellular matrix of cartilage.
This study was performed on a limited number of subjects (19 controls and 28 OA with different KL scores), but preliminary results show that machine learning could be a promising method for assessing diseases (OA in joint cartilage in our case) from MRI data in an automatic and objective manner. More controls and OA patients with different symptoms (and KL grade) must be scanned with sodium MRI alongside proton MRI (DTI, dGEMRIC, T2, T1ρ) for further validating the technique. As multiparametric MRI data might be long to acquire, we expect to improve the speed of acquisition for sodium MRI with compressed sensing reconstruction of undersampled data [50,52].
In conclusion, either Min[STD] alone or in combination with Mean[MEAN] or Min[MEAN], all from fluid suppressed data, were the best predictors with an accuracy >74%, mainly with linear LGR and linear SVM. Other good classifiers include DIA, LNR and NAB. A robust and accurate machine learning classification method based on sodium MRI data could therefore help detecting early signs of OA, assessing treatment follow-ups from cartilage repair procedures [53] or disease modifying osteoarthritis drug (DMOAD) [54] under test. On the long term, sodium MRI could be implemented as a complement to other imaging methods (radiography, proton MRI) associated with machine learning for increasing the objectivity of the decision making process by radiologists.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grant nos. 1R01AR053133, 1R01AR056260, 1R01AR060238, and 1R03AR065763.
References
- 1.Borthakur A, Mellon E, Niyogi S, Witschey W, Kneeland JB, Reddy R. Sodium and T1ρ MRI for molecular and diagnostic imaging of articular cartilage. NMR in Biomedicine. 2006;19(7):781–821. doi: 10.1002/nbm.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith HE, Mosher TJ, Dardzinski BJ, Collins BG, Collins CM, Yang QX, Schmithorst VJ, Smith MB. Spatial variation in cartilage T2 of the knee. Journal of Magnetic Resonance Imaging. 2001;14(1):50–55. doi: 10.1002/jmri.1150. [DOI] [PubMed] [Google Scholar]
- 3.Akella SV, Reddy Regatte R, Gougoutas AJ, Borthakur A, Shapiro EM, Kneeland JB, Leigh JS, Reddy R. Proteoglycan-induced changes in T1ρ-relaxation of articular cartilage at 4T. Magnetic resonance in medicine. 2001;46(3):419–423. doi: 10.1002/mrm.1208. [DOI] [PubMed] [Google Scholar]
- 4.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proceedings of the National Academy of Sciences. 2008;105(7):2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashir A, Gray ML, Burstein D. Gd-DTPA2- as a measure of cartilage degradation. Magnetic resonance in medicine. 1996;36(5):665–673. doi: 10.1002/mrm.1910360504. [DOI] [PubMed] [Google Scholar]
- 6.Filidoro L, Dietrich O, Weber J, Rauch E, Oerther T, Wick M, Reiser M, Glaser C. High-resolution diffusion tensor imaging of human patellar cartilage: Feasibility and preliminary findings. Magnetic Resonance in Medicine. 2005;53(5):993–998. doi: 10.1002/mrm.20469. [DOI] [PubMed] [Google Scholar]
- 7.Reddy R, Insko EK, Noyszewski EA, Dandora R, Kneeland JB, Leigh JS. Sodium MRI of human articular cartilage in vivo. Magnetic resonance in medicine. 1998;39(5):697–701. doi: 10.1002/mrm.1910390505. [DOI] [PubMed] [Google Scholar]
- 8.Gold GE, Chen CA, Koo S, Hargreaves BA, Bangerter NK. Recent advances in MRI of articular cartilage. AJR American journal of roentgenology. 2009;193(3):628. doi: 10.2214/AJR.09.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madelin G, Regatte RR. Biomedical applications of sodium MRI in vivo. J Magn Reson Imaging. 2013;38:511–529. doi: 10.1002/jmri.24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro EM, Borthakur A, Gougoutas A, Reddy R. 23Na MRI accurately measures fixed charge density in articular cartilage. Magnetic Resonance in Medicine. 2002;47(2):284–291. doi: 10.1002/mrm.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesperance LM, Gray ML, Burstein D. Determination of fixed charge density in cartilage using nuclear magnetic resonance. Journal of orthopaedic research. 1992;10(1):1–13. doi: 10.1002/jor.1100100102. [DOI] [PubMed] [Google Scholar]
- 12.Madelin G, Lee JS, Inati S, Jerschow A, Regatte RR. Sodium inversion recovery MRI of the knee joint in vivo at 7T. Journal of Magnetic Resonance. 2010;207(1):42–52. doi: 10.1016/j.jmr.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madelin G, Babb J, Xia D, Chang G, Krasnokutsky S, Abramson SB, Jerschow A, Regatte RR. Articular cartilage: evaluation with fluid-suppressed 7.0-T sodium MR imaging in subjects with and subjects without osteoarthritis. Radiology. 2013;268(2):481–491. doi: 10.1148/radiol.13121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sajda P. Machine learning for detection and diagnosis of disease. Annu Rev Biomed Eng. 2006;8:537–565. doi: 10.1146/annurev.bioeng.8.061505.095802. [DOI] [PubMed] [Google Scholar]
- 15.Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RS. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: a tutorial overview. Neuroimage. 2009;45(1):S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, Davatzikos C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magnetic Resonance in Medicine. 2009;62(6):1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin PC, Reiter DA, Spencer RG. Classification of degraded cartilage through multiparametric MRI analysis. Journal Of Magnetic Resonance. 2009;201(1):61–71. doi: 10.1016/j.jmr.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin PC, Irrechukwu O, Roque R, Hancock B, Fishbein KW, Spencer RG. Multivariate analysis of cartilage degradation using the support vector machine algorithm. Magnetic Resonance in Medicine. 2012;67(6):1815–1826. doi: 10.1002/mrm.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy KP. Machine learning: a probabilistic perspective. MIT Press; 2012. [Google Scholar]
- 21.Barber D. Bayesian reasoning and machine learning. Cambridge University Press; 2012. [Google Scholar]
- 22.Hastie T, Tibshirani R, Friedman J. Data mining, inference and prediction. Vol. 2. Springer; 2009. The elements of statistical learning. [Google Scholar]
- 23.Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. MIT Press; 2012. [Google Scholar]
- 24.James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning. Springer; 2013. [Google Scholar]
- 25.Kuhn M, Johnson K. Applied predictive modeling. Springer; 2013. [Google Scholar]
- 26.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- 27.Cover T, Hart P. Nearest neighbor pattern classification. Information Theory, IEEE Transactions on. 1967;13(1):21–27. [Google Scholar]
- 28.Lewis DD. In Machine learning: ECML-98. Springer; 1998. Naive (Bayes) at forty: The independence assumption in information retrieval; pp. 4–15. [Google Scholar]
- 29.Fisher RA. The use of multiple measurements in taxonomic problems. Annals of eugenics. 1936;7(2):179–188. [Google Scholar]
- 30.McCullagh P. Generalized linear models. European Journal of Operational Research. 1984;16(3):285–292. [Google Scholar]
- 31.McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. The bulletin of mathematical biophysics. 1943;5(4):115–133. [PubMed] [Google Scholar]
- 32.De Boer PT, Kroese DP, Mannor S, Rubinstein RY. A tutorial on the cross-entropy method. Annals of operations research. 2005;134(1):19–67. [Google Scholar]
- 33.Safavian SR, Landgrebe D. A survey of decision tree classifier methodology. IEEE transactions on systems, man, and cybernetics. 1991;21(3):660–674. [Google Scholar]
- 34.Breiman L. Bagging predictors. Machine learning. 1996;24(2):123–140. [Google Scholar]
- 35.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke T, Greenwald R, Hochberg M, et al. Development of criteria for the classification and reporting of osteoarthritis: classification of osteoarthritis of the knee. Arthritis & Rheumatism. 1986;29(8):1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 36.Kellgren J, Lawrence J. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16(4):494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown R, Madelin G, Lattanzi R, Chang G, Regatte RR, Sodickson DK, Wiggins GC. Design of a nested eight-channel sodium and four-channel proton coil for 7T knee imaging. Magnetic Resonance in Medicine. 2013;70(1):259–268. doi: 10.1002/mrm.24432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielles-Vallespin S, Weber MA, Bock M, Bongers A, Speier P, Combs SE, Wöhrle J, Lehmann-Horn F, Essig M, Schad LR. 3D radial projection technique with ultrashort echo times for sodium MRI: clinical applications in human brain and skeletal muscle. Magnetic resonance in medicine. 2007;57(1):74–81. doi: 10.1002/mrm.21104. [DOI] [PubMed] [Google Scholar]
- 39.Dutt A, Rokhlin V. Fast Fourier transforms for nonequispaced data. SIAM Journal on Scientific computing. 1993;14(6):1368–1393. [Google Scholar]
- 40.Kupce E, Freeman R. Adiabatic pulses for wideband inversion and broadband decoupling. Journal of Magnetic Resonance, Series A. 1995;115(2):273–276. [Google Scholar]
- 41.Madelin G, Jerschow A, Regatte RR. Sodium relaxation times in the knee joint in vivo at 7T. NMR in Biomedicine. 2012;25(4):530–537. doi: 10.1002/nbm.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borthakur A, Shapiro EM, Akella SV, Gougoutas A, Kneeland JB, Reddy R. Quantifying sodium in the human wrist in vivo by using MR imaging. Radiology. 2002;224(2):598–602. doi: 10.1148/radiol.2242011039. [DOI] [PubMed] [Google Scholar]
- 43.Wheaton AJ, Borthakur A, Shapiro EM, Regatte RR, Akella SV, Kneeland JB, Reddy R. Proteoglycan loss in human knee cartilage: quantitation with sodium MR imaging-feasibility study. Radiology. 2004;231(3):900–905. doi: 10.1148/radiol.2313030521. [DOI] [PubMed] [Google Scholar]
- 44.Kim JH. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Computational Statistics & Data Analysis. 2009;53(11):3735–3745. [Google Scholar]
- 45.Blum A, Kalai A, Langford J. Beating the hold-out: Bounds for k-fold and progressive cross-validation; In Proceedings of the twelfth annual conference on Computational learning theory. ACM; 1999. pp. 203–208. [Google Scholar]
- 46.Raya JG, Horng A, Dietrich O, Krasnokutsky S, Beltran LS, Storey P, Reiser MF, Recht MP, Sodickson DK, Glaser C. Articular cartilage: in vivo diffusion-tensor imaging. Radiology. 2012;262(2):550–559. doi: 10.1148/radiol.11110821. [DOI] [PubMed] [Google Scholar]
- 47.Domingos P. A few useful things to know about machine learning. Communications of the ACM. 2012;55(10):78–87. [Google Scholar]
- 48.Pipe JG, Zwart NR, Aboussouan EA, Robison RK, Devaraj A, Johnson KO. A new design and rationale for 3D orthogonally oversampled k-space trajectories. Magnetic Resonance in Medicine. 2011;66(5):1303–1311. doi: 10.1002/mrm.22918. [DOI] [PubMed] [Google Scholar]
- 49.Boada FE, Gillen JS, Shen GX, Chang SY, Thulborn KR. Fast three dimensional sodium imaging. Magnetic Resonance in Medicine. 1997;37(5):706–715. doi: 10.1002/mrm.1910370512. [DOI] [PubMed] [Google Scholar]
- 50.Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed sensing MRI. Signal Processing Magazine, IEEE. 2008;25(2):72–82. [Google Scholar]
- 51.Hunter D, Arden N, Conaghan P, Eckstein F, Gold G, Grainger A, Guermazi A, Harvey W, Jones G, Hellio Le Graverand M, et al. Definition of osteoarthritis on MRI: results of a Delphi exercise. Osteoarthritis and Cartilage. 2011;19(8):963–969. doi: 10.1016/j.joca.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madelin G, Chang G, Otazo R, Jerschow A, Regatte RR. Compressed sensing sodium MRI of cartilage at 7T: preliminary study. Journal of Magnetic Resonance. 2012;214:360–365. doi: 10.1016/j.jmr.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang G, Sherman O, Madelin G, Recht M, Regatte R. MR imaging assessment of articular cartilage repair procedures. Magnetic resonance imaging clinics of North America. 2011;19(2):323. doi: 10.1016/j.mric.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qvist P, Bay-Jensen AC, Christiansen C, Dam EB, Pastoureau P, Karsdal MA. The disease modifying osteoarthritis drug (DMOAD): Is it in the horizon? Pharmacological Research. 2008;58(1):1–7. doi: 10.1016/j.phrs.2008.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.