Abstract
In March 2013, a patient infected with a novel avian influenza A H7N9 virus was reported in China. Since then, there have been 458 confirmed infection cases and 177 deaths. The virus contains several human-adapted markers, indicating that H7N9 has pandemic potential. The outbreak of this new influenza virus highlighted the need for the development of universal influenza vaccines. Previously, we demonstrated that a tetrameric peptide vaccine based on the matrix protein 2 ectodomain (M2e) of the H5N1 virus (H5N1-M2e) could protect mice from lethal infection with different clades of H5N1 and 2009 pandemic H1N1 influenza viruses. In this study, we investigated the cross-protection of H5N1-M2e against lethal infection with the new H7N9 virus. Although five amino acid differences existed at positions 13, 14, 18, 20, and 21 between M2e of H5N1 and H7N9, H5N1-M2e vaccination with either Freund's adjuvant or the Sigma adjuvant system (SAS) induced a high level of anti-M2e antibody, which cross-reacted with H7N9-M2e peptide. A mouse-adapted H7N9 strain, A/Anhui/01/2013m, was used for lethal challenge in animal experiments. H5N1-M2e vaccination provided potent cross-protection against lethal challenge of the H7N9 virus. Reduced viral replication and histopathological damage of mouse lungs were also observed in the vaccinated mice. Our results suggest that the tetrameric H5N1-M2e peptide vaccine could protect against different subtypes of influenza virus infections. Therefore, this vaccine may be an ideal candidate for developing a universal vaccine to prevent the reemergence of avian influenza A H7N9 virus and the emergence of potential novel reassortants of influenza virus.
Keywords: influenza A virus, H7N9, M2e, peptide vaccine, cross protection
INTRODUCTION
Since the first human infection by a novel avian influenza A H7N9 virus was reported in March 2013, a total of 458 confirmed cases with 177 deaths in China had been reported by December 2014.1 After a relatively silent period from July to October 2013, in which only four cases with one death were reported, the virus has reemerged since November 2013, resulting in the second outbreak in China.2 This novel influenza virus can bind to both avian (alpha 2,3-linked sialic acid) and human (alpha 2,6-linked sialic acid) receptors.3 It also contains human-adapted amino acid markers.4,5 These adaptations may explain why the virus can cause outbreaks in the human population.6 Patients infected with the H7N9 virus typically show symptoms such as fever, cough, opacities, and consolidation on chest radiography, and some severe cases can progress to acute respiratory distress syndrome (ARDS) and multiorgan failure.4 Although the lethality of H7N9 influenza is comparatively lower than that of highly pathogenic H5N1 viral infection, it is much higher than that of 2009 pandemic H1N1 influenza, reaching approximately 30%.7 Furthermore, high-pathogenicity markers for human-adapted influenza virus, such as an E637K amino acid substitution in the PB2 gene and Q226L in the haemagglutinin (HA) gene, have been identified in recent isolates of the H7N9 virus,4 suggesting that the virus might become more virulent in humans. Thus, the pandemic potential of lethal H7N9 influenza has raised public concern.
Although early treatments with oseltamivir and peramivir were effective,3,8,9 drug-resistant mutants of the virus were found soon after the patients received the anti-influenza therapy.10 Candidates of inactivated virus vaccines and virus-like particle (VLP) vaccines of H7N9 virus have been reported.11 However, the use of inactivated virus vaccines or VLP may not provide protection against mutated or reassorted influenza virus. In fact, a surveillance study has suggested that the novel influenza A H7N9 virus is a new reassortant of several influenza viruses, including H7, N9, and H9N2.6 Therefore, development of universal influenza vaccines is an attractive goal for scientists around the world. In this regard, the ectodomain of matrix protein 2 (M2e) of influenza viruses may be a promising target for the development of a universal influenza vaccine. This is because M2e is relatively conserved in different subtypes of influenza virus,12,13 and animal experiments have demonstrated that M2e vaccination can provide cross-protection against infection with different subtypes of influenza virus. H5N1-M2e-specific antibodies could react with different subtypes of influenza virus, such as H5N2, H9N2, H7N7, and H11N6.14 Various M2e-based vaccines have been developed, such as recombinant protein vaccines,15,16,17,18 plasmid DNA vaccines,19 and peptide vaccines.20,21 In our previous studies, we constructed a tetrameric H5N1-M2e vaccine candidate and demonstrated that it could provide cross-protection against lethal infections with different clades of H5N1 and 2009 pandemic H1N1 viruses.20,21 In this study, we extend our investigation to evaluate cross-protection of H5N1-M2e against lethal infection by the novel avian influenza A H7N9 virus in mice.
MATERIALS AND METHODS
Mice
Six- to eight-week-old female BALB/c mice were provided by the Laboratory Animal Unit of the University of Hong Kong. Mice were maintained in cages and provided with sterilized food and water in the animal facility. The animal study was approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR) of the University of Hong Kong.
Peptide
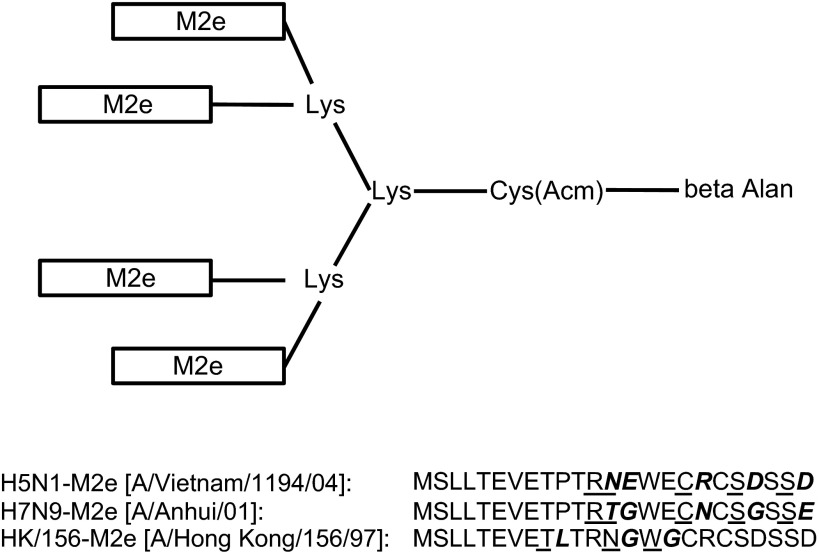
Tetra-branched peptides encoding the M2e of A/Vietnam/1194/04 (H5N1), A/Anhui/01(H7N9) and A/Hong Kong/156/97 (Figure 1) were synthesized by GL Biochem Ltd. (Shanghai, China).
Figure 1.
Structure of the tetrameric M2e peptide vaccine. The peptide vaccine was synthesized on [Fmoc-Lys(Fmoc)]2-Lys-Cys(Acm)-βAla-Wang Resin in a tetrameric form carrying four copies of H5N1-M2e or H7N9-M2e. Each rectangle represents a copy of M2e. The amino acid sequences of M2e were determined according to two virus strains, A/Vietnam/1194/04 (H5N1), A/Anhui/01/13 (H7N9), and A/Hong Kong/156/97 (H5N1), which are shown at the bottom.
Virus
Mouse-adapted A/H7N9/Anhui/01 virus was inoculated in 10-day-old specific pathogen-free (SPF) eggs and the allantoic fluid was collected after three-day incubation at 35 °C. The allantoic fluid was aliquoted and stored at −80 °C before use. The virus titer was detected by 50% tissue culture infective dose (TCID50) assay, while the 50% lethal dose (LD50) was determined by challenging BALB/c mice with serial dilutions of the virus as described previously.20 All experiments involving H7N9 virus were performed in biosafety level 3 (BSL-3) facilities as described previously.22
Animal experiment
Two groups of BALB/c mice (15 mice/group) were subcutaneously (s.c.) vaccinated with H5N1-M2e tetramer peptide pre-mixed, respectively, with Freund's adjuvant (FA, Sigma, St. Louis, SO, USA) and the Sigma adjuvant system (SAS) (Sigma, St. Louis, SO, USA) as described previously.23 Three other groups of mice (15 mice/group) were s.c. injected with FA, SAS or phosphate-buffered saline (PBS) alone as negative controls. Briefly, the mice were primary-immunized with 10 µg of H5N1-M2e pre-mixed with complete FA or SAS, and then boosted with 10 µg of the peptide with incomplete FA or SAS twice in a three-week interval. Mice sera were collected 1 day prior to each vaccination and 10 days after the second booster vaccination for detection of specific antibodies. Ten days after the last vaccination, the mice were intranasally (i.n.) inoculated with 10 LD50 of the mouse-adapted strain of A/Anhui/01 (H7N9) after anesthetization with ketamine and xylazine. The challenged mice were observed for 21 days or till death. Lung tissues were collected from 5 mice/group at day 6 post-challenge for virological and pathological investigation.
Enzyme-linked immunosorbant assay (ELISA)
The titers of M2e-specific antibodies were detected by ELISA as described previously.24 Briefly, a 96-well micro-titer plate (Sigma, St. Louis, SO, USA) was coated with either H5N1–, H7N9–, or HK/156-M2e (GL Biochem Ltd, Shanghai, China) at a concentration of 1 µg/well and incubated overnight at 4 °C. After the coated plate was blocked with 3% bovine serum albumin in PBS for 2 h at room temperature (RT), serially diluted sera were added to the plate and incubated at RT for 2 h. Horseradish peroxide-conjugated goat anti-mouse immunoglobulin G antibody (Dako, Glostrup, Denmark) was added to the plate and incubated at RT for 1 h. Substrate 3,3′,5,5′-tetramethylbenzidine (Life Technologies, Carlsbad, CA, USA) was added to the plate and incubated at RT for 0.5 h. The reaction was stopped by adding 1 M H2SO4, and the results were measured at absorbance of 450 nm using an ELISA reader (Beckman Coulter, Brea, CA, USA).
Virological tests
Virus titers were detected by TCID50 assay as described previously.20 Briefly, the infected allantoic fluid and mouse lung homogenates were serially diluted 10-fold in minimum essential medium (Invitrogen, Waltham, MA, USA) containing a cocktail of antibiotics. The diluted samples were added to PBS-prewashed Madin–Darby canine kidney cell monolayer in a 96-well plate. The cell cultures were incubated at 37 °C for 1 h, and the supernatants were replaced with fresh minimum essential medium containing a cocktail of antibiotics. The cytopathic effect that appeared in the infected Madin–Darby canine kidney cells was observed daily and recorded on the third-day post-infection.
Viral RNA copies in mouse lung homogenates were measured by quantitative reverse-transcription polymerase chain reaction (QRT-PCR) as described previously.23 Briefly, viral RNA of mouse lung homogenate was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The extracted viral RNA was reverse-transcribed to cDNA using Superscript II (Invitrogen, Waltham, MA, USA) and primer Flu12 (AGC AAA AGC). The viral RNA copy numbers in the samples were determined using a Lightcycler 96 (Roche Applied science, Penzberg, Germany) with SYBR green I master (Roche Applied science, Penzberg, Germany). Primers specific for influenza viral M gene (forward primer: 5-CTT CTA ACC GAG GTC GAA ACG-3; reverse primer: 5-GGC ATT TTG GAC AAA KCG TCT A-3) were used in the QRT-PCR assay.
Histopathological analysis
The lung tissues collected from infected mice were fixed with 10% formalin. The fixed tissues were embedded in paraffin wax, and the samples were cut to 6-µm-thick sections. The sections were mounted on the slide and examined by H&E staining as described previously.25
M2e 3D structure prediction
The 3D structures of the A/Vietnam/1194/04 (H5N1), A/Hong Kong/156/97 (H5N1), A/Anhui/01/13 (H7N9), and A/Beijing/501/09 (H1N1) M2e were predicted using the online software PEP-FOLD (http://bioserv.rpbs.univ-paris-diderot.fr/PEP-FOLD/) as described previously.26,27,28 The predicted model with the highest score was chosen for discussion purposes.
Statistical analysis
The results are presented as the mean ± standard deviation (SD). Statistical significance between different vaccination groups was calculated by Student's t-test and Kaplan–Meier analysis using the statistical package for the social sciences (International Business Machine Corporation, Armonk, NY, USA) statistical software. P-values less than 0.05 were considered significant.
RESULTS
High level of cross-reacted antibody is induced by H5N1-M2e vaccination
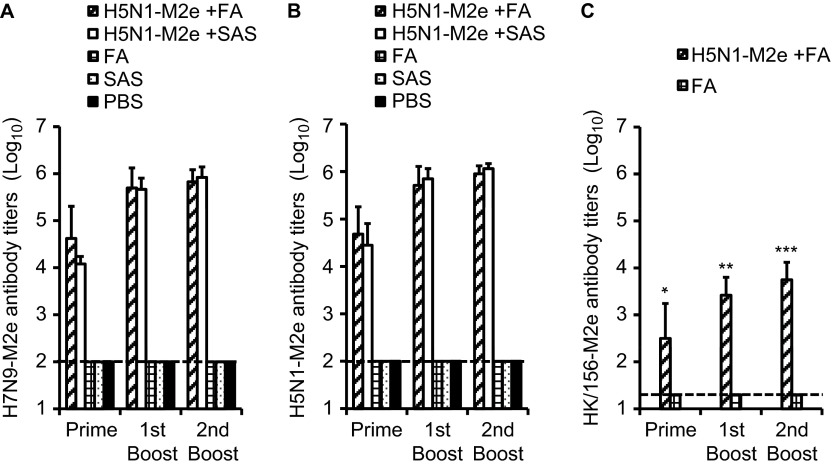
As shown in Figure 1, a difference of five amino acids was observed between the sequences of H5N1-M2e and H7N9-M2e. To examine whether these variations would affect the reaction of the antibody induced by the H5N1-M2e vaccine and H7N9-M2e, we tested the antibody titers in serum samples collected from H5N1-M2e-vaccinated mice using ELISA with plates coated with H5N1-M2e and H7N9-M2e, respectively. The titers of immunoglobulin G antibody specific to H7N9-M2e (Figure 2A) and H5N1-M2e (Figure 2B) were gradually increased in mouse serum samples collected after each vaccination, reaching very high levels (>1:105) 10 days after the last booster vaccination. By contrast, M2e-specific antibody was undetectable in serum samples collected from mice vaccinated with FA, SAS, or PBS alone. The adjuvant effect of SAS seemed to be better than that of FA because H5N1-M2e plus SAS induced a higher level of antibody responses. Importantly, the antibody titers specific to H5N1-M2e and H7N9-M2e were similar (P > 0.05). These results indicate that H5N1-M2e vaccination induced a high level of M2e-specific antibody that effectively cross-reacted with H7N9-M2e, although a difference of approximately 21% (5/24) amino acids was identified between the sequences of H5N1-M2e and H7N9-M2e.
Figure 2.
Cross-reacting M2e-specific antibody responses induced by H5N1-M2e vaccination. Mice were vaccinated three times with tetrameric H5N1-M2e peptide mixed with FA (H5N1−M2e+FA) or SAS (H5N1−M2e+SAS). Mice vaccinated with PBS, FA, or SAS were used as control. Mice sera were sampled 21 days after priming and the first booster vaccination and 10 days after the second booster vaccination for the ELISA detection. The end point titer of each sample was determined as the highest dilution that yielded an OD450 nm absorbance higher than the blank OD450 nm absorbance plus 2 SD. The data are shown as the geometric mean of 10 mice per group with its corresponding SD on a log10 scale. (A) H7N9-M2e tetrameric peptide was used as coating antigen in the ELISA. (B) H5N1-M2e tetrameric peptide was used as coating antigen in the ELISA. (C) HK/156-M2e tetrameric peptide was used as coating antigen in the ELISA. The detection limits (1:100 and 1:20) are indicated by the dotted line. *, **, and *** indicate significant differences (P < 0.05) between the H5N1-M2e and HK/156 M2e cross-reactive antibody titers detected on 21 days after priming, 21 days after first booster and 10 days after second booster H5N1−M2e+FA vaccinations, respectively. Experiments were repeated five times.
Furthermore, M2e of another H5N1 virus strain A/Hong Kong/156/97, which is nonreactive with H5N1-M2e (A/Vietnam/1194/04)-induced antibody,19 was included as a negative control in the ELISA experiments. As shown in Figure 2C, the titer of HK/156-M2e cross-reactive antibody was approximately 2 logs lower than that of H7N9-M2e (P < 0.05), indicating that H5N1-M2e vaccination did not induce sufficient antibody toward HK/156-M2e.
H5N1-M2e vaccination provides potent cross-protection against lethal challenge with H7N9 influenza virus
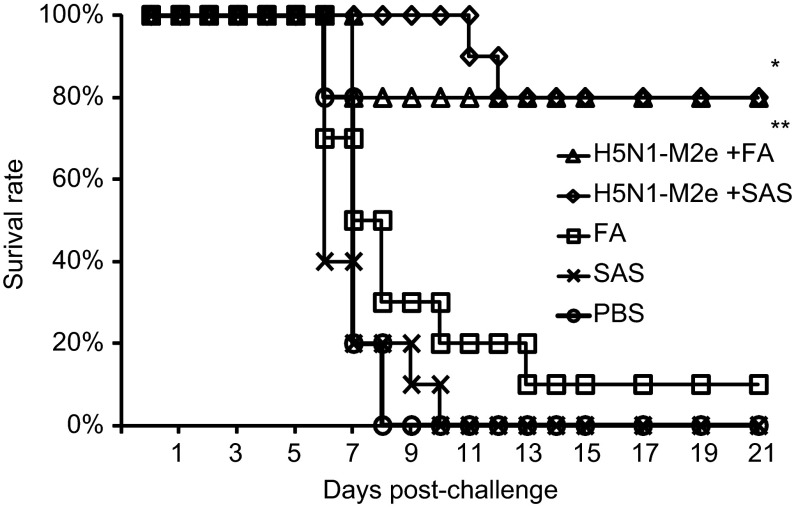
To evaluate whether H5N1-M2e vaccination could provide effective cross-protection against lethal infection of H7N9 virus, the H5N1-M2e-vaccinated mice were challenged with 10 LD50 of mouse-adapted A/Anhui/01/13 (H7N9) virus. As shown in Figure 3, vaccination of H5N1-M2e with either FA or SAS, could protect 80% (8/10) of mice from lethal challenge of H7N9 virus. The protective effect was similar to the results from experiments using lethal challenge with homogenous H5N1 virus.20 By contrast, the survival rate of mice vaccinated with SAS or PBS was 0% (0/10). Interestingly, 1 out of 10 mice vaccinated with FA also survived the lethal challenge (survival rate 10%). The survival rate of H5N1-M2e-vaccinated mice were significantly higher than that of mice vaccinated with FA, SAS, or PBS (P < 0.01). These results show that H5N1-M2e vaccination could provide satisfactory cross-protection against the lethal infection with the novel H7N9 virus.
Figure 3.
Survival rate in mice challenged with lethal dose of H7N9 virus. H5N1-M2e+FA- and H5N1−M2e+SAS-vaccinated mice were challenged with 10 LD50 of A/Anhui/01/13 (H7N9) influenza virus. FA, SAS, and PBS alone were used as negative controls. The survival of mice (10/group) was observed for 21-day post-challenge. The survival rates (%) were calculated corresponding to the mice survival. * and ** indicate significant differences (P < 0.01) between the survival rates of the H5N1−M2e+FA group and the FA group and between the survival rates of the H5N1−M2e+SAS group and the SAS group, respectively.
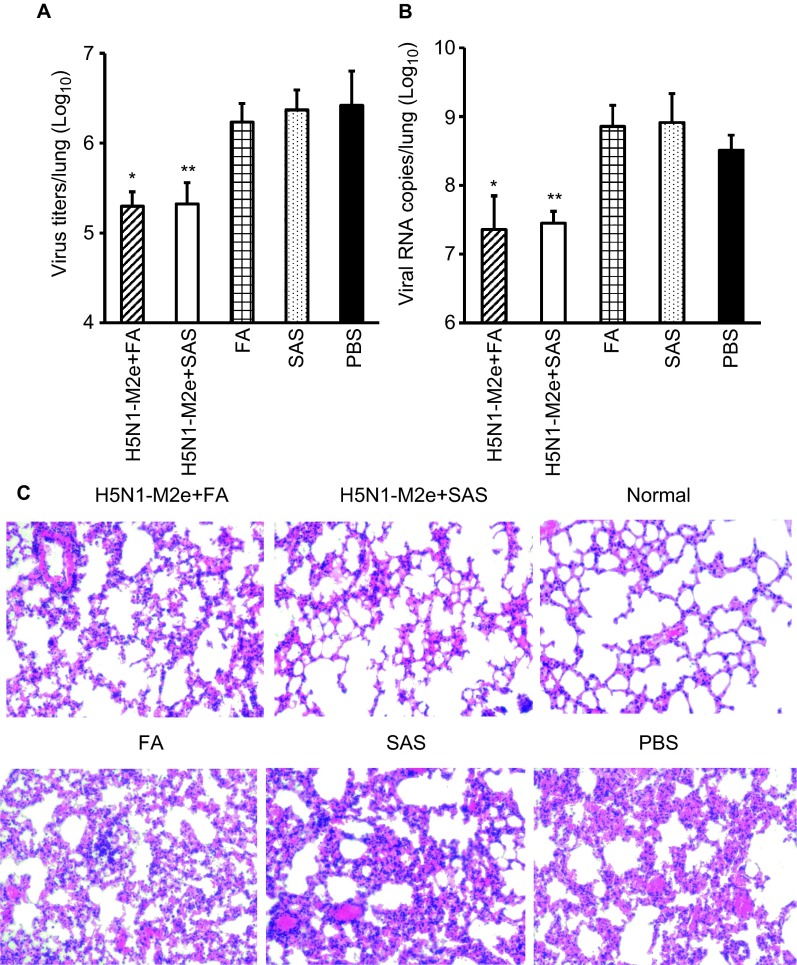
H5N1-M2e vaccination reduces viral load and tissue damage in lungs of mice with lethal infection with the H7N9 virus
To examine whether H5N1-M2e vaccination could inhibit viral infection and tissue damage in the lungs of mice challenged with a lethal dose of H7N9 virus, six mice per group were sacrificed at day 6 post-challenge and their lungs were collected for detection of viral load and tissue damage. As detected by TCID50 assay and QRT-PCR, virus titers (Figure 4A) and viral RNA copies (Figure 4B) in the lungs of mice vaccinated with H5N1-M2e were significantly reduced, which was approximately 2 logs lower than that in the mice vaccinated with FA, SAS, or PBS (P < 0.01). Consistent with this result, H&E staining showed that the lung pathology of mice vaccinated with H5N1-M2e was less severe than mice vaccinated with adjuvant or PBS. Severe damage of lung tissue was observed in adjuvant- or PBS-vaccinated mice, which included damage to the bronchial epithelium with denatured pneumocytes, neutrophils, and mononuclear cells, marked infiltration of lymphocytes, and fused alveoli wall with focal hemorrhage (Figure 4C).
Figure 4.
Virus titers and pathological changes in mouse lungs challenged with H7N9 virus strain A/Anhui/01/13. Mice were challenged with 10 LD50 of A/Anhui/01/2013 (H7N9) 10 days after the second booster vaccination. Mice lungs were sampled at day 6 post-challenge, and the virus titers in the lungs were determined by (A) TCID50 assay and (B) QRT-PCR. The data are presented as geometric mean +SD of (A) TCID50 per mouse lung and (B) viral RNA copies per mouse lung on a Log10 scale. * and ** indicate significant difference (P < 0.01) between the virus titer of the H5N1−M2e+FA group and the FA group and between the SAS H5N1−M2e+SAS group and the SAS group, respectively. (C) Histopathological changes in mouse lungs collected at day 6 post-challenge were detected by H&E staining. The histopathological changes in representative images of lungs from mice vaccinated with H5N1−M2e+FA, H5N1−M2e+SAS, FA, SAS, or PBS are shown. A representative image of normal mouse lung is included as negative control (magnification ×100).
H5N1-M2e and H7N9-M2e 3D structure analysis
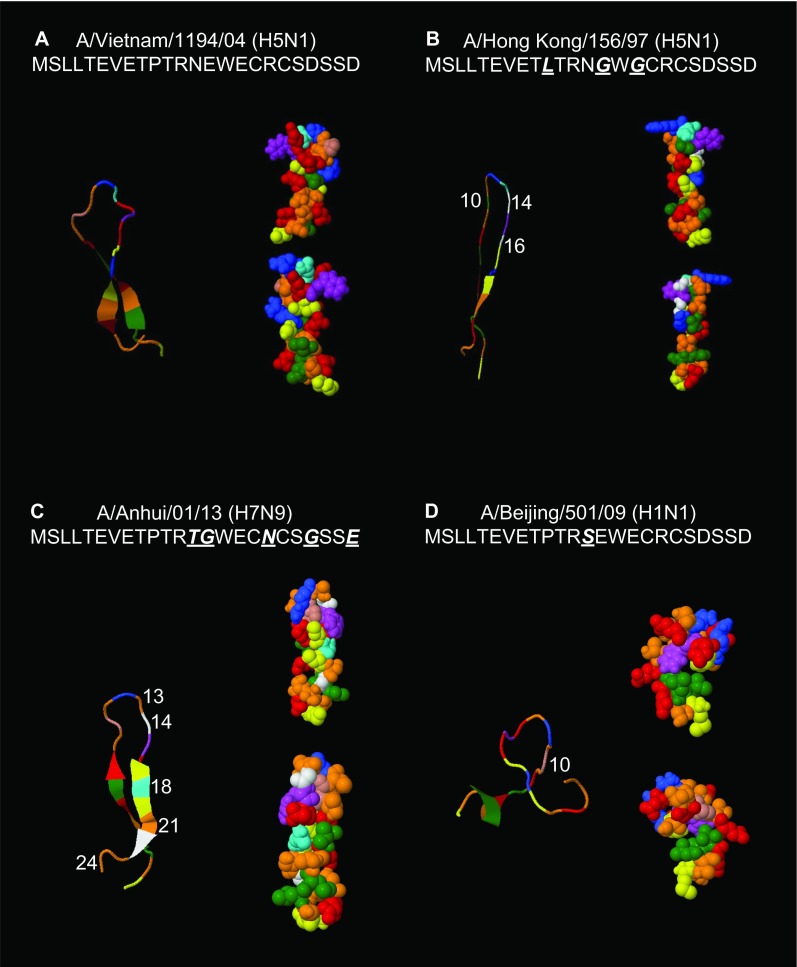
We analyzed monomer sequences of H5N1-M2e (A/Vietnam/1194/04) and H7N9-M2e (A/Anhui/01/13) using PEP-FOLD and compared the structural differences between the predicted 3D models. M2e of A/Hong Kong/156/97 (H5N1), which is non-reactive with H5N1-M2e (A/Vietnam/1194/04) induced antibody,19 and M2e of A/Beijing/501/09 (H1N1), which is cross-reactive with H5N1-M2e-induced antibody,21 were included for analysis. As shown in Figure 5, all four M2e were folded as hairpin shapes. The U-shaped regions of M2e-A/Vietnam/1194/04 (H5N1), M2e-A/Anhui/01/13 (H7N9), and M2e-A/Beijing/501/09 (H1N1) were more exposed than that of M2e-A/Hong Kong/97 (H5N1). The structural differences between these four M2e peptides might be due to the properties of the substituted amino acids, and they might affect the antigenic site accessibility.
Figure 5.
The 3D structure prediction of the M2e structures of H5N1 and H7N9 subtypes. The 3D structures of M2e of (A) A/Vietnam/1194/04 (H5N1), (B) A/Anhui/01/13 (H7N9), (C) A/Hong Kong/156/97 (H5N1), and (D) A/Beijing/501/09 were predicted using the online software PEP-FOLD. The skeleton structure prediction model is shown on the left, and the amino acid spherical structure prediction model is shown on the right with two different angles. Each amino acid is shown with its specific color according to the software. The amino acid variations between A/Vietnam/1194/04 (H5N1), A/Anhui/01/13 (H7N9), A/Hong Kong/156/97 (H5N1), and A/Beijing/501/09 (H1N1) are indicated with white numbers on the skeleton prediction models.
DISCUSSION
To control potential new influenza pandemics, the best measure may be to develop a universal influenza vaccine because no existing vaccines can provide effective cross-protection against a mutated or reassorted novel influenza virus. Current efforts to develop universal vaccines include the development of M2e-based vaccines, HA stem vaccines 29 and M1 VLPs.30 M2e-based vaccine candidates can provide cross-protection against infections of different subtypes of influenza virus.20,21,31 In our previous studies, we showed that an H5N1-M2e-based tetrameric peptide vaccine exhibited promising protection against lethal challenge with different clades of H5N1 virus and other subtypes of influenza virus.20,21 In this study, we further demonstrated that this tetrameric peptide vaccine could also offer potent protection against infection with a novel H7N9 influenza virus, which caused a first outbreak from March to June 2013 and a second outbreak from November 2013 to December 2014 in China.
Our results showed that the H5N1-M2e tetrameric peptide vaccine, together with either FA or SAS, induced a high antibody response that was able to effectively cross-react with H7N9-M2e (Figure 2). The vaccinations thus protected the mice from a lethal challenge with the H7N9 virus (Figure 3). The survival rate of H5N1-M2e-vaccinated mice against a H7N9 lethal challenge was 80%, which was the same as that of H5N1-M2e-vaccinated mice against a H5N1 lethal challenge.20 Consistently, viral load and lung damage in H5N1-M2e-vaccinated mice were much lower and less severe than that in adjuvant- or PBS-injected mice (Figure 4). Although T-cell responses induced by an M2e-based vaccine have been reported previously, the protection by the M2e vaccination was mainly attributed to the induced antibodies but not the T cells.19 Similarly, our results showed that the cross-protection of the H5N1-M2e vaccination was directly related to the levels of cross-reactive antibodies toward the M2e of challenge viruses. Unlike vaccinations with inactivated viruses, HA-subunit vaccines and VLP vaccines, which may provide complete protection against infection by homogenous viruses but poor cross-protection against infection by heterogeneous viruses, our results illustrate that the H5N1-M2e tetrameric peptide vaccine can provide satisfactory cross-protection against infection with a novel influenza virus that has newly emerged in humans.
Amino acid mutations at positions 10 and 11 of M2e reduce the monoclonal antibody binding affinity,32 whereas virus mutants at M2e position 10 can escape the protection of M2e antibodies.33 Moreover, M2e peptide variants at positions 10, 14, and 16 significantly reduce reactive antibody-binding titer.19 These findings indicate that although M2e is relatively conserved among different subtypes of influenza virus, amino acid mutations at certain positions could indeed affect the efficacy of M2e vaccination. Notably, there are five amino acid differences at positions 13, 14, 18, 21, and 24 between H5N1-M2e and H7N9-M2e, which accounted for approximately 21% (5/24) of the total amino acids of M2e (Figure 1). However, these amino acid variations did not abolish the efficacy of the vaccination. The protection against lethal challenge by H7N9 virus (Figures 3 and 4) was not affected, despite the fact that the amino acid variations between H5N1-M2e and H7N9-M2e included a mutation at position 14, which has been reported to affect the antibody cross-reaction.19 To examine how the amino acid variations may affect the structure of the M2e peptide, the 3D structures of M2e-A/Vietnam/1194/04 (H5N1), M2e-A/Hong Kong/156/97 (H5N1), M2e-A/Anhui/01/13 (H7N9), and M2e-A/Beijing/501/09 (H1N1) were analyzed using the online software PEP-FOLD (Figure 5). The 3D structures of M2e-A/Vietnam/1194/04 (H5N1) and M2e-A/Anhui/01/13 (H7N9) showed that the middle regions were similarly folded as an “open” structure, although there were five amino acid differences between these two M2e sequences (Figure 1). Although there was only one amino acid difference between M2e-A/Vietnam/1194/04 (H5N1) and M2e-A/Beijing/501/09 (H1N1), the 3D structure of M2e-A/Beijing/501/09 (H1N1) showed a relatively more “open” structure (Figure 5). However, M2e-A/Hong Kong/156/97 (H5N1), which contained amino acid variations at positions 10, 14, and 16 (Figure 1) and had low cross-reactivity with the H5N1-M2e-induced antibody (Figure 2C), had the same region folded as a hairpin structure. According to the 3D prediction model, the 12th amino acid, arginine, and the 15th amino acid, tryptophan, in the structure of M2e-A/Hong Kong/156/97 (H5N1) are bulky and are located at the top of the hairpin, which may provide steric hindrance that blocks the access to the lower region of the structure. This structure may limit the antibody-binding ability and the antigen processing by the immune system. By contrast, the 12th-position arginine and the 15th-position tryptophan of the other three M2e structures orientate in such a way that they would not block the lower access. These observations may explain why the H5N1-M2e vaccine-induced antibody could still react with M2e that contained variations at positions 13, 14, 18, 21, and 24, but it could not react with M2e containing amino acid variations at positions 10, 14, and 16, because their 3D structure were not compatible.
A slight protection (10% survival) was observed in mice vaccinated with FA alone (Figure 3). A similar phenomenon was also reported in other studies, in which vaccination with FA alone provided a slight protection (10% survival) against lethal challenge by enterovirus 71 and group A Streptococcus, respectively.34,35 A possible explanation is that the adjuvant alone may induce innate immune responses that can provide a certain level of protection against viral infections.36 Nevertheless, the survival rate of the mice vaccinated with H5N1-M2e plus FA was significantly higher than that of the mice vaccinated with FA alone (P < 0.01), indicating that it was H5N1-M2e but not the adjuvant that provided the key protection against lethal challenge with the H7N9 virus.
In summary, this study has illustrated that H5N1-M2e may provide potent cross-protection against lethal challenge from a novel avian influenza A H7N9 virus, even though approximately 21% amino acids were different existed between H5N1-M2e and H7N9-M2e. These results suggest that the M2e tetrameric peptide may provide broad spectrum of cross-protection against infections by heterogeneous influenza viruses. The M2e tetrameric peptide may be a promising candidate for the development of a universal vaccine. To improve the vaccine's protection, the M2e vaccine may be used together with inactivated virus, HA-subunit and/or other types of vaccines. Complete protection has been reported in the combined use of an M2e-based vaccine and inactivated virus vaccine.37
Acknowledgments
This research is supported by grant from the Control of Infectious Diseases, Welfare and Food Bureau of the Hong Kong SAR Government (09080812).
References
- European Center For Disease Prevention and Control Communicable disease threats report, week 50, 7–13 December 2014 Stockholm; ECDC; 2014. Available at http://www.ecdc.europa.eu/en/publications/Publications/communicable-disease-threats-report-13-dec-2014.pdf [Google Scholar]
- World Health Organization Background and summary of human infection with avian influenza A(H7N9) virus – as of 31 January 2014 Geneva; WHO; 2014. Available at http://www.who.int/influenza/human_animal_interface/20140131_background_and_summary_H7N9_v1.pdf?ua=1 [Google Scholar]
- Zhou J, Wang D, Gao R, et al. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499:500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT, Wang J, Shen Y, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling BJ, Jin L, Lau EH, et al. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet. 2013;382:129–137. doi: 10.1016/S0140-6736(13)61171-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Emergence of avian influenza A(H7N9) virus causing severe human illness – China, February–April 2013. MMWR Morb Mortal Wkly Rep. 2013;62:366–371. [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu S, Song Z, et al. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013;381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- Klausberger M, Wilde M, Palmberger D, et al. One-shot vaccination with an insect cell-derived low-dose influenza A H7 virus-like particle preparation protects mice against H7N9 challenge. Vaccine. 2014;32:355–362. doi: 10.1016/j.vaccine.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W, De Filette M, Birkett A, et al. A “universal” human influenza A vaccine. Virus Res. 2004;103:173–176. doi: 10.1016/j.virusres.2004.02.030. [DOI] [PubMed] [Google Scholar]
- Liu W, Zou P, Ding J, et al. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005;7:171–177. doi: 10.1016/j.micinf.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Hemmatzadeh F, Sumarningsih S, Tarigan S, et al. Recombinant M2e protein-based ELISA: a novel and inexpensive approach for differentiating avian influenza infected chickens from vaccinated ones. PLoS ONE. 2013;8:e56801. doi: 10.1371/journal.pone.0056801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huleatt JW, Nakaar V, Desai P, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008;26:201–214. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- Neirynck S, Deroo T, Saelens X, et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. 1999;5:1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- Zhao G, Du L, Xiao W, et al. Induction of protection against divergent H5N1 influenza viruses using a recombinant fusion protein linking influenza M2e to Onchocerca volvulus activation associated protein-1 (ASP-1) adjuvant. Vaccine. 2010;28:7233–7240. doi: 10.1016/j.vaccine.2010.08.049. [DOI] [PubMed] [Google Scholar]
- De Filette M, Martens W, Roose K, et al. An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J Biol Chem. 2008;283:11382–11387. doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins SM, Zhao ZS, Lo CY, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007;13:426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Lin Y, Du L, et al. An M2e-based multiple antigenic peptide vaccine protects mice from lethal challenge with divergent H5N1 influenza viruses. Virol J. 2010;7:9. doi: 10.1186/1743-422X-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Sun S, Du L, et al. An H5N1 M2e-based multiple antigenic peptide vaccine confers heterosubtypic protection from lethal infection with pandemic 2009 H1N1 virus. Virol J. 2010;7:151. doi: 10.1186/1743-422X-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng BJ, Chan KW, Lin YP, et al. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci USA. 2008;105:8091–8096. doi: 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Leung VH, Zhang X, et al. A recombinant vaccine of H5N1 HA1 fused with foldon and human IgG Fc induced complete cross-clade protection against divergent H5N1 viruses. PLoS ONE. 2011;6:e16555. doi: 10.1371/journal.pone.0016555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhao G, He Y, et al. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25:2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhao G, Lin Y, et al. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J Immunol. 2008;180:948–956. doi: 10.4049/jimmunol.180.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet P, Shen Y, Maupetit J, et al. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37:W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J, Derreumaux P, Tuffery P. A fast method for large-scale de novo peptide and miniprotein structure prediction. J Comput Chem. 2010;31:726–738. doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- Bommakanti G, Citron MP, Hepler RW, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci USA. 2010;107:13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan FS, Kim MC, Lee BJ, et al. Influenza M1 VLPs containing neuraminidase induce heterosubtypic cross-protection. Virology. 2012;430:127–135. doi: 10.1016/j.virol.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Lee JH, Pascua PN, et al. Prokaryote-expressed M2e protein improves H9N2 influenza vaccine efficacy and protection against lethal influenza A virus in mice. Virol J. 2013;10:104. doi: 10.1186/1743-422X-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zou P, Ding J, et al. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005;7:171–177. doi: 10.1016/j.micinf.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Zharikova D, Mozdzanowska K, Feng J, et al. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. J Virol. 2005;79:6644–6654. doi: 10.1128/JVI.79.11.6644-6654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, He D, Li Z, et al. Protection against lethal enterovirus 71 challenge in mice by a recombinant vaccine candidate containing a broadly cross-neutralizing epitope within the VP2 EF loop. Theranostics. 2014;4:498–513. doi: 10.7150/thno.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CN, Lin YC, Fann C, et al. Protection against lethal enterovirus 71 infection in newborn mice by passive immunization with subunit VP1 vaccines and inactivated virus. Vaccine. 2001;20:895–904. doi: 10.1016/s0264-410x(01)00385-1. [DOI] [PubMed] [Google Scholar]
- Billiau A, Matthys P. Modes of action of Freund's adjuvants in experimental models of autoimmune diseases. J Leukoc Biol. 2001;70:849–860. [PubMed] [Google Scholar]
- Wu F, Yuan XY, Huang WS, et al. Heterosubtypic protection conferred by combined vaccination with M2e peptide and split influenza vaccine. Vaccine. 2009;27:6095–6101. doi: 10.1016/j.vaccine.2008.11.037. [DOI] [PubMed] [Google Scholar]