Abstract
De novo somatic mutations in focal areas are well documented in diseases such as neoplasia but are rarely reported in malformation of the developing brain. Hemimegalencephaly (HME) is characterized by overgrowth of either one of the two cerebral hemispheres. The molecular etiology of HME remains a mystery. The intractable epilepsy that is associated with HME can be relieved by the surgical treatment hemispherectomy, allowing sampling of diseased tissue. Exome sequencing and mass spectrometry analysis in paired brain-blood samples from individuals with HME (n = 20 cases) identified de novo somatic mutations in 30% of affected individuals in the PIK3CA, AKT3 and MTOR genes. A recurrent PIK3CA c.1633G>A mutation was found in four separate cases. Identified mutations were present in 8–40% of sequenced alleles in various brain regions and were associated with increased neuronal S6 protein phosphorylation in the brains of affected individuals, indicating aberrant activation of mammalian target of rapamycin (mTOR) signaling. Thus HME is probably a genetically mosaic disease caused by gain of function in phosphatidylinositol 3-kinase (PI3K)-AKT3-mTOR signaling.
Malformations of cortical development (MCD) are increasingly recognized as an important cause of epilepsy and developmental delay. Disruption at various critical stages of cortical development—neuronal proliferation, migration and organization—leads to characteristic MCD1,2. HME is a clinically devastating pediatric MCD characterized by enlargement of one cerebral hemisphere. Individuals with HME typically present with epilepsy, psychomotor disability, contralateral hemiparesis and hemianopsia3. Although most affected individuals have isolated forms of the disease, HME is occasionally associated with neurocutaneous syndromes, including tuberous sclerosis, hypomelanosis of Ito (HI) and Proteus syndrome (PS)4. Most affected individuals display progressive and catastrophic epilepsy, which is medically intractable and inevitably requires radical hemispherectomy (removal of the affected cerebral hemisphere) as a surgical treatment to relieve epilepsy and psychomotor disability4. HME occurs sporadically with no association with a specific ancestry group or gender. In addition, focal involvement of one affected hemisphere and discordance in monozygotic twins imply that somatic mutations occurring in cells within the affected area may contribute to HME during early cerebral development3,5. However, the genetic etiology and pathogenesis of HME remain poorly understood.
The histopathological abnormalities in HME brain are described as cortical dyslamination, dysmorphic immature neurons and neuronal heterotopia, indicating primary defects in neuroglial differentiation, migration and cellular growth6. These pathological features are often shared with tuberous sclerosis complex (TSC), which is caused by mutations in TSC1 and TSC2 (ref. 7). In fact, the link between TSC and mTOR signaling suggests the hyperactivation of mTOR signaling in HME8, but the wide range of altered genes in expression profiles9 and the phenotypic variability and heterogeneity of diseased brain have hampered the identification of genetic causes.
Hemispheric surgical resection samples were available for 20 cases that met clinical and pathological criteria for HME3, and pre-surgical magnetic resonance imaging (MRI) scans were available for a subset of these (Supplementary Fig. 1 and Supplementary Table 1). Peripheral blood or saliva was obtained for probands and their parents and siblings in accordance with ethical approval. Due to the association of HME with Proteus syndrome10 and the recent discovery in Proteus syndrome of a recurrent AKT1 c.49G>A (p.Glu17Lys) activating mosaic mutation11, we first assessed each available brain sample for the AKT1 c.49G>A mutation. No mosaicism was detected in AKT1 (Supplementary Fig. 2), suggesting that the connection between HME and Proteus syndrome may have other genetic origins.
We next considered the possibility of a common, large de novo copy-number variation (CNV) such as those that have been reported in various neurodevelopmental disorders, including in epilepsy12. Using SNP-Genotyping arrays13, we analyzed five pairs of HME brain-blood genomic DNA samples. In four of the five cases, we detected identical common CNVs in both brain and blood. In one case, HME-1573, we detected a de novo CNV on chromosome 1, but this CNV has been reported previously to be a normal variant14, suggesting that de novo CNVs are unlikely to be a major cause of HME (Supplementary Fig. 2). We additionally considered the possibility of germline mutation, with silencing of the mutation occurring in healthy tissue due to mitotic recombination, akin to the mutation patterns observed for other recently reported mosaic phenotypes15–17. However, neither brain nor blood samples showed evidence of loss of heterozygosity (LOH), which is consistent with a previous finding of a lack of LOH in PTEN hamartoma tumor syndrome associated with an HME-like condition18 (data not shown).
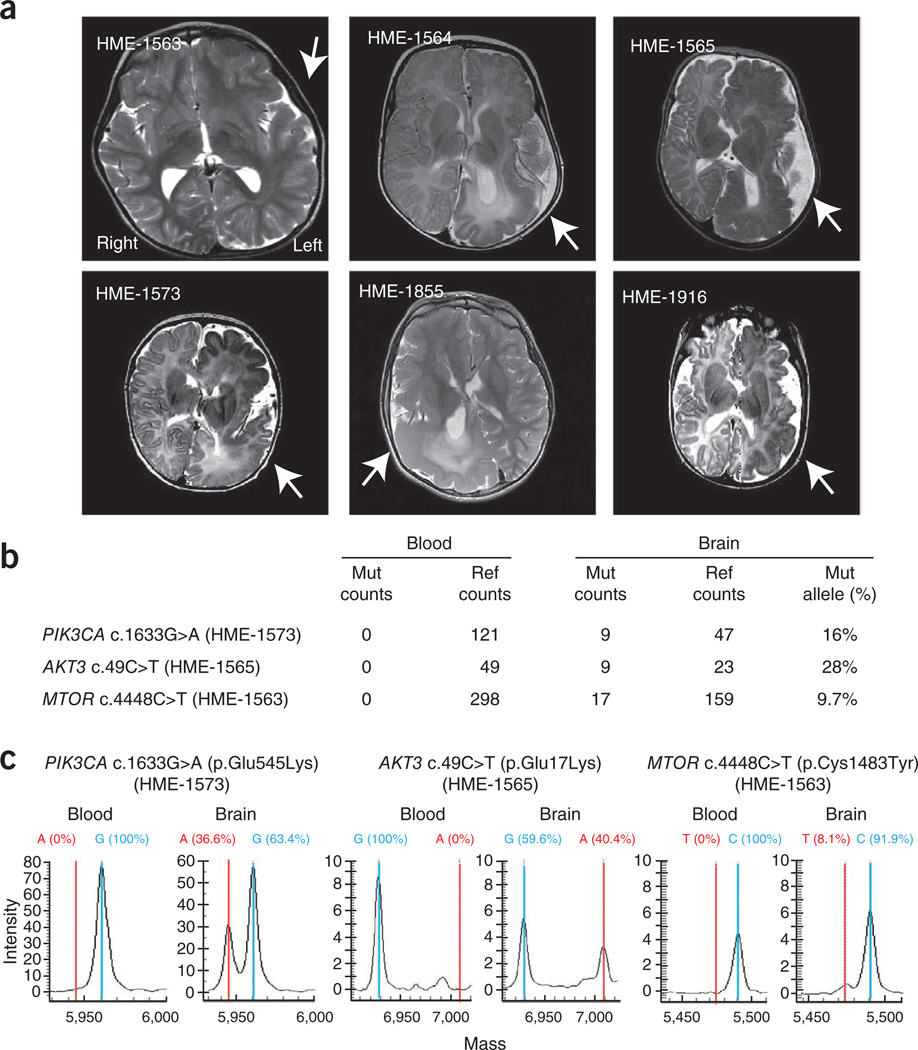
These results are consistent with the idea that HME is caused by de novo point mutations in critical regulatory gene(s). To test this model, we performed whole-exome sequencing on brain and peripheral blood DNA from five HME cases, with 93% exonic recovery at >30-fold coverage. To maximize the possibility of detecting somatic mutations at low copy in cellular heterogeneous brain tissues from HME cases, we used the recently developed JointSNVMix computational algorithm for the detection of somatic mutations in next-generation sequencing to simultaneously analyze sequencing data from blood-brain pairs and generate probabilities for de novo mutations at all detected alleles19. Using stringent criteria (Supplementary Fig. 3), we identified 26 candidate somatic mutations specific to HME brain. We then prioritized variants for those likely to substantially alter function by considering the biological importance of evolutionarily conserved sequences (Online Methods). With this prioritization, the top three missense mutations were found in PIK3CA (c.1633G>A; p.Glu545Lys) in HME-1573, AKT3 (c.49C>T; p.Glu17Lys) in HME-1565 and MTOR (c.4448C>T; p.Cys1483Tyr) in HME-1563 (Fig. 1), suggesting these as candidate genes involved in HME. None of these mutations were found in any blood sample (with a range of 49–298 reads per allele) but were identified in 9–17 reads from the affected hemisphere compared with 23–159 reference reads (9.7–28% of reference reads). Visualization of the reads in the Integrative Genomics Viewer suggested that the base quality of the mutant alleles was similar to that of the reference alleles, and there was no evidence that the variant calls were due to misalignment of the reads (Supplementary Fig. 3). In the remaining two cases, the evidence for somatic variants was less compelling, and none of the possible variants were confirmed using complementary methods. The data suggest that a fraction of the cells in the three samples contain de novo somatic mutations encoding altered amino acids.
Figure 1.
MRI and mutation analysis in hemimegalencephaly. (a) Axial T2-weighted brain MRI of cases identified with mutations. Arrows indicate the affected hemispheres, showing thickened cortical mantle, changes in white matter signal and alterations in ventricular shape, resulting in increased hemispheric size and midline shift of falx cerebri. (b) Sequencing counts from exome sequencing of each of three brain-blood paired samples. Mut, mutation; ref, reference. (c) Mass spectrometry validation of mutations. Wild-type sequences (blue) and de novo mutations (red) are correlated with results from next-generation sequencing.
As capillary (Sanger) sequencing cannot reliably detect somatic mutations in heterogeneous cell mixtures20, we instead used a modified single base–extension protocol followed by mass spectrometry analysis, which is able to detect somatic mutations at a frequency of as low as 3% in genetically heterogeneous samples21 (Supplementary Fig. 3). Reanalysis of the same DNA samples used for whole-exome sequencing again showed the absence of the mutant allele in blood but its presence in the brain, with similar mutation burden as that detected with Illumina sequencing (Fig. 1). These somatic mutations were detected at a frequency of 36.6%, 40.4% and 8.1% in each brain sample (Fig. 1). Using this technology, we further screened these mutations in the remaining 15 HME cases and identified 3 additional cases (HME-1855, HME-1916 and HME-1564) carrying the PIK3CA c.1633G>A (p.Glu545Lys) variant, each with a mutation burden of ~30% (Table 1 and Supplementary Fig. 3). Therefore, we identified de novo somatic mutations in the PIK3CA, AKT3 and MTOR genes in 6 out of our total cohort of 20 HME cases (30%), with the PIK3CA c.1633G>A (p.Glu545Lys) variant being recurrent.
Table 1.
Clinical and molecular data from individuals with HME carrying mutations in the PI3K-AKT3-mTORpathway
| Subject ID | Age at surgery | Sex | Lesion locationa | Brain pathology | Other pathology | Gene | Nucleotide change | Protein change |
|---|---|---|---|---|---|---|---|---|
| HME-1565 | 6 months | F | Left | CD, CN, EN, PMG | – | AKT3 | c.49C>T | p.Glu17Lys |
| HME-1563 | 5 years | M | Left | CD, CN, EN | Hypomelanosis of Ito | MTOR | c.4448C>T | p.Cys1483Tyr |
| HME-1573 | 4 months | M | Left | CD, CN, EN, PMG | – | PIK3CA | c.1633G>A | p.Glu545Lys |
| HME-1916 | 4 months | M | Left | CD, CN, EN, PMG | – | PIK3CA | c.1633G>A | p.Glu545Lys |
| HME-1564 | 6 months | M | Left | CD, EN, PMG | – | PIK3CA | c.1633G>A | p.Glu545Lys |
| HME-1855 | 4 years | M | Right | CD, CN, EN, PMG | Hemihypertrophy of right hand and foot | PIK3CA | c.1633G>A | p.Glu545Lys |
M, male; F, female; CD, cortical dyslamination; CN, cytomegalic neuron; EN, ectopic neurons; PMG, polymicrogyria. Results are ordered by gene name and age of the subject at the time of surgery.
Affected cerebral hemisphere.
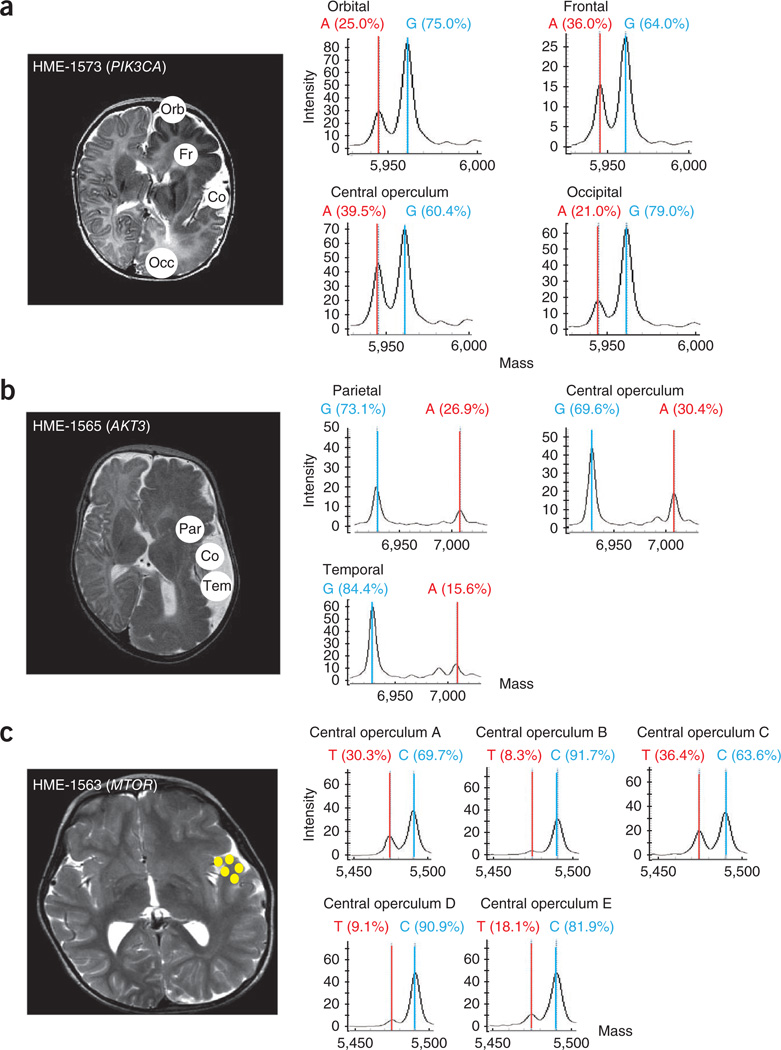
The differences in mutation burden between HME cases prompted us to examine this burden in anatomically defined brain regions in available samples from the cases. In HME-1573 with the PIK3CA mutation, we found a mutation burden of 21–40% in orbital, frontal, central operculum and occipital regions (Fig. 2). In HME-1565 with the AKT3 mutation, we found a mutation burden of ~16–30% in parietal, central operculum and temporal regions. Notably, in HME-1563, where tissue was available only from multiple distinct regions of the central operculum, we found the MTOR mutation burden to be quite variable, ranging from 8–36%, even within this one region. These data indicate that HME harbors admixtures of mutant alleles that make up between 8–40% of the alleles within affected brain regions. As the human genome is diploid and because we expect these mutations to be present in only one of the two alleles, we infer that a substantially higher percentage of cells carry the HME-associated mutations in these various brain regions than in other tissues, although the exact percentages remain to be determined.
Figure 2.
Consistent de novo mutation burden across affected hemispheres. (a) HME-1573 sampled during surgery in left orbital (Orb), frontal (Fr), central operculum (Co) and occipital (Occ) regions. (b) HME-1656 sampled in parietal (Par), central operculum and temporal (Tem) regions. (c) HME-1563 was sampled multiple independent times in the central operculum region (yellow circles), but samples could not be precisely aligned anatomically. Left, MRI scans; circles indicate brain areas examined by mass spectrometry analysis. Right, each sample aliquot was analyzed by mass spectrometry for mutation burden, describing variation in mutation burden across anatomical locations.
The PIK3CA, AKT3 and MTOR genes encode well-known regulators of the mTOR signaling pathway (Fig. 3), which is involved in cell growth and metabolism. Whereas PIK3CA and MTOR have widespread expression, AKT3 is most strongly expressed in the nervous system. Akt3-null mice have smaller brains than wild-type mice, as a result of reduced cell number and size22,23, and an activating point mutation in Akt3 results in seizure susceptibility24. The PIK3CA gene encodes the catalytic domain of the PI3K protein known as p110α, which uses ATP to phosphorylate phosphatidylinositol (PtdIns), PtdIns 4-phosphate (PtdIns(4)P) and PtdIns 4,5-bisphosphate (PtdIns(4,5)P2). Upon stimulation with extracellular signals, such as insulin and platelet-derived growth factor, the resultant production of PtdIns 3,4,5-trisphosphate (PtdIns(3,4,5)P3), one of the most potent bioactive lipids, functions to activate proteins in the AKT family by binding to the pleckstrin homology (PH) domain. AKT family proteins can directly phosphorylate the mTOR protein, which is the product of the MTOR gene. The identification of de novo mutations in these three genes, which are central to the mTOR pathway, is evidence that disrupted signaling underlies HME.
Figure 3.
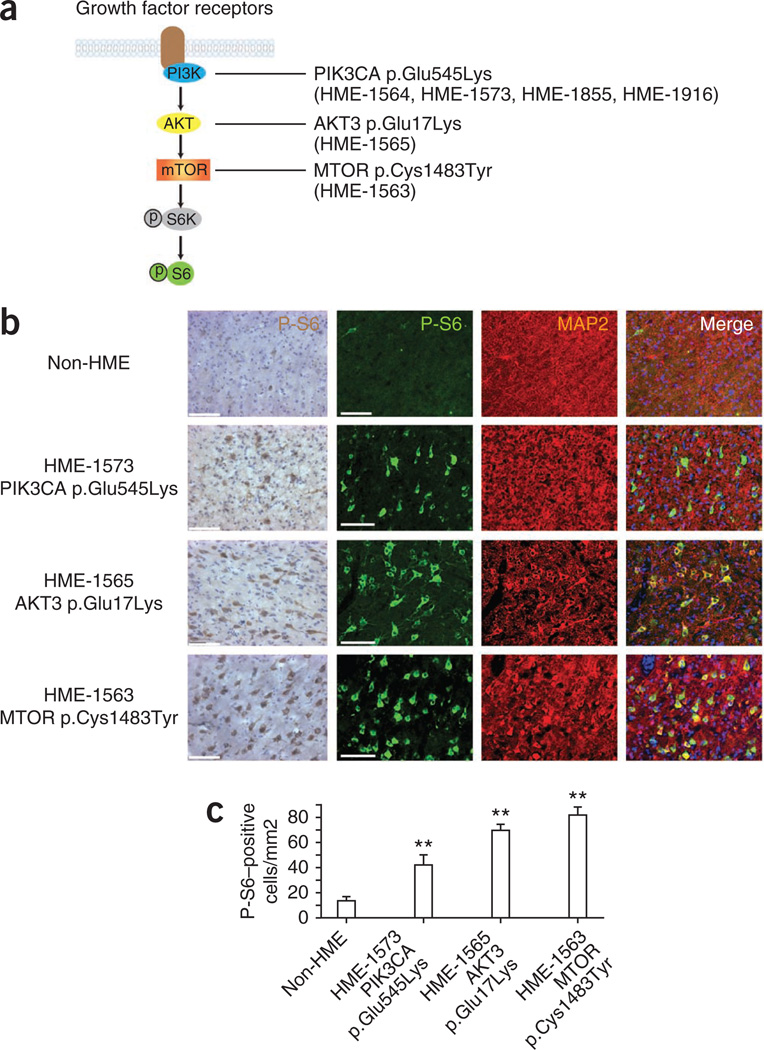
The de novo mutations identified in HME correlate with hyperactive mTOR signaling. (a) Schematic of the PI3K-AKT3-mTOR pathway and downstream phosphorylated ribosomal protein S6 kinase (P-S6K) and phosphorylated ribosomal protein S6 (P-S6). (b) HME pathological samples show an increased percentage of cells with positive staining for P-S6 and MAP2. Left P-S6, biotin-streptavidin DAB staining; right, P-S6, fluorescence-conjugated staining. MAP2 was used to identify neurons. Scale bars, 100 µm. (c) Quantification of P-S6–positive cells from 5–7 representative cortical areas per case. **P < 0.01 (relative to non-HME samples, one-way ANOVA with Bonferroni posttest, n > 50 cells per region). Error bars, s.e.m.
A large body of work has implicated somatic mutations in these genes in the pathogenesis of solid malignancies, and two of the three mutant alleles that we identified were previously reported in cancers. The PIK3CA p.Glu545Lys variation within the helical domain of the p110α protein was identified in tumors of the colon and brain25,26 and constitutively activates the mTOR pathway27. The AKT3 p.Glu17Lys variation within the PH domain was identified in melanomas and results in increased AKT3 phosphorylation in transfected cells28. Although the transversion we identified in MTOR has not been encountered previously in human disease, other activating point mutations in this gene have been found in cancer29,30. Nevertheless, it remains to be determined whether the mTOR p.Cys1483Tyr variation in HME leads to increased signaling.
To determine whether the affected individuals in our study had aberrant mTOR signaling, we immunostained brain sections of HME cases (HME-1573, HME-1565 and HME-1563) with an antibody specific to the phosphorylated epitope of the S6 protein in a standard assay for the activation of mTOR signaling. Cells with the morphology of cytomegalic neurons were strongly labeled for phosphorylated S6 in the 3′-diaminobenzidine (DAB) staining of HME brains (Fig. 3). In addition, we co-immunostained for the neuronal marker MAP2, comparing samples with age-matched, similarly processed non-HME cortical hemisphere, and we found a marked increase in the number of cells that were positive for phosphorylated S6 and greater intensity of staining for phosphorylated S6 in cytomegalic neurons of HME cases (Fig. 3). We conclude that the mutations identified here are associated with increased mTOR signaling in affected brain regions.
On the basis of these data and those previously published, we propose a model of HME involving somatic de novo gain of function in the mTOR pathway. Mutations were identified in 6 of the 20 samples using exome data from the first 5 cases, so it is possible that there are other mutations in these genes or in completely different genes in the remaining cases. No clear genotype-phenotype correlation was observed in the affected individuals identified to date (Supplementary Table 1), which is not unexpected, given the potential for heterogeneity in the genetic cause and mosaicism rates and the potential for allelic heterogeneity, any of which might influence phenotype. In addition, the connection to TSC suggests the possibility of using mTOR inhibitors to treat individuals with HME who have aberrant mTOR signaling because the TSC-encoded proteins Hamartin and Tuberin function as negative regulators of the mTOR pathway31–34. Brain pathology from TSC tubers shows hyperactivation of downstream mTOR signaling, and inhibition of mTOR with rapamycin (also known as sirolimus) is of proven benefit in the treatment of TSC-related brain lesions35.
We note that subject HME-1563 with the MTOR mutation also carries a diagnosis of hypomelanosis of Ito, a dermatological condition caused by segmental depletion of melanocytes. The co-occurrence of HME and hypomelanosis of Ito has been observed repeatedly36–38, suggesting a common genetic origin, but the basis of hypomelanosis of Ito remains unknown. We speculate that activating mutations in the mTOR pathway may account for at least some cases and suggest that these somatic mutations may extend to other areas of the body and result in loss of mutation-carrying melanocytes in the affected areas. Similarly, the PIK3CA mutation in HME-1855 may extend to hypertrophic regions (the right hand and foot), thereby causing the overgrowth of affected tissues. In both cases, it might be possible to test this model by assessing mutation burden in affected regions.
Our results are consistent with at least three potential explanations for the strict unilateral hemisphere affectation in HME. The first model involves a substantial somatic mutation burden in a subset of progenitors contributing to one hemisphere, without such burden in progenitors in the contralateral hemisphere. The second model involves approximately equal mutation burden in the two hemispheres, which is exacerbated by an unidentified spatially restricted change to signaling, such as epileptic seizures. The third model involves a combination of these two models, with uneven mutation burden in the two hemispheres that is exacerbated by unilateral signaling. Although access to tissue from the uninvolved hemisphere in HME is lacking, it should be possible to test these models using animal models of HME. Accessing brain tissue from humans with neurological disease will be increasingly important for future research.
ONLINE METHODS
Subject ascertainment
Individuals with HME (n = 20) were identified through the UCLA Pediatric Epilepsy Surgery Program database. Affected individuals underwent extensive presurgical evaluation, including structural MRI and video electroencephalography (EEG) telemetry, to localize the anatomic lesions. The affected individuals had surgery between 2003 and 2011; the presurgical and surgical protocols have been published previously3,39,40 and since 1997 have involved a modified lateral hemispherectomy. Twenty sequentially enrolled affected individuals met study entry criteria for HME, defined as those surgical cases where most (at least three lobes) or all of one cerebral hemisphere was larger than the opposite hemisphere when visualized by MRI. Both fresh-frozen and formalin-fixed tissues with anatomical coordinates were available from most cases. Institutionally approved informed consent was obtained to use clinical data and study genetic etiology. None of the individuals in this cohort had concordant solid malignancies. The study was performed in accordance with the ethics boards of UCLA and UCSD.
PCR restriction enzyme assays for the c.49G>A mutation in AKT1
A modified PCR–restriction endonuclease assay was performed as described previously11. The assay can detect mosaicism in as little as 1% of sampled cells and included a Proteus syndrome sample with AKT1 c.49G>A mutation as a control. We tested each brain DNA sample in the cohort. Briefly, a 6-carboxyfluorescein (FAM)-labeled forward primer and an unlabeled modified reserve primer were used to amplify the target sequence containing the AKT1 c.49G site. The modified reverse primer created an MboII restriction site in the presence of the c.49G>A mutation. MboII digestion of a mutant allele gave a 122-bp fragment, and uncut wild-type alleles gave a 141-bp fragment. Fragments were analyzed using an Applied Biosystems 3130 instrument. Skin genomic DNA from an individual with Proteus syndrome (a gift from L. Biesecker, US National Institutes of Health) was used as a positive control.
SNP genotyping analysis for CNVs
DNA from brain and blood samples of five cases was analyzed for structural variations using the HumanCytoSNP-12 DNA BeadChip kit (Illumina) according to the manufacturer’s recommendations. Briefly, labeled DNA was hybridized to the BeadChip containing a whole-genome panel of >200,000 SNPs probes with the highest tagging power. Hybridized arrays were scanned and analyzed with Illumina’s BeadStudio software. To identify high-confidence CNVs, raw data were analyzed with both PennCNV41 and Illumina’s BeadStudio software. CNVs identified with both methods were tabulated.
DNA extraction and whole-exome sequencing
Genomic DNA from frozen brain and blood was extracted using Qiagen DNA methodology. Saliva samples were collected using Oragene kits and genomic DNA was isolated. Whole-exome capture was performed with the SureSelect All Exon Target Enrichment System (Agilent Human All Exon 50 Mb kit). Libraries underwent paired-end sequencing (2 × 100 bp) on an Illumina HiSeq 2000 instrument according to manufacturer’s protocol. For each case sample, >90% of the exome was covered at >30×.
Analysis of blood-brain paired exome sequencing
We used the JointSNVMix model19 to jointly analyze blood-brain paired sequencing data sets to generate probabilities that any given allele occurred de novo (not in the germline). In a paired genotype gN × gT, where gN is the genotype of the blood sample and gT is that of the brain sample, JointSNVMix calculated the probabilities for nine possible permutations; each sample can have three genotypes: AA, AB and BB, where A denotes a nucleotide matched to the reference genome and B denotes a mismatched nucleotide. To calculate the probability of observing allele counts a and b given a prior joint genotype, JointSNVMix uses a mixture of binomial densities, which consists of AA, AB and BB. We used a more general version (JointSNVMix2) that incorporates base- and mapping-quality information to model conditional densities. To extract only somatic mutations, a cutoff for the sum of two joint genotype probabilities of AA_AB and AA_BB (P(AA_AB) + P(AA_BB)) was applied.
To generate a candidate mutation set, we applied the following filters to our blood-brain data set: (i) read depth of ≥15 and (ii) joint genotype probabilities of greater than 0.5 (AA_AB, AA_BB, AB_BB), with AB_BB added to include the possibility of alleles with LOH. A total of 7,036 variants were identified with these filters on the basis of a probability value threshold of 0.5. These were further filtered by (i) setting a threshold of 0.9, (ii) excluding intronic or synonymous variants and (iii) excluding annotated SNPs, resulting in 861 variants. These were filtered by (i) excluding those identified in our in house exome database of >1,000 individuals without HME, (ii) excluding those in more than 2% of reads from blood samples (to exclude germline mutations), (iii) excluding those in ≤3 reads from brain (to exclude error calls in brain samples) and (iv) excluding those with Phastcon score of ≤0.50, resulting in 26 variants. These were prioritized on the basis of (i) missense mutations (representing alleles likely to impart gain of function), (ii) previous publications for the variant and (iii) functional impact score42. From this list, the top three candidate mutations emerged.
Mutation burden analysis using single–base pair extension
Mutation burdens were analyzed with the Sequenom MassARRAY platform. The general workflow for this is shown in Supplementary Figure 3. Briefly, MassARRAY Designer software (Sequenom) was used to design the PCR assay and iPLEX single-base extension primers for analysis. Using designed primers and the iPLEX Pro kit, PCR and subsequent single–base pair extension reactions were performed according to the manufacturer’s guide. Extension products (5–12 nl) were arrayed on silicon chips. Allele-specific differences in mass between extension products were detected by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Mutation burdens were scored by Mass ARRAY Typer Analyzer software.
Immunohistochemistry
Non-HME brain was collected in the operating room from the normal, uninvolved brain of an individual with Rasmussen syndrome (chronic inflammation of one side of the brain) as part of a planned hemispherectomy. This sample was processed in the same way as the HME cases. Surgical tissue blocks were fixed in freshly prepared phosphate-buffered 4% paraformaldehyde for 24–48 h, cryoprotected overnight in 20% buffered sucrose and stored at −80 °C. Cryostat-cut sections (30-µm thick) were collected and placed on glass slides, blocked in 0.2% gelatin and 0.1% Triton X-100 in PBS and stained with the following antibodies: rabbit antibody to phosphorylated S6 ribosomal protein (Ser240/Ser244) (1:50 dilution; 5364, Cell Signaling Technology) and mouse antibody to MAP2 (1:100 dilution; MAB3418, Millipore). Samples were then washed in PBS and stained with the following secondary antibodies: Alexa Fluor 594–conjugated donkey antibody to mouse (1:200 dilution; A21203, Invitrogen), Alexa Fluor 488–conjugated donkey antibody to rabbit (1:200 dilution; A21206, Invitrogen). Hoechst (Molecular Probes, H3570) was used for nuclear staining. We acquired and quantified images using a DeltaVision deconvolution microscope. The number of cells positive for phosphorylated S6 was determined using the 20× objective lens; four fields were acquired per subject within the neuron-rich regions, and >100 cells were scored per region. For DAB staining, immunostaining was performed on free-floating 30-µm cryostat-cut sections by first blocking in a Tris-buffered solution with 2% normal goat serum (Vector S-1000) and 0.2% Triton X-100 for 1 h. Sections were then incubated in rabbit antibody to phosphorylated S6 ribosomal protein for 1 h at 37 °C and incubated in biotinylated secondary antibody to rabbit IgG (1:300 dilution, Vector) for 35 min at 37 °C. Immunoreactivity was visualized using peroxidase-conjugated avidin (Vectastain Elite ABC Kit, Vector) with the addition of DAB substrate (MP Biomedicals). Sections were mounted on slides and counterstained with hematoxylin (Fisher), and coverslips were applied using Cytoseal XYL (Thermo Scientific). Images were acquired using an Olympus BX-51 microscope.
Supplementary Material
Acknowledgments
We thank J. Meerloo at the UCSD Neurosciences Core for microscopy services (supported by the US National Institute of Neurological Disorders and Stroke; P30NS047101), the Genomics Core at Cedars-Sinai Medical Center, the Broad Institute (supported by the US National Human Genome Research Institute; U54HG003067 to E. Lander) for sequencing support and analysis, D. Neelam (Sequenom) for technical support and W.B. Dobyns and M.L. Warman for sharing unpublished results. This work was supported by grants from the Daland Fellowship from the American Philosophical Society (to J.H.L.), the US National Institutes of Health (R01 NS038992 to G.W.M. and R01 NS048453, R01 NS052455, R01 NS041537 and P01 HD070494), the Simons Foundation Autism Research Initiative and the Howard Hughes Medical Institute (to J.G.G.).
Footnotes
Accession codes. The whole-exome sequencing data from individuals in this study have been deposited in the database of Genotypes and Phenotypes (dbGAP). Reference sequences for the affected genes include NM_006218 (PIK3CA), NM_181690 (AKT3) and NM_004958 (MTOR).
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.H.L. and J.L.S. organized the project and performed genetic studies. K.J.H., A.C. and J.H.L. recruited subjects. J.H.L., M.H. and T.D.-S. performed immunostaining. C.R. and S.B.G. generated and interpreted exome results. S.K., A.H., E.S., V.B. and J.H.L. performed JointSNVMix analysis. V.F. and J.H.L. oversaw SNP genotyping and analyzed CNVs. G.W.M. performed surgeries and managed samples along with M.H. G.W.M. and J.G.G. conceived of the project and oversaw data collection and manuscript preparation.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Blümcke I, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52:158–174. doi: 10.1111/j.1528-1167.2010.02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain. 2012;135:1348–1369. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salamon N, et al. Contralateral hemimicrencephaly and clinical-pathological correlations in children with hemimegalencephaly. Brain. 2006;129:352–365. doi: 10.1093/brain/awh681. [DOI] [PubMed] [Google Scholar]
- 4.Di Rocco C, Battaglia D, Pietrini D, Piastra M, Massimi L. Hemimegalencephaly: clinical implications and surgical treatment. Childs Nerv. Syst. 2006;22:852–866. doi: 10.1007/s00381-006-0149-9. [DOI] [PubMed] [Google Scholar]
- 5.Crino PB. mTOR: a pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 2011;17:734–742. doi: 10.1016/j.molmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Palmini A, et al. Terminology and classification of the cortical dysplasias. Neurology. 2004;62:S2–S8. doi: 10.1212/01.wnl.0000114507.30388.7e. [DOI] [PubMed] [Google Scholar]
- 7.Jozwiak J, Jozwiak S, Skopinski P. Immunohistochemical and microscopic studies on giant cells in tuberous sclerosis. Histol. Histopathol. 2005;20:1321–1326. doi: 10.14670/HH-20.1321. [DOI] [PubMed] [Google Scholar]
- 8.Boer K, et al. A neuropathological study of two autopsy cases of syndromic hemimegalencephaly. Neuropathol. Appl. Neurobiol. 2007;33:455–470. doi: 10.1111/j.1365-2990.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, et al. Targeted gene expression analysis in hemimegalencephaly: activation of beta-catenin signaling. Brain Pathol. 2005;15:179–186. doi: 10.1111/j.1750-3639.2005.tb00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths PD, Welch RJ, Gardner-Medwin D, Gholkar A, McAllister V. The radiological features of hemimegalencephaly including three cases associated with proteus syndrome. Neuropediatrics. 1994;25:140–144. doi: 10.1055/s-2008-1073012. [DOI] [PubMed] [Google Scholar]
- 11.Lindhurst MJ, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mefford HC, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann. Neurol. 2011;70:974–985. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 15.Jonkman MF, Pasmooij AM. Revertant mosaicism—patchwork in the skin. N. Engl. J. Med. 2009;360:1680–1682. doi: 10.1056/NEJMc0809896. [DOI] [PubMed] [Google Scholar]
- 16.Choate KA, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330:94–97. doi: 10.1126/science.1192280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jongmans MC, et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am. J. Hum. Genet. 2012;90:426–433. doi: 10.1016/j.ajhg.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merks JH, et al. PTEN hamartoma tumour syndrome: variability of an entity. J. Med. Genet. 2003;40:e111. doi: 10.1136/jmg.40.10.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth A, et al. JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next generation sequencing data. Bioinformatics. 2012;28:907–913. doi: 10.1093/bioinformatics/bts053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 21.Oeth P, del Mistro G, Marnellos G, Shi T, van den Boom D. Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY) Methods Mol. Biol. 2009;578:307–343. doi: 10.1007/978-1-60327-411-1_20. [DOI] [PubMed] [Google Scholar]
- 22.Tschopp O, et al. Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development. 2005;132:2943–2954. doi: 10.1242/dev.01864. [DOI] [PubMed] [Google Scholar]
- 23.Easton RM, et al. Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol. Cell. Biol. 2005;25:1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokuda S, et al. A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Hum. Mol. Genet. 2011;20:988–999. doi: 10.1093/hmg/ddq544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broderick DK, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- 26.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 27.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies MA, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardt M, Chantaravisoot N, Tamanoi F. Activating mutations of TOR (target of rapamycin) Genes Cells. 2011;16:141–151. doi: 10.1111/j.1365-2443.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 33.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 34.Ito N, Rubin GM. gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell. 1999;96:529–539. doi: 10.1016/s0092-8674(00)80657-1. [DOI] [PubMed] [Google Scholar]
- 35.Bissler JJ, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N. Engl. J. Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S, Sankhyan N, Kabra M, Kumar A. Hypomelanosis of Ito with hemimegalencephaly. Dermatol. Online J. 2009;15:12. [PubMed] [Google Scholar]
- 37.Chapman K, Cardenas JF. Hemimegalencephaly in a patient with a neurocutaneous syndrome. Semin. Pediatr. Neurol. 2008;15:190–193. doi: 10.1016/j.spen.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Auriemma A, et al. Hemimegalencephaly in hypomelanosis of Ito: early sonographic pattern and peculiar MR findings in a newborn. Eur. J. Ultrasound. 2000;12:61–67. doi: 10.1016/s0929-8266(00)00099-9. [DOI] [PubMed] [Google Scholar]
- 39.Hemb M, et al. Improved outcomes in pediatric epilepsy surgery: the UCLA experience, 1986–2008. Neurology. 2010;74:1768–1775. doi: 10.1212/WNL.0b013e3181e0f17a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonas R, et al. Cerebral hemispherectomy: hospital course, seizure, developmental, language, and motor outcomes. Neurology. 2004;62:1712–1721. doi: 10.1212/01.wnl.0000127109.14569.c3. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.