Abstract
Background
MicroRNAs are small non-coding RNAs that are involved in various biological processes including insect development. Anopheles stephensi serves as primary vector of malaria parasite in Asia and exhibits holometabolous life cycle that involves four different stages of development. Regulation and role of mosquito miRNAs during various stages of mosquito development remain largely unknown.
Methods
High throughput small RNA sequencing was employed for identification and profiling of miRNAs across immature and adult stages of malaria vector, which were further validated using Northern hybridization and real time PCR. Target prediction and pathway analysis was carried out to understand the role of regulated miRNAs in insect development. Degradome sequencing was employed to identify cleaved targets of some regulated miRNAs. Loss of function strategy was employed for miR-989 to understand its probable role in female reproductive process.
Results
Small RNA sequencing and data analysis revealed 111 and 14 known and novel miRNAs respectively across all stages of Anopheles stephensi. Nine miRNAs showed gender specific regulation across different stages of mosquito development. Analysis of miRNAs revealed regulation of 24 and 26 miRNAs across different stages of male and female mosquito development respectively. mRNA targets and significant pathways targeted by regulated miRNAs were identified for each stage of mosquito development. Degradome sequencing revealed twenty nine cleaved targets of insect miRNAs. MicroRNA-989 showed significant up-regulation in the adult female as compared to adult male mosquito. Knockdown of miR-989 expression in adult female using miRNA specific antagomir affected targets playing roles in protein binding, proteolysis and nucleic acid binding in ovary tissue of female mosquito post blood feeding.
Conclusions
This is the first comprehensive effort to understand regulation of Anopheles stephensi miRNAs across developmental stages of male and female mosquito. Preliminary role of regulated miRNAs in mosquito development was revealed by target prediction and pathway analysis. MicroRNA-989 emerged to have important roles in adult female mosquitoes showing significant up-regulation which was further studied using miR-989 specific antagomir. This study provides insights into mosquito development and reproductive process and has implications for effective control of mosquito population required for reducing spread of mosquito-borne infectious diseases.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-015-0772-y) contains supplementary material, which is available to authorized users.
Keywords: Mosquito development, microRNA, NGS, Antagomir, Degradome sequencing
Background
Mosquitoes exhibit holometabolous life cycle that proceeds through four different life stages. Eggs laid by blood fed female mosquitoes hatch into larvae that metamorphosis into pupae and finally to imago or adult mosquitoes. These stages exhibit distinctive morphological and physiological differences as depicted by their choice of different ecological niches. After hatching, larva grows and periodically sheds its old cuticle by the process of ecdysis. This process results in larval growth through successive instar to reach fourth instar stage of development. Larva metamorphosis into pupa involves almost complete histolysis and phagocytosis of the larval tissues. Pupa is a non feeding inactive stage during which reconstruction of histolysed tissue results in formation of organs of an adult mosquito.
Such complete metamorphosis involves complex interplay of ecdysteroid and juvenile hormone activities and regulation of transcriptional events [1,2].
MicroRNAs are non protein coding small RNAs of length 18–24 nucleotides that are produced by plants, animals and viruses [3-5]. They regulate numerous biological processes such as growth and development, differentiation, disease progression, apoptosis and immunity [6]. Such regulation is mediated by miRNA binding to 3’UTR sequence of target gene and regulating their expression either by target cleavage or translation repression [7]. Repertoire of miRNAs in different mosquito species has been reported [8-10]. Few of them have been functionally characterized and were shown to play critical role in insect reproduction and immunity against pathogens [11-13]. Few studies have focused on understanding the regulation of miRNAs across developmental stages of mosquito [14,15]. Previous studies have identified miRNAs in An.stephensi and studied miRNA regulation in blood fed and post Plasmodium infected female mosquito [9,16]. Understanding the regulation of miRNAs across immature stages of An.stephensi, an important disease vector in Asia remains largely unfulfilled.
In this study, we employed next generation small RNA sequencing to identify miRNAs that are regulated across Anopheles stephensi immature stages. We studied miRNA differences between male and female mosquito during larval, pupal and adult stages of their development, as differences between the genders render the female mosquito fit to serve as vector for parasite transmission. MicroRNAs differentially expressed during metamorphosis from larval to pupal and then from pupal to adult stages were identified. Characterization of these miRNAs may offer insight into vital processes such as ecdysis, histolysis and generation of adult organs during mosquito life cycle. Additionally, a number of novel mosquito-specific miRNAs were also discovered. Further, mRNA targets were predicted to understand their role in mosquito development. Antagomir injections and degradome sequencing were employed for the first time in insects to identify mRNA targets cleaved by regulated miRNAs in ovary tissue of female mosquito. Ovary specific cleavage of targets highlights towards their role in insect reproduction. Understanding the functions of these regulated miRNAs will provide useful insights in mosquito biology and is capable of deciphering ways to control mosquito-borne infectious diseases.
Methods
Ethics statement
Animal experiments were performed in accordance with National animal ethics guidelines of the Government of India after approval by Institutional Animal Ethics Committees of International Centre for Genetic Engineering & Biotechnology, New Delhi (Permit number: ICGEB/AH/2011/01/IR-8).
Rearing of mosquito
All developmental stages of Anopheles stephensi including egg, larva, pupa and adult mosquitoes were reared under optimum temperature at 28 ± 2°C and 70 -75% humidity in an insectary. Laid eggs were transferred to the enamel trays and were allowed to hatch into a first instar larvae. Larvae were fed on fish food and were allowed to grow from first instar to fourth instar larvae. Fourth instar larvae transforms into pupae, which were collected and kept into cloth cages. Emerging adult mosquitoes were maintained in same cages and were fed on water soaked raisins and 1% glucose soaked cotton pads. To obtain next generation of eggs, 5–6 days old female mosquitoes were fed for two hours on mice as a source of blood meal. Female mosquitoes developed and laid eggs three days post blood feeding in a bowl of sterilized water.
Sample collection and RNA isolation
Mosquitoes at different stages of their development, namely fourth instar larvae, pupae and 5–6 days old adult mosquitoes, with a minimum sample size of 100 numbers in each group for both genders were collected. The samples were collected a minimum of three times during different rearing cycles for each stage of development. Total RNA enriched in small RNA population was extracted using miRNeasy kit (Qiagen) as per the manual’s protocol. Total RNA from all biological replicates were pooled during small RNA library preparation. Collecting samples from different cycles of mosquito rearing would help in nullifying changes induced by possibly altered rearing conditions and hence served as biological replicates. Quality and quantity of RNA was checked by using Agilent 2100 Bioanalyzer RNA Nano 6000 kit.
Small RNA sequencing library preparation, sequencing and data analysis
Small RNA libraries were prepared from total RNA extracted from all six samples viz., male and female fourth instar larva, male and female pupa and 5–6 days old male and female adult mosquitoes. Small RNA library preparation and post sequencing data analysis was carried out as described in [9]. Briefly, Illumina Trueseq libraries were prepared by ligating adaptors to both side of the RNA sequence followed by reverse transcription. Adaptors ligated fragments were PCR amplified and were 14–160 bps fragments were purified using 6% TBE PAGE gel. Prepared libraries were then sequenced using Illumina Genome Analyzer II.
In-house developed data analysis pipeline was used for analysing post sequencing raw data as described in [8]. Briefly, raw sequences were filtered and sequences with length >18 nt were selected. Total number of unique reads were identified in each library, which were then aligned against mature miRNA sequence downloaded from mirbase database using bowtie with zero mismatch as a parameter [17,18]. Unmatched sequences were used for novel miRNAs identification. Unmatched sequences were mapped on to pre-miRNA, ncRNA database [19] and coding region of genes of different mosquito species downloaded from vectorbase [20]. Unmatched sequences were then matched to An.stephensi genome. Matched sequences as well as 75 nt sequences flanking on either side of the mapped sequences were extracted. These sequences were folded into secondary structures using RNAfold [21] and their folding energies were calculated using RNAplot [21]. Sequences folding into hairpin loop structure, folding energy < −20 kcal/mol and small sequence mapping on the arm of secondary structure were classified as novel miRNAs [8].
Differential expression of miRNAs and statistical analysis
Total number of reads for each miRNA in all the six libraries were obtained by using in house Perl script. Reads were normalised by calculating tags per million of total RNA reads (TPM) for each miRNA in all the six libraries. TPM data obtained was then fed into edgeR module for identification of differentially expressed genes between different developmental stages. The P value cut off was performed on the data with the significance threshold selected as 0.05. The final set of miRNAs that were considered to be significantly regulated were selected on the basis of three criteria, namely, > two fold change in expression between compared stages, P value ≤ 0.05, and TPM >10.
Antagomir injections in mosquitoes
Antagomir for miR-989 was synthesized complementary to mature miRNA sequence with 2’-O-methyl (2’-OMe) group at each base and also with 3’ cholesterol group (Dharmacon, USA). Scrambled RNA was synthesized with same modifications and was used as a negative control. 4–5 days old female mosquitoes were divided into three batches of 200 mosquitoes each. First batch was injected with 69 nl of PBS. Second and third batch of mosquitoes were injected with 69 nl of 100 uM scrambled RNA and miR-989 antagomir each. Mosquitoes were allowed to recover for two days and were fed on uninfected mice blood. Ovaries were dissected out of female mosquitoes 24 hours post blood feeding and were then stored in Trizol at −80°C till RNA extraction. Knockdown of miRNA expression post injection was checked by miRNA qRT-PCR.
Quantitative RT-PCR
Expression profiling of miRNAs across three immature stages in both genders was carried out by quantitative RT-PCR. Ten ng of RNA was reverse transcribed using cDNA synthesis kit (Exiqon). Real time reactions were set up using 1:80 diluted cDNA, custom miRNA LNA PCR primer sets (Exiqon) and SYBR green master mix (Exiqon) following manufacturer’s instructions in ABI one step detection system. Experiments were conducted a minimum of two times, with each experiment set up in triplicates for all developmental stages. 5.8 s rRNA was used as an endogenous control for miRNA expression profiling. Expression levels were then calculated against adult female mosquito as a calibrator using 2-ΔΔCT method.
Northern hybridization
Digoxigenin-labelled antisense miRCURY LNA probes (Exiqon) were used for detection of miRNAs by Northern hybridization. 10ug of total RNA isolated from 5–6 days old female mosquito was run using 15% denaturing polyacrylamide gels (PAGE). After the run was completed, gels were stained with ethidium bromide and RNA quality was checked using transilluminator. RNA was then transferred to the nylon membrane using semi-dry transfer apparatus. Nylon membrane with the transferred RNA was subjected to EDC crosslinking. Crosslinked membranes were pre-hybridized in a rotating hybridization oven for 30 min at 37 deg in hybridization buffer. Overnight hybridization was done in same buffer at 37 deg with the final concentration of 0.5 nM miRCURY LNA probe (Exiqon). The membranes were washed twice for 10 min each in a low stringent buffer (2X SSC, 0. 1% SDS) at RT and then once in washing buffer (1X SSC) at RT. The membranes were incubated for 3 h in blocking buffer (Roche) followed by 30 min incubation with Anti-DIG-alkaline phosphatase fragment antibody (Roche) in blocking buffer. The membranes were then washed in DIG washing buffer at RT for 15 min each and then incubated for 5 min in development buffer. CSPD substrate (1:100 diluted in development buffer) was applied on to the membranes and incubated in dark for 10 min. Chemiluminescence signal was then measured in a Fluorchem machine (Protein Simple) to detect miRNA on the membrane.
MicroRNA target prediction and identification of enriched pathways
mRNA targets of miRNAs regulated across developmental stages were predicted using RNAhybrid software [22]. 3’UTR sequence of An.stephensi genes were downloaded from vector base. Mature miRNA sequence (fasta) and downloaded 3’UTR sequence were used as input data files in RNAhybrid. Targets were predicted using following parameters (i) Perfect miRNA seed complementarity with 3’UTR sequence (ii) P value < 0.05 (iii) miRNA:mRNA binding energy < −20 kcal/mol. Targets predicted using these criteria’s were selected for pathway analysis. Orthologs of these targets present in An.gambiae were fetched from vector base. Targets ortholog of each miRNA cluster were analysed by NIH DAVID resource [23]. mRNA targets of miRNA cluster were analysed using functional annotation clustering with EASE score of 0.1. Enrichment score for each cluster was calculated to highlight enriched role of group members in the given study.
Degradome sequencing for miRNA target identification
Total RNA was extracted from ovaries of mosquitoes injected with PBS, scrambled RNA and miR-989 antagomir as described before. Degradome library construction and sequencing was performed by LC Sciences (Houston, USA). Data analysis was performed in the lab using an in-house developed pipeline. Raw reads obtained after sequencing were processed to remove low quality reads. Adaptor sequences were then trimmed from the raw reads. Resulting 20–25 nt sequences were selected for further analysis. Potentially cleaved miRNA target were identified using Parallel Analysis of RNA Ends (PAREsnip) [24]. Degradome sequences, cDNA of An. stephensi downloaded from Vectorbase and mature miRNA sequences were provided as an input to the PAREsnip software. Target plots (T-plots) were generated showing relative abundance of fragments mapping at the miRNA target site relative to the abundance of fragments found at other sites on the transcript. Depending on this, identified targets were grouped into five categories. In category 0, maximum number of degraded sequences (>1) were present at one site on the transcript. In category 1, two or more sites were present on the transcripts where degraded sequences (>1) map with the same abundance. If abundance at a site was less than maximum but greater than median abundance for the transcript, target was classified as category 2. In category 3, abundance at a position was less or equal to the median value for that transcript. Category 4 was classified with only one raw read at the position.
Results
MicroRNA dynamics was studied during metamorphosis of both male and female An.stephensi mosquito. Six small RNA libraries enriched in miRNAs were constructed from larva male (LM), larva female (LF), pupa male (PM), pupa female (PF) adult male (AM) and adult female (AF) mosquitoes. Constructed libraries were sequenced using sequencing by synthesis technology, (Illumina Inc.). Raw data generated for all six libraries was analysed using in-house data analysis pipeline [8].
Deep sequencing of small RNA libraries
Sequencing by synthesis of six different libraries yielded 7.2 × 107 raw reads. After adaptor trimming, total of 5.9 × 107 reads remained for further analysis (Table 1). Size distribution of reads was studied in each library. Size distribution of reads in each library showed two distinct peaks. One was observed at 20–23 nt corresponding to miRNAs and another at 32–35 nt representing piRNA like small RNAs (Figure 1). A total of 3.3 × 106, 2.3 × 106, 4.0 × 106, 6.7 × 106, 9.6 × 106 and 2.8 × 106 reads were found to lie between size range of 18-30 nt in LM, LF, PM, PF, AM and AF respectively. These sequences were mapped on to the known miRNA database for miRNA identification in various developmental stages of An. stephensi (Table 1). Unmatched sequences were mapped on the ncRNA database and coding sequences from different mosquito species (Table 1). The remaining unmatched sequences were used for novel miRNAs identification.
Table 1.
Composition of small RNA in deep sequenced libraries from larva male (LM), larva female (LF), pupa male (PM), pupa female (PF), adult male (AM) and adult female (AF) mosquito
| Sample name | Raw reads | Reads post adaptor trimming | 18-30 nt reads | Reads mapped to ncRNAs | Reads mapped to known miRNAs | Reads mapped to coding region of Anopheles | Reads mapped to An.stephensi genome |
|---|---|---|---|---|---|---|---|
| LM | 3.5 × 106 | 3.3 × 106 | 3.3 × 106 | 5.6 × 105 | 1.1 × 104 | 2.9 × 104 | 2.9 × 105 |
| LF | 2.7 × 106 | 2.6 × 106 | 2.3 × 106 | 5.4 × 105 | 6.3 × 104 | 4.7 × 104 | 1.8 × 105 |
| PM | 1.6 × 107 | 1.2 × 107 | 4.0 × 106 | 7.5 × 105 | 6.2 × 105 | 1.1 × 105 | 4.0 × 105 |
| PF | 1.9 × 107 | 1.2 × 107 | 6.7 × 106 | 1.3 × 106 | 8.3 × 105 | 1.5 × 105 | 7.0 × 105 |
| AM | 2.5 × 107 | 2.4 × 107 | 9.6 × 106 | 7.9 × 105 | 2.6 × 106 | 5.8 × 104 | 9.9 × 105 |
| AF | 5.4 × 106 | 4.4 × 106 | 2.8 × 106 | 1.9 × 105 | 7.8 × 105 | 1.9 × 104 | 5.2 × 105 |
| Total | 7.2 × 107 | 5.9 × 107 | 2.8 × 107 | 4.1 × 106 | 4.9 × 106 | 4.2 × 105 | 3.1 × 106 |
Figure 1.
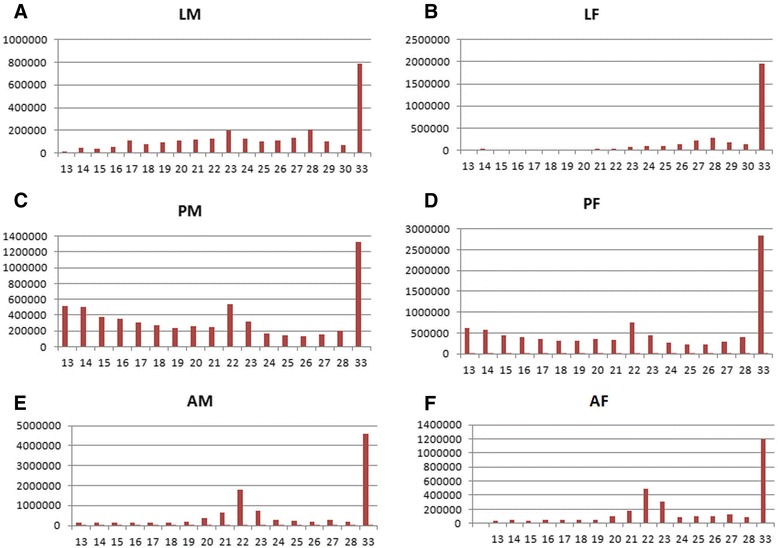
Size distribution of small RNA reads in deep sequenced libraries. Column graphs showing different sizes of small RNA reads in (A) larva male (LM), (B) larva female (LF), (C) pupa male (PM), (D) pupa female (PF), (E) adult male (AM) and (F) adult female (AF) mosquito.
Complete repertoire of miRNAs in immature and mature stages of An.stephensi 18-30 nt length sequences in each of the libraries were mapped on to known miRNA sequences fetched from mirbase database. A total of 4.9×106 reads from all six libraries mapped with 100% match with known miRNA sequences from eight different insect species (Table 1). After removing mapped redundancy, 111 total known miRNAs were identified across all stages of An.stephensi. Maximum and minimum number of known miRNAs were identified in AM (n = 109) and LF (n = 70) respectively (Table 2). Remaining stages LM, PM, PF, AF contained 85, 104, 106 and 101 known miRNAs respectively. Sixty nine miRNAs were expressed in all the stages of development of the mosquito. Remaining 42 were absent in one or more developmental stages of the mosquito (Table 2). Many miRNAs were absent in larval stage of mosquito although they showed expression with TPM >10 in other stages of development. Five miRNAs (miR-1891, miR-190-3p, miR-285, miR-988-3p and miR-989) were not expressed in both male and female larva. Whereas nine miRNAs (miR-bantam-5p, miR-137, miR-184b, miR-193, miR-71-5p, miR-929-5p, miR-980, miR-998 and miR-9c-3p) were found absent only in female larva. Out of these 111 miRNAs, three miRNAs namely miR-8-3p, miR-bantam-3p and miR-281-5p were found most abundant in developmental stages. MicroRNA-8-3p was the most abundant miRNA in larval stages of the mosquito while bantam-3p was most abundant in both male and female pupal stages of development. Most abundant miRNA in AM and AF was miR-281-5p and miR-bantam-3p respectively (Table 2). Differential expression of known miRNAs between the genders and during metamorphosis is described in following sections of the manuscript.
Table 2.
List of known miRNAs expressed and regulated across mosquito developmental stages
| S. No. | miRNA | Length | Sequence | Tags Per Million (TPM) | |||||
|---|---|---|---|---|---|---|---|---|---|
| LM | LF | PM | PF | AM | AF | ||||
| 1 | ast-bantam-3p | 22 | TGAGATCACTTTGAAAGCTGAT | 2199.4 | 245.4 | 5410.0 | 6599.4 | 9907.7 | 25521.7 |
| 2 | ast-bantam-5p | 23 | CCGGTTTTCATTTTCGATCTGAC | 3.3 | 5.9 | 7.5 | 24.0 | 43.4 | |
| 3 | ast-let-7 | 21 | TGAGGTAGTTGGTTGTATAGT | 65.8 | 5.1 | 384.4 | 538.1 | 983.3 | 1301.1 |
| 4 | ast-miR-100 | 22 | AACCCGTAGATCCGAACTTGTG | 254.1 | 30.9 | 2226.8 | 2173.4 | 3732.3 | 8010.1 |
| 5 | ast-miR-1000 | 21 | ATATTGTCCTGTCACAGCAGT | 8.1 | 0.6 | 14.1 | 19.8 | 45.6 | 54.0 |
| 6 | ast-miR-10-3p | 23 | CAAATTCGGTTCTAGAGAGGTTT | 21.7 | 7.1 | 41.1 | 60.8 | 67.8 | 102.0 |
| 7 | ast-miR-10-5p | 22 | ACCCTGTAGATCCGAATTTGTT | 614.5 | 151.3 | 2437.9 | 3984.4 | 25072.2 | 9310.7 |
| 8 | ast-miR-11 | 22 | CATCACAGTCTGAGTTCTTGCT | 218.1 | 23.8 | 370.2 | 574.2 | 626.4 | 1269.3 |
| 9 | ast-miR-1174 | 22 | TCAGATCTACTTCATACCCATG | 18.0 | 2.0 | 7.9 | 7.4 | 6.9 | 20.6 |
| 10 | ast-miR-1175-3p | 21 | TGAGATTCTACTTCTCCGACT | 41.6 | 4.0 | 50.8 | 65.1 | 81.6 | 66.8 |
| 11 | ast-miR-1175-5p | 22 | AAGTGGAGTAGTGGTCTCATCG | 9.2 | 3.2 | 4.9 | 3.4 | 4.2 | |
| 12 | ast-miR-12 | 23 | TGAGTATTACATCAGGTACTGGT | 107.4 | 12.8 | 128.2 | 109.3 | 373.3 | 438.8 |
| 13 | ast-miR-124 | 20 | TAAGGCACGCGGTGAATGCC | 1.5 | 0.3 | 7.2 | 9.7 | 40.1 | 45.1 |
| 14 | ast-miR-125-3p | 21 | ACAAGTTTTGATCTCCGGTAT | 1.1 | 0.3 | 3.6 | 4.9 | 3.5 | 11.0 |
| 15 | ast-miR-125-5p | 22 | TCCCTGAGACCCTAACTTGTGA | 10.3 | 1.1 | 66.7 | 80.3 | 228.9 | 194.0 |
| 16 | ast-miR-133-3p | 22 | TTGGTCCCCTTCAACCAGCTGT | 9.2 | 0.3 | 30.4 | 42.8 | 138.3 | 129.4 |
| 17 | ast-miR-133-5p | 20 | AGCTGGTTGACATCGGGTCA | 0.2 | 0.1 | ||||
| 18 | ast-miR-137 | 22 | TATTGCTTGAGAATACACGTAG | 1.8 | 5.8 | 6.4 | 19.0 | 39.6 | |
| 19 | ast-miR-13a | 22 | CTCCTCAAAGGGTTGTGAAATG | 0.4 | 0.4 | 0.6 | 1.3 | 1.3 | |
| 20 | ast-miR-13b-3p | 23 | TATCACAGCCATTTTGACGAGTT | 44.1 | 9.6 | 135.1 | 190.7 | 138.4 | 124.9 |
| 21 | ast-miR-13b-5p | 22 | TCGTAAAAATGGTTGTGCTGTG | 3.7 | 2.3 | 6.9 | 14.3 | 5.5 | 18.6 |
| 22 | ast-miR-1-3p | 22 | TGGAATGTAAAGAAGTATGGAG | 19.5 | 2.8 | 243.3 | 259.6 | 428.2 | 197.9 |
| 23 | ast-miR-14 | 22 | TCAGTCTTTTTCTCTCTCCTAT | 89.0 | 13.6 | 551.5 | 690.8 | 1779.2 | 4857.7 |
| 24 | ast-miR-1-5p | 18 | TACTTCTTTACATTCCAT | 0.2 | 0.4 | 0.2 | |||
| 25 | ast-miR-184a | 22 | TGGACGGAGAACTGATAAGGGC | 1360.2 | 161.5 | 4415.3 | 4174.1 | 13173.6 | 16430.4 |
| 26 | ast-miR-184b | 22 | TGGACGGAGAACTGATAAAGGA | 22.8 | 12.0 | 151.6 | 76.9 | 115.5 | |
| 27 | ast-miR-1889 | 20 | CACATTACAGATTGGGATTA | 0.1 | 0.2 | 1.1 | 0.9 | ||
| 28 | ast-mir-1890 | 20 | TGAAATCTTTGATTAGGTCT | 7.7 | 1.7 | 126.0 | 133.4 | 34.3 | 38.7 |
| 29 | ast-miR-1891 | 22 | TGAGGAGTTAATTTGCGTGTTT | 0.1 | 0.3 | 129.2 | 85.4 | ||
| 30 | ast-miR-190-3p | 22 | CCCAGGAATCAAACATATTATT | 0.5 | 2.8 | 13.1 | 23.4 | ||
| 31 | ast-miR-190-5p | 24 | AGATATGTTTGATATTCTTGGTTG | 9.2 | 4.5 | 23.0 | 30.5 | 57.7 | 56.8 |
| 32 | ast-miR-193 | 20 | TACTGGCCTACTAAGTCCCA | 0.7 | 8.4 | 15.9 | 0.3 | 1.1 | |
| 33 | ast-miR-210-3p | 21 | CTTGTGCGTGTGACAACGGCT | 4.0 | 0.6 | 6.0 | 7.5 | 107.6 | 291.1 |
| 34 | ast-miR-210-5p | 21 | AGCTGCTGACCACTGCACAAG | 0.1 | 0.2 | 0.5 | |||
| 35 | ast-miR-219 | 20 | TGATTGTCCAAACGCAATTC | 0.2 | 0.7 | 2.4 | 0.4 | ||
| 36 | ast-miR-252-3p | 21 | CTGCTGCCCAAGTGCTTATCG | 0.4 | 0.5 | 0.7 | 3.0 | 4.4 | |
| 37 | ast-miR-252-5p | 22 | CTAAGTACTAGTGCCGCAGGAG | 60.7 | 5.7 | 128.4 | 166.9 | 950.6 | 1111.8 |
| 38 | ast-mir-263a | 22 | AATGGCACTGGAAGAATTCACG | 2272.9 | 575.0 | 831.3 | 887.0 | 1205.4 | 3564.4 |
| 39 | ast-miR-263b-3p | 21 | TGGATCTTTTCGTGCCATCGT | 0.1 | |||||
| 40 | ast-miR-263b-5p | 23 | CTTGGCACTGGGAGAATTCACAG | 5.5 | 2.3 | 17.3 | 25.8 | 133.4 | 137.4 |
| 41 | ast-miR-275 | 22 | TCAGGTACCTGAAGTAGCGCGC | 135.0 | 11.9 | 286.9 | 451.9 | 341.0 | 711.3 |
| 42 | ast-miR-276-3p | 22 | TAGGAACTTCATACCGTGCTCT | 1118.3 | 136.9 | 2720.4 | 3464.7 | 3892.3 | 4702.5 |
| 43 | ast-miR-2765 | 22 | TGGTAACTCCACCACCGTTGGC | 157.0 | 21.5 | 956.6 | 858.5 | 2.1 | 0.5 |
| 44 | ast-miR-276-5p | 22 | AGCGAGGTATAGAGTTCCTACG | 12.9 | 2.6 | 29.6 | 39.5 | 31.2 | 29.2 |
| 45 | ast-miR-277 | 22 | TAAATGCACTATCTGGTACGAC | 36.8 | 6.0 | 605.0 | 657.4 | 3277.0 | 5776.7 |
| 46 | ast-miR-278 | 21 | TCGGTGGGACTTTCGTCCGTT | 15.1 | 2.6 | 12.7 | 22.9 | 25.6 | 24.3 |
| 47 | ast-miR-279 | 20 | TGACTAGATCCACACTCATT | 44.1 | 5.7 | 174.4 | 264.0 | 172.4 | 176.1 |
| 48 | ast-miR-2796-3p | 20 | GTAGGCCGGCGGAAACTACT | 2.6 | 0.6 | 3.2 | 4.3 | 20.1 | 25.7 |
| 49 | ast-miR-2796-5p | 22 | AGGGGTTTCTTTCGGCCTCCAG | 0.1 | 0.1 | 0.1 | 0.2 | ||
| 50 | ast-mir-281-3p | 22 | TGTCATGGAATTGCTCTCTTTA | 36.0 | 4.0 | 31.3 | 38.1 | 74.2 | 29.0 |
| 51 | ast-miR-281-5p | 22 | AAGAGAGCTATCCGTCGACAGT | 3986.2 | 233.8 | 4066.0 | 3725.7 | 14959.7 | 13394.8 |
| 52 | ast-miR-282 | 22 | TAGCCTCTTCTAGGCTTTGTCT | 8.1 | 0.3 | 6.8 | 10.2 | 0.3 | 0.9 |
| 53 | ast-miR-283 | 19 | AAATATCAGCTGGTAATTC | 1.8 | 0.6 | 2.4 | 2.7 | 6.8 | 5.5 |
| 54 | ast-miR-285 | 22 | TAGCACCATTCGAAATCAGTAC | 51.4 | 14.2 | 313.7 | 4.2 | ||
| 55 | ast-miR-286 | 25 | TGACTAGACCGAACACTCGCGTCCT | 0.1 | 0.1 | ||||
| 56 | ast-miR-2944a-3p | 22 | TATCACAGTAGTTGTACTTTAA | 0.1 | 0.2 | ||||
| 57 | ast-miR-2944a-5p | 22 | GAAGGAACTTCTGCTGTGATCT | 0.4 | 1.3 | 0.8 | 0.4 | ||
| 58 | ast-miR-2944b-3p | 22 | TATCACAGCAGTAGTTACCTGA | 0.1 | 0.0 | ||||
| 59 | ast-miR-2944b-5p | 23 | GAAGGAACTCCCGGTGTGATATA | 0.1 | 0.1 | 0.1 | 0.2 | ||
| 60 | ast-miR-2945 | 19 | TGACTAGAGGCAGACTCGT | 0.4 | 1.6 | 2.5 | 3.5 | 4.7 | |
| 61 | ast-miR-2a-3p | 23 | TATCACAGCCAGCTTTGAAGAGC | 110.3 | 13.3 | 280.0 | 375.7 | 475.8 | 601.6 |
| 62 | ast-miR-2a-5p | 21 | ACTCTCAAAGTGGTTGTGAAA | 14.0 | 2.0 | 79.0 | 35.4 | 8.8 | 36.5 |
| 63 | ast-miR-2b | 24 | TATCACAGCCAGCTTTGATGAGCT | 51.5 | 8.5 | 139.2 | 307.4 | 295.8 | 225.8 |
| 64 | ast-miR-2c | 18 | TCACAGCCAGCTTTGATG | 7.4 | 0.2 | 0.8 | 4.3 | ||
| 65 | ast-miR-305-3p | 22 | CGGCACATGTTGGAGTACACTT | 5.1 | 0.3 | 8.8 | 8.8 | 20.8 | 21.9 |
| 66 | ast-miR-305-5p | 21 | ATTGTACTTCATCAGGTGCTC | 95.2 | 9.9 | 189.8 | 288.3 | 168.4 | 277.6 |
| 67 | ast-miR-306 | 22 | TCAGGTACTGGATGACTCTCAG | 410.0 | 39.4 | 556.8 | 792.3 | 845.3 | 2160.1 |
| 68 | ast-miR-307-3p | 20 | TCACAACCTCCTTGAGTGAG | 0.3 | 0.6 | 1.2 | 1.5 | ||
| 69 | ast-miR-307-5p | 20 | ACTCACTCAACCTGGGTGTG | 0.1 | 0.1 | 0.1 | 0.2 | ||
| 70 | ast-miR-308 | 18 | AATCACAGGAGTATACTG | 32.4 | 2.6 | 22.7 | 30.4 | 13.0 | 24.5 |
| 71 | ast-miR-309 | 22 | TCACTGGGCAAAGTTTGTCGCA | 0.1 | |||||
| 72 | ast-miR-31 | 21 | TGGCAAGATGTTGGCATAGCT | 14.0 | 2.8 | 48.6 | 53.6 | 104.0 | 63.7 |
| 73 | ast-miR-315 | 23 | TTTTGATTGTTGCTCAGAAAGCC | 129.1 | 34.6 | 75.8 | 129.8 | 382.3 | 980.6 |
| 74 | ast-miR-317 | 25 | TGAACACATCTGGTGGTATCTCAGT | 111.8 | 17.6 | 60.2 | 47.6 | 480.5 | 1900.9 |
| 75 | ast-miR-33 | 21 | GTGCATTGTAGTTGCATTGCA | 0.3 | 0.2 | 0.2 | 0.6 | 0.5 | |
| 76 | ast-miR-34 | 23 | TGGCAGTGTGGTTAGCTGGTTGT | 18.0 | 2.8 | 8.5 | 8.0 | 789.4 | 657.3 |
| 77 | ast-miR-375 | 22 | TTTGTTCGTTTGGCTCGAGTTA | 72.1 | 6.0 | 8.5 | 11.6 | 55.5 | 145.1 |
| 78 | ast-miR-7 | 21 | TGGAAGACTAGTGATTTTGTT | 8.5 | 2.3 | 1.9 | 2.4 | 13.6 | 2.9 |
| 79 | ast-miR-71-3p | 22 | TCTCACTACCTTGTCTTTCATG | 26.5 | 2.8 | 64.8 | 80.9 | 97.0 | 98.0 |
| 80 | ast-miR-71-5p | 22 | AGAAAGACATGGGTAGTGAGAT | 1.1 | 5.6 | 5.6 | 15.3 | 8.4 | |
| 81 | ast-miR-79-3p | 23 | ATAAAGCTAGATTACCAAAGCAT | 0.4 | 0.6 | 1.5 | 3.0 | 2.3 | 3.8 |
| 82 | ast-miR-79-5p | 23 | GCTTTGGCGCTTTAGCTGTATGA | 0.3 | 0.6 | 0.2 | 0.5 | ||
| 83 | ast-miR-8-3p | 23 | TAATACTGTCAGGTAAAGATGTC | 4190.3 | 577.3 | 3906.2 | 5280.5 | 8065.6 | 24812.4 |
| 84 | ast-miR-8-5p | 22 | CATCTTACCGGGCAGCATTAGA | 130.2 | 13.6 | 230.3 | 285.7 | 901.1 | 2144.5 |
| 85 | ast-miR-87 | 19 | GTGAGCAAATATTCAGGTG | 0.4 | 0.2 | 0.2 | 1.3 | 1.3 | |
| 86 | ast-miR-927-3p | 22 | CAAAGCGTTTGGATTCTGAAAC | 4.0 | 0.9 | 11.8 | 15.3 | 50.7 | 126.7 |
| 87 | ast-miR-927-5p | 22 | TTTAGAATTCCTACGCTTTACC | 22.8 | 7.1 | 146.7 | 97.7 | 866.4 | 934.0 |
| 88 | ast-miR-929-3p | 20 | TCCCTAACGGAGTCAGATTG | 0.2 | 0.4 | ||||
| 89 | ast-miR-929-5p | 21 | AAATTGACTCTAGTAGGGAGT | 0.4 | 4.2 | 4.3 | 13.5 | 15.5 | |
| 90 | ast-miR-92a | 20 | TATTGCACTTGTCCCGGCCT | 36.4 | 1.7 | 33.1 | 21.3 | 29.7 | 91.3 |
| 91 | ast-miR-92b | 22 | AATTGCACTTGTCCCGGCCTGC | 192.0 | 11.9 | 283.7 | 339.2 | 147.6 | 218.3 |
| 92 | ast-miR-932 | 23 | TCAATTCCGTAGTGCATTGCAGT | 4.8 | 0.9 | 22.7 | 28.1 | 55.7 | 80.9 |
| 93 | ast-miR-957 | 22 | TGAAACCGTCCAAAACTGAGGC | 68.4 | 24.4 | 268.1 | 366.7 | 898.2 | 819.6 |
| 94 | ast-miR-965 | 22 | TAAGCGTATAGCTTTTCCCATT | 1.1 | 2.0 | 2.8 | 0.8 | 2.0 | |
| 95 | ast-miR-970-3p | 21 | TCATAAGACACACGCGGCTAT | 43.4 | 3.4 | 75.9 | 131.4 | 313.4 | 453.4 |
| 96 | ast-miR-980 | 19 | TAGCTGCCTAGTGAAGGGC | 0.4 | 1.6 | 1.9 | 13.3 | 37.8 | |
| 97 | ast-miR-981 | 22 | TTCGTTGTCGACGAAACCTGCA | 4.8 | 0.9 | 8.2 | 17.3 | 63.0 | 49.1 |
| 98 | ast-miR-988-3p | 22 | CCCCTTGTTGCAAACCTCACGC | 2.5 | 8.3 | 12.1 | 27.6 | ||
| 99 | ast-miR-988-5p | 21 | GTGTGCTTTGTGACAATGAGA | 0.1 | 0.2 | ||||
| 100 | ast-miR-989 | 21 | TGTGATGTGACGTAGTGGTAC | 4.5 | 2.1 | 0.5 | 55.1 | ||
| 101 | ast-miR-993-3p | 24 | GAAGCTCGTTTCTATAGAGGTATC | 4.4 | 2.3 | 6.7 | 9.2 | 14.7 | 28.5 |
| 102 | ast-miR-993-5p | 22 | TACCCTGTAGTTCCGGGCTTTT | 1.1 | 0.9 | 3.4 | 6.7 | 10.6 | 15.7 |
| 103 | ast-miR-996 | 20 | TGACTAGATTACATGCTCGT | 68.8 | 6.5 | 149.5 | 199.4 | 175.1 | 315.8 |
| 104 | ast-miR-998 | 21 | TAGCACCATGAGATTCAGCTC | 7.4 | 14.3 | 10.1 | 33.9 | 38.3 | |
| 105 | ast-miR-999 | 22 | TGTTAACTGTAAGACTGTGTCT | 210.0 | 24.9 | 440.1 | 609.1 | 2221.6 | 3492.3 |
| 106 | ast-miR-9a | 23 | TCTTTGGTTATCTAGCTGTATGA | 2025.1 | 406.1 | 1594.9 | 2663.9 | 321.4 | 1049.7 |
| 107 | ast-miR-9c-3p | 22 | TAAAGCTTTAGTACCAGAGGTC | 2.6 | 2.1 | 3.1 | 3.9 | 11.1 | |
| 108 | ast-miR-9c-5p | 22 | TCTTTGGTATTCTAGCTGTAGA | 840.6 | 92.1 | 669.4 | 1056.2 | 366.8 | 1565.7 |
| 109 | ast-miR-iab-4-3p | 24 | CGGTATACCTTCAGTATACGTAAC | 0.1 | |||||
| 110 | ast-miR-iab-4-5p | 22 | ACGTATACTGAATGTATCCTGA | 3.3 | 0.3 | 2.3 | 2.6 | 1.7 | 2.2 |
| 111 | ast-miR-iab-8-5p | 20 | TTACGTATACTGAAGGTATA | 0.1 | 0.2 | 0.2 | 0.2 | ||
Several novel miRNAs were identified using high throughput deep sequencing data in adult mosquito. Novel miRNAs identified in adult female mosquito were reported previously [9]. In this study, we focused on identification of novel miRNAs in adult male mosquito. Their expression was then checked in other stages of mosquito development (Table 3). Fourteen small RNA sequences folded into a perfect hairpin structure with folding energy < −20 kcal/mol (Additional file 1: Figure S1). Seven of these were present on 5’ arm of the precursor and hence were annotated as miR-5p whereas rest seven sequences were present on 3’arm and were annotated as miR-3p. Nomenclature of these novel miRNAs was given in a specific order (Table 3). Novel miRNA sequences were blasted against known miRNAs in mirBase database. These sequences did not show similarity with any of the known miRNAs indicating towards identification of novel class of miRNAs in mosquito.
Table 3.
List of Novel miRNAs expressed and regulated across mosquito developmental stages
| S. No | Length | Mature miRNA sequence | Pre-miRNA sequence | Novel miRNA | Tags Per Million (TPM) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| LM | LF | PM | PF | AM | AF | |||||
| 1 | 20 | AGTCATTAATGATATTAGAC | AUUUAUGGGAAAUAAUCUCUUUAAUCAGGCUUUAUAGUCAUUAAUGAUAUUAGACUGCAAUUCUAAAGGA AUAAUUUAAUUUAUUAAGGCUUAAGAAUUAAAAGAUUAAUACAAAUAUUUUCA | ast-Novel-1-3p | 0.37 | 0.00 | 0.06 | 0.05 | 0.08 | 0.00 |
| 2 | 21 | ATAATTTAATTTATTAAGGCT | AUUUAUGGGAAAUAAUCUCUUUAAUCAGGCUUUAUAGUCAUUAAUGAUAUUAGACUGCAAUUCUAAAGGA AUAAUUUAAUUUAUUAAGGCUUAAGAAUUAAAAGAUUAAUACAAAUAUUUUCA | ast-Novel-1-5p | 0.18 | 0.16 | 2.84 | 0.18 | ||
| 3 | 23 | GTATTCAATTTGTATATCGTCGT | GUUGGACCUUACUUAAAUUUGUAUUCAAUUUGUAUAUCGUCGUCAUCAGAAUAUAUUAUAAGAUUAAUAAUUUUCUAAAUAUUUUAUUAAAUAAUAUGUCAGGUCAAGGUGCAGUUUAUGUUUAAGUAG | ast-Novel-2-3p | 0.18 | 0.10 | 0.08 | |||
| 4 | 19 | ATTGATAATCCACGTTGGA | GUUGGACCUUACUUAAAUUUGUAUUCAAUUUGUAUAUCGUCGUCAUCAGAAUAUAUUAUAAGAUUAAUAAUUUUCUAAAUAUUUUAUUAAAUAAUAUGUCAGGUCAAGGUGCAGUUUAUGUUUAAGUAG | ast-Novel-2-5p | 0.37 | 0.54 | 0.16 | 0.11 | ||
| 5 | 24 | TTAAGATTGGTATGATTAGCGTCT | UUUCCUUUAAGAUUGGUAUGAUUAGCGUCUUGUUUAGCAGAAACUAAUCGUACUCCUUUU | ast-Novel-3-3p | 0.16 | 0.12 | 0.24 | |||
| 6 | 24 | AAACTAATCGTACTCCTTTTGATT | UUUCCUUUAAGAUUGGUAUGAUUAGCGUCUUGUUUAGCAGAAACUAAUCGUACUCCUUUU | ast-Novel-3-5p | 0.16 | |||||
| 7 | 18 | AATGGGTAGTCGAAGATT | AAUUGCUCAUGGUUUAUGUUCAUCUGGAUUAUUUUGUUUAGCUAAUAUUUCUUACGAACGAAUGGGUAGUCGAAGAUUAUUAAUUAAUAA | ast-Novel-4-3p | 6.00 | 0.47 | 0.16 | |||
| 8 | 22 | AATTGCTCATGGTTTATGTTCA | AAUUGCUCAUGGUUUAUGUUCAUCUGGAUUAUUUUGUUUAGCUAAUAUUUCUUACGAACGAAUGGGUAGUCGAAGAUUAUUAAUUAAUAA | ast-Novel-4-5p | 0.3 | |||||
| 9 | 21 | CGTAGTTTCTACATTAGGAGT | UUUUACUUUUUUUGUAGGAUCAAUAUGAUUUAUACCCGUAGUUUCUACAUUAGGAGUAAU | ast-Novel-5-3p | 0.12 | 0.10 | 0.04 | |||
| 10 | 21 | ACTTTTTTTGTAGGATCAATA | UUUUACUUUUUUUGUAGGAUCAAUAUGAUUUAUACCCGUAGUUUCUACAUUAGGAGUAAU | ast-Novel-5-5p | 0.60 | 0.42 | 0.56 | 0.18 | ||
| 11 | 25 | TTTCGGATATGAATCAAAGTAATTT | ACCACUUAAAUUUCGGAUAUGAAUCAAAGUAAUUUUCAUCAAUUCCCUCUACCCUCAUGAGCGAGUGAUGUAAUGUACUCUCGUUCUGGACGAUUUUCAACCACUUAAAU | ast-Novel-6-3p | 100.76 | 44.78 | 95.03 | 141.73 | 135.20 | 796.74 |
| 12 | 21 | CGTTCTGGACGATTTTCAACC | ACCACUUAAAUUUCGGAUAUGAAUCAAAGUAAUUUUCAUCAAUUCCCUCUACCCUCAUGAGCGAGUGAUGUAAUGUACUCUCGUUCUGGACGAUUUUCAACCACUUAAAU | ast-Novel-6-5p | 0.37 | 0.6 | ||||
| 13 | 23 | AGCCGAACCAGCAGTACGAGTTT | GACCCGACCGAGCCGAACCAGCAGUACGAGUUUUAACGCACGUUUCUUCGACUGGGAGCGUGGCGUCUCCUGUAACGCGGCUACUCGUGG | ast-Novel-7-3p | 0.04 | |||||
| 14 | 22 | CGTCTCCTGTAACGCGGCTACT | GACCCGACCGAGCCGAACCAGCAGUACGAGUUUUAACGCACGUUUCUUCGACUGGGAGCGUGGCGUCUCCUGUAACGCGGCUACUCGUGG | ast-Novel-7-5p | 0.28 | 0.30 | 0.31 | 0.04 | 0.18 | |
Validation of known miRNAs by Northern hybridization
MicroRNAs identified by small RNA sequencing was verified by northern hybridization. Total RNA from sugar fed female mosquito was probed using locked nucleic acid (LNA) probes (Exiqon) for representative miRNAs. Expression of eight miRNAs namely miR-277, miR-14, miR-34, miR-285, miR-13b-3p, miR-989, miR-1174 and mir-219 was validated in An.stephensi (Figure 2). Out of these eight miRNAs, miR-277 and miR-14 were highly abundant in female mosquito with reads more than 2×104. Hence, these two miRNAs showed distinct and specific band in northern hybridization. Two MicroRNA miR-285 and miR-219 were low abundance miRNAs with reads less than 25 in female mosquito. Nevertheless, such low expressing miRNAs was detectable by specific LNA based probes in the mosquito (Figure 2) thereby validating the sensitivity of this assay.
Figure 2.
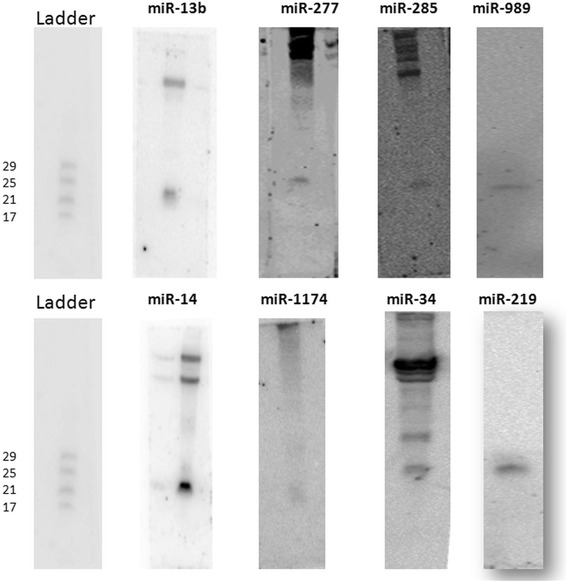
Northern hybridization based detection of mosquito miRNAs. 10ug total RNA from adult female was used for detection of (A) miR-13b, (B) miR-277, (C) miR-285, (D) miR-989, (E) miR-14, (F) miR-1174, (G) miR-34 and (H) miR-219 via DIG based Northern hybridization. Ladder was run along with total RNA with oligos ranging from 17 nt to 29 nt.
Differential expression of miRNAs across different stages of mosquito development
To study differential expression of miRNAs, tags per million (TPM) of individual miRNAs was calculated in all the six libraries. MicroRNAs with TPM >10 in any of the developmental stages were subjected to EdgeR analysis. Regulation of miRNAs was studied between two developmental stages and miRNAs showing fold change >2 with P value ≤ 0.05 were classified as differentially expressed. Heat map of all known miRNAs is shown in Figure 3. A total of 36 miRNAs were found differentially expressed between different stages of mosquito development.
Figure 3.
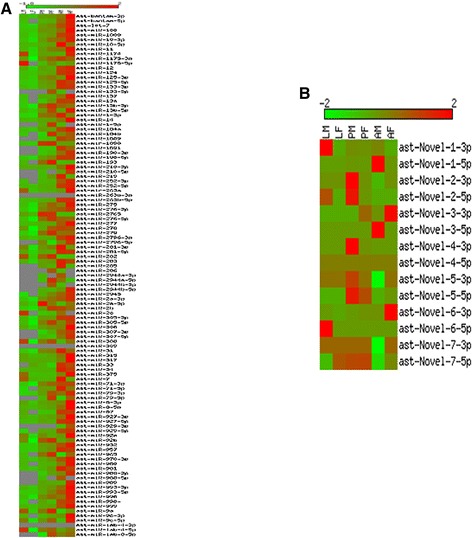
Heat map of miRNAs differentially expressed across developmental stages. Expression profiles of (A) known miRNAs and (B) Novel miRNAs in small RNA libraries prepared from larva male (LM), larva female (LF), pupa male (PM), pupa female (PF), adult male (AM) and adult female (AF) mosquito. Colour gradation from light green to dark red represents relative increase in miRNA expression.
Gender regulated microRNAs
We studied difference in miRNAs expression between both genders during larval, pupa and adult mosquito stages. Minimum number of miRNA differences was identified during larval stages as it is the early stage of mosquito life cycle. Only two miRNAs namely miR-184b and miR-1175-5p were significantly down-regulated in female larvae when compared with male larvae (Additional file 2: Figure S2). Both genders during pupa stage showed difference in the expression of three miRNAs. MicroRNA-285 was down-regulated whereas miR-190-3p and miR-184b were up-regulated in female pupa as compared to male pupa stage (Additional file 2: Figure S2). Maximum differences in miRNA expression between genders were observed during adult stages of the mosquito. Only one miRNA miR-989 was found significantly up-regulated in adult female mosquito when compared to male mosquito. Whereas, five miRNAs namely miR-2c, miR-285, miR-219, miR-7 and miR-2765 were found down-regulated in female compared with male mosquito (Additional file 2: Figure S2).
MicroRNA regulation during metamorphosis of male mosquito from larval to pupa and to adult stages
MicroRNA regulation was observed during transition of male mosquito from larva to pupa stage and during metamorphosis from pupa to adult stage of mosquito life cycle. Many miRNAs were found differentially expressed during these two transition phases. Seven miRNAs (miR-2c, miR-375, miR-7, miR-1175-5p, miR-263a, miR-1174 and miR-34) were down-regulated whereas cluster of seven miRNAs (miR-100, miR-193, miR-1-3p, miR-1890, miR-277, miR-989 and miR-285) were up-regulated in pupa stage when compared to the larval male mosquito (Additional file 3: Figure S3). Metamorphosis from pupa to adult stage also resulted in differential expression of miRNAs. We identified seven miRNAs (miR-10-5p, miR-219, miR-2c, miR-210-3p, miR-190-3p, miR-34 and miR-1891) up-regulated in male adult mosquito when compared to pupal stage (Additional file 3: Figure S3). Transition to adult mosquito resulted in down-regulation of eight miRNAs namely miR-2765, miR-193, miR-282, miR-989, miR-2a-5p, miR-9a, miR-1890 and miR-965 (Additional file 3: Figure S3). Out of these regulated miRNAs, five miRNAs are of significant importance as they showed differential expression during larval to pupa as well as during pupa to adult metamorphosis. Two microRNAs, miR-2c and miR-34 were down-regulated from larval to pupa transition whereas its expression increases again in adult stages. Three miRNAs, miR-193, miR-1890 and miR-989 were up-regulated in pupa stage. Their expression down-regulates in adult male mosquito when compared to its pupa stage (Additional file 3: Figure S3).
MicroRNA regulation during metamorphosis of female mosquito from larval to pupa and to adult stages
Regulation of miRNAs was also observed during metamorphosis of female mosquito from larva to pupa and to adult stages of development. Eight miRNAs (miR-1-3p, miR-let-7, miR-277, miR-133-3p, miR-285, miR-193, miR-998 and miR-184b) were up-regulated in pupa female when compared to larva female mosquito (Additional file 4: Figure S4). Another set of eight miRNAs (miR-7, miR-263a, miR-375, miR-317, miR-34, miR-1174, miR-315 and miR-993-3p) were found down-regulated in pupa female compared with larval female mosquito (Additional file 4: Figure S4). Metamorphosis of female pupa to adult also resulted in differential expression of miRNAs. We identified eight miRNAs (miR-927-5p, miR-375, miR-980, miR-989, miR-210-3p, miR-317, miR-34 and miR-1891) up-regulated in adult female mosquito compared to pupa stage (Additional file 4: Figure S4). A set of eight miRNAs (miR-2765, miR-2c, miR-1-5p, miR-193, miR-282, miR-2944a-5p, miR-285 and miR-9a) were down-regulated in adult mosquito when compared with pupa female mosquito (Additional file 4: Figure S4). Five miRNAs were differentially expressed in all developmental stages of female mosquito. Two miRNAs, miR-285 and miR-193 were up-regulated in pupa stages whereas gets down-regulated in adult stages. Three miRNAs namely miR-375, miR-317 and miR-34 were down-regulated in pupa female whereas adult stage resulted in up-regulation of these miRNAs when compared to pupa stage (Additional file 4: Figure S4).
Validation of miRNA expression profiling by real-time PCR
Differential expression of miRNAs across immature and mature stages in both male and female mosquitoes was validated by qRT-PCR. Two miRNAs, miR-989 and miR-219 were up-regulated in adult female and adult male mosquito respectively when compared to other stages of development (Figure 4A and B). Significant down-regulation was observed for miR-277, miR-210 and miR-285 in larval stages compared to pupa and adult stages of mosquito (Figure 4C, D and H). MicroRNA-34 was down-regulated in male and female pupa compared to larva and adult mosquito (Figure 4E). MicroRNA-1174 expression was down-regulated in both genders of pupa stage and male mosquito (Figure 4F). MicroRNA -9a was the only miRNA that showed up-regulated expression in larva and pupa stages when compared to adult mosquito (Figure 4G). Similar expression pattern of miRNAs was observed in small RNAs sequencing and real time data analysis, thereby validating our analysis.
Figure 4.
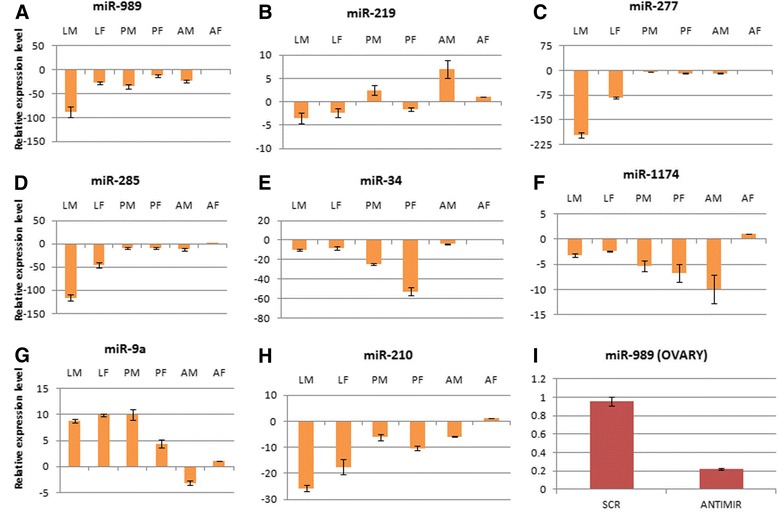
Expression profiling of regulated miRNAs by Real time PCR. (A) miR-989, (B) miR-219, (C) miR-277, (D) miR-285, (E) miR-34, (F) miR-1174, (G) miR-9a and (H) miR-210 were profiled in larva male (LM), larva female (LF), pupa male (PM), pupa female (PF), adult male (AM) and adult female (AF) mosquito. Y axis depicts fold change in miRNA expression in samples compared with adult female mosquito (AF), fold change in which was taken as 1. (I) Fold change in miRNA-989 expression in ovaries from scrambled (SCR) and antagomir (ANTIMIR) injected mosquitoes. Y axis depicts fold change in miRNA expression in samples compared with PBS injected ovaries, fold change in which was taken as 1. 5.8 s RNA was taken as an endogenous control.
MicroRNA target prediction using RNA hybrid and degradome sequencing
Regulation of gene expression is brought about by binding of miRNAs on 3’UTR sequence of the target genes and in silico analysis for identifying miRNA binding regions could be employed for better validation [25]. In the present study, we employed bioinformatic analysis and in vivo assays to identify targets of regulated miRNAs. Targets binding to miRNAs were predicted first by identifying miRNAs seed binding sites on 3’UTR of An. stephensi genes using RNAhybrid. Further, degree of binding complementarity determines the mechanism of target regulation, either by mRNA cleavage or by translational repression. As ovary is an important organ for insect reproduction, we aimed to identify mRNAs cleaved by miRNAs in ovary tissue of blood fed female mosquito by degradome sequencing.
Target prediction of differentially expressed miRNAs and enriched pathway analysis
Targets of miRNAs regulated in gender-specific manner and during metamorphosis from larva to pupa and to adult stages in both genders were predicted using RNAhybrid (P value < 0.05). mRNA targets were predicted for 36 miRNAs that were regulated during different stages of mosquito development (as described above) (Additional file 5: Table S1). Maximum number of targets were identified for miR-34 (n = 524) whereas minimum targets were predicted for miR-190-3p (n = 24). Orthologs of all the mRNA targets known in An.gambiae were retrieved from vector-base. MicroRNAs were divided into five clusters depending upon developmental stage at which they were found differentially expressed. MicroRNAs that showed gender differential expression during adult stage were grouped as a cluster. MicroRNAs regulated between the developmental stages in both genders were analysed separately (Table 4). Orthologs of target genes known in An. gambiae were retrieved from vector base and analysed using functional annotation clustering tool of DAVID resource. Analysis was carried out using An.gambiae gene IDS as An.stephensi database is not linked to the DAVID resource. Annotation term groups playing more enriched role in present study were identified. Targets of miRNAs showing gender differential expression were analysed which resulted in identification of five enriched annotation clusters. These clusters were involved in cellular polysaccharide biosynthetic process, chaperone, hexose metabolic pathway, heme binding and phagocytotic pathways (Table 4). Targets of miRNAs regulated from larva to pupa stage in male mosquito were enriched in genes functional in phagocytosis and membrane organization, polysaccharide metabolic process, protease activity, peptidase activity and nuclear transport. Female larva to pupa metamorphosis involved genes functional in enriched pathways such as endocytosis, polysaccharide metabolic process, cell-redox homeostasis, peptidase and amino acid metabolic pathway (Table 4). Transition of pupa to adult male mosquito is mediated by enriched pathways such as gelsolin–actin binding protein, polysaccharide biosynthetic process, zinc finger, vesicle mediated transport, protein biosynthesis, translation, vitamin and cofactor binding, nucleoside binding, protein catabolic process and insect pheromone /odorant binding protein phBP (Table 4). In female mosquitoes, pathways functional in phagocytosis, cellular carbohydrate biosynthetic process, protein biosynthesis, vitamin binding, cyclin, golgi membrane, ATP binding, hydrolase activity and transcription were enriched during pupa to adult metamorphosis (Table 4).
Table 4.
List of enriched pathways functional during different stages of mosquito development
| (I) | miRNAs differentially expressed in male and female adult mosquitoes | |
| Down-regulated miRNAs in females: miR-2c, miR-285, miR-219, miR-7 and miR-2765 | ||
| Up-regulated miRNAs in females: miR-989 | ||
| Significant clusters identified | Enrichment score | |
| 1 | Cellular polysaccharide biosynthetic process | 2.1 |
| 2 | Chaperone | 1.8 |
| 3 | Hexose metabolic pathway | 1.3 |
| 4 | Heme binding | 1.1 |
| 5 | Phagocytotic pathways | 1.09 |
| (II) | miRNAs regulated from larval to pupa transition of male mosquito | |
| Down-regulated miRNAs in pupa male: miR-2c, miR-375, miR-7, miR-1175, miR-263a, miR-1174 and miR-34 | ||
| Up-regulated miRNAs in pupa male: miR-100, miR-193, miR-1-3p, miR-1890, miR-277, miR-989 and miR-285 | ||
| Significant clusters identified | Enrichment score | |
| 1 | phagocytosis and membrane organization | 2.6 |
| 2 | polysaccharide metabolic process | 2.1 |
| 3 | protease activity | 1.9 |
| 4 | peptidase activity | 1.6 |
| 5 | nuclear transport | 1.1 |
| (III) | miRNAs regulated from transition of pupa to adult male mosquito | |
| Down-regulated miRNAs in adult male: miR-2765, miR-193, miR-282, miR-989, miR-2a-5p, miR-9a, miR-1890 and miR-965 | ||
| Up-regulated miRNAs in adult male: miR-10-5p, miR-219, miR-2c, miR-210-3p, miR-190-3p, miR-34 and miR-1891 | ||
| Significant clusters identified | Enrichment score | |
| 1 | gelsolin –actin binding protein | 2.2 |
| 2 | polysaccharide biosynthetic process | 2 |
| 3 | zinc finger | 1.8 |
| 4 | vesicle mediated transport | 1.7 |
| 5 | protein biosynthesis | 1.6 |
| 6 | translation | 1.5 |
| 7 | vitamin and cofactor binding | 1.4 |
| 8 | nucleoside binding | 1.3 |
| 9 | protein catabolic process | 1.2 |
| 10 | insect pheromone/odorant binding protein phBP | 1.1 |
| (IV) | miRNAs regulated from larval to pupa transition of female mosquito | |
| Down-regulated miRNAs in pupa female: miR-7, miR-263a, miR-375, miR-317, miR-34, miR-1174, miR-315 and miR-993-3p | ||
| Up-regulated miRNAs in pupa female : miR-1-3p, miR-let-7, miR-277, miR-133-3p, miR-285, miR-193, miR-998 and miR-184b | ||
| Significant clusters identified | Enrichment score | |
| 1 | endocytosis | 3 |
| 2 | polysaccharide metabolic process | 1.9 |
| 3 | cell-redox homeostasis | 1.7 |
| 4 | peptidase | 1.5 |
| 5 | amino acid metabolic pathway | 1.4 |
| (v) | miRNAs regulated from transition of pupa to adult female mosquito | |
| Down-regulated miRNAs in adult female: miR-2765, miR-2c, miR-1-5p, miR-193, miR-282, miR-2944a-5p, miR-285 and miR-9a | ||
| Up-regulated miRNAs in adult female : miR-927-5p, miR-375, miR-980, miR-989, miR-210-3p, miR-317, miR-34 and miR-1891 | ||
| Significant clusters identified | Enrichment score | |
| 1 | phagocytosis | 2.4 |
| 2 | cellular carbohydrate biosynthetic process | 2.1 |
| 3 | protein biosynthesis | 1.6 |
| 4 | vitamin binding | 1.5 |
| 5 | golgi membrane | 1.1 |
| 6 | ATP binding | 1.1 |
| 7 | hydrolase activity | 1.1 |
| 8 | transcription | 1.1 |
These pathways were identified using NIH DAVID functional annotation clustering tool with E score =0.1.
Target identification using antagomirs and degradome sequencing
Loss of function strategy was employed for identification of miRNA targets in ovary of female mosquito. PBS, scrambled RNA and miRNA specific antagomirs were Nano-injected in female mosquito. RNA was extracted and subjected to degradome sequencing. Degradome libraries were constructed from ovary tissue of PBS, scrambled RNA and miR-989 antagomir injected mosquitoes respectively dissected at 24 hours post blood feeding. Analysis of data from PBS injection revealed cleaved products of several miRNAs identified in our study (Data not shown). We restricted our analysis to those significantly regulated miRNAs between the genders, namely, miR-2c, miR-285, miR-219, miR-7, miR-989 and miR-2765. These miRNAs showed distinct regulation in their expression pattern between the two genders at their reproductively active mature stages. Such difference in expression between the genders highlights their role in reproductive processes. mRNA targets of these miRNAs were identified by sequencing the degraded mRNA due to miRNA cleavage using PARE sequencing. Cleaved targets were identified for four miRNAs (miR-285, miR-219, miR-989 and miR-7). Twenty nine mRNAs cleaved by these miRNAs as visualised by target plots were identified in ovary of female mosquito. mRNAs (n = 6) were cleaved by miR-219 and miR-7 whereas miR-285 cleaved only one mRNA in ovary tissue of female mosquito (Table 4). Maximum number if targets were found to be cleaved by miR-989 (n = 16).
In an attempt to validate these targets and to understand the role of miR-989 in mosquito reproduction, expression of miR-989 was knocked down by injecting miR-989 specific antagomir in female mosquito. Four fold decreases in miRNA expression was observed in antagomir injected tissue compared to PBS and scrambled injected ovaries (Figure 4I). Of the sixteen mRNAs that were found to be cleaved in the ovary of PBS injected mosquitoes, ten mRNAs were found regulated in miR-989 antagomir injected ovaries (Table 5). Four of the cleaved targets (ASTE002227, ASTE008381, ASTE003701 and ASTE005749) were also predicted as miR-989 targets by RNAhybrid (Table 5).
Table 5.
List of miRNA targets identified by degradome sequencing in ovary tissue of female mosquito
| S. No. | miRNA | Target | OVARY_PBS (Category) | OVARY_SCR (Category) | OVARY_989 (Category) | GO term name | Cleavage position | Ortholog in An. gambiae |
|---|---|---|---|---|---|---|---|---|
| 1 | miR-989 | ASTE002227* | yes (2) | yes (4) | yes (2) | protein binding | 500 | AGAP003558 |
| 2 | miR-989 | ASTE014169 | yes (0) | yes (0) | yes (0) | 151 | ||
| 3 | miR-989 | ASTE004090 | yes (3) | yes (3) | yes (4) | signal transduction | 3429 | AGAP002261 |
| 4 | miR-989 | ASTE010155 | yes (0) | DNA binding | 840 | AGAP003982 | ||
| 5 | miR-989 | ASTE008099 | yes (4) | protein binding | 553 | AGAP003439 | ||
| 6 | miR-989 | ASTE003651 | yes (4) | aminoacyl-tRNA ligase activity | 1680 | AGAP004708 | ||
| 7 | miR-989 | ASTE007120 | yes (4) | yes (2) | yes (2) | 2092 and 1931 | AGAP000879 | |
| 8 | miR-989 | ASTE014120 | yes (2) | yes (4) | yes (2) | 275 | ||
| 9 | miR-989 | ASTE010290 | yes (4) | yes (2) | yes (2) | 1682 | AGAP005504 | |
| 10 | miR-989 | ASTE002911 | yes (2) | yes (4) | yes (2) | nucleic acid binding | 731 | AGAP004361 |
| 11 | miR-989 | ASTE008381* | yes (4) | yes (2) | 984 | AGAP010075 | ||
| 12 | miR-989 | ASTE004431 | yes (4) | proteolysis | 1824 | AGAP007280 | ||
| 13 | miR-989 | ASTE003701* | yes (4) | 795 | AGAP010226 | |||
| 14 | miR-989 | ASTE011654 | yes (2) | yes (3) | yes (2) | 2304 | ||
| 15 | miR-989 | ASTE005749* | yes (4) | 2347 | AGAP001150 | |||
| 16 | miR-989 | ASTE003996 | yes (2) | yes (4) | integral component of membrane | 1681 | AGAP004486 | |
| 17 | miR-219 | ASTE006120 | yes (4) | 1650 | AGAP004892 | |||
| 18 | miR-219 | ASTE006196 | yes (4) | proteolysis | 4239 | AGAP006203 | ||
| 19 | miR-219 | ASTE011346 | yes (4) | 1764 | AGAP004892 | |||
| 20 | miR-219 | ASTE007196 | yes (4) | 1442 | AGAP001357 | |||
| 21 | miR-219 | ASTE002692 | yes (4) | GTP binding | 1763 | AGAP009441 | ||
| 22 | miR-219 | ASTE009304 | yes (4) | protein binding | 1268 | AGAP011439 | ||
| 23 | miR-285 | ASTE005579 | yes (2) | acid phosphatase activity | 337 | AGAP002387 | ||
| 24 | miR-7 | ASTE008161 | yes (4) | 428 | AGAP010789 | |||
| 25 | miR-7 | ASTE001260 | yes (4) | 677 | AGAP008905 | |||
| 26 | miR-7 | ASTE008160 | yes (4) | 383 | AGAP010789 | |||
| 27 | miR-7 | ASTE010939 | yes (4) | 2608 | AGAP012493 | |||
| 28 | miR-7 | ASTE002046 | yes (4) | ATP binding | 211 | AGAP002996 | ||
| 29 | miR-7 | ASTE003640 | yes (2) | metabolic process | 1342 | AGAP004721 |
*Targets predicted by RNAhybrid.
Out of 29 total targets cleaved by all six miRNAs, 19 were classified under category 4, one under category 3, seven under category 2 and two targets were classified under category 0. None of these targets were classified under category 1. T-plots of targets falling under category 2, 1 and 0 are shown in Figure 5. GO terms of these targets are provided in Table 5.
Figure 5.
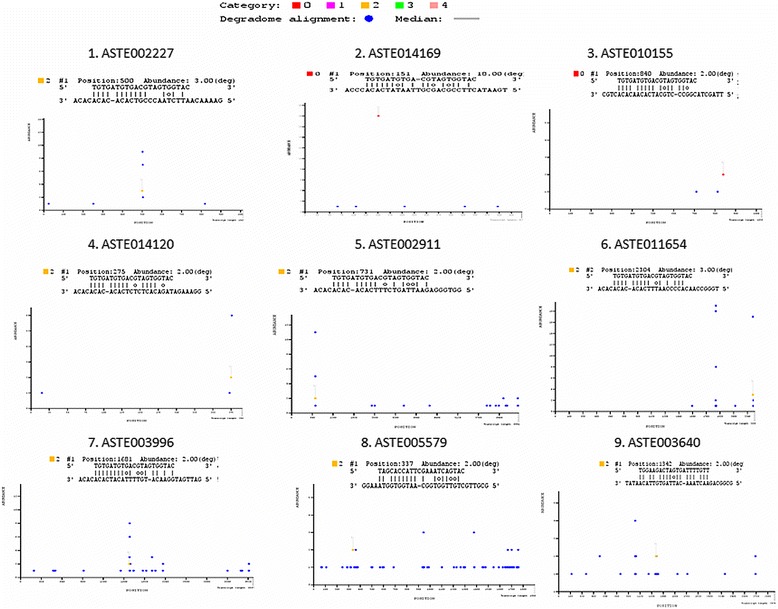
miRNA targets identification by Degradome sequencing. T-plots of miRNA targets classified under category 2, 1 and 0. X-axis represents transcript length whereas Y-axis represents number of degradome read that were mapped on to specific position on the target sequence. Blue dot depicts position on the transcript where degraded read was mapped. Coloured dots represent the position on the transcript where degraded fragment (s) and miRNA seed sequence mapped on the transcript.
Out of 29 targets identified, GO term name were identified for 14 mRNAs (Table 5). GO terms of these mRNAs were enriched in pathways related to protein binding and proteolysis. These targets might be important for insect reproduction as protein deposition in the ovary results in maturation of the developing egg. Another GO term significantly represented in this cluster of targets was related to nucleic acid binding. Nucleic acid binding might be responsible proper transcription and translation of genes required for maternal to zygotic transition in developing eggs.
Discussion
An.stephensi is an important vector of malaria parasite and is distributed throughout the Middle East and South Asia region. Various strategies were planned to control malaria spread, few of which were directed towards controlling the expansion of vector population. Such strategies require detailed understanding of mosquito biology pertaining to its developmental stages. Mosquito being a holometabolous insect goes through four stages of development. All four stages show characteristic morphological and physiological differences, resulting from differential gene expression at each stage of development. To understand molecular mechanisms mediating such differences, we studied the stage specific differences in miRNA expression which are known regulators of gene expression.
In this study, we employed deep sequencing technology to identify miRNAs expressed during mosquito development. Many of the 111 known miRNAs were same as those identified in our previous study [9]. Nevertheless, few more known miRNAs were added to the list that were not identified earlier in blood fed and parasitized mosquito [9]. Three microRNAs (miR-8, miR-bantam and miR-281) were abundantly expressed across all developmental stages of development indicating house-keeping function of these miRNAs. In Drosophila, miR-bantam functions in maintaining circadian rhythm and controls cell proliferation [26,27]. MicroRNA-8 regulates PI3K activity in larval fat boy and controls apoptosis/neurodegeneration in insects [28,29]. MicroRNA-281 was shown to be involved in regulating dengue virus replication in Aedes albopictus [30]. Efforts were also taken to identify novel mosquito-specific miRNAs in An. stephensi. Seven pairs of small RNA reads (−5p and -3p) fulfilled set criteria’s to be classified as mature miRNAs [31]. Expression values of each miRNA in all six libraries were normalised and TPM vales were compared to study miRNAs regulation across mosquito developmental stages. Understanding the differences between male and female mosquito is the most significant as these differences make female mosquitoes fit for reproduction and parasite transmission. Therefore, we identified miRNAs that were differentially regulated between genders during larval, pupal and adult stages of development. Few miRNAs were differentially expressed between genders during larval and pupal stages as these stages are early stages of development and differences between genders are not very apparent. On the other hand, six miRNAs were found differentially expressed between male and female adult mosquito. Out of these six miRNAs, miR-989 was of significant importance as it was reported to get abundantly expressed in ovary of female mosquito [16]. Its expression levels were up-regulated in female mosquito post blood feeding indicating towards its role in mosquito reproduction [9]. Two microRNAs, miR-989 and miR-7 also showed gender difference expression in Anopheles anthropophagus [14].
This is the first extensive study wherein efforts were taken to understand miRNA regulation during complete metamorphosis in male and female mosquito. MicroRNA differences were studied during larva to pupa transition and from pupa to adult metamorphosis in both male and female mosquito. Larva undergoes molting or ecdysis resulting in its growth and development from first to fourth instar larva. Histolysis and phagocytosis of larval tissues results in larval transition to pupa stage. Many miRNAs were found differentially expressed between these two stages. Few miRNAs were common in male and female metamorphosis from larva to pupa stage. While, few miRNAs regulated differently during male and female metamorphosis were identified. The role of few of these regulated miRNAs has been shown in Drosophila. Let-7 was required for neuromuscular remodeling and temporal organization during metamorphosis [32,33]. Another regulated miRNA, miR-7 was involved in notch signalling and photoreceptor differentiation in Drosophila eye [34,35]. Metamorphosis of pupa to adult mosquito involves tissue lysis and re-organization resulting in formation of adult organs. This process involves complex interplay between molecular factors as indicated by significant number of miRNAs found regulated during pupa to adult mosquito metamorphosis. MicroRNA-989 was identified up-regulated in female mosquito whereas it was down-regulated in male mosquito during the transition. It was found specifically up-regulated in female mosquito when compared to other stages of mosquito development. Such observation highlights its function in physiological activities related specifically with adult female mosquito. Regulated miRNAs such as miR-34 and miR-9a were involved in various processes involved in insect development [36-39].
Development of complex organisms requires controlled balance between mRNA degradation and translation. This fine balance is maintained by many factors including miRNAs. To understand the role of regulated miRNAs in mosquito development, it is necessary to identify their mRNA targets. As miRNAs binds to 3’UTR of targets sequence and regulate their expression, efforts were taken to identify miRNA seed binding sites on 3’UTR sequence of An. stephensi genes. Many targets of each miRNAs were identified using RNA hybrid. Targets of miRNA regulated at specific stage were analysed together and enriched annotation clusters playing significant role at specific stage of development were identified. Pathways involved in metabolic processes, phagocytosis and membrane organization were found enriched during larval to pupal transition. These pathways might be important for histolysis and phagocytosis of larval tissue which results in its transition to pupal stage. Metamorphosis from pupal to adult stage involves pathways such as vesicle mediated transport, phagocytosis, protein and polysaccharide biosynthetic processes. Genes functional in translation and enzymatic activity were also found enriched during mosquito metamorphosis. These pathways clearly are required for pupa tissue re-organization and re-formation of adult mosquito organs.
In our study, we identified miRNAs that were differentially expressed in male and female adult mosquitoes. Several studies have emphasised the role of some of these regulated miRNAs in female mosquitoes especially upon blood feeding [9,16]. In an attempt to identify targets of miRNAs regulated in our study that could have a possible role in female mosquitoes, ovary tissues were analysed upon blood feeding to identify targets, which could further be studied using miRNA specific antagomirs. This will be helpful to elucidate miRNA biological functions in an organism [12,25]. In this study, we inhibited miR-989 expression by injecting antagomir in mosquitoes. MicroRNA-989 up-regulates in female mosquito compared to male mosquito. Its expression further increases in ovary tissue of mosquito post blood feeding highlighting towards its significant role in mosquito reproduction [16]. Inhibition of miRNA expression via antagomir was combined with degradome sequencing to identify and validate miR-989 targets in ovary of blood fed female mosquito. Cleaved miRNA targets have been identified in plant kingdom using degradome sequencing [40,41]. This is the first report to have used degradome sequencing for miRNA targets identification in insects. Degradome sequencing has been employed mainly for identification of miRNA targets in plants as these miRNAs bind to their target sequence with perfect complementarity resulting in their cleavage [40,41]. Therefore, we identified very few cleaved targets in mosquitoes as miRNAs in animal kingdom generally regulate gene expression by mRNA translational repression. Targets were identified in ovary tissue of mosquito by degradome sequencing for miRNAs differentially expressed between both genders. Differentially expressed miRNAs resulted in regulation of twenty nine targets by mediating their cleavage in ovary of female mosquito. Many targets have GO term related to protein binding and proteolysis which might be important for egg development that requires massive protein deposition in developing embryo. Also targets with GO term related to metabolic processes, nucleic acid binding and signal transduction were identified. Functional characterizations of these targets are underway to understand their role in mosquito developmental processes. Deciphering function of many cleaved targets identified in ovary tissue would provide us with a better idea about their role in mosquito reproduction.
Conclusions
In conclusion, this study provides comprehensive account of miRNA regulated across different stages of mosquito development. Efforts were taken to understand targets of these miRNAs which can provide better understanding of their biological function in mosquito development. Identification of miRNA targets in ovary of blood fed female mosquito could provide insights in mosquito reproductive process and has implications for effective control of mosquito population required for reducing spread of mosquito-borne infectious diseases.
Acknowledgements
The study was funded through Science and Engineering Research Board (SERB) Young Scientist grant (SR/FT/LS-109/2011) awarded to SS and through ICGEB internal funds. SS is a recipient of Ramanujan Fellowship, Department of Science and Technology, Government of India and their support is duly acknowledged. SJ and VR are recipients of senior research fellowship from the Council of Scientific and Industrial Research, India. Small RNA Truseq sequencing was performed at NxGenBio Life Sciences, New Delhi. Degradome sequencing was performed at LC Sciences, Hangzhou, China. We acknowledge the assistance of Jatin Shrinet in bioinformatic data analysis. We are grateful to the technical assistance of all NIMR staff in mosquito rearing and collection. We acknowledge assistance of Dr. Anil Sharma in mosquito collection. Tara Ram’s assistance in maintenance of mosquitarium at ICGEB is duly acknowledged.
Abbreviations
- miRNA
microRNA
- Pre-miRNA
Precursor microRNA
- AF
Adult female
- AM
Adult male
- LM
Larva male
- LF
Larva female
- PM
Pupa male
- PF
Pupa female
- piRNA
Piwi-interacting RNA
- nt
Nucleotide
- UTR
Untranslated region
- NGS
Next generation sequencing
- TPM
Tags per million
- LNA
Locked nucleic acid
- bp
Base pair
- mRNA
Messenger RNA
- rRNA
Ribosomal RNA
- tRNA
Transfer RNA
- snoRNA
Small nucleolar RNA
- RT-PCR
Real time polymerase chain reaction
- EDC
l-ethyl-3-(3-dimethylaminopropyl) carbodiimide
- SSC
Saline sodium citrate
Additional files
Predicted stem-loop structures of Novel miRNAs. Precursor structures of mature miRNAs were folded using RNA fold. Sequences forming stem-loop structures with folding energy < −20 kcal/mol and with mature miRNAs sequences lying on the stem region were selected as Novel miRNAs.
miRNAs regulated between two genders across different stages of their development. (A) Heat map of miRNAs differentially expressed between larva male (LM) and larva female (LF), pupa male (PM) and pupa female (PF), adult male (AM) and adult female (AF) mosquito. Colour gradation from light green to dark red represents relative increase in miRNA expression. (B) Column graph showing fold change in miRNAs expression between larva male (LM) and larva female (LF) mosquito. (C) Column graph showing fold change in miRNAs expression between pupa male (PM) and pupa female (PF) mosquito. (D) Column graph showing fold change in miRNAs expression between adult male (AM) and adult female (AF) mosquito.
miRNAs regulated across different stages of male mosquito development. (A) Heat map of miRNAs differentially expressed between larva male (LM), pupa male (PM) and adult male (AM) mosquito. Colour gradation from light green to dark red represents relative increase in miRNA expression. (B) Column graph showing fold change in miRNAs expression between larva male (LM) and pupa male (PM) mosquito. (C) Column graph showing fold change in miRNAs expression between pupa male (PM) and adult male (AM) mosquito.
miRNAs regulated across different stages of female mosquito development. (A) Heat map of miRNAs differentially expressed between larva female (LF), pupa female (PF) and adult female (AF) mosquito. Colour gradation from light green to dark red represents relative increase in miRNA expression. (B) Column graph showing fold change in miRNAs expression between larva female (LF) and pupa female (PF) mosquito. (C) Column graph showing fold change in miRNAs expression between pupa female (PF) and adult female (AF) mosquito.
List of miRNA targets identified by RNAhybrid on 3’UTR of An.stephensi genes.
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
SJ and VR performed the experiments. SJ analysed the data. AT and SS were involved in mosquito maintenance and sample collection. SS and RKB conceptualised the study. SJ, SS and RKB drafted the manuscript. All authors read and approved the final version.
Contributor Information
Shanu Jain, Email: shanujain07@gmail.com.
Vandita Rana, Email: rana.vandita@gmail.com.
Adak Tridibes, Email: adak.mrc@gmail.com.
Sujatha Sunil, Email: sujatha@icgeb.res.in.
Raj K Bhatnagar, Email: raj@icgeb.res.in.
References
- 1.Harker BW, Behura SK, de Bruyn BS, Lovin DD, Mori A, Romero-Severson J, et al. Stage-specific transcription during development of Aedes aegypti. BMC Dev Biol. 2013;13:29. doi: 10.1186/1471-213X-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Hedo M, Goodman WG, Schafellner C, Martini A, Sehnal F, Eizaquirre M, et al. Control of larval-pupal-adult molt in the moth Sesamia nonagrioides by juvenile hormone and ecdysteroids. J Insect Physiol. 2011;57(5):602–7. doi: 10.1016/j.jinsphys.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–99. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;82(18):9164–70. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 6.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Shrinet J, Jain S, Jain J, Bhatnagar RK, Sunil S. Next generation sequencing reveals regulation of distinct Aedes microRNAs during chikungunya virus development. PLoS Negl Trop Dis. 2014;8(1):e2616. doi: 10.1371/journal.pntd.0002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain S, Rana V, Shrinet J, Sharma A, Tridibes A, Sunil S, et al. Blood feeding and Plasmodium infection alters the miRNome of Anopheles stephensi. PLoS One. 2014;9(5):e98402. doi: 10.1371/journal.pone.0098402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skalsky RL, Vanlandingham DL, Scholle F, Cullen BR HSa. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter F, Edaye S, Hüttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35(20):6953–62. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2010;107(52):22391–8. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puthiyakunnon S, Yao Y, Li Y, Gu J, Peng H, Chen X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasit Vectors. 2013;6(1):230. doi: 10.1186/1756-3305-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Huang H, Xing C, Li C, Tan F, Liang S. Identification and characterization of the expression profile of microRNAs in Anopheles anthropophagus. Parasit Vectors. 2014;7:159. doi: 10.1186/1756-3305-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu W, Criscione F, Liang S, Tu Z. MicroRNAs of two medically important mosquito species: Aedes aegypti and Anopheles stephensi. Insect Mol Biol, 2014 [DOI] [PMC free article] [PubMed]
- 16.Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, et al. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35(Database issue):D145–8. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, et al. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40(Database issue):D729–34. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms. Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Folkes L, Moxon S, Woolfenden HC, Stocks MB, Szittya G, Dalmay T, et al. PAREsnip: a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res. 2012;40(13):e103. doi: 10.1093/nar/gks277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci U S A. 2014;111(40):14460–5. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 27.Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23(18):2179–91. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139(6):1096–108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131(1):136–45. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Liu Y, Yan H, Li Y, Zhang H, Xu J, et al. miR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication. Parasit Vectors. 2014;7(1):488. doi: 10.1186/s13071-014-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18(13):943–50. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22(12):1591–6. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123(7):1267–77. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–82. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259(1):9–18. doi: 10.1016/S0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 37.Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–23. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol. 2010;338(1):63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 2009;327(2):487–96. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 40.Liu N, Tu L, Tang W, Gao W, Lindsey K, Zhang X. Small RNA and degradome profiling reveals a role for miRNAs and their targets in the developing fibers of Gossypium barbadense. Plant J. 2014;80(2):331–44. doi: 10.1111/tpj.12636. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Qin C, Chen Z, Zuo T, Yang X, Zhou H, et al. Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics. 2014;15:25. doi: 10.1186/1471-2164-15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]


