Abstract
A common strategy in mass spectrometry analyses of complex protein mixtures is to digest the proteins to peptides, separate the peptides by microcapillary liquid chromatography and collect tandem mass spectra (MS/MS) on the eluting, complex peptide mixtures, a process commonly termed “shotgun proteomics”. For years, the most common way of data collection was via data-dependent acquisition (DDA), a process driven by an automated instrument control routine that directs MS/MS acquisition from the highest abundant signals to the lowest, a process often leaving lower abundant signals unanalyzed and therefore unidentified in the experiment. Advances in both instrumentation duty cycle and sensitivity allow DDA to probe to lower peptide abundance and therefore enable mapping proteomes to a more significant depth. An alternative to acquiring data by DDA is by data-independent acquisition (DIA), in which a specified range in m/z is fragmented without regard to prioritization of a precursor ion or its relative abundance in the mass spectrum. As a consequence, DIA acquisition potentially offers more comprehensive analysis of peptides than DDA and in principle can yield tandem mass spectra of all ionized molecules following their conversion to the gas-phase. In this work, we evaluate both DDA and DIA on three different linear ion trap instruments: an LTQ, an LTQ modified in-house with an electrodynamic ion funnel, and an LTQ-Velos. These instruments were chosen as they are representative of both older (LTQ) and newer (LTQ-Velos) ion trap designs i.e., linear ion trap and dual ion traps, respectively, and allow direct comparison of peptide identification using both DDA and DIA analysis. Further, as the LTQ-Velos has an improved “S-lens” ion guide in the high-pressure region to improve ion flux, we found it logical to determine if the former LTQ model could be leveraged by improving sensitivity by modifying with an electrodynamic ion guide of significantly different design to the S-lens. We find that the ion funnel enabled LTQ identifies more proteins in the insoluble fraction of a yeast lysate than the other two instruments in DIA mode, while the faster scanning LTQ-Velos performs better in DDA mode. We explore reasons for these results, including differences in scan speed, source ion optics, and linear ion trap design.
Introduction
Shotgun proteomics by liquid chromatography tandem mass spectrometry (LC-MS/MS) is the common paradigm for characterizing proteins in complex mixtures [1]. Proteins are commonly digested to peptides and the complex peptide mixtures separated by microcapillary nanoflow liquid chromatography followed by the acquisition of tandem mass spectra of the eluting peptides using an approach known as data-dependent acquisition (DDA). In DDA, the most abundant peptides in a mass spectrum are selected for fragmentation automatically by the instrument data system using an instrument control language [2]. The approach is very powerful, especially when coupled with numerous offline and online sample fractionation approaches, and the advent of robust, commercially available, fast-scanning and sensitive mass spectrometers. However, there are some disadvantages to using DDA methods. For example, while DDA offers a superficially logical and directed way of acquiring many peptide tandem mass spectra in a complex mixture, it is accepted that this method results in a stochastic random sampling of peptides, due largely to biases caused by poor control of ion selection with respect to order, retention time, and ion abundance at the point of selection [3]. With complex peptide mixtures, where under sampling of eluting peptides is common, intra-assay variation of peptide identification can change by at least 50% from replicate to replicate injections (i.e. technical replicates), often leading to performing the same experiment on the same sample multiple times to raise repeatability of the results and confidence for the analyst. While this is not necessarily a problem for a purely discovery-based experiment, in which the number and identity of constituent proteins are the most important results, it is a major problem for quantitative proteomics experiments and investigators seeking precise measurement of differential protein expression. Furthermore, while DDA methods are always improving, the analyses are rarely comprehensive with respect to sample dynamic range often leaving low abundant peptides in the sample uncharacterized [4]. Overall, there is a need to improve the throughput of comprehensive proteomics analyses which still remain slow relative to comparable genomics technologies [5].
As an alternative to DDA, data-independent acquisition (DIA) methods are based on activation of all ions within a defined mass-to-charge (m/z) isolation window regardless of their detected relative ion abundance [6]. This unique way of acquiring data departs from DDA approaches and seeks to improve the depth and coverage of proteomic analyses as well as the overall reproducibility of the experiment. The term “data-independent acquisition” was first coined by Venable et al. who used a Thermo Scientific LTQ ion trap mass spectrometer employing collision-induced dissociation (CID) isolation windows of 10 m/z acquired successively across a desired m/z range [7]. Peptides were identified by database search by considering the center of the isolation window as the peptide parent mass. Earlier, the concept of activating and fragmenting the entire m/z range was explored by Purvine et al. whose “shotgun CID” method demonstrated feasibility to perform parallel peptide sequencing using a quadrupole time-of-flight (Q-TOF) instrument [8]. This concept was further developed by Silva et al. and commercially developed by Waters Corporation on their Q-TOF instrument platforms [9, 10]. Coined MSE, the method employs alternating acquisitions of mass spectra followed by full spectrum activation to produce CID mass spectra. Fragment ions are related to precursor ions for peptide identification through reconstruction and alignment of ion chromatograms using a proprietary software algorithm. Currently, the approach has been further advanced by combining MSE data acquisition with ion mobility separations [11], a technique pioneered by Clemmer and co-workers and previously demonstrated for peptide separations to reduce chemical noise and improve sample dynamic range [12, 13]. A similar parallel peptide sequencing technique called All Ion Fragmentation (AIF) was recently reported for specific use with Thermo’s Orbitrap Exactive [14].
More recently Panchaud et al. modified the concept described by Venable by leaving the systematic data-independent isolation of m/z windows intact, but by using m/z isolation windows that are typical of those found in DDA methods i.e. < 3.0 Th [15]. A primary motivation was to reduce the extent of chimeric precursor ions selected during the MS/MS event as to allow the use of common, established database search engines for peptide identification. This approach, coined Precursor Acquisition Independent from Ion Count (PAcIFIC), benefited under the strict constraint that the number of MS/MS events per m/z range interrogated needed to fall within a typical peptide chromatographic peak width and hence necessitated a large amount of instrument time. The approach has benefited greatly from the development of instruments with faster duty cycles, with notable improvements moving from the linear ion trap (LTQ) to the dual ion trap (Velos) [16] and quadrupole orbitrap hybrid (Q-Exactive) [17] instruments. Recently, Aebersold and coworkers took the DIA concept and developed a targeted informatics approach for data analysis [18]. The method, coined SWATH and currently adapted to AB Sciex Triple-TOF instrument platforms, acquires tandem mass spectra over moderately large m/z windows (e.g. 25 Th) but uses reference library spectra (e.g. PeptideAtlas) to extract peptide identifications from the acquired spectra. Similarly, Bruce and co-workers developed another combined DIA informatics approach for use with high mass accuracy data called FT-All Reaction Monitoring (FT-ARM) [19]. A notable difference over SWATH, AIF, and MSE is that FT-ARM does not utilize peptide chromatographic elution profiles in the analysis.
Concurrent with the developments in data acquisition strategies have been improvements in ion source transmission efficiency. A large percentage of molecules in solution are lost during the process of ionizing at atmospheric pressure and subsequent transmission into the high vacuum region of the mass spectrometer [20–22]. A significant improvement to this problem has been the emergence of the electrodynamic ion funnel, an ion guide that specifically addresses and corrects ion losses that occur while transmitting ions formed at atmospheric pressure through the pressure gradient of the first differentially-pumped vacuum region of the mass spectrometer [23, 24]. The ion funnel replaces more traditional interfaces, usually consisting of a nozzle/skimmer device, with a stacked-ring ion guide with large ion acceptance area. By utilizing co-application of RF and DC electric fields, the expanding ion plume can be captured and refocused to a collimated ion beam ensuring near 100% transmission of ions from the first vacuum region of the mass spectrometer to downstream ion optics and detector [24–26].
While improving ion transmission efficiency has obvious advantages and sensitivity improvements to beam-based instruments (e.g. triple quadrupole mass spectrometers), advantages for pulsed-source ion trap instruments are less clear. A particular advantage of Thermo linear ion trap instruments is the time during which ions are allowed to accumulate in the trap, the ion injection time, can be varied to keep the ion population constant from scan to scan [27]. This has the effect of producing high quality tandem mass spectra from both low and high abundance molecular precursor ions with the only significant difference reflected in the corresponding ion injection times. The improvement offered by installing an ion funnel on an ion trap instrument is therefore manifested as a reduction in time taken to fill the trap [28]. Moreover, since DIA strategies focus on acquiring data regardless of precursor signal intensity, improvements in source efficiency would likely provide a more measurable benefit to identifying signals of low relative ion abundance than do DDA approaches which prioritize exclusively on the most abundant signals. The Thermo LTQ-Velos instrument incorporates technology improvements over the previous LTQ design, including using a stacked-ring ion guide called the “S-lens” which replaced the LTQ’s tube lens-skimmer configuration [29]. The RF-driven S-lens is similar in principle to the ion funnel but with the notable difference is that it lacks a DC field component. Software improvements to the LTQ-Velos enable elimination of the MS/MS prescan and instead predict the ion injection time using the signal from the MS precursor scan, thus saving time for each MS/MS spectrum acquired. Most significantly, the LTQ-Velos incorporates a dual-pressure ion trap which enables decoupling of ion accumulation and fragmentation, effectively analyzing ions at higher speeds than in previous single-trap instruments. Overall, these improvements result in an increase in instrument duty cycle of at least 50%, enabling significant improvements in the context of proteomics experiments on complex protein mixtures [30].
In this work we use three different Thermo Scientific linear ion trap mass spectrometers of different configuration to explore the effects of instrumentation differences and data acquisition strategies within the context of shotgun proteomics. These instruments include an unmodified LTQ, an LTQ modified in-house with an electrodynamic ion funnel, and an unmodified LTQ-Velos. We employ both data-dependent and data-independent acquisition strategies on each instrument. The merits of different data acquisition approaches are compared and placed into context with the respective instrument hardware. Note that it is not our intention to offer an exhaustive comparison of DDA and DIA; rather, this work is focused on evaluating our chosen hardware platforms in the context of these two data acquisition strategies to offer data to those seeking alternatives to DDA for comprehensive protein identification and quantitative proteomics applications.
Experimental
This work is divided into two parts. In the first part (Part A), we performed a typical shotgun proteomics experiment, using DIA and DDA methods on each of three different instrument types. In the second part (Part B), we performed direct infusion experiments on each of the instrument platforms to help explain and explore the results of the comparisons in Part A.
Part A: Sample preparation and liquid chromatography-tandem mass spectrometry (LC-MS/MS)
The ammonium bicarbonate-insoluble protein fraction of a whole-cell lysate of wild-type yeast (Saccharomyces cerevisiae) was denatured with 0.1% RapiGest, reduced with 5 mM dithiothreitol, and incubated at 60°C for 30 minutes. After cooling, the lysate was alkylated with 15 mM iodoacetamide. The alkylated sample was digested to peptides using trypsin, with an enzyme-substrate ratio of 1:50, for one hour at 37°C with shaking. RapiGest was cleaved by the addition of 200 mM HCl, at 37°C for 45 minutes with shaking. The resulting peptide solution was desalted and particulates removed by passing it through a mixed-mode cation exchange column (Oasis MCX, Waters) according to the manufacturer’s instructions. Liquid chromatography was performed using 40 cm microcapillary fused-silica columns (75 µm ID) packed with Jupiter C12 resin (Phenomenex), pulled in-house to a tip of ∼5 µm using a laser puller (Sutter Instruments). Samples were run on a linear gradient of 9% to 32% acetonitrile for 95 minutes, using an Agilent 1100 binary pump, at flow rate of ∼300 nL/min.
LC-MS/MS experiments were run in DDA and DIA mode (see below) on three different instruments: (1) an unmodified linear ion trap (LTQ, Thermo Scientific); (2) a linear ion trap (LTQ) modified with the addition of an electrodynamic ion funnel; (3) a dual linear ion trap (LTQ Velos, Thermo Scientific). The front-end optics of each instrument were tuned using the 3+ ion (m/z ∼ 433) of a 1 µM solution of human angiotensin I (Sigma) in 50:49.9:0.1 (v/v/v) water:methanol:formic acid. The MS/MS ion target was set to 2,000 ions for all experiments. The ESI voltage was applied via liquid junction electrode, in a source arrangement built in-house and available through the University of Washington’s Proteomics Resource (details found at http://proteomicsresource.washington.edu/nsisource.php).
Part A: Data-independent and data-dependent acquisition
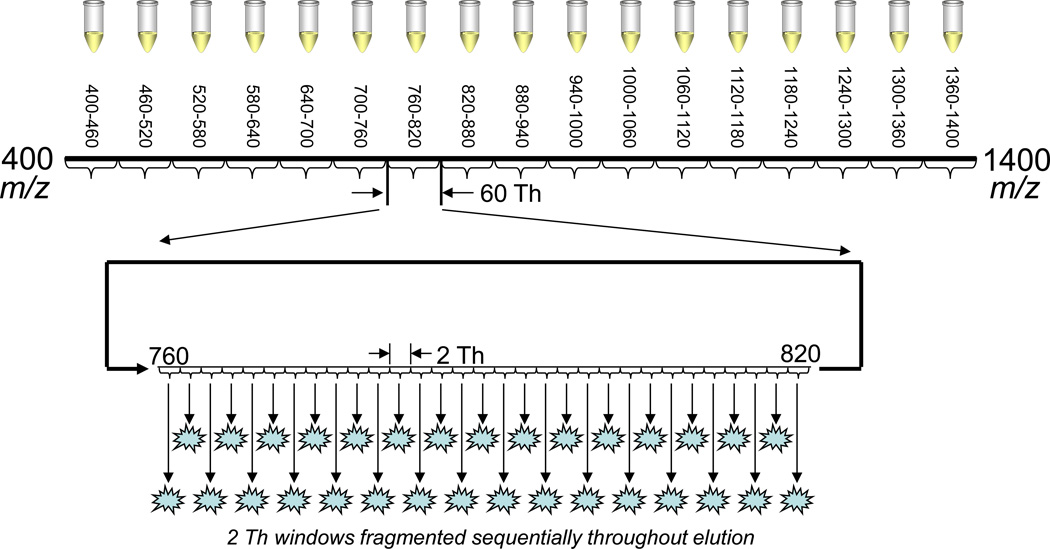
Data-independent acquisition (DIA) was implemented using a fragmentation window of 2 Th and designed according to the PAcIFIC method [15]. The 2-Th window size yielded ∼30 successive mass windows in < 6 seconds (depending on the instrument) and covering a total m/z range of 60 Th, as shown in Figure 1. These 30 windows were sampled sequentially and repeatedly throughout a single sample injection and chromatographic run. In all, 17 separate sample injections were required to cover the m/z range 400–1400. All dynamic exclusion features were turned off in DIA mode. Data-dependent acquisition (DDA) was implemented as a “top 5” experiment: one precursor scan covering m/z 400–1400 followed by CID spectra targeting the top five most intense ions in the precursor spectrum. Sampled peaks were added to a dynamic exclusion list of 30 second duration, repeat count 1, and list length of 50. DDA experiments were performed in 17 replicates to compare with the DIA experiments. In both DDA and DIA mode, CID event parameters were (on all instruments): isolation width, 2 Th; collision energy, 35%; activation q, 0.25; activation time, 30 ms.
Figure 1.
Data-independent acquisition (DIA) scheme. The full mass range is split into seventeen windows of 60 m/z range. Each of these windows is subjected to DIA with 2 Th windows throughout one chromatographic run. Thus each 60 Th window represents one sample injection, and 17 separate injections are required to cover the whole mass range from m/z 400– 1400.
Part A: Data analysis
Instrument data files were converted to the MS2 format [31] with MakeMS2, an in-house software program (available at http://proteome.gs.washington.edu/software.html). MS2 files were searched against a fasta file containing the S. cerevisiae open reading frames (Saccharomyces Genome Database, downloaded March 25, 2009) using SEQUEST [32]. SEQUEST output was processed with Percolator which computed q-values and posterior error probabilities at the peptide-spectrum match level [33]. Percolator output was loaded into MSDaPl (Mass Spectrometry Data Platform), an in-house Java-based platform for storing, viewing, and analyzing the output of shotgun proteomics experiments [34]. Peptides were assembled into proteins using a modified version of the IDPicker algorithm [35]. The entire data pipeline was also processed using a concatenated forward-reverse database, and false discovery rates at the protein level were estimated after setting a threshold based on a target q-value on the peptide spectrum match level.
Part B: Infusion MS
In addition to the three instruments described above, three additional instruments were used and configured similarly: an LTQ (the quadrupole ion trap portion of an LTQ-Orbitrap), an LTQ modified in-house with an ion funnel (the quadrupole ion trap portion of an LTQ-FT), and another standalone LTQ-Velos. Glu[1]-fibrinopeptide B (Sigma) was dissolved in 50:49.9:0.1 (v/v/v) water:methanol:formic acid to a concentration of 1 µM. The peptide solution was infused using the IonMax ESI probe (metal needle kit) and source housing (Thermo) with nitrogen sheath gas, ESI voltage, and source parameters optimized for the particular instrument in use. A separate syringe pump (Harvard Apparatus) supplied a sample flow rate of 3 µL/min. The same source, probe, tubing, syringe pump, and flow rate were used for all infusion experiments. The front-end optics of all instruments were tuned using the 2+ charge state of Glu[1]-fibrinopeptide B (m/z ∼ 786). The MS/MS ion target was set at 10,000 ions, and CID parameters were the same for all instruments.
Part B: Data analysis
Data acquired in Part B were analyzed as described below (see “Comparing Instrument Platforms").
Parts A and B: Ion funnel
An electrodynamic ion funnel was constructed based on the design reported by Page et al. [28]. The hardware (electrodes, vacuum housing, and fittings) was built at the University of Washington (Department of Physics Machine Shop), as were the associated electronics and power supplies (Department of Chemistry Electronics Shop). The funnel itself consists of a set of 100 brass electrodes with the first 58 electrodes having a fixed inner diameter of 25.4 mm and the remaining electrodes decreasing linearly to a final inner diameter of 2 mm. Electrode 20 was replaced with a “jet disrupter,” a 5 mm brass plate centered in the x-y plane of the electrode, serving to block neutral species from entering the downstream vacuum regions [36, 37]. RF waveforms (∼825 kHz, 75 Vpp) were applied to the electrodes via a capacitor network with each adjacent electrode alternating in RF phase by 180 degrees. A linear DC gradient was supplied to the electrodes via a resistor network with the entrance electrode set at ∼200 V and the exit electrode set to ∼5 V. A final conductance-limiting plate was biased at ∼2 V and was not part of the resistor gradient. The jet disrupter was biased at ∼175V and carried no RF voltage. The mass spectrometer's stainless steel transfer tube was biased at ∼210V and maintained at a temperature of 275°C. The pressure in the ion funnel region was approximately 1 Torr at all times with no additional pumping added to the system.
Results and Discussion
Comparing data acquisition approaches
The intent of this work was to evaluate three different hardware platforms in the context of two different data acquisition methods. As described above and in Figure 1, the DIA method developed required 17 separate sample injections to cover the peptide-rich m/z 400–1400 range. The window size of 2 Th was chosen to simplify the data analysis process [15], as the wider windows used in some previous DIA work introduce complications in traditional analysis methods due to the presence of multiple peptides in the same isolation window [7, 8]. As a comparison we chose to perform 17 separate sample injections using DDA covering the same mass range. Another comparison would be to perform DDA on the same windows as used for each of the DIA sample injections. While a gas-phase fractionation (GPF) approach [38] is indeed a possible comparison, it carries an important disadvantage, namely that restricting DDA to a short mass range while retaining dynamic exclusion parameters will lead to a situation in which the mass spectrometer spends large amounts of time without triggering any MS/MS events [39]. This effect, sometimes called “acquisition blackout”, can be reduced by removing the dynamic exclusion parameters to maximize the number of CID spectra acquired but will broadly lead to an increase in data redundancy (i.e. spectral counts) as the instrument control routine will focus on the signals of highest spectral abundance. Early in this study, we performed several such gas-phase fractionation experiments on smaller m/z windows, with the common result that DIA always did better than GPF in terms of number of identifications (data not shown). Based on these experiments, we chose instead to perform replicates of DDA experiments utilizing the whole mass range, exploiting DDA to maximize protein identification.
A typical metric in shotgun proteomics experiments is the number of protein identifications obtained in the context of a defined false-discovery rate. Table 1 displays the number of tandem mass spectra acquired, protein identifications, and percent overlap for each dataset: the composite of all runs for each instrument using either DDA or DIA. The protein identification data illustrate a clear trend for the data acquired by DIA mode where the ion funnel enabled LTQ (designated IF-LTQ) provided the largest number of identifications, 2275, as compared to 2091 for the LTQ-Velos and 2051 for the LTQ. Across all instruments, the DIA approach produced more protein identifications than DDA. The primary reason for the larger number of protein identifications for the IF-LTQ in DIA mode is that the ion funnel delivered improved ion transmission and therefore more ion flux as compared to the ion interfaces of the other instruments. In instruments with automatic gain control (AGC), as discussed in the next section, improved ion transmission will directly result in shorter ion injection times and will overall produce a higher proportion of tandem mass spectra with improved spectral quality compared to source designs with lower relative ion transmission efficiencies. Numerous ion funnel prototypes have been shown to transmit over ten times the ion currents over instruments with tube lens-skimmer configurations [26, 28]. Sensitivity claims for the S-lens has been reported at five times transmission improvement over the LTQ tube lens-skimmer design [30]. Taken collectively, the data support that the electrodynamic ion funnel > S-lens > tube lens-skimmer configuration in terms of ion transmission, ion flux, and overall sensitivity. This reflects the order of total protein identifications for data acquired by DIA by the three instruments and supports our conclusion that overall ion transmission (i.e. sensitivity) is the most important factor for improving proteome coverage using DIA methods.
Table 1.
Protein identification results from Saccharomyces cerevisiae datasets. Column 1 refers to the number of MS/MS spectra that were matched to a peptide below a 1% false-discovery rate, as estimated by Percolator. Column 2 lists the number of protein identifications for each instrument/method, along with a false-discovery rate at the protein level, estimated by a reverse-database strategy. Column 3 lists the percentage of all identified proteins found by a particular instrument/method combination. The group of columns marked “Percent overlap” lists percent of proteins in instrument/method row found by instrument/method column. For example, 74.3% of the proteins found in Velos DDA were also found in LTQ DDA.
| Number of matched MS/MS spectra |
Number of proteins identified; protein FDR (Total = 3335) |
Percentage of all identified proteins |
Percent overlap | ||||||
|---|---|---|---|---|---|---|---|---|---|
| LTQ DDA |
IF- LTQ DDA |
Velos DDA |
LTQ DIA |
IF- LTQ DIA |
Velos DIA |
||||
| LTQ DDA |
74900 | 1791; 6.6% |
53.7 | 77.2 | 78.7 | 81.3 | 83.8 | 81.8 | |
| IF-LTQ DDA |
92246 | 1783; 8.2% |
53.5 | 77.1 | 78.8 | 81.0 | 84.2 | 81.3 | |
| Velos DDA |
142135 | 1895; 11.7% |
56.8 | 74.3 | 74.5 | 77.4 | 80.7 | 78.2 | |
| LTQ DIA |
20028 | 2094; 3.2% |
62.8 | 69.0 | 68.9 | 69.7 | 86.7 | 79.8 | |
| IF-LTQ DIA |
25600 | 2321; 3.5% |
69.6 | 64.2 | 64.6 | 65.6 | 78.3 | 79.4 | |
| Velos DIA |
33913 | 2146; 4.3% |
64.3 | 68.1 | 67.8 | 69.1 | 78.2 | 86.2 | |
In contrast to the DIA results, DDA mode using the LTQ Velos produced the most protein identifications, 1847, as compared with 1742 for the IF-LTQ and 1735 for the LTQ. The main reason for the larger number of protein identifications for the LTQ Velos in DDA mode is that the dual-trap configuration nearly doubles the speed at which spectra are collected, directly leading to more sampled peptides per unit time and resulting in more protein identifications [30]. Indeed, the LTQ Velos in DDA mode collected a total of 142,135 tandem mass spectra as compared to 92,246 for the IF-LTQ and 74,900 for the LTQ. Strikingly, data from DIA experiments collected less than a third the number of spectra as compared to DDA experiments collected on the same respective instrument, but overall produced more protein identifications. The difference was greatest for the LTQ Velos, where the DIA experiment acquired only 33,913 spectra as compared to 142,135 spectra in the DDA experiment, an overall 76% reduction in spectra acquired. It is also worthy to note that the lower percentages in the DIA-overlap rows in Table 1, when compared to the DDA columns, indicate that there are more proteins uniquely identified by DIA than by DDA. This supports the general conclusion the DIA methods can consistently identify peptides at greater proteome depth relative to DDA methods.
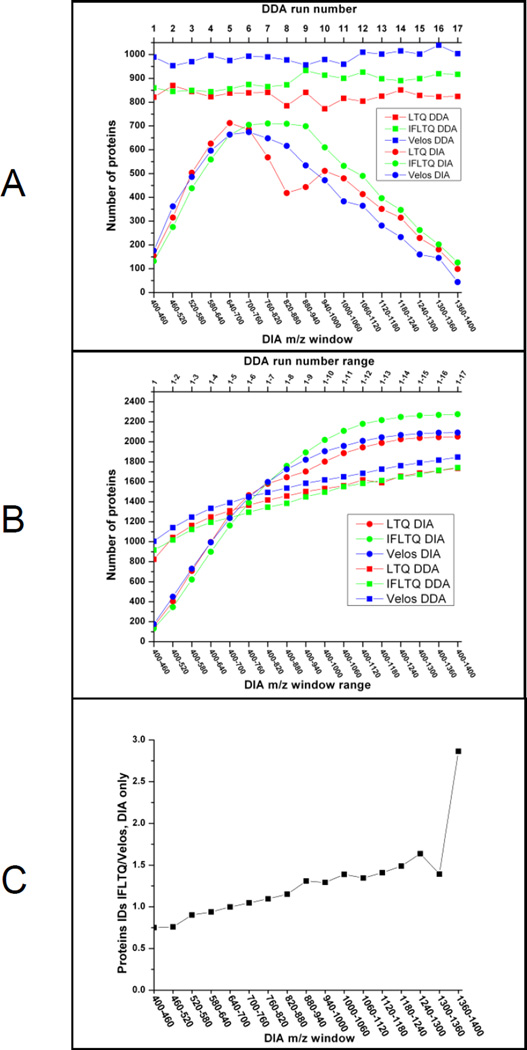
The numbers of protein identifications per LC-MS/MS run as a function of mass range (in the case of DIA) and run number (in the case of DDA) are displayed in Figure 2. In Figure 2A, the majority of identifications in DIA originate in the high ion density m/z range between 500 and 1000 Th where 2+ and 3+ charge states of tryptic peptides are typically detected. In the DDA data, the LTQ-Velos consistently returned more identifications than either the IF-LTQ or LTQ DDA runs. The cumulative identification of peptides and proteins is displayed in Figure 2B. This figure illustrates that the protein identifications from cumulative DDA runs increase rapidly and then achieve a relatively consistent slope; the continual increase through later runs is likely in part due to the addition of false positives. By contrast, the DIA curves increase sharply through the first half of the m/z range, consistent with the expected ion density, and cross the DDA curve relatively early between m/z 700–760. The cumulative protein identifications plateau at the latter part of the m/z range, consistent with the lower ion density at that region. In Figure 2C, identifications from the IF-LTQ and LTQ Velos are compared directly for data acquired by DIA. The ratio of number of protein identifications (IF-LTQ/LTQ Velos) are plotted versus mass range. The resulting line crosses 1.0 in the m/z 600–640 range and increases throughout the higher m/z range, suggesting better ion transmission for the higher m/z ions for the ion funnel configuration over that of the S-lens.
Figure 2.
A, Protein identifications by mass range (DIA) and run number (DDA); B, Accumulation of protein IDs versus mass range (DIA) and run number (DDA); C, Ratio of protein identifications for the IF-LTQ to the LTQ Velos.
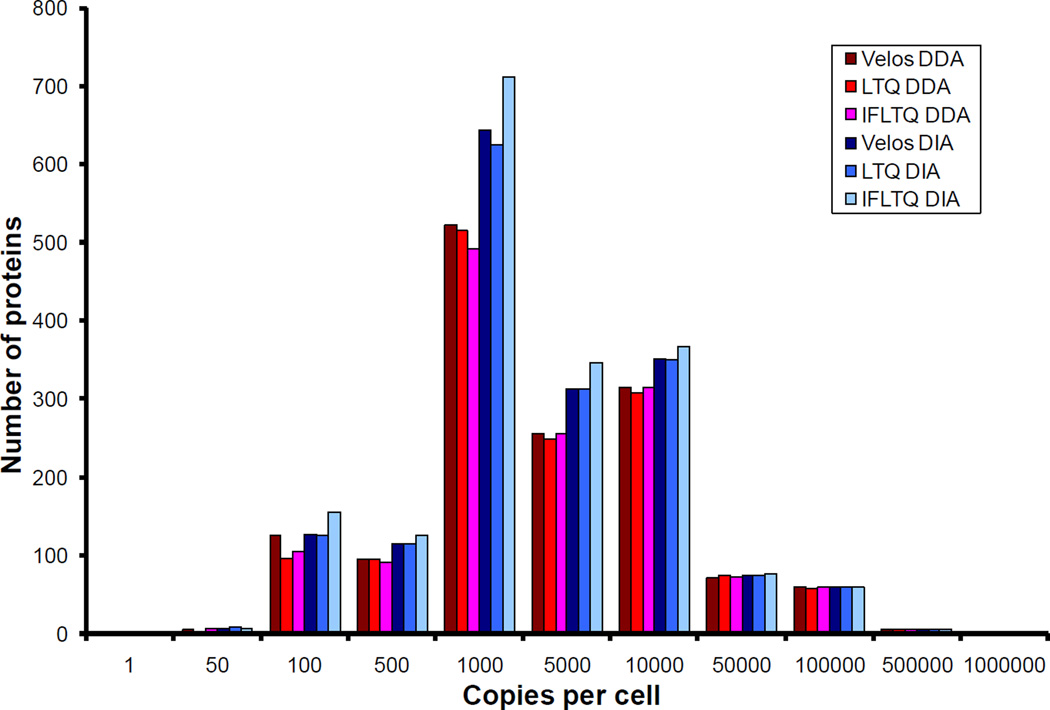
Yeast was chosen as the sample of interest to take advantage of protein copy number information reported previously [40]. Histograms depicting the numbers of proteins identified versus copy number are shown in Figure 3. The IF-LTQ collecting data using DIA performed significantly better than the other instruments and data acquisition strategies. However, when collecting data using DDA, the LTQ Velos performed the best. The additional protein identifications using DIA or DDA are a direct result of increased sampling in the lower half of the copy number range. The improved identification of low copy number proteins using the LTQ Velos by DDA confirms analyses reported previously and is directly attributed to the improved sensitivity of the S-lens ion guide and increased scan speed of the dual ion trap over the former LTQ design [30]. More significantly, our data indicate that DIA outperforms DDA in all cases, especially in the lower half of the copy number range where the copies per cell falls below 10,000. This is a direct illustration of the DIA approach and its ability to identify proteins at wider dynamic range and proteome depth. The IF-LTQ gave the greatest characterization of the yeast proteome in this study and can be directly attributed to the increased performance of the electrodynamic ion funnel which provided the most robust ion transmission to the mass analyzer. Both acquisition techniques were able to detect the most highly abundant species, regardless of instrument.
Figure 3.
Number of identified proteins versus protein copies per cell (Saccharomyces cerevisiae).
Overall, the results in Table 1 and Figure 2 confirm what has been reported previously for DIA and the PAcIFIC method [15]. In particular, DIA is able to identify more proteins than DDA, and these gains are apparent in the lower abundance range of the yeast proteome, as shown in Figure 3. However, we anticipated that the LTQ Velos would perform better overall than either of the other two instruments, because it has an RF ion guide (the S-lens) aimed at improving the sensitivity in a manner similar to the electrodynamic ion funnel. In addition, the LTQ Velos has almost twice the instrument duty cycle as either the IF-LTQ or LTQ ion trap designs, as shown by the greater numbers of MS/MS spectra collected per unit time by the Velos in both modes of acquisition (Table 1). However, the DIA data on the IF-LTQ suggests that scan speed is not the dominant factor in influencing the number of protein identifications using the DIA strategy. We therefore continued the investigation into a more direct comparison of the instrumentation platforms used in our study.
Comparing instrument platforms
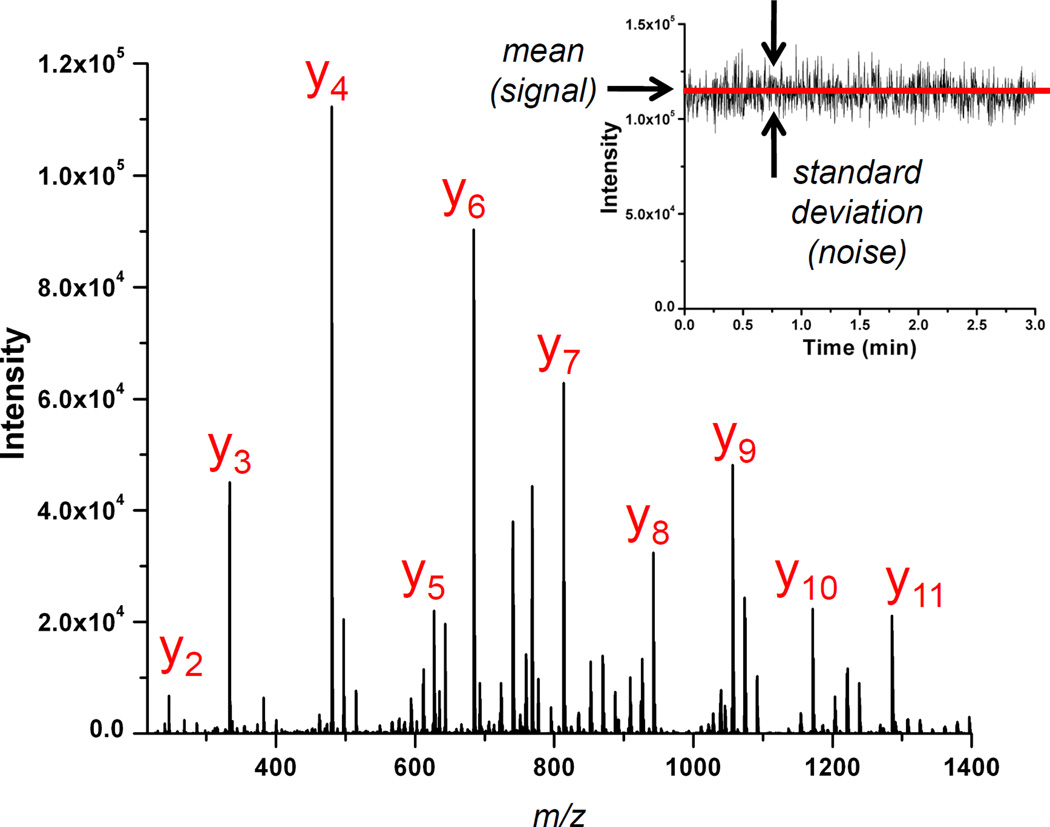
To improve our understanding of the LC-MS/MS experiments, we approached comparison of all three instrument platforms by collecting data by direct infusion of a peptide standard. We included measurements on three additional instruments that were similar in design and modification to the three instrument platforms described above to provide replicate measurements and increase the confidence of the reported results. We performed a simple experiment that enabled the general characterization of all six instruments in terms of signal-to-noise, where noise is defined as the precision obtained over many measurements of a nominally constant signal, as shown in Figure 4 (inset). This experiment involved infusing a peptide standard, collecting tandem mass spectra, and performing an analysis similar to that which the instrument vendor uses to determine the number of charges (i.e. ions) in the mass analyzer (i.e. ion trap) [41, 42]. This analysis parallels previous work reported on gas chromatography-mass spectrometry instruments using selected ion monitoring [43].
Figure 4.
A signal-averaged CID spectrum of Glu[1]-fibrinopeptide-B (y2-y11 labeled). Signal-to-noise measured as signal precision (inset).
The analysis proceeds as follows. We assume that instrument noise sources can be divided into two groups: (1) noise dependent solely on the statistics of the arrival of ions at the detector, i.e., shot noise, described formally by Poisson statistics [44]; and (2) all other noise sources, including noise due to the electron multiplier itself, pulse counting electronics and associated circuitry, and digitization errors. The signal variance due to these two groups can be written as
| (1) |
where σT2 is the total variance in a repeated measurement, σP2 is the variance due to Poisson (shot) noise, and σO2 contains the variance due to all other noise sources.
We acquire many nominally identical spectra (1000 spectra in this case) and compute intensity ratios for several of the most prominent ions relative to a single reference ion. Assuming the mass analyzer and attendant ion optics and detection systems perform adequately, by far the greatest source of instability in electrospray mass spectrometry is that of the electrospray ionization source itself. Measuring the ratio of the intensities of two different ions in the same spectrum ensures that the multiplicative contribution of noise sources which affect all peaks in a spectrum, the common mode noise which includes electrospray instability, will effectively cancel.
In the instruments under consideration here, automatic gain control (AGC) was used to keep the number of charges (i.e. ions) in the trap approximately constant by varying the time during which ions are allowed to accumulate in the trap [27]. This time is called the ion injection time as discussed in the Introduction. The measured peak area is therefore A = α(N/t), where N is the number of ions in the peak, and t is the ion injection time, which is the same for all ions in a given spectrum. The factor α is included to account for inaccuracies in the instrument calibration. Because α and t are the same for all ions in a given spectrum, the ratio of peak areas for any two ions a and b in a spectrum is R = Na/Nb.
The variance σR2 in repeated measurements of R can be computed theoretically (for small variations in Na and Nb) by using the propagation of errors [44]:
| (2) |
Inserting R = Na/Nb into Eq (2) gives:
| (3) |
This equation can now be simplified by assuming that the variance in the intensity measurements is dominated by Poisson (shot) noise. The variance of any set of Poisson distributed events is equal to the number of events, σ2 = N. Substituting this relation for the variances on the right-hand-side of Eq (3) results in the Poisson limited noise:
| (4) |
Since the number of ions in the peak is N = At/α (as defined above), Eq (4) can be rewritten as
| (5) |
where the bars indicate averages over repeated measurements. Inserting Eq (5) into Eq (1) gives:
| (6) |
In this equation, the measured quantities are R (ratio of peak areas), σT2 (variance in the ratio of peak areas), Aa (average peak area of ion a over many measurements), Ab (average peak area of ion b over many measurements), and t (ion injection time). The unknown quantities are α and σo2. Thus this equation is of the form y = mx + b, with m = α and b = σ O2. Taking measurements of many different ratios over many mass spectra allows us to perform a linear regression to obtain α and σO2. Because the instruments under consideration here use AGC, we anticipate that α will be near 1, indicating good instrument calibration. We further anticipate that σo2 will be small, indicating that instrument noise is limited by Poisson (shot) noise.
To carry out the analysis described above, we infused a solution of Glu[1]-fibrinopeptide-B into each of six mass spectrometers (see Methods) and continuously acquired 1000 tandem mass spectra of the 2+ charge state. An example spectrum is shown in Figure 4, with the y-ion series labeled. We monitored the top five most abundant fragment ions (y3, y4, y6, y7, and y9) and computed the peak areas, the ratios of peak areas relative to y9, and the variance in the ratio measurements. We then performed a linear regression on the resulting data (Table 2); each regression resulted in a R2 value in excess of 0.96. Four of the six instruments had an α value between 1.1 and 1.4, while the other two differ from these values. Interestingly, the latter two instruments were the only two of the six that were not calibrated immediately prior to performing these experiments. Because only one of the six is close to 1, which would indicate perfect calibration, and the four that were calibrated immediately prior to performing the experiments had more consistent values of α, we speculate that there may be some systematic error in calibration or in the analysis leading to a value of α different from 1. In particular, we note that it is known that variation in electrospray does not fully cancel in the types of analyses presented above, and can only be considered to cancel when ion populations are large. In our case, we estimate ion populations over all instruments and all ions of between 200 and 900 ions per peak. The lower ion counts can help explain the deviation from the expected value of α=1, both in terms of non-cancellation of ESI variation and in terms of overall fit. Nevertheless, results from different instruments are consistent with each other, and in all cases, the magnitude of σ O2 is small compared to other terms in Eq (6). This result indicates that non-Poisson noise sources are small in magnitude and strongly suggests that noise in each instrument is dominated by Poisson limited noise. We therefore conclude that all instruments perform according to expectations and that our results indicating fewer protein identifications in DIA mode for the LTQ Velos over the IF-LTQ are not due to any non-optimum parameters employed in the LTQ Velos. Additionally, we note that this procedure and analysis could be utilized on a routine basis to check the quality and consistency of the calibration routine, as well as to check for the presence of any non-Poisson limited noise which could indicate problems with the instrument hardware or detection electronics.
Table 2.
Results of linear regression as described in the Results and Discussion section. Values of α are consistent across instruments that were recently calibrated, while the two instruments that were not calibrated immediately prior to the experiments had different values (as noted by *). The magnitude of σO2 is small compared to other terms in Eq. (6). Regressions were obtained with R2 > 0.96 in all cases.
| Instrument | α | σO2 | R2 |
|---|---|---|---|
| Velos 1 | 1.32 | 0.0007 | 0.997 |
| LTQ 1 | 1.38 | 0.0005 | 0.965 |
| IF-LTQ 1 | 1.29 | 0.0002 | 0.994 |
| Velos 2 | 1.16 | 0.0001 | 0.997 |
| LTQ 2 | 1.81* | 0.0006 | 0.999 |
| IF-LTQ 2 | 0.994* | −0.0011 | 0.992 |
In a nanoflow LC-MS/MS experiment, typical peptide chromatographic peak widths (FWHM) usually fall somewhere between 5 and 30 seconds. Under typical operating conditions, the LTQ (with or without the ion funnel) is capable of acquiring a CID spectrum every 200 milliseconds with the LTQ Velos acquiring at roughly twice this rate. In DDA mode, this enables the LTQ Velos to acquire CID spectra on more molecular species, leading to more protein identifications as observed in Table 1 and Figure 2. By contrast, in DIA mode extra acquisition speed results in extra spectra collected in a given amount of time for each mass window, however the increase in spectra will generally not lead to additional protein identifications but rather an increase in redundant matched spectra. Indeed, we note that duty cycle improvements do not account for the trends in numbers of protein identifications for DIA reported in this study. For example, the LTQ Velos in DIA mode offers a large improvement in duty cycle due to the dual-pressure ion trap configuration and returns 32% more matching tandem mass spectra than IF-LTQ operating in DIA mode, but overall returned 8% fewer protein identifications than the IF-LTQ. It is worthy to note, however, that adding more m/z windows per sample injection would be a better use of the extra speed of the LTQ Velos in DIA mode and would provide the distinct advantage of completing the experiment in a fewer number of injections.
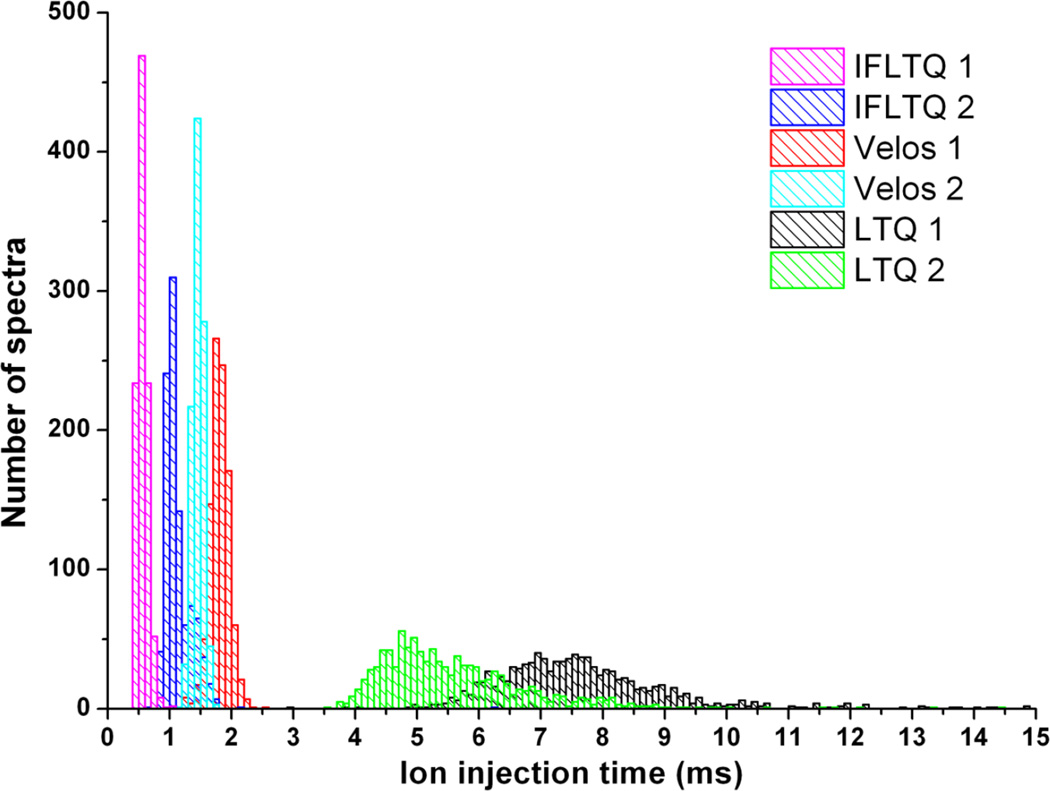
More important than the number of spectra collected in DIA mode is the quality of the collected spectra; only one or two good spectra are needed across a chromatographic peak to produce a high quality spectrum that can lead to confident peptide identification. In instruments with AGC, the ion injection time allows for accumulation of a specified number of ions in the trap, thus allowing lower abundance ions to produce spectra with similar quality to higher abundance ions. The AGC time is subject to a user-defined maximum which prevents unproductive accumulation of extremely low abundance or non-existent precursor ions. If the ion flux is too small, the ion injection time will reach the maximum more often. Conversely, when the ion flux is larger, the ion injection times will be smaller and less likely to reach the maximum. Thus a higher proportion of good-quality spectra will be obtained on an instrument with a source configuration that can deliver a larger ion flux. With this in mind, we compared the ion injection times for all spectra examined in the signal-to-noise infusion measurements described above. A histogram of these ion injection times is shown in Figure 5. The two IF-LTQ instruments have overall shorter ion injection times than the two LTQ Velos instruments. The previous generation unmodified LTQ instruments have much longer, broader distribution of ion injection times. The reason for the observed increase in protein identifications in the IF-LTQ in DIA mode is that the ion funnel delivers more analytically useful ions to the trap in a shorter amount of time. Referring to the percent overlap for protein identifications for DIA (Table 1), the LTQ Velos identified 79.4% of the same proteins as the IF-LTQ, whereas for DIA IF-LTQ identified 86.2% of the same proteins as the LTQ Velos. In other words, there is significant overlap between the two, but the IF-LTQ identified significantly more proteins. One additional reason for the increased identifications in the IF-LTQ could be a broader m/z transmission range for the IF-LTQ compared to the LTQ Velos. Referring to the number of protein identifications as a function of m/z range in Figure 2A, the identifications from the IF-LTQ instrument are greater than the other two instruments for higher values of m/z, while the LTQ Velos performs better at the lower m/z values. This is likely reflective of the broad m/z transmission range of the ion funnel as well as differences in front-end tuning between the instruments.
Figure 5.
Histogram of ion injection times for six linear ion trap instruments: two ion funnel-modified LTQs, two LTQ Velos, and two LT Qs.
Conclusions
We have presented a systematic study examining two different modes of data acquisition for proteomics on three different instrument platforms. In these experiments we find that the ion funnel modified LTQ instrument outperforms the previous generation LTQ and the current generation LTQ Velos for experiments acquired in DIA mode. Having analyzed for instrumental sources of non-Poisson limited noise, with additional instruments of similar configuration used for corroboration, we conclude that the improvement is primarily due to the improved ion flux and transmission of the electrodynamic ion funnel. The ion funnel delivered a higher flux of ions to the trap than either the S-lens or tube lens-skimmer configurations of the other two instruments. Because mass spectrometer speed only provides a time advantage in DIA mode, the technology advances in the LTQ Velos do not lead automatically to an improvement for proteomics in this mode of operation. In DIA mode, instrument sensitivity as measured by the ion injection time is the more important factor and the ion funnel modified LTQ holds a distinct advantage. However, the LTQ Velos outperformed the other instruments in DDA mode. In DDA mode, the time advantage gained through an improved acquisition duty cycle is evident, resulting in the acquisition of more spectra per unit time for the LTQ Velos. Although the IF-LTQ yielded more peptide identifications in DIA mode than the LTQ Velos, the LTQ Velos has the ability to perform the analysis faster by adding more DIA windows per sample injection, while continuing to obtain tandem mass spectra within the typical peptide chromatographic time scale. Throughput and optimization improvement for DIA will form the basis of future work.
Acknowledgements
We thank Jason Page, Keqi Tang, and Richard D. Smith at PNNL for assistance with the ion funnel designs. We thank Jae Schwartz, Michael Senko, Jean-Jacques Dunyach, and Philip Remes of Thermo Scientific for helpful discussions. We acknowledge Larry Stark and Jim Greenwell in the Department of Physics Machine Shop and Jim Gladden, Lon Buck, and Roy Olund in the Department of Chemistry Electronics Shop at the University of Washington. We thank Vagisha Sharma and Mike Riffle at the Proteomics Resource at the University of Washington for help with the yeast proteome analysis. This work was supported by NIH R01 DK069386 and the Yeast Resource Center at the University of Washington.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Stahl DC, Swiderek KM, Davis MT, Lee TD. Data-controlled automation of liquid chromatography/tandem mass spectrometry analysis of peptide mixtures. J. Am. Soc. Mass Spectrom. 1996;7:532–540. doi: 10.1016/1044-0305(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 3.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2010;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, Grant SF. Advances in whole genome sequencing technology. Curr. Pharm. Biotechnol. 2011;12:293–305. doi: 10.2174/138920111794295729. [DOI] [PubMed] [Google Scholar]
- 6.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 2013 doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 7.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 8.Purvine S, Eppel JT, Yi EC, Goodlett DR. Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics. 2003;3:847–850. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 9.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 10.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Bond NJ, Shliaha PV, Lilley KS, Gatto L. Improving qualitative and quantitative performance for MS(E)-based label-free proteomics. J. Proteome Res. 2013;12:2340–2353. doi: 10.1021/pr300776t. [DOI] [PubMed] [Google Scholar]
- 12.Myung S, Lee YJ, Moon MH, Taraszka J, Sowell R, Koeniger S, Hilderbrand AE, Valentine SJ, Cherbas L, Cherbas P, Kaufmann TC, Miller DF, Mechref Y, Novotny MV, Ewing MA, Sporleder CR, Clemmer DE. Development of high-sensitivity ion trap ion mobility spectrometry time-of-flight techniques: a high-throughput nano-LC-IMS-TOF separation of peptides arising from a Drosophila protein extract. Anal. Chem. 2003;75:5137–5145. doi: 10.1021/ac030107f. [DOI] [PubMed] [Google Scholar]
- 13.Sowell RA, Hersberger KE, Kaufman TC, Clemmer DE. Examining the proteome of Drosophila across organism lifespan. J. Proteome Res. 2007;6:3637–3647. doi: 10.1021/pr070224h. [DOI] [PubMed] [Google Scholar]
- 14.Geiger T, Cox J, Mann M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics. 2010;9:2252–2261. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panchaud A, Scherl A, Shaffer SA, von Haller PD, Kulasekara HD, Miller SI, Goodlett DR. Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal. Chem. 2009;81:6481–6488. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panchaud A, Jung S, Shaffer SA, Aitchison JD, Goodlett DR. Faster, quantitative, and accurate precursor acquisition independent from ion count. Anal. Chem. 2011;83:2250–2257. doi: 10.1021/ac103079q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, Wu CC, MacCoss MJ. Multiplexed MS/MS for improved data-independent acquisition. Nat. Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11O111:016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisbrod CR, Eng JK, Hoopmann MR, Baker T, Bruce JE. Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. J. Proteome Res. 2012;11:1621–1632. doi: 10.1021/pr2008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kebarle P, Tang L. From Ions in Solution to Ions in the Gas-Phase - the Mechanism of Electrospray Mass-Spectrometry. Anal. Chem. 1993;65:A972–A986. [Google Scholar]
- 21.Cech NB, Enke CG. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 22.Page JS, Kelly RT, Tang K, Smith RD. Ionization and transmission efficiency in an electrospray ionization-mass spectrometry interface. J. Am. Soc. Mass Spectrom. 2007;18:1582–1590. doi: 10.1016/j.jasms.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Shaffer SA, Tang KQ, Anderson GA, Prior DC, Udseth HR, Smith RD. A novel ion funnel for focusing ions at elevated pressure using electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1997;11:1813–1817. [Google Scholar]
- 24.Shaffer SA, Prior DC, Anderson GA, Udseth HR, Smith RD. An ion funnel interface for improved ion focusing and sensitivity using electrospray ionization mass spectrometry. Anal. Chem. 1998;70:4111–4119. doi: 10.1021/ac9802170. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer SA, Tolmachev A, Prior DC, Anderson GA, Udseth HR, Smith RD. Characterization of an improved electrodynamic ion funnel interface for electrospray ionization mass spectrometry. Anal. Chem. 1999;71:2957–2964. doi: 10.1021/ac990346w. [DOI] [PubMed] [Google Scholar]
- 26.Kelly RT, Tolmachev AV, Page JS, Tang K, Smith RD. The ion funnel: theory, implementations, and applications. Mass Spectrom. Rev. 2010;29:294–312. doi: 10.1002/mas.20232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz JC, Zhou X-G, Bier ME. Method and apparatus of increasing dynamic range and sensitivity of a mass spectrometer. US Patent. 1996;5(572):022. [Google Scholar]
- 28.Page JS, Tang K, Smith RD. An electrodynamic ion funnel interface for greater sensitivity and higher throughput with linear ion trap mass spectrometers. Int. J. Mass Spectrom. 2007;265:244–250. [Google Scholar]
- 29.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR. 3rd: MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 32.Yates JR, Eng JK, 3rd, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 33.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 34.Sharma V, Eng JK, Maccoss MJ, Riffle M. A mass spectrometry proteomics data management platform. Mol. Cell. Proteomics. 2012;11:824–831. doi: 10.1074/mcp.O111.015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang KQ, Tolmachev AV, Nikolaev E, Zhang R, Belov ME, Udseth HR, Smith RD. Independent control of ion transmission in a jet disrupter dual-channel ion funnel electrospray ionization MS interface. Anal. Chem. 2002;74:5431–5437. doi: 10.1021/ac0202583. [DOI] [PubMed] [Google Scholar]
- 37.Page JS, Bogdanov B, Vilkov AN, Prior DC, Buschbach MA, Tang K, Smith RD. Automatic gain control in mass spectrometry using a jet disrupter electrode in an electrodynamic ion funnel. J. Am. Soc. Mass Spectrom. 2005;16:244–253. doi: 10.1016/j.jasms.2004.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23:3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 39.Scherl A, Shaffer SA, Taylor GK, Kulasekara HD, Miller SI, Goodlett DR. Genome-specific gas-phase fractionation strategy for improved shotgun proteomic profiling of proteotypic peptides. Anal. Chem. 2008;80:1182–1191. doi: 10.1021/ac701680f. [DOI] [PubMed] [Google Scholar]
- 40.Ghaemmaghami S, Huh W, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 41.Kaur P, O'Connor PB. Use of statistical methods for estimation of total number of charges in a mass spectrometry experiment. Anal. Chem. 2004;76:2756–2762. doi: 10.1021/ac035334w. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz JC. Measuring ion number and detector gain. US Patent. 2006;7(109):474. [Google Scholar]
- 43.MacCoss MJ, Toth MJ, Matthews DE. Evaluation and optimization of ion-current ratio measurements by selected-ion-monitoring mass spectrometry. Anal. Chem. 2001;73:2976–2984. doi: 10.1021/ac010041t. [DOI] [PubMed] [Google Scholar]
- 44.Bevington PR, Robinson DK. Data reduction and error analysis for the physical sciences. 2nd ed. New York: McGraw-Hill; 1992. [Google Scholar]