Abstract
Long non-coding RNAs (lncRNAs) are emerging as important regulators of tissue physiology and disease processes including cancer. In order to delineate genome-wide lncRNA expression, we curated 7,256 RNA-Seq libraries from tumors, normal tissues, and cell lines comprising over 43 terabases of sequence from 25 independent studies. We applied ab initio assembly methodology to this dataset, yielding a consensus human transcriptome of 91,013 expressed genes. Over 68% (58,648) of genes were classified as lncRNAs, of which 79% (48,952) were previously unannotated. About 1% (597) of the lncRNAs harbored ultraconserved elements and 7% (3,900) overlapped disease-associated single nucleotide polymorphisms (SNPs). To prioritize lineage-specific, disease-associated lncRNA expression we employed non-parametric differential expression testing and nominated 7,942 lineage- or cancer-associated lncRNA genes. The lncRNA landscape characterized here may shed light into normal biology and cancer pathogenesis, and be valuable for future biomarker development.
Suggested Keywords: cancer, long noncoding RNA (lncRNA), high-throughput RNA sequencing (RNA-Seq), transcriptome, transcriptome assembly, Sample Set Enrichment Analysis (SSEA), MiTranscriptome
Introduction
Cancers are a leading cause of morbidity and mortality worldwide, with over 14 million new cases and 8 million deaths in 20121. To improve our understanding of cancer pathogenesis, ongoing large-scale efforts led by The Cancer Genome Atlas (TCGA) consortium are using high-throughput molecular profiling strategies to characterize genetic, epigenetic, and transcriptional changes2, 3. However, efforts to interpret these data have mainly focused on protein-coding genes, despite definitive evidence that transcription of the non-coding genome produces functional RNAs4. In particular, long non-coding RNAs (lncRNAs) have been implicated in biological, developmental, and pathological processes and act through mechanisms such as chromatin reprogramming, cis-regulation at enhancers, and post-transcriptional regulation of mRNA processing5, 6.
The emergence of high-throughput RNA sequencing (RNA-Seq) technology provides a revolutionary means for systematic discovery of transcriptional units. Indeed, RNA-Seq has led to a deeper appreciation of the intricate nature of transcription by revealing a milieu of lncRNAs both located in intergenic ‘gene deserts’ and overlapping protein-coding loci4. The aligned sequence data generated by RNA-Seq experiments can be used to predict full-length transcripts in silico with ab initio transcriptome assembly7, 8. Ab initio assembly provides an unbiased modality for gene discovery, and has been successful in pinpointing novel cancer-associated lncRNAs9. Despite such efforts to catalog human lncRNAs, several lines of evidence suggest that our current knowledge of lncRNAs remains inadequate. First, reported discrepancies between independent lncRNA cataloguing efforts suggest that lncRNA annotations are fragmented or incomplete10. Second, previous studies largely avoided the annotation of monoexonic transcripts and intragenic lncRNAs due to the added complexity of transcriptional reconstruction in these regions11. Third, the rapid co-evolution of high-throughput sequencing technologies and bioinformatics algorithms now enables more accurate transcript reconstruction compared to previous efforts8. Fourth, high-throughput cataloguing efforts have thus far been confined to select cell lines, individual cancer types, or relatively small cohorts4,9,11. However, cancers possess highly heterogeneous gene expression patterns and detecting recurrent expression of subtype-specific lncRNAs will likely require analysis of much larger tumor cohorts. Here, we utilized a compendium of 7,256 RNA-Seq libraries to comprehensively interrogate the human transcriptome, identifying 58,648 lncRNA genes. Moreover, we leveraged our dataset to identify myriad lncRNAs associated with 27 tissue and cancer types. By uncovering this expansive landscape of tissue- and cancer-associated lncRNAs, we provide the scientific community a powerful starting point to begin investigating their biological relevance.
Results
An expanded landscape of human transcription
We attempted to capture the spectrum of human transcriptional diversity by curating 25 independent datasets totaling 7,256 poly-A+ RNA-Seq libraries, including 5,847 from TCGA, 928 from the Michigan Center for Translational Pathology (MCTP), 67 from the Encyclopedia of DNA Elements (ENCODE), and 414 from other public datasets (Supplementary Fig. 1a and Supplementary Tables 1, 2). We developed an automated transcriptome assembly pipeline and employed it to process the raw sequencing datasets into ab initio transcriptome assemblies (Supplementary Fig. 1b, Supplementary Table 3, and Methods). This bioinformatics pipeline utilized approximately 1,870 core-months (average 0.26 core-months per library) on high-performance computing environments.
Collectively the RNA-Seq data constituted 493 billion fragments; individual libraries averaged 67.9M total fragments and 55.5M successful alignments to human chromosomes. On average 86% of aligned bases from individual libraries corresponded to annotated RefSeq exons, while the remaining 14% fell within introns or intergenic space12. We applied coarse quality control measures to account for variations in sequencing throughput, run quality, and RNA content by removing 753 libraries with (1) fewer than 20 million total fragments, (2) fewer than 20 million total aligned reads, (3) read length less than 48bp, or (4) fewer than 50% of aligned bases corresponding to RefSeq genes (Supplementary Fig. 1c, d). After coarse filtration, we obtained approximately 391 billion aligned fragments (43.69 terabases of sequence) to use for subsequent analysis. The set of 6,503 libraries passing quality control filters included 6,280 datasets from human tissues and 223 samples from cell lines. Of the tissue libraries, 5,298 originated from primary tumor specimens, 281 from metastases, and 701 from normal or benign adjacent tissues (Supplementary Fig. 1e). We subsequently refer to this set of samples as the MiTranscriptome compendium.
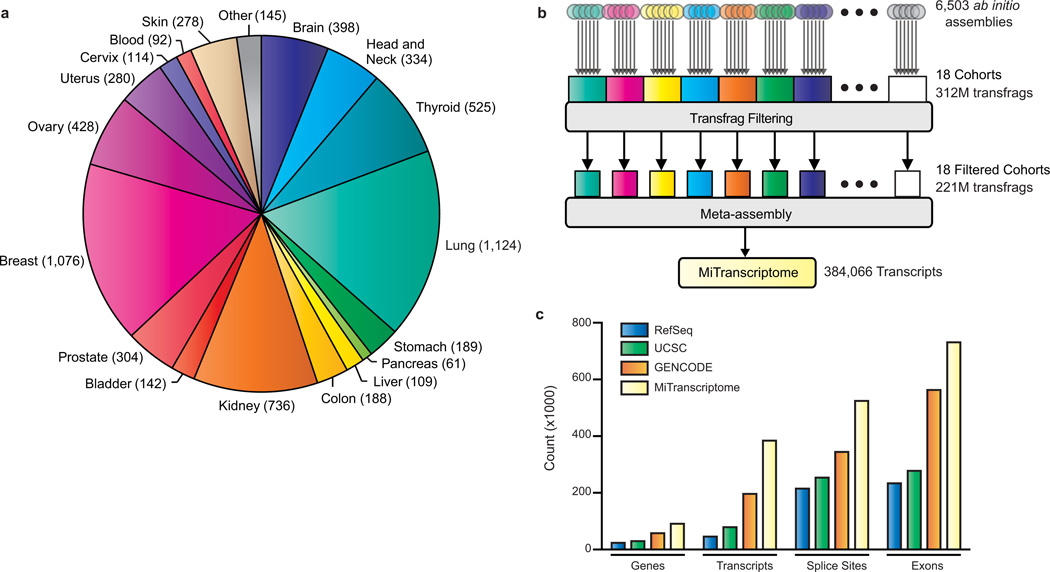
To permit sensitive detection of lineage-specific transcription we partitioned the libraries into 18 cohorts by organ system (Fig. 1a, Supplementary Table 2), performed cohort-wise filtering and meta-assembly, before re-merging the data (Fig. 1b). We developed and employed computational methods to filter library-specific background noise and predict the most likely isoforms from the assemblies of transcript fragments (transfrags) (Fig. 1b). Our filtering approach utilized transcript abundance and recurrence information to differentiate robust transcription from incompletely processed RNA or genomic DNA contamination4 (Methods). This stringent approach eliminated the vast majority (>96%) of unannotated transfrags in the compendium (Methods, Supplementary Fig. 2a–f). The remaining transfrags were collapsed into full-length transcript predictions using a greedy dynamic programming algorithm (Methods, Supplementary Fig. 3a,b). For example, in the chromosome 12 locus containing HOTAIR and HOXC11, the algorithm consolidated 7,471 raw transfrags into 17 transcripts, including ones that accurately matched annotated HOTAIR and HOXC11 isoforms (Supplementary Fig. 3c). After merging meta-assemblies from 18 organ system cohorts, we established a consensus set of 384,066 predicted transcripts that we designated as the MiTranscriptome assembly (Fig. 1b).
Figure 1. Ab initio transcriptome assembly reveals an expansive landscape of human transcription.
(a) Pie chart showing composition and cohort sizes for transcriptome reconstruction. The 6,503 RNA-Seq libraries were categorized into 18 cohorts by organ system. Organ systems with relatively few libraries were grouped together as ‘other’.
(b) Workflow diagram for transcriptome reconstruction. Ab initio assembly was carried out on each RNA-Seq library yielding transcript fragments (transfrags) predictions that may represent full or partial length transcripts. Ab initio assemblies were grouped by cohort and filtered to remove unreliable transfrags. Meta-assembly was performed on filtered transfrags for each cohort. Finally, transcripts from individual cohorts were merged to produce a consensus MiTranscriptome assembly.
(c) Bar chart comparing exons, splice sites, transcripts, and genes in the MiTranscriptome assembly with the RefSeq (Dec, 2013), UCSC (Dec, 2013) and GENCODE (release 19) catalogs.
To characterize the MiTranscriptome we compared it to reference catalogs from RefSeq (Dec, 2013)12, UCSC (Dec, 2013)13, GENCODE (Release 19)10, and intergenic lncRNA predictions from the previous cataloguing study by Cabili et al.11. We observed increases in exons, splice sites, transcripts, and genes of 29%, 52%, 95%, and 57%, respectively, relative to GENCODE, the largest of the reference catalogs (Fig. 1c and Methods). In terms of well-annotated genes, the assembly demonstrated high sensitivity at the nucleotide and splice site level, recovering 94% and 93% of RefSeq nucleotides and splice sites, respectively (Supplementary Fig. 4a,b). However, detection of precise RefSeq splicing patterns, an ongoing challenge for in silico transcriptome reconstruction methods8, was just 31%. Unannotated transcripts were defined as lacking strand-specific nucleotide overlap with reference transcripts (RefSeq, UCSC, and GENCODE). While the fraction of transcripts overlapping annotated genes was high in individual cohorts (range 62–88%, mean 75%), the fraction of annotated genes within the entire MiTranscriptome was just 46%, alluding to the presence of much unannotated transcription unique to specific lineages (Supplementary Fig. 4c).
To assess the robustness of the MiTranscriptome we stratified transcripts into confidence tiers based on annotation status, the presence of annotated splice junctions, and mono- or multi-exonic structure (Supplementary Table 4). Using the empirical cumulative distribution function derived from annotated transcript expression levels, we assigned confidence scores to unannotated transcripts (Supplementary Fig. 5a). Next, we performed qRT-PCR validations of 100 unannotated transcripts (38 mono-exonic, 62 multi-exonic) with modest expression (i.e. FPKM > 1.0) in at least one of the lung, prostate, or breast cancer cell lines A549, LNCaP, or MCF7, respectively (Methods). To assess false positives arising from background levels of genomic DNA control reactions without reverse transcriptase were also included. Of the 100 lncRNAs tested, 95 had significantly higher expression in the appropriate cell line relative to control (Student’s t-test, p-value < 0.05, Supplementary Fig. 6), and showed high correlation between qRT-PCR and RNA-Seq expression profiles (Supplementary Fig. 7a). In addition we also performed independent Sanger sequence verification of 18 amplicons that were highly expressed in the three cell lines (Supplementary Table 5, Supplementary Fig. 7b,c).
Coding potential assessment of long RNA transcripts
To facilitate further study of the assembly we classified transcripts into one of five categories: (1) Protein-coding, (2) Read-through (implying a transcript overlapped multiple separate annotated genes), (3) Pseudogene, (4) lncRNA, and (5) Transcript of Unknown Coding Potential (TUCP) (Supplementary Fig. 8a). The TUCP classification was originally suggested by Cabili et al.11 and pertains to long RNAs with in silico evidence of coding potential. The ability to predict coding potential from sequence features alone has important implications for ab initio transcript annotation studies (Supplementary Note). Here, we predicted TUCPs by incorporating two methods: (1) predictions from the Coding Potential Assessment Tool (CPAT)14, which analyzes the sequence features of transcript open reading frames (ORFs), and (2) presence of a known Pfam domain15 within a transcript ORF (Supplementary Note and Supplementary Fig. 8b–h).
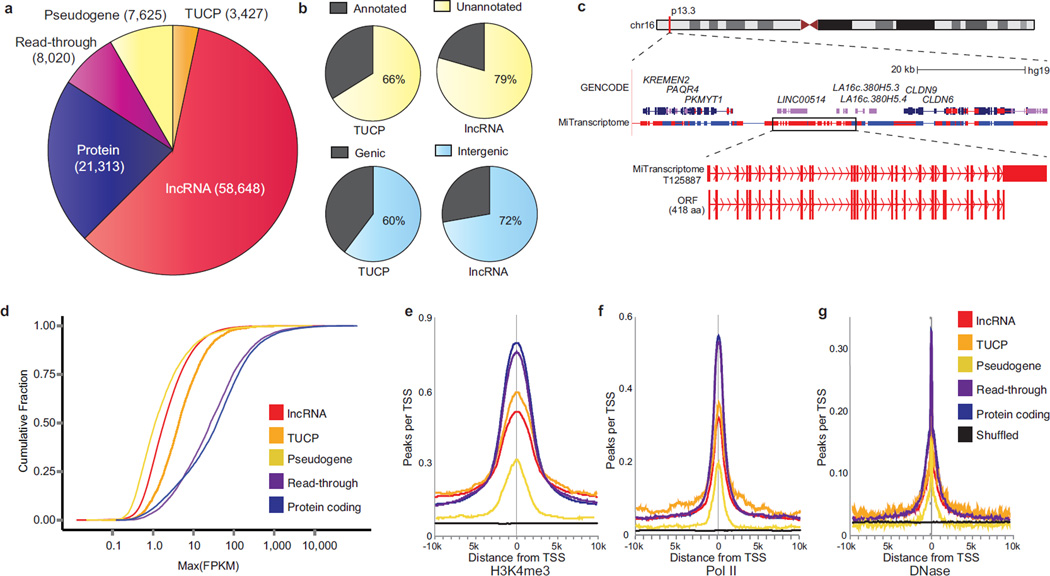
Remarkably, over sixty percent of MiTranscriptome genes were classified as either lncRNAs or TUCPs (59% lncRNAs, 3.5% TUCPs, Fig. 2a). The majority of lncRNAs and TUCPs were unannotated relative to RefSeq, UCSC, and GENCODE genes (79% and 66%, respectively) and located within intergenic regions (72% and 60%, respectively) (Fig. 2b). Interestingly, 5,248 transcripts overlapping annotated lncRNAs were flagged as TUCPs, suggesting that previous annotation attempts identified ostensibly non-coding fragments of transcripts possessing robust ORFs. For example, in a chromosome 16 intergenic locus we detected transcripts harboring a 418 amino acid ORF containing 29 exons overlapping three independent genes annotated by GENCODE as lncRNAs (LINC00514, LA16c-380H5.3, LA16c-380H5.4), suggesting that some annotated lncRNAs may in fact be inaccurate partial representations of a larger protein-coding gene (Fig. 2c). To further investigate coding potential we searched a large human proteomics dataset derived from benign tissue samples16 for peptides uniquely mapping to TUCP ORFs and noted 268 such genes (Supplementary Table 6). Given these intriguing results we anticipate that future integration of proteomics data from tumor tissues will strengthen our TUCP predictions.
Figure 2. Characterization of the MiTranscriptome assembly.
(a) Pie chart of composition and quantities of lncRNA, transcripts of unknown coding potential (TUCP), expressed pseudogene, read-through, and protein-coding genes in the MiTranscriptome assembly.
(b) Pie charts of number of lncRNAs and TUCP genes (top) unannotated versus annotated relative to reference catalogs and (bottom) intragenic versus intergenic.
(c) Genomic view of the chromosome 16p13.3 locus. Protein coding genes (PKMYT1 to CLDN9) border an intergenic region containing GENCODE lncRNA genes LINC00514 and LA16c.380H5. MiTranscriptome transcripts encompassing these genes are shown in a dense view, and (bottom) an individual isoform containing a 29-exon, 418aa ORF is highlighted. This ORF spans multiple GENCODE lncRNAs.
(d) Empirical cumulative distribution plot comparing the maximum expression (FPKM) of the major isoform of each gene across gene categories.
(e, f, and g) Plots of aggregated ENCODE ChIP-Seq data from 13 cell lines at 10kb intervals surrounding expressed transcription start sites (FPKM > 0.1) for (e) H3K4me3, (f) RNA polymerase II (Pol II), and (g) DNase hypersensitivity.
Characterization of long RNAs
LncRNA and TUCP genes tended to have fewer exons than read-through or protein coding genes, but we nevertheless observed appreciable alternative splicing for all classes of transcripts11, 17 (Supplementary Fig. 5b). Furthermore, we observed that lncRNAs and TUCPs were expressed at lower levels than read-through or protein-coding transcripts, which is consistent with previous studies9, 11, 17, 18 (Fig. 2d). To further corroborate active transcription of the lncRNAs and TUCPs, we intersected intervals surrounding the TSSs with ECNODE ChipSeq data for histone 3 lysine 4 trimethylation (H3K4me3) ChIP-Seq, RNA polymerase II (PolII) binding sites, and DNase hypersensitivity data from 13 cell lines19, 20 (Methods). Maximal enrichment of these marks at their TSSs, but not in randomly shuffled control regions, suggested that the assembled lncRNA and TUCP transcripts possess actively regulated promoters (Fig. 2e–g).
LncRNAs harboring conserved elements
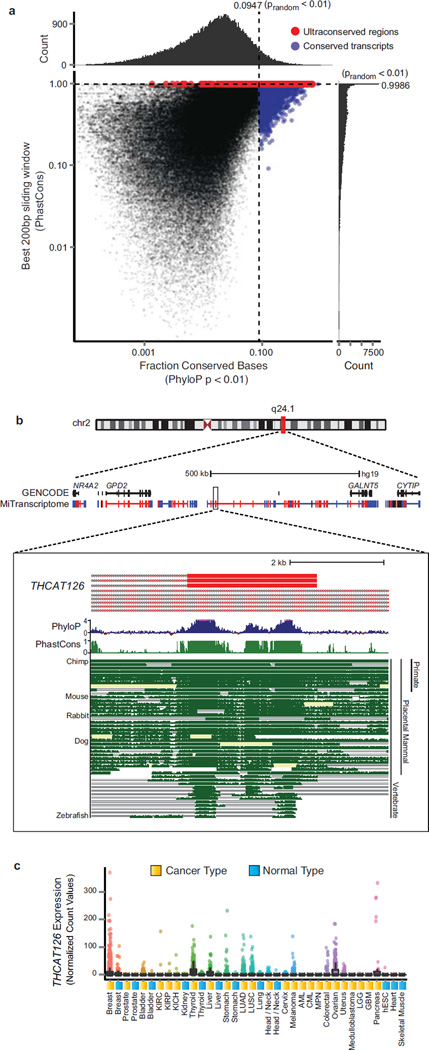
The evolutionary conservation of lncRNAs has been a topic of ongoing conversation, with several reports suggesting that lncRNAs are modestly conserved11, 17, 18, 21. In agreement with previous reports we observed increases in both transcript and promoter conservation levels for lncRNAs and TUCPs relative to random control regions (Supplementary Fig. 5c–f and Methods). Shifts in the cumulative distributions of lncRNA and TUCP transcripts were greater for annotated transcripts relative to unannotated transcripts. This difference may reflect discovery bias towards highly conserved genes detectable across multiple model systems. Moreover, the subtle conservation increases we observe for lncRNAs suggest, at least in humans, that lncRNA conservation might be an exceptional phenomenon rather than a general one. Therefore, we specifically delineated 3,309 lncRNAs (5.6% of all lncRNAs) harboring markedly higher base-wise conservation relative to random intergenic regions to enable focused study of these transcripts (Fig. 3a, Supplementary Fig. 5e and Methods). In addition, an intriguing aspect of the non-coding genome includes ultraconserved elements (UCE), which are stretches of DNA >200nt with nearly perfect sequence identity across multiple organisms22, 23. We delineated 597 intergenic lncRNAs (1.2% of all intergenic lncRNAs) harboring UCEs and designated these as Highly Conserved Long Intergenic Non-Coding RNAs (HICLINCs) (Supplementary Fig. 5h and Methods). For example, THCAT126, a previously unannotated intergenic lncRNA on chromosome 2q24, contains elements in its final exons that are conserved in nearly all vertebrates including zebrafish (Fig. 3b). Moreover, THCAT126 is expressed widely across many tissue types, including thyroid cancer (Fig. 3c). Highly conserved lncRNAs such as THCAT126 (and other cancer-associated HICLINCs described below) provide an exciting avenue for in vivo study of the role of lncRNAs in development and carcinogenesis.
Figure 3. Analysis of conservation in lncRNAs.
(a) Scatter plot with marginal histograms depicting the distribution of full transcript conservation levels (x axis) and maximal 200bp window conservation levels (y axis) for lncRNA and TUCP transcripts. Full transcript conservation levels were measured using the fraction of conserved bases (PhyloP p < 0.01). Sliding window conservation levels were measured using the average PhastCons score across 200bp regions along the transcript. Blue points indicate transcripts that were conserved relative to random non-transcribed intergenic control regions (false positive rate < 0.01). Red points indicate transcripts with 200bp windows that meet the criteria for ‘ultraconserved’ regions. Marginal histograms depict the distribution of scores along both axes. Scores of zero were omitted from the plot.
(b) Genomic view of chromosome 2q24.1 locus. Protein coding genes GALNT5 and GPD2 flank an intergenic region with no annotated transcripts. MiTranscriptome transcripts are shown in a dense view populating this intergenic space. Blue and red color represents positive and negative strand transcripts, respectively (color scheme applies to all subsequent genomic views). Most zoomed view (bottom) depicts a highly conserved exon from the lncRNA THCAT126. Multiz alignment of 46 vertebrate species depicted as well as the per base PhyloP and PhastCons conservation score.
(c) Expression data for THCAT126 across all MiTranscriptome cancer and normal tissue type cohorts.
LncRNAs overlapping disease-associated SNPs
To investigate the relationship of the MiTranscriptome assembly with disease-associated regions of the genome, we assessed overlap of transcripts in the assembly with 11,194 unique disease associated single nucleotide polymorphisms (SNPs) from a catalog of genome-wide association studies (GWAS)24. MiTranscriptome exons and transcripts overlapped 2,586 and 9,770 GWAS SNPs compared to just 1,096 and 7,050 SNPs overlapping reference transcripts, respectively (Supplementary Fig. 9a,b). Altogether transcripts in the assembly overlapped 2,881 formerly intergenic SNPs located within ‘gene deserts’, and only lacked 161 GWAS SNPs overlapping annotated genes. We tested for the possibility that the increased overlap with GWAS SNPs occurred at a rate above chance and observed that both MiTranscriptome transcripts and exons were significantly enriched for GWAS SNPs relative to random SNPs chosen from the same chip platform (paired t-test, p-value, 5.25e-135 and 1.15e-199, respectively, Supplementary Fig. 9c, Methods and Supplementary Note). Moreover, unannotated intergenic lncRNAs and TUCPs were also significantly enriched for disease-associated regions, with exons more highly enriched than full-length transcripts (paired t-test, p-values 9.90e-78 and 5.50e-50, for whole transcript and exon, respectively, Supplementary Fig. 9d). These data argue that a rigorous reevaluation of allele-specific gene expression regulation in regions proximal to GWAS SNPs may yield informative biological associations with the new lncRNAs identified in this study.
Differential Expression Analysis
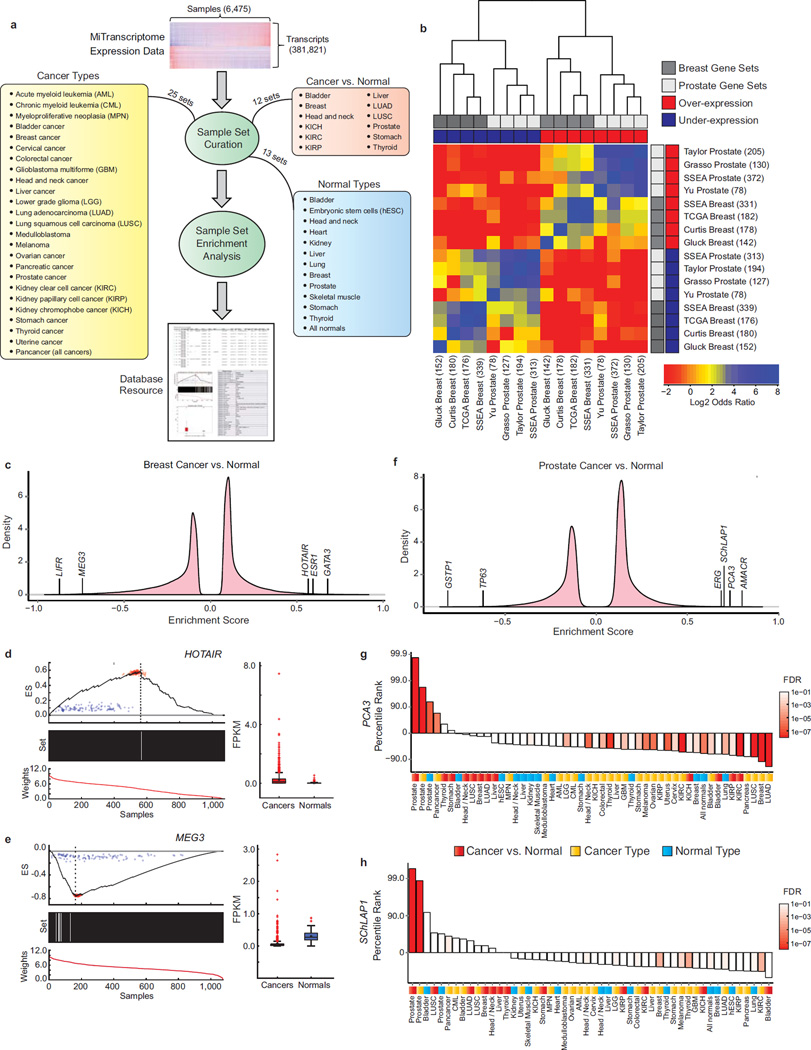
Our large-scale transcriptome reconstruction process unveiled tremendous transcriptional complexity highlighted by the presence of thousands of uncharacterized lncRNAs and TUCPs. To prioritize disease-associated and lineage-specific transcription, we developed a non-parametric method for differential expression testing called Sample Set Enrichment Analysis (SSEA) (Methods and Supplementary Note). SSEA adapts the weighted Kolmorgorov-Smirnoff-like tests used by Gene Set Enrichment Analysis (GSEA)25 to discover transcript expression changes between two groups of samples. The non-parametric nature of this method permits sensitive detection of differential expression within heterogeneous sample populations (e.g., tumor sub-types). We performed 50 differential expression analyses including various cancer or normal lineage types (i.e., one cancer or lineage type versus all other MiTranscriptome samples), and cancer versus normal comparisons within a single tissue type (Fig. 4a and Methods). Collectively, SSEA detected over two million significant associations (FDR < 1e-3 for cancer versus normal analyses and FDR < 1e-7 for lineage analyses) involving 267,726 MiTranscriptome transcripts (Supplementary Table 7 and Methods). To validate the enrichment testing approach we assessed its ability to rediscover known biomarkers up-regulated and down-regulated in prostate and breast cancers. We assessed the concordance between the top 1% positively and negatively enriched genes from each cancer type with cancer gene signatures obtained from the Oncomine database of microarray studies26–32 (Supplementary Table 8 and Methods). A heatmap of the odds ratios of the gene signature associations revealed striking agreement between SSEA and the other studies, with SSEA often demonstrating equal or better concordance to each microarray study than comparison between microarray studies (Supplementary Table 9, Fig. 4b). Thus, isoform-level differential expression testing from the MiTranscritpome ab initio assembly of RNA-Seq data recapitulated the results from cancer microarray gene expression studies, supporting the SSEA method as a viable tool for detection of differential expression.
Figure 4. Methodology for discovering cancer-associated lncRNAs.
(a) Samples were grouped into 50 different sample sets in three categories: (1) cancer type, (2) normal type, and (3) cancer versus normal. Enrichment testing was performed using SSEA, and significant transcripts were imported into an online resource.
(b) Heatmap showing concordance of SSEA algorithm with prostate and breast cancer gene signatures obtained from the Oncomine database. The top 1% over-expressed and under-expressed genes from each analysis were compared using Fisher’s Exact Tests.
(c) Enrichment score density plots for breast cancers versus normal samples.
(d and e) Enrichment and expression plots for lncRNAs (d) HOTAIR and (e) MEG3. Subplots include: (top) running ES across all samples (dotted line: max/min ES, red points: Poisson resamplings of fragment counts, blue points: random permutations of the sample labels). (middle) Black bars (cancers) or white bars (normals). (bottom) Rank-ordered normalized expression values. Adjacent boxplots (interquartile range and median shown by box and whiskers) depict transcript expression (FPKM) in cancers and normals. 967 and 109 patients in the breast cancer and normal groups, respectively.
(f) Enrichment score density plots for prostate cancers versus normal samples.
(g and h) Bar plots of percentile ranks for prostate cancer-specific lncRNAs (g) PCA3 and (h) SChLAP1 across Cancer vs. Normal (red), Cancer Type (gold) and Normal Type (blue) sample sets. Bar colors depict statistical significance (FDR).
To further credential the enrichment testing approach, we assessed the ability to detect positive control lncRNAs and protein-coding genes in breast and prostate cancers. For example, SSEA correctly identified the oncogenic lncRNA HOTAIR, estrogen receptor 1 (ESR1), and GATA binding protein 3 (GATA3) as highly positively enriched in breast cancers, and accurately nominated the tumor suppressor lncRNA MEG3 and the metastasis suppressor LIFR36 as highly negatively enriched30, 31, 33, 34 (Fig. 4c–e). Similarly, in prostate cancers SSEA detected differential expression of lncRNAs and protein-coding genes consistent with the literature (Fig. 4f). Notably, the known prostate cancer lncRNAs Prostate Cancer Antigen-3 (PCA3) and SChLAP1 were strikingly enriched in a cancer-specific and prostate-specific manner relative to all other sample set analyses (Fig. 3g,h)28, 35. Overall the ability of the enrichment testing approach to rediscover known cancer genes in an unbiased fashion indicates its utility for analysis of the cancer association and lineage specificity within the panorama of uncharacterized transcription unveiled by MiTranscriptome.
Characterization of differentially expressed lncRNAs
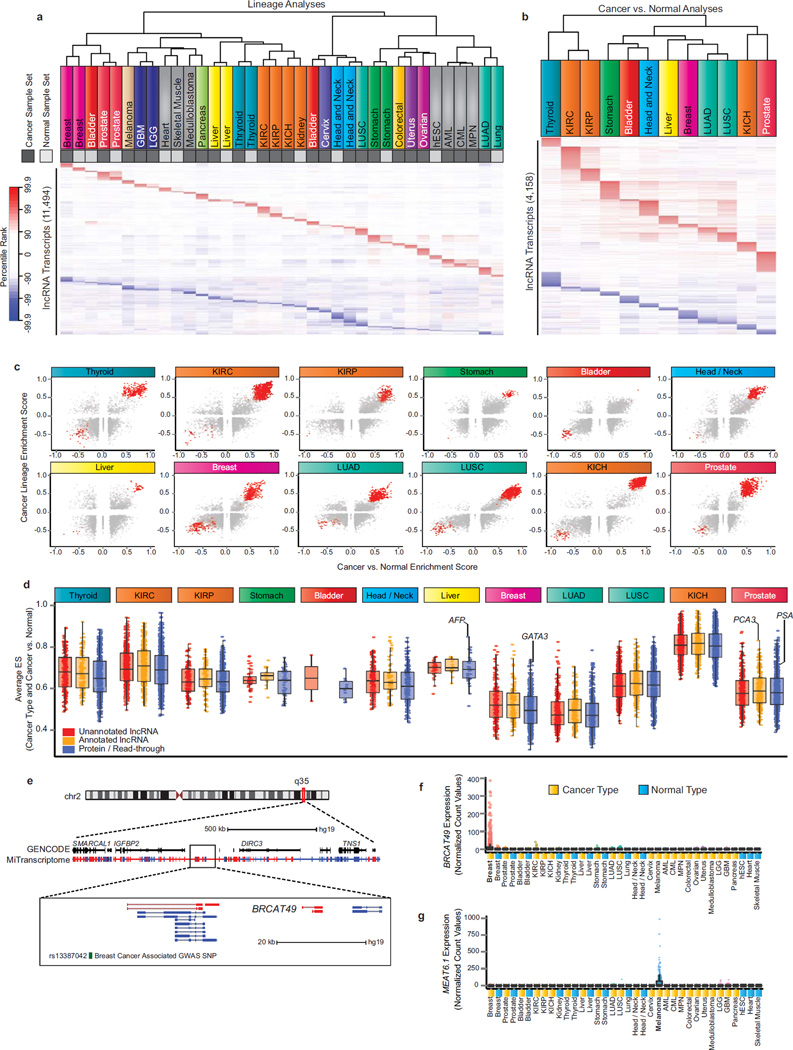
To extend our study beyond known cancer genes, we mined the enrichment test results for lineage-specific and cancer-specific transcripts in an unbiased manner. Lineage specificity was assayed using sample sets for each cancer or tissue type compared to all other samples in the MiTranscriptome compendium (Figure 4a, “Cancer Types/Normal Types”), and SSEA results were utilized to determine the degree of enrichment for each transcript in the various cancer and tissue types. Unsupervised clustering of transcript percentile ranks for the top 1% of transcripts in each lineage demonstrated distinct lineage specific signatures while also suggesting relationships among lineages and between cancer and normal sets from the same lineage (Supplementary Fig. 10a, and Methods). Examples of closely related lineage clusters include blood cancers (acute myeloid leukemia (AML), chronic myeloid leukemia (CML), and myeloproliferative neoplasia (MPN)), brain cancers (lower grade glioma (LGG) and glioblastome multiforme (GBM)), and muscle tissue (cardiac and skeletal). Additionally, a cluster comprising cervical cancer, head and neck cancer and normal lineages, lung squamous cell cancer, and bladder cancer emerged and suggested that primarily squamous (and transitional) cell carcinomas from distant primary sites share important gene expression relationships. Intriguingly, unsupervised clustering of only the lncRNAs in the top 1% of the SSEA analysis for lineage association recapitulated all of these relationships, indicating the capacity for lncRNAs to independently identify cancer and normal lineages (Fig. 5a).
Figure 5. Discovery of lineage-associated and cancer-associated lncRNAs in the MiTranscriptome compendia.
(a) Heatmap of lineage-specific lncRNAs. Each column represents a sample set from one of 25 cancer (dark grey) and normal (light grey) lineages and each row represents an individual lncRNA transcript. All transcripts were statistically significant (FDR < 1e-7) and ranked in the top 1% most positively or negatively enriched transcripts within at least one sample set. The heatmap color spectrum corresponds to percentile ranks, with under-expressed transcripts (blue) and over-expressed transcripts (red).
(b) Heatmap of cancer-specific lncRNAs nominated by SSEA Cancer vs. Normal analysis of 12 cancer types (columns). All transcripts were statistically significant (FDR < 1e-3) and ranked in the top 1% most positively or negatively enriched transcripts within at least one sample set.
(c) Scatter plots showing enrichment score for Cancer vs. Normal (x axis) and Cancer Lineage (y axis) for all lineage-specific and cancer-associated lncRNA transcripts across 12 cancer types. Red points indicate transcripts meeting the percentile cutoffs for cancer- and lineage-association.
(d) Boxplot comparing the performance of cancer- and lineage-associated lncRNAs across 12 cancer types. The average of the lineage and cancer versus normal ES is plotted on the y axis.
(e) Genomic view of chromosome 2q35 locus. Most zoomed view (bottom) depicts BRCAT49, a breast lineage and breast cancer specific lncRNA. Breast cancer associated GWAS SNP, rs13387042, is depicted in green.
(f) Expression data for BRCAT49 across all MiTranscriptome cancer and normal tissue type cohorts.
(g) Expression data for MEAT6 across all MiTranscriptome cancer and normal tissue type cohorts.
Next, we investigated the dimension of cancer-specific transcriptional dynamics in twelve tissues with ample numbers of both cancer and normal samples (Figure 4a, “Cancer vs. Normal”). Similar to above, unsupervised clustering of the top 1% cancer-associated lncRNAs demonstrated highly specific signatures for each cancer type, with the exception of lung and kidney cancers (Fig. 5b and Supplementary Fig. 10b). Lung squamous cell carcinomas (LUSC) and adenocarcinomas (LUAD) clustered together and shared numerous transcripts with cancer association. Similarly, renal clear cell (KIRC) and papillary cell (KIRP) carcinomas exhibited highly overlapping signatures, while renal chromophobe carcinomas (KICH) remained distinct from KIRC and KIRP.
Finally, we intersected results from lineage and cancer analyses. With extensive further evaluation, such transcripts may have translational potential for use in non-invasive clinical tests, particularly for cancers that lack reliable biomarkers. Notable examples included the prostate-specific lncRNAs PCA3 and SChLAP1 presented earlier (Fig. 4g,h). A myriad of lncRNAs were detected as being lineage and cancer associated (i.e. in the top 5% of both analyses) for each of the cancer types analyzed (Fig. 5c, Supplementary Fig. 11a). A direct comparison of lncRNAs and protein-coding transcripts revealed that both annotated and unannotated lncRNAs have the potential to perform at a comparable level to protein-coding genes, supporting a role for lncRNAs in augmenting tissue and cancer specificity gene signatures (Fig. 5d and Supplementary Fig. 11b,c).
We applied stringent statistical cutoffs to nominate 7,942 lncRNA or TUCP genes (11,478 transcripts) with as cancer associated, lineage associated, or both (Methods, Supplementary Table 10). Transcripts meeting the stringent cutoffs in the cancer versus normal analyses were designated as having “cancer association”. Those transcripts meeting stringent cutoffs for lineage specificity in non-cancerous tissue (e.g. heart, skeletal muscle, embryonic stem cells) and in cancers lacking RNA-Seq data for benign tissue were designated as “lineage associated”. Moreover, transcripts meeting the cutoffs for both the cancer versus normal and lineage specificity analyses were designated as having “cancer and lineage association” (Table 1). Transcripts with significant association in just one tissue type were given names according to that tissue type (Table 1), and transcripts with associations in multiple tissues were named “Cancer Associated Transcripts” (CATs). An additional 545 lncRNA genes (1634 transcripts) that possessed ultraconserved elements but did not meet the stringent lineage and cancer association criteria were designated as HICLINCs (Highly Conserved Long Intergenic Non-Coding RNA). Of these 8,487 lncRNAs, 7,804 did not possess an official gene symbol according to the HUGO Gene Nomenclature Committee36, and were thus named according to the convention described in Table 1.
Table 1. Summary of lineage and/or cancer- specific lncRNAs nominated in this study.
For each of the 27 tissue types (rows), the table lists numbers of lncRNA genes associated with tissue and/or cancer type, enriched for conserved nucleotides, containing ultraconserved elements, or classified as TUCP. Cancer and tissue specific lncRNAs only delineated for tissue types for which there was a sufficient number of matched normal to perform a cancer versus normal analysis (NA reported otherwise).
| Tissue/Cancer Type (Naming Convention) |
Total Associated Non-Coding Transcripts |
# Cancer- & Tissue- Specific |
# Conserved | # Containing Ultraconserved Element |
# Classified as TUCP |
|---|---|---|---|---|---|
| Acute Myelogenous Leukemia Associated Transcripts (AMATs) | 373 | NA | 29 | 13 | 26 |
| Bladder Cancer Associated Transcripts (BLCATs) | 61 | 0 | 9 | 2 | 5 |
| Breast Cancer Associated Transcripts (BRCATs) | 1115 | 134 | 82 | 27 | 76 |
| Cervical Cancer Associated Transcripts (CVATs) | 162 | NA | 12 | 2 | 13 |
| Chronic Myelogenous Leukemia Associated Transcripts (CMATs) | 157 | NA | 16 | 3 | 11 |
| Colorectal Cancer Associated Transcripts (CRATs) | 163 | NA | 29 | 4 | 17 |
| Glioblastoma Multiforme Associated Transcripts (GBATs) | 161 | NA | 11 | 2 | 22 |
| Head and Neck Cancer Associated Transcripts (HNCATs) | 766 | 5 | 45 | 15 | 68 |
| Heart Tissue Associated Transcripts (HRATs) | 170 | NA | 16 | 1 | 12 |
| Human Embryonic Stem Cells Associated Transcripts (ESATs) | 205 | NA | 10 | 0 | 20 |
| Chromophobe Renal Cell Carcinoma Associated Transcripts (KCHCATs) | 1050 | 52 | 64 | 20 | 92 |
| Renal Clear Cell Carcinoma Associated Transcripts (KCCATs) | 1429 | 215 | 84 | 26 | 123 |
| Renal Papillary Cell Carcinoma Associated Transcripts (KPCATs) | 474 | 0 | 41 | 8 | 38 |
| Low Grade Glioma Associated Transcripts (LGATs) | 265 | NA | 31 | 10 | 23 |
| Liver Cancer Associated Transcripts (LVCATs) | 250 | 0 | 18 | 1 | 20 |
| Lung Adenocarcinoma Associated Transcripts (LACATs) | 953 | 19 | 64 | 19 | 61 |
| Lung Squamous Cell Carcinoma Associated Transcripts (LSCATs) | 1014 | 10 | 70 | 23 | 58 |
| Medulloblastoma Associated Transcripts (MBATs) | 312 | NA | 26 | 3 | 33 |
| Melanoma Associated Transcripts (MEATs) | 339 | NA | 24 | 2 | 34 |
| Myeloproliferative Neoplasia Associated Transcripts (MPATs) | 101 | NA | 12 | 1 | 8 |
| Ovarian Cancer Associated Transcripts (OVATs) | 163 | NA | 37 | 12 | 30 |
| Pancreatic Cancer Associated Transcripts (PNATs) | 247 | NA | 27 | 4 | 22 |
| Prostate Cancer Associated Transcripts (PRCATs) | 727 | 38 | 49 | 14 | 62 |
| Skeletal Muscle Tissue Associated Transcripts (SMATs) | 123 | NA | 5 | 1 | 11 |
| Stomach Cancer Associated Transcripts (STCATs) | 95 | 0 | 10 | 1 | 10 |
| Thyroid Cancer Associated Transcripts (THCATs) | 1289 | 80 | 73 | 21 | 111 |
| Uterine Endometrial Carcinoma Associated Transcripts (UTATs) | 183 | NA | 31 | 1 | 16 |
To infer putative roles for cancer or lineage associated lncRNAs in oncogenesis, we curated 2,078 MSigDB gene sets into categories corresponding to biological function (angiogenesis/hypoxia, metastasis, proliferation/cell-cycle, cell adhesion, DNA damage/repair) or signatures from gene expression profiling studies (Supplementary Table 11)25. We constructed an expression correlation matrix between lncRNAs and protein-coding genes and employed a “guilt by association” analysis whereby the correlation data was processed by GSEA to generate a matrix of the association of each lncRNA with each gene set, capturing over 14,000 transcripts with significant associations (Methods and Supplementary Tables 12, 13)37.
To allow the scientific community to explore our discoveries, we developed an online portal featuring detailed characteristics of the nominated transcripts (see URLs), and present several examples of intriguing lncRNAs here. First, the lncRNA Breast Cancer Associated Transcript-49 (BRCAT49) is a breast cancer- and lineage-associated lncRNA (Fig. 5d) located ~45kb downstream of the intergenic breast cancer SNP rs13387042 that has been implicated by multiple GWAS studies (Fig. 5e,f)38–42. BRCAT49 provides a possible target for explaining the breast cancer association of this genomic region, and would be a candidate for intergenic expression quantitative trait loci (eQTL) analysis. Further interrogation of the relationship with GWAS SNPs was also performed, and all transcripts within 50kb of a GWAS SNP implicated in a disease site for which the lncRNAs was identified as having a significant association are reported in Supplementary Table 14. Second, the lncRNA we termed Melanoma Associated Transcript-6 (MEAT6) was found to be in the 99.8th percentile in the melanoma lineage SSEA analysis (Fig. 5a). Genomic investigation delineated MEAT6 as a partially annotated transcriptional variant of the lncRNA AK090788 on chromosome 6q26 (Supplementary Fig. 12a). However, MEAT6 utilizes an alternative start site and upstream exons absent from reference catalogs. Expression of MEAT6 isoforms using the novel start site were highly specific to the melanoma samples in the MiTranscriptome cohort (Fig. 5g); however, isoforms lacking the MEAT6 start site had a dramatically different pan-cancer expression profile with almost no expression in melanoma (Supplementary Fig. 12b). Additional examples of expression profiles for cancer- or lineage-specific lncRNAs in other tissue types are displayed in Supplementary Fig. 12c,d. The examples shown here are indeed representative, and we anticipate that an abundance of uncharacterized transcription with biological and translational potential can be leveraged using our discoveries here and our online resource (see URLs, Supplementary Tables 10, 11).
Discussion
Here, we discovered and characterized an expanded landscape of transcription via unbiased transcriptome reconstruction from thousands of tumors, normal tissues, and cell lines. Our work utilizes several orders-of-magnitude more RNA-seq data (~100 fold) than previous RNA-seq lncRNA discovery efforts and vastly increases the universe of known transcripts in both normal tissues and cancer. The unprecedented breadth (6,503 samples) and depth (>43 Terabases of sequence) of our compendia enabled sensitive detection of robust transcription and specific filtration of background noise. The lncRNAs in our assembly (58,648 genes, often with multiple isoforms) far outnumber entries in current lncRNA databases (<16,000 genes), implying that reference transcript annotations may be fragmented or otherwise incomplete11, 17, 43–46. Moreover, our assembly indicates that the genomic diversity of lncRNAs eclipses coding transcripts (i.e., nearly 60,000 lncRNA genes versus approximately 30,000 protein coding genes), a disparity that may grow as additional diseases and cell types are sequenced and more lncRNAs are discovered.
Multiple lines of in silico evidence support the biological and functional relevance of MiTranscriptome transcripts, including robust expression, protein-coding potential (for TUCPs), high conservation, active regulation at promoters, proximity to disease-associated genomic polymorphisms, correlation with protein-coding gene signatures, lineage-specificity, and cancer-specificity. Moreover, many lncRNAs independently identified by this study have been previously validated and mechanistically linked to carcinogenesis (Supplementary Table 15)34, 35, 47–49. Regardless of their functional contributions, uncharacterized MiTranscriptome transcripts could serve as future cancer biomarkers.
Although the central dogma remains a core tenet of cellular and molecular biology, the appreciation of lncRNAs as functional genomic elements that defy the central dogma may be essential for fully understanding biology and disease. Taken together, our results indicate that the vastness and complexity of lncRNA transcription has been grossly underappreciated, and that myriad lncRNAs are associated with carcinogenesis. We anticipate that the MiTranscriptome assembly and lncRNAs identified by this study, as well as the computational tools developed herein, will provide a foundation for lncRNA genomics, biomarker development, and the delineation of cancer disease mechanisms.
Online Methods
High performance computing
Computational analysis was performed using the Flux high-performance computer cluster hosted by the Advanced Research Computing (ARC) at the University of Michigan.
RNA-Seq Data Processing
A comprehensive RNA-Seq analysis pipeline was employed on all samples (Supplementary Fig. 1b). The analysis pipeline provided sequence quality metrics, filtering of contaminant reads, fragment size estimation, strand-specific library type estimation, spliced alignment of reads to the human reference genome (version hg19/GRCh37), alignment performance metrics, generation of visualization tracks for genome browsers, and ab initio transcript assembly. The third-party tools used to process RNA-Seq data were selected based on computational performance, ease-of-use, user and community support, and experience (Supplementary Table 3). Further details are described in Supplementary Note.
Overview of transcriptome reconstruction
To merge ab initio assembled transcript fragments (transfrags) into a consensus transcriptome we developed and utilized a bioinformatics method that (1) classifies and filters sources of background noise in individual libraries and (2) reassembles transfrags weighted by their expression levels from multiple libraries into a consensus transcriptome. More details in Supplementary Note.
Filtration of noise contamination
We controlled for alignment artifacts and poorly assembled transcripts by clipping very short first or last exons (< 15bp) and excluding short transfrags (≤ 250bp). We removed noise due genomic DNA contamination and incompletely processed RNA using a machine learning method. The method models the empirical distributions of relative transcript abundance and recurrence (number of independent samples in which the transcript was observed). From this model the method determines optimal library-specific thresholds for distinguishing annotated from unannotated transcription as a proxy for signal versus background noise, respectively. Further details are described in Supplemental Note.
Transcriptome meta-assembly
We created directed acyclic splicing graphs where nodes in the graph reflected contiguous exonic regions and edges corresponded to splicing possibilities (Supplementary Fig. 3a). Nodes in the splicing graph with relatively low abundance were then pruned. We then incorporated partial path information inherent in transfrags spanning multiple exons by building splicing pattern graphs that subsumed the original splice graphs (Supplementary Fig. 3b). The splicing pattern graph is a type of De Bruijn graph where each node represents a contiguous path of length k through the splice graph, and edges connect paths with k-1 nodes in common. The algorithm finds and reports a set of highly abundant transcripts by iteratively traversing the graph using dynamic programming in a greedy fashion. Further details are described in Supplementary Note.
Merging of meta-assemblies
To merge meta-assemblies from 18 cohorts we used the Cuffmerge tool50, which produced a final transcriptome GTF file.
Comparisons of MiTranscriptome with reference catalogs
The exons, splice sites, and splicing patterns of all assembled transcripts were compared to RefSeq, UCSC, GENCODE (version 19), and the merged union of all three reference catalogs using custom python scripts. Sensitivity and precision values were computed using the number of shared strand-specific transcribed bases, introns, and splicing patterns. Precision was also computed for the subset of ab initio transcripts that overlapped any part of a reference transcript. Transcripts that overlapped a reference transcript on the same strand were designated annotated. When an ab initio transcript matched multiple reference transcripts, a best match was chosen using the following criteria: (1) matching splicing pattern, (2) fraction of shared introns, and (3) fraction of shared transcribed bases. The biotype (protein, read-through, pseudogene, or lncRNA) for annotated transcripts was imputed from the best matching reference transcript. Annotated lncRNAs and unannotated transcripts were reclassified as either lncRNAs or TUCPs.
Prediction of transcripts of unknown coding potential (TUCP)
We predicted coding potential by integrating two sources of evidence: (1) predictions from the alignment-free Coding Potential Assessment Tool (CPAT)14 and (2) searches for Pfam 27.0 matches15. CPAT determines the coding probability of transcript sequences using a logistic regression model built from ORF size, Fickett TESTCODE statistic51, and hexamer usage bias. We chose a CPAT probability cutoff by repeatedly randomly sampling 100,000 each of putative non-coding and protein-coding transcripts and optimizing on the balanced accuracy (average of sensitivity and specificity) metric (Supplementary Fig. 8b,c). The average area-under-the-curve (AUC) across 100 iterations was 0.9310 (minimum 0.9302, maximum 0.9320), and the average optimal probability cutoff was 0.5242 (minimum 0.5090, maximum 0.5482). This cutoff value achieved accurate discrimination of lncRNAs and protein-coding genes (sensitivity: 0.84, specificity: 0.95, FDR: 0.076). Of the putative non-coding transcripts 9,903 (5.3%) exceeded the CPAT cutoff and met the criteria for TUCP. As additional evidence of coding potential we scanned all transcripts for Pfam A or B domains across the three translated reading frames for stranded transcripts and six frames for monoexonic transcripts of unknown strand (Supplementary Note). We designated putative non-coding transcripts with either a Pfam domain or a positive CPAT prediction as TUCP.
Proteomics analysis
We obtained the following Thermo files (in the RAW format) from a recent study mapping the human proteome52: Adult_Kidney_Gel_Elite_55, Adult_Liver_Gel_Elite_56, Adult_Pancreas_Gel_Elite_60, Adult_Rectum_Gel_Elite_63, Adult_Urinarybladder_Gel_Elite_40, Fetal_Brain_Gel_Velos_16, Adult_Lung_Gel_Elite_56, and Adult_Prostate_Gel_Elite_62. The Thermo files were transformed into mzXML using MSConverter53 and interrogated against human UniProt database V.15.11 using the X!tandem search engine. The database was concatenated with all possible open reading frames longer than 7 amino acids from the lncRNAs and with reversed sequences for determination of false discovery rate (FDR). The X!Tandem search parameters were: fully tryptic cleavage, parent mass error 5 ppm, fragment mass error 0.5 Da, 2 allowed missed cleavages. Fixed modifications: Cys carbamidomethylation. Variable modifications: Met oxidation. X!Tandem output files were processed by PeptideProphet and ProteinProphet. Data was filtered at peptide probability 0.5 and protein probability 0.9 to ensure protein FDR < 1%.
Confidence scoring system
After assembly of the MiTranscriptome, transcripts were subjected to an additional confidence evaluation. lncRNAs in the MiTranscriptome were categorized into tiers based on their annotation status and the degree of matching of splice junctions to the reference annotation (Supplementary Table 4). Tier 1 transcripts are all annotated and tier 2 transcripts are unannotated. An empirical cumulative distribution function (eCDF) was developed by profiling the second highest expression value (across all 6,503 samples) for each tier 1 transcript. The second highest value was used to control for outlier expression. The eCDF was using to compute confidence scores for tier 2 transcripts using the same expression summary statistic.
Validation of lncRNA transcript by qRT-PCR
We chose 150 lncRNAs with at least 1 FPKM expression in either A549, LNCaP, or MCF7 cells for biological validation. For each transcript, primer pairs were designed using the Primer-BLAST tool54. Primer pairs with the following parameters were selected: (1) amplicon length between 80–140 bp (2) primer GC content between 35–65%, and (3) primer length greater than 20 bp. Primers were blasted against the human genome to ensure specificity to our target gene, and primers designed against multi-exonic transcripts spanned exon junctions. Regions of any transcript that directly overlapped an exon on the antisense strand were avoided. Primer pairs meeting these criteria could be designed for 100 out of 150 lncRNAs (38 monoexonic and 62 multiexonic). All oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA) and are listed in Supplementary Table 5.
RNA was isolated from A549, LNCaP and MCF7 cells in Trizol (Invitrogen) using the RNeasy Mini Kit (Qiagen). Equal amount of RNA was converted into cDNA using random primers and the Superscript III reverse transcription system (Invitrogen). Quantitative real-time PCR (qPCR) was performed using Power SYBR Green Mastermix (Applied Biosystems, Foster City, CA) on an Applied Biosystems 7900HT Real-Time PCR System. The housekeeping genes, CHMP2A, EMC7, GPI, PSMB2, PSMB4, RAB7A, REEP5, SNRPD3 were used as loading controls55. Data was normalized first to housekeeping genes and then to the median value of all samples using the delta-delta Ct method and plotted as fold change over median. To ensure the specificity of the primers, 20 amplicons were further analyzed by Sanger sequencing.
Cell lines and reagents
All cell lines were obtained from the American Type Culture Collection (Manassas, VA). Cell lines were maintained using standard conditions. Specifically, A549 were grown in F-12K plus 10% fetal bovine serum (FBS), LNCaP in RMPI1640 (Invitrogen) plus 10% FBS and 1% penicillin-streptomycin, and MCF7 in Eagle’s Minimum Essential Media (EMEM) plus 10% FBS. All of the cell lines were grown at 37°C degrees in a 5% CO2 cell culture incubator. To ensure identity, cell lines were genotyped at the University of Michigan Sequencing Core using Profiler Plus (Applied Biosystems) and compared with the short tandem repeat (STR) profiles of respective cell lines available in the STR Profile Database (ATCC). All of the cell lines were routinely tested and found to be free of Mycoplasma contamination.
Evidence for active regulation of transcriptional start sites
To conduct analysis of TSS intervals ENCODE project datasets were downloaded from the UCSC Genome Browser13. For H3K4me3 analysis we used the Encode Project Broad Institute H3K4me3 ChIP-Seq peaks for cell lines GM12878, H1-hESC, HeLa-S3, HepG2, HMEC, HSMM, HSMMtube, HUVEC, K562, NH-A, NHDF-Ad, NHEK, and NHLF56. For RNA polymerase II analysis we used POL2RA binding sites from the Encode Project Uniform TFBS master file version 3 for any of the cell lines with H3k4me3 data19. Finally, for the DNase hypersensitivity analysis the Encode Project combined UW and Duke DNaseI hypersensitivity regions were downloaded as a master file from EMBL-EBI, and filtered for any of the cell lines with H3k4me3 data. Peak enrichment files (BED format) were aggregated across all cell lines. Intervals of +/− 10 kilobases surrounding unique MiTranscriptome TSSs were generated using BEDTools ‘slop’ tool57. To control for expression, TSSs were filtered for transcripts not expressed in any of the cell lines (FPKM < 0.1). Basewise peak coverage was generated for each TSS interval using the BEDTools ‘coverage’ function and summarized across subsets of TSSs. Summed per-base coverage histograms were normalized by dividing by the number of expressed TSSs.
Conservation analysis
The evolutionary conservation of transcripts in our assembly was studied using two metrics: (1) the fraction of significantly conserved bases (p ≤ 0.01, phyloP algorithm), and (2) the maximally conserved 200nt sliding window (phastCons scores averaged within each window). The former captures independently conserved elements within a transcript regardless of position, and the latter captures contiguous regions of high conservation. The 200nt sliding window size was chosen to aid in discovery of putative ultraconserved elements23. As a negative control we measured the conservation of non-transcribed regions using these metrics by randomly sampling contiguous length-matched intervals from intergenic and intronic space. Non-transcribed interval sampling was restricted to regions with valid 46-way conservation data.
The fractional basewise conservation and contiguous window conservation metrics were used to nominate highly conserved and ultraconserved transcripts, respectively. In both cases cutoffs for significant transcripts were determined by controlling the rate of observing elements with similar conservation levels within non-transcribed intergenic space at a level of 0.01. For fractional basewise conservation a score of 0.0947 (9.5% of transcript bases conserved at phyloP p-value < 0.01) corresponded to a false discovery rate < 0.01. At this cutoff the sensitivity for detecting protein-coding transcripts was 0.67. For contiguous sliding window conservation an average PhastCons probability of 0.9986 corresponded to a false discovery rate < 0.01. At this cutoff the sensitivity for detecting true positive ultraconserved non-coding elements downloaded from UCNEbase was 0.6922. Applying these criteria to our assembly yielded 6,034 lncRNAs (3.4%) and 541 TUCPs (4.7%) with significant basewise conservation levels. Additionally, 1,686 lncRNAs (0.96%) and 121 TUCPs (0.01%) harbored contiguous ultraconserved regions.
GWAS analysis
A list of GWAS SNPs was obtained from the National Human Genome Research Institute’s GWAS catalog (accessed Jan 6, 2014)24. SNP haplotypes were excluded from the SNP overlap analysis, and a list of 11,194 unique SNPs was obtained. The merged union of the RefSeq, UCSC, and GENCODE catalogs was used as a reference for comparison with MiTranscriptome. Please refer to Supplementary Methods for a description of the GWAS overlap enrichment testing analysis.
Transcript expression estimation
Expression levels (FPKM) of the transcripts in the assembly were determined using Cufflinks (version 2.02 and 2.1.1)58. Normalized abundance estimates (FPKM) were computed for all MiTranscriptome transcripts, converted into approximate fragment count values, and aggregated into a matrix of expression data (Fig. 4a and Supplementary Methods). Library size factors for expression normalization were computed by applying the geometric normalization method described by Anders and Huber59.
Transcript expression enrichment analysis
To analyze differential expression of transcripts relative to sample phenotypes we developed a method called Sample Set Enrichment Analysis (SSEA). SSEA performs weighted KS-tests using normalized count data vectors as weights. To convert count values into weights for a single KS-test the following steps are performed: (1) raw count values are normalized by library-specific size factors, (2) normalized count values are “resampled” from a Poisson distribution (lambda equals the observed count value) to mimic the effect of technical replication, and (3) random Poisson noise (by default, lambda equals 1) is added to the normalized, resampled count values to destabilize zero-valued counts and break ties. A power transform (exponential or logarithmic) is then applied to the weights (by default, a log-transformation is applied after incrementing normalized count values by 1). The choice of power transformation influences the relative importance of precision versus recall during enrichment testing. For example, users aiming to discover genes new in molecular subtypes of a disease would prioritize precision over sensitivity, whereas a user aiming to discover ideal biomarkers may value sensitivity over precision. Following count data normalization and power transformation, SSEA performs the weighted KS-test procedure described in GSEA25, 60. The resulting enrichment score (ES) statistic describes the strength of association between the weights and the sample set.
To control for random sampling bias in count values (e.g. “shot noise”) SSEA performs repeated enrichment tests using resampled count values to mimic observations from technical replicates and uses the median enrichment score (by default, 100 tests are performed). The basis for Poisson resampling as a legitimate model for technical replication was established by Marioni et al.60 To test for significance, SSEA performs enrichment tests using randomly shuffled sample labels to derive a set of null enrichment scores with the same sign as the observed score (by default, 1000 null enrichment scores are computed). The nominal p value reported is the relative rank of the observed enrichment score within the null enrichment scores. To control for multiple hypothesis testing, SSEA maintains the null normalized enrichment score (NES) distributions for all transcripts in a sample set, and uses the null NES distribution to compute FDR q values in the same manner as proposed by Subramanian et al.25
Benchmarking SSEA performance using microarray gene signatures
Gene signatures for the top 1% of overexpressed and underexpressed genes from three prostate cancer26–28 and three breast cancer29–31 microarray studies were obtained using Oncomine32 (Supplementary Table 8). The top 1% gene signatures as detected by SSEA in the MiTranscriptome breast and prostate cohorts were determined using prostate cancer versus normal and breast cancer versus normal sample sets (Fig. 4a). Given that the MiTranscriptome was produced from an ab initio assembly, transcript identity was assigned to the annotated reference gene with the greatest degree of concordance, where degree of splicing agreement was prioritized over degree of exonic same-stranded overlap. The most-enriched isoform for each gene was used to produce a gene signature.
Degree of overlap for all combinations of the 16 gene sets tested (3 published breast up-regulated sets, 3 published breast down-regulated sets, 3 published prostate up-regulated sets, 3 published prostate down-regulated sets, 1 SSEA-determined prostate up-regulated set, 1 SSEA-determined prostate down-regulated set, 1 SSEA-determined breast up-regulated and 1 SSEA-determined breast down-regulated set) was determined by calculating an odds ratio and performing a Fisher’s exact test for each gene set pair (Supplementary Table 9). Each comparison was restricted to the set of genes assessed by both profiling platforms. Microarray chip annotation files were downloaded from the Molecular Signatures Database (MSigDB)61 . The set of all annotated genes (relative to RefSeq, UCSC, and GENCODE) was used as the annotation file for MiTranscriptome. Unsupervised hierarchical clustering of the heatmap data was performed using the ‘euclidean’ distance measure and the ‘complete’ agglomeration method.
Discovery of lineage-specific and cancer-specific transcripts
To generate enrichment test data for unsupervised clustering, we ranked transcripts within each SSEA sample set by normalized enrichment score (NES) and assigned fractional ranks (e.g. a fractional rank of 0.95 implies the transcript ranked in the top 5th percentile of all transcripts in the sample set). Only significant results (FDR < 1e-7 for lineage analysis and FDR < 1e-3 for cancer versus normal analysis) were used. Unsupervised clustering was performed using Pearson correlation of log-transformed fractional ranks as a distance metric and Ward’s method. Transcripts that were significantly associated with multiple sample sets were grouped with the most strongly associated sample set. Heatmaps were produced using the ‘heatmap.2’ function from the ‘gplots’ package in R.
Guilt-by-association GSEA analysis
For each cancer and/or lineage associated lncRNA (Supplemental Table 10), expression levels of the target lncRNA were correlated to the expression of all protein-coding genes across all samples in the associated tissue cohort. For cancer cohorts (e.g. breast, prostate), correlations were performed (Spearman) using only the cancer samples (i.e. normal samples were excluded). The protein-coding genes were then ranked by the Rho value, and used in a weighted, pre-ranked GSEA analysis against a collection of cancer associated gene sets from MSigDB (Supplementary Table 11). Significant associations were determined for any gene set having an FWER p-value below 0.001.
Supplementary Material
Acknowledgements
We thank Brock Palen and Jeremy Hallum for technical assistance with the high performance computing cluster, Sameek Roychowdhury for reviewing the manuscript, the University of Michigan DNA Sequencing Core for Sanger sequencing, and K. Giles for critically reading manuscript and submission of documents. This work was supported in part by the NIH Prostate Specialized Program of Research Excellence grant P50CA69568, the Early Detection Research Network grant UO1 CA111275, US National Institutes of Health R01CA132874, RO1 CA154365 (D.G.B, A.M.C.), and the Department of Defense grant PC100171 (A.M.C.). A.M.C. is supported by the Prostate Cancer Foundation and the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor and a Taubman Scholar of the University of Michigan. R.M. was supported by a Prostate Cancer Foundation Young Investigator Award and the Department of Defense Post-doctoral Fellowship W81XWH-13-1-0284. Y.S.N. is supported by a University of Michigan Cellular & Molecular Biology NIGMS Training Grant.
Footnotes
URLs
MiTranscriptome Online Portal: mitranscriptome.org
Accession codes
Transcript sequences have been submitted to GenBank (accession number pending).
Author Contributions
M.K.I., Y.S.N. and A.M.C. conceived the study and analyses. M.K.I. processed RNA-seq data and performed ab initio assembly. M.K.I. and Y.S.N. performed data processing and data analysis with assistance from T.R.B., R.M., A.S., Y.H., J.E., S.Z., J.R.P., and F.Y.F.; R.M., U.S., A.S., and Y.H. performed qPCR validations; M.K.I. and Y.S.N. developed SSEA with help of H.K.I.; D.G.B. contributed primary samples. D.R.R., W.Y. and S.M.D. generated RNA-seq libraries and X.C. performed the sequencing. M.K.I., Y.S.N., and A.S. developed the web resource; T.R.B. provided systems administration, data storage, high-performance computing, and networking support. A.P. performed the proteomics analysis. M.K.I, Y.S.N., and A.M.C. wrote the manuscript. All authors discussed results and commented on the manuscript.
Competing financial interests
Oncomine is supported by ThermoFisher, Inc. (Previously Life Technologies and Compendia Biosciences). A.M.C was a co-founder of Compendia Biosciences and served on the scientific advisory board of Life Technologies before it was acquired.
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer. 2014 doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nature genetics. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nature methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruitt KD, et al. RefSeq: an update on mammalian reference sequences. Nucleic acids research. 2014;42:756. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic acids research. 2014;42:764. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic acids research. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:222. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenbloom, et al. ENCODE Data in the UCSC Genome Browser: year 5 update. Nucleic Acids Research. 2012;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrieva S, Bucher P. UCNEbase--a database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic acids research. 2013;41:101. doi: 10.1093/nar/gks1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 24.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:1001. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 28.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gluck S, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/− trastuzumab. Breast cancer research and treatment. 2012;132:781–791. doi: 10.1007/s10549-011-1412-7. [DOI] [PubMed] [Google Scholar]
- 30.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nature medicine. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Research. 2014 doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nature genetics. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nature genetics. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 40.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature genetics. 2013;45:353–361. doi: 10.1038/ng.2563. 361e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nature genetics. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, et al. A combined analysis of genome-wide association studies in breast cancer. Breast cancer research and treatment. 2011;126:717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- 43.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic acids research. 2011;39:146. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volders PJ, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic acids research. 2013;41:246. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park C, Yu N, Choi I, Kim W, Lee S. lncRNAtor: a comprehensive resource for functional investigation of long noncoding RNAs. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu325. [DOI] [PubMed] [Google Scholar]
- 46.Hangauer Vaughn, McManus Rinn. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. The Journal of biological chemistry. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 48.Tomlins SA, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Science translational medicine. 2011;3 doi: 10.1126/scitranslmed.3001970. 94ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prensner JR, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer research. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fickett JW. Recognition of protein coding regions in DNA sequences. Nucleic acids research. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chambers, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnology. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye, et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends in genetics : TIG. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liberzon, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.