SUMMARY
Gene inactivation by transposon insertion or allelic exchange is a powerful approach to probe gene function. Unfortunately, many microbes, including Chlamydia, are not amenable to routine molecular genetic manipulations. Here we describe an arrayed library of chemically-induced mutants of the genetically-intransigent pathogen Chlamydia trachomatis, in which all mutations have been identified by whole genome sequencing, providing a platform for reverse genetic applications. An analysis of possible loss-of-function mutations in the collection uncovered plasticity in the central metabolic properties of this obligate intracellular pathogen. We also describe the use of the library in a forward genetic screen that identified InaC as a bacterial factor that binds host ARF and 14-3-3 proteins to modulate F-actin assembly and Golgi redistribution around the pathogenic vacuole. This work provides a robust platform for reverse and forward genetic approaches in Chlamydia and should serve as a valuable resource to the community.
INTRODUCTION
Advances in genome sequencing technologies enable the cost effective sequencing of microbial genomes (Markowitz et al., 2012), many of which are recalcitrant to genetic analysis. As a result, the function of individual genes in these microbes is often inferred based on their homology to genes in model organisms where molecular genetic approaches, such as insertional mutagenesis or gene replacement, are standard. However, the development of a robust system for DNA transformation and the molecular tools to perform targeted mutagenesis in many microbes can be a lengthy process or may be unattainable. For instance, Chlamydia trachomatis, an intracellular bacterial pathogen responsible for blinding trachoma and sexually transmitted infections (Haggerty et al., 2010), was only recently reported to be amenable to DNA transformation and limited molecular genetic manipulation (Heuer et al., 2007; Kari et al., 2011; Wang et al., 2011; Nguyen and Valdivia., 2012; Johnson and Fisher., 2013), even though the organism was first cultured more than half a century ago.
The Chlamydia life cycle (Figure S1) alternates between an infectious elementary body (EB), and an intracellular replicative reticulate body (RB). Upon internalization, the EB modifies its membrane bound vacuole to generate a compartment termed the inclusion (Hatch., 1999). Within the inclusion EBs differentiate into RBs, replicate, and eventually differentiate back into EBs, which are released to initiate new rounds of infections (Dautry-Varsat et al., 2005). From within the inclusion, Chlamydia manipulates host cellular pathways to ensure its proliferation and survival (Bastidas et al., 2013), including changes to the organization of the host cell’s internal architecture such as the redistribution of organelles and cytoskeletal elements around the inclusion (Kokes and Valdivia., 2012). Given the lack of robust tools for molecular genetic manipulation in C. trachomatis, the bacterial genes underlying these cellular changes, their contribution to Chlamydia pathogenesis, and the ensuing metabolic adaptations to the intracellular environment remain poorly understood.
In this work we generated and characterized a collection of chemically mutagenized C. trachomatis strains in which all induced gene variants were identified by whole genome sequencing. In addition to providing a robust framework for reverse genetic applications, an analysis of variant alleles led to insights into the metabolic requirements of Chlamydia during infection of mammalian cells. Finally, we implemented a microscopy-based forward genetic screen and identified a bacterial factor important for regulating cytoskeletal rearrangements at the periphery of the inclusion. We find that this ARF and 14-3-3-recruiting factor also mediates Golgi reorganization yet is dispensable for trafficking of Golgi derived sphingolipids to the inclusion. Overall, our work illustrates the value of combining standard chemical mutagenesis and whole genome sequencing as a platform for reverse and forward genetics applications.
RESULTS
A collection of chemically mutagenized and sequenced strains provides a broad array of mutant alleles in C. trachomatis
To facilitate a dissection of C. trachomatis traits important for infection and manipulation of host cellular targets, we subjected a rifampin-resistant (RifR) lymphogranuloma venereum (LGV) C. trachomatis L2 strain (L2/434/Bu) to ethyl methyl sulfonate (EMS) or N-ethyl-N-nitrosourea (ENU) mutagenesis. We isolated variants that exhibited a small plaque phenotype as these mutants are more likely to have been exposed to high mutagenic doses. From initial whole genome sequencing (WGS) of 43 mutant strains, we determined that the number of chemically induced genetic lesions per genome ranged from 7–25 and 6–22 transitions for EMS and ENU treated strains, respectively. We expanded each clonal isolate in Vero cells and arrayed them into a collection of 934 strains (Figure 1A). Because plaque isolation and clonal expansion requires a complete infectious cycle (Figure S1), these mutants are unlikely to be biased for defects in any one specific stage of infection.
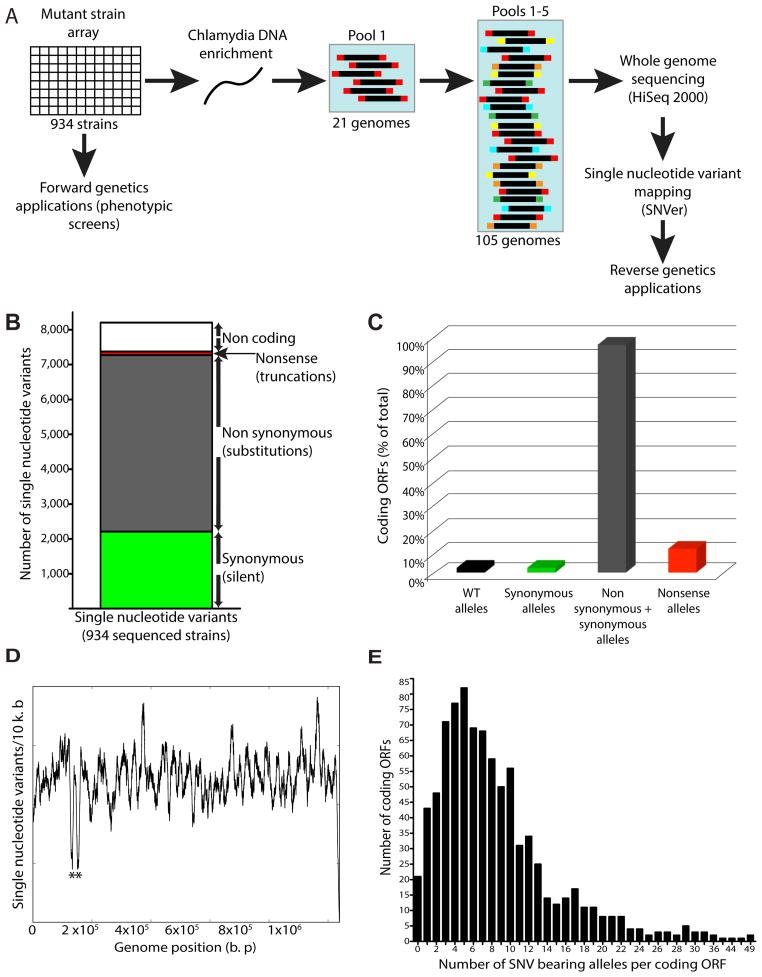
Figure 1. Generation of an ordered array of sequenced C. trachomatis mutants for use in genetic analyses.
(A) Chemically mutagenized C. trachomatis (C. t) LGV-L2 434/Bu strains were clonally isolated by plaque purification and arrayed in 96 well plates. Genomic sequencing libraries from a pool of 20 mutant strains and 1 reference strain were tagged with a unique barcode, combined with four other pools, and sequenced on an Illumina NGS platform. (B) Distribution of synonymous (green), non synonymous (gray), nonsense (red), and non coding (white) single nucleotide variants (SNVs) identified among mutagenized C. t strains. (C) Distribution of ORFs without variant alleles (2%, black), with alleles harboring only synonymous SNVs (2%, green), containing both synonymous and non synonymous SNVs (96%, gray), and with nonsense SNVs (10%, red). (D) Density of SNVs (number of SNVs/10 k. b of DNA sequence) across the C. t LGV-L2 genome. * denotes rRNA paralogues (CTL_r01_2 CTL_r02_2). Underrepresentation of SNVs is due to an inability to unambiguously map sequencing reads to these loci. (E) Distribution of the number of variant alleles identified per ORF.
This strain collection would constitute a useful platform for reverse genetics applications if all mutagen-induced single nucleotide variants (SNVs) could be identified and mapped. We enriched Chlamydia DNA from infected Vero cells, pooled DNA from 20 strains (Table S1), and sequenced five barcoded pools totaling 100 strains in an Illumina HiSeq 2000 Next Generation Sequencing (NGS) platform (Figure 1A) leading to an average of 14X-94X coverage per genome (data not shown). We used SNVer (Single Nucleotide Variant caller), a program developed to identify variants from pooled NGS data (Wei et al., 2011), and identified 8,205 SNVs (Table S2). Among these variants, 2,212 SNVs (27%) were not predicted to incur amino acid substitutions in the corresponding protein sequences (synonymous) (Figure 1B). Variants mapping to non-protein coding regions of the genome (intergenic regions, rRNA, tRNA loci) were underrepresented (10%) (Figure 1B), likely due to the high coding density of the C. trachomatis L2 genome (Thomson et al., 2008). In contrast, non-synonymous variants were predominant (5,061 SNVs, 62%) (Figure 1B) which can lead to production of “neutral” protein variants that are functionally indiscernible from their wild type counterparts or to “non-neutral” amino acid substitutions that can be deleterious for protein function, folding, or stability. We assessed the impact of these variants on gene product function by applying SNAP (Screening for Non-Acceptable Polymorphisms), a neural network based method that measures the likelihood that non-synonymous amino acid substitutions have a neutral or non-neutral impact on protein function (Bromberg and Rost., 2007). We identified 2,842 neutral (35%) and 2,219 non-neutral (27%) non-synonymous SNVs (Table S2). In addition, we identified 99 mutations (1%) (Figure 1B, Table S3) that lead to nonsense codons and presumably non-functional truncated proteins.
The C. trachomatis 434/Bu genome encodes 889 ORFs (protein coding genes), and 54 non-protein coding genes (tRNAs, rRNAs, and pseudogenes) (Thomson et al., 2008). We find that for 21 ORFs (21/889 (2%)) we did not identify alleles harboring SNVs and that for 18 ORFs (18/889 (2%)) we only identified alleles harboring synonymous SNVs (Figure 1C). Nonetheless, since we identified alleles with non-synonymous SNVs for 96% (853/889) of ORFs in the genome, including alleles with nonsense mutations for 84 protein coding genes (Figure 1C), our collection of mutants provides a broad representation of the possible mutational landscape of C. trachomatis. SNVs were widely distributed throughout the chromosome at a density of ~ 8 SNVs/kb with the exception of the orthologous rRNA operons (CTL_r01/CTL_r02), which were excluded from our SNV calling workflow due to the inability to unambiguously map sequence reads to this region (Figure 1D). We generated more than one distinct mutant allele for the majority of coding genes in the C. trachomatis chromosome with mutant allele frequencies ranging from 1 to 49 variant alleles/ORF (Figure 1E, Table S2). This collection of SNVs allows us to quickly identify strains bearing mutant alleles in any gene of interest. These strains can be then phenotyped to define the function of the mutated gene (“reverse genetics”) as we have reported for the effectors CPAF and TepP (Snavely and Kokes et al., 2014; Chen et al., 2014).
The C. trachomatis LGV L2 biovar tolerates perturbations in metabolic and DNA damage repair pathways
Based on 99 nonsense mutations in 84 ORFs, we can predict physiological properties inherent to C. trachomatis, such as the ability to tolerate alterations in central metabolism, amino acid metabolism and transport, membrane transport and stability, DNA processing, and transcription (Table S3). In addition, these nonsense alleles suggest that several Chlamydia proteins postulated to interact with host factors also appear to be dispensable for completion of its life cycle (Table S3). These include putative inclusion membrane proteins and effectors such as the mammalian histone methyltransferase NUE (Pennini et al., 2010), the early-translocated scaffolding phosphoprotein TepP (Chen et al., 2014), the lipid droplet associated protein Lda2 (Kumar et al., 2006), and the protease CPAF (Zhong., 2009; Zhong., 2011). The recovery of three CPAF null alleles in our collection is consistent with findings indicating that the function of this protease is more nuanced during infection than previously appreciated (Chen et al., 2012; Snavely and Kokes et al., 2014). While every nonsense allele we have characterized has led to a loss of gene product production (data not shown), we cannot exclude the possibility that some alleles can still code for partially functional proteins.
Based on these nonsense mutations we inferred which biological pathways are not necessary for Chlamydia to complete its infectious cycle in a cell culture model of infection. For instance, recovery of nonsense mutations in glgB and glgX provide genetic confirmation that Chlamydia does not require the synthesis and break down of branched glycogen in cell culture (Figure 2A) as previously suggested (Matsumoto et al., 1998; Nguyen and Valdivia., 2012). Similarly, the breakdown of glucose-6-phosphate into pyruvate by the Embden-Meyerhoff-Parnas (EMP or Glycolysis) pathway is expendable as indicated by a loss-of-function mutation in glucose-6 phosphate isomerase (pgi), an enzyme that channels glucose-6 phosphate into the EMP pathway (Figure 2B). Based on metabolic pathways inferred from the C. trachomatis L2 genome we hypothesize that the breakdown of glucose-6-phosphate in a pgi null strain can be restored by the non-oxidative branch of the Pentose Phosphate Pathway (Figure 2B).
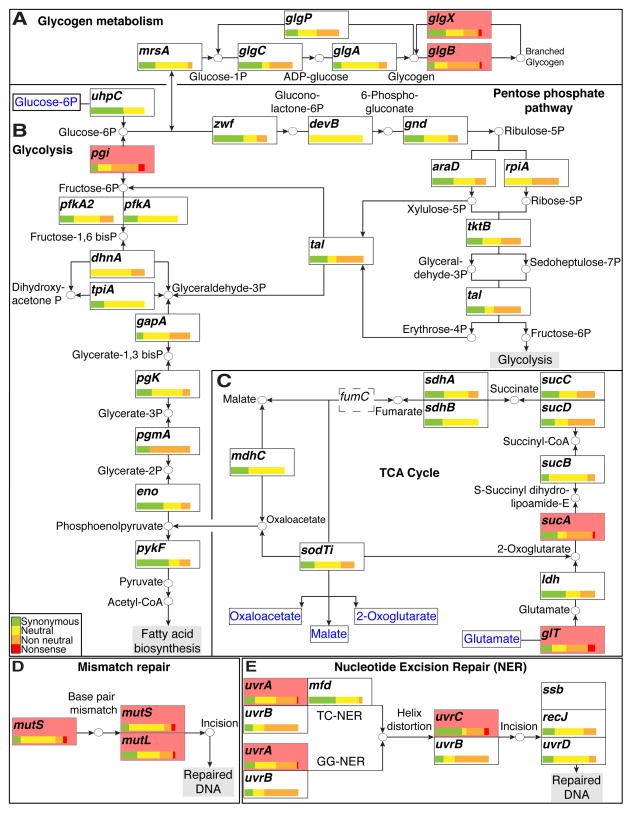
Figure 2. C. trachomatis tolerates perturbations in the tricarboxylic cycle and DNA repair networks.
(A–C) The LGV C. t central carbon metabolic network. The distribution of all synonymous (green), neutral (yellow), non-neutral (orange), and nonsense (red) bearing alleles is depicted for each ORF in the network. Distributions are normalized to total number of SNV bearing alleles identified per ORF. All ORFs with observed nonsense alleles are shaded in light red. (A) Production and break down of branched glycogen is not necessary as LGV C. t tolerates nonsense mutations in glgB and glgX. (B) The presence of a nonsense mutation in pgi suggests that the pentose phosphate pathway is sufficient to circumvent loss of fructose-6-phosphate production when the EMP pathway (Glycolysis) is blocked. (C) Nonsense mutations in glt and sucA suggest that LGV biovars can sustain metabolic activity with a severely curtailed TCA cycle. The TCA cycle is further truncated due the presence of a fumC (dashed box) pseudogene. All compounds imported from the host are shown in blue. (D) LGV C. t biovars can tolerate defects in their DNA mismatch repair machinery as suggested by the presence of nonsense bearing alleles in mutS and mutL. (E) Defects in the nucleotide excision repair pathway are also tolerated as evidenced by the presence of nonsense bearing alleles in uvrA and uvrC.
In most bacteria, the TCA cycle is important for the generation of reductive potential, energy generation, and the synthesis of metabolic precursors. The TCA cycle in C. trachomatis is incomplete given the lack of citrate synthase (gltA), aconitase (acn), and isocitrate dehydrogenase (icd) (McClarty., 1999). In the absence of a complete TCA cycle, production of oxaloacetate is likely fueled either by the import of 2-oxoglutarate from the host by the Chlamydia homolog of a dicarboxylate translocator (SodTi) (Weber et al., 1995), or by deamination of glutamate into 2-oxoglutarate (Weiss., 1967). Under cell culture conditions, glutamate does not appear to be necessary since we isolated a strain containing a nonsense mutation in the putative glutamate transporter-encoding gene (gltT) (Figure 2C). Interestingly, the TCA cycle is further constrained in C. trachomatis LGV biovars, since the fumarate hydratase enzyme that catalyzes the conversion of fumarate to malate is encoded by a pseudogene (Thomson et al., 2008), suggesting that a continuous cycle beginning with 2-oxoglutarate and ending with oxaloacetate is not required. Consistent with this, we identified a nonsense mutation in sucA (Figure 2C), encoding the E1 subunit of the 2-oxoglutarate-dehydrogenase complex that catalyzes the decarboxylation of 2-oxoglutarate to succinate. Therefore, in addition to being unable to synthesize malate, the Chlamydia LGV L2 biovar can also survive without succinate and fumarate (Figure 2C).
C. trachomatis can also propagate in cultured cells without a functional DNA mismatch repair system or with an impaired nucleotide excision DNA repair pathway (Figures 2 D and E). C. trachomatis is hypothesized to employ a methylation-independent mismatch repair pathway (reviewed in (Larrea et al., 2010) and (Pillon et al., 2010)) as they lack dam and mutH (Stephens et al., 1998; Carlson et al., 2005; Thomson et al., 2008). Instead mismatch repair is postulated to be mediated by the mismatch-sensing protein MutS and the repair protein MutL (reviewed in (Schofield and Hsieh., 2003) and (Kunkel and Erie., 2005)). Interestingly, we identified nonsense mutations in mutS and mutL indicating that under normal growth conditions in cultured cells there is not sufficient DNA damage such that the sensing and repair functions provided by MutS and MutL are required (Figure 2E).
Nucleotide excision repair (NER) allows for repair of drug and UV-induced lesions to DNA by the UvrABC excinuclease complex (reviewed in (Morita et al., 2010)). NER repairs damage in both transcribed and untranscribed DNA strands (Global genomic NER (GG-NER)), and repairs damaged DNA when RNA polymerase stalls at a lesion in DNA (Transcription coupled repair (TC-NER)) (Selby and Sancar., 1994; Svejstrup., 2002). As observed with mismatch repair, the identification of uvrA and uvrC nonsense mutations imply that both GG-NER and TC-NER are dispensable for C. trachomatis in epithelial cells (Figure 2D).
The inclusion membrane protein InaC is necessary for F-actin assembly at the inclusion periphery
An arrayed C. trachomatis mutant library is also a valuable resource for forward genetic approaches to identify mutants impaired for specific cellular traits. To illustrate this application, we implemented a microscopy-based screen to identify bacterial factors mediating rearrangement of filamentous actin (F-actin) at the periphery of the inclusion in infected cells (Kumar and Valdivia., 2008).
We screened for mutants that failed to promote F-actin assembly at the inclusion and identified two strains, CTL2M0407 (Figure 3A) and CTL2M0448 (data not shown, see below) that displayed a reproducible defect in this cytoskeletal reorganization. Since the genomes of these mutants were sequenced in pools and the exact number of SNVs in each strain was unknown, we re-sequenced the genomes of both strains and determined them to be identical (Table S4). Consequently we chose CTL2M0407 (herein referred to as M407) for further characterization. M407 harbors 12 non-synonymous variants as compared to the reference genome (Table S4). By performing genetic crosses between M407 (RifR) and a spectinomycin resistant (SpcR) wild type strain (Nguyen and Valdivia., 2012), we identified a nonsense mutation in CTL0184 (C307T/Q103*) as responsible for the failure to promote F-actin assembly at inclusions (Figure S2). Western blot analysis and immunofluorescence microscopy with antibodies raised against the inclusion membrane protein CT813 (homolog of CTL0184 in C. trachomatis Serotype D (Chen et al., 2006; Li et al., 2008)) confirmed the absence of CTL0184 in M407 (Figure 3B and C). Given the genetic link between the CTL0184C307T allele, the lack of detectable CTL0184 gene product, and the loss of F-actin assembly to the inclusion, we renamed CTL0184 as InaC (Inclusion membrane protein for actin assembly), to reflect its role in F-actin re-programming.
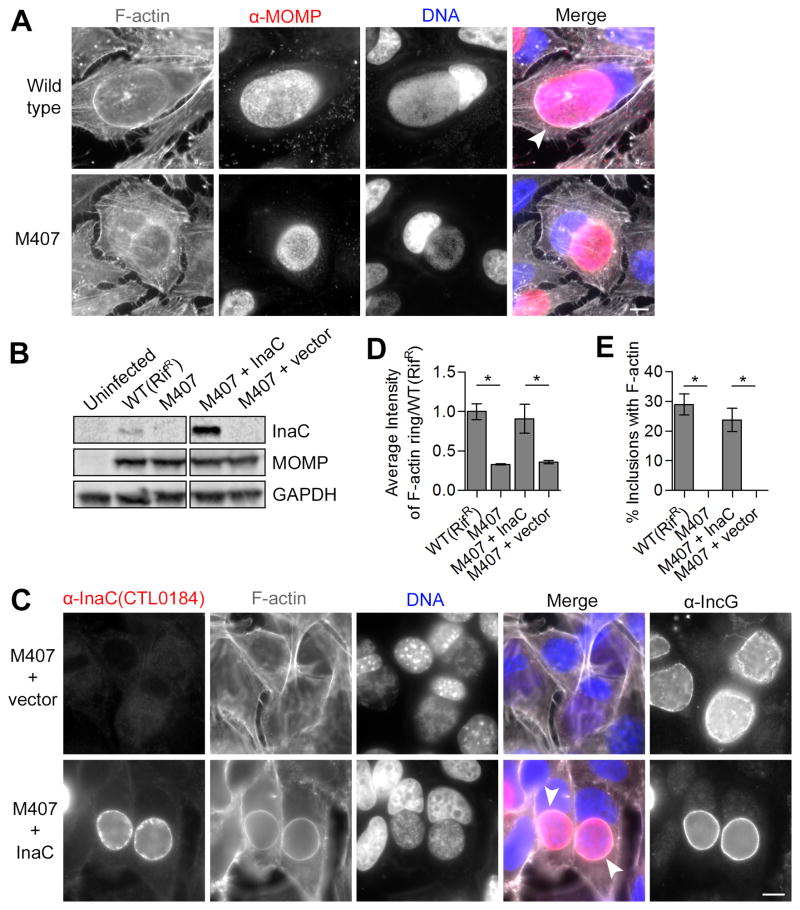
Figure 3. The inclusion membrane protein InaC is required for F-actin assembly at the inclusion.
(A) F-actin assembly at inclusions (arrowhead) is absent in HeLa cells infected with M407. F-actin (grayscale) and C. t (red) were detected by indirect immunofluorescence at 30 hpi using rhodamine-phalloidin and anti-MOMP antibodies respectively. DNA (blue) was stained with Hoechst. (B–E) F-actin assembly in M407 inclusions is rescued upon expression of wild type InaC. InaC expression is restored in M407 mutants transformed with a plasmid expressing wild type inaC from its own promoter as assessed by immunoblot (C) and immunofluorescence (red, D) analysis and without affecting the distribution of another inclusion membrane protein, IncG. F-actin assembly at the inclusion (white arrowheads, (C)) is restored in M407 transformed with a plasmid encoding inaC but not an empty vector. Note that the average intensity (D) and frequency (E) of F-actin around complemented inclusions is comparable to wild type. Mean ± SEM for three independent experiments is shown. At least 50 (F) or 300 (E) inclusions were enumerated in each triplicate for each experiment. * indicates P < 0.05 by one-way ANOVA and Newman-Keuls post hoc. Scale bars represent 10μm.
To confirm InaC’s role in F-actin rearrangements, we transformed M407 with a plasmid shuttle vector encoding wild type inaC under the control of its endogenous promoter. Transformation restored InaC production in M407 (Figure 3B and 3C) and F-actin assembly at the inclusion (Figure 3C). We found that both the amounts of F-actin (Figure 3D) and the frequency of F-actin structures at inclusions in M407 (Figure 3E) were restored to wild type levels after transformation with an InaC expressing plasmid but not an empty vector control. We conclude that InaC is required for F-actin assembly around the C. trachomatis inclusion.
InaC recruits ARF and 14-3-3 proteins to the inclusion
To define the function of InaC we identified interacting host proteins by immunoprecipitating (IP) a GFP-tagged full-length InaC transiently expressed in HEK 293T cells. Ten of the 19 interacting proteins identified by mass spectrometry (Table S5) belong to two protein families – human ADP-ribosylation factors (ARF1, 4 and 5), and 14-3-3 proteins (Figure 4A). We confirmed the co-precipitation of GFP-InaC with endogenous 14-3-3β, 14-3-3ε, and ARF1 by immunoblot analysis (Figure 4B). ARFs are small GTPases that function in vesicular traffic and secretory organelle function (Donaldson and Jackson., 2011) and 14-3-3s are a family of conserved regulatory proteins that bind and regulate signaling proteins in numerous cellular pathways (Morrison., 2009). Given that GFP-ARF1 and 14-3-3β localize to the inclusion periphery (Moorhead et al., 2010; Scidmore and Hackstadt., 2001), we tested if InaC mediates recruitment of ARFs and 14-3-3s by assessing the localization of GFP-ARFs (ARF1, 4, 5, and 6) and endogenous 14-3-3β and 14-3-3ε in cells infected with wild type C. trachomatis or InaC mutants (Figure 4C). 14-3-3β and 14-3-3ε are enriched around wild type inclusions yet are barely detectable around inclusions lacking InaC. Similarly, GFP-ARF1, -ARF4, and -ARF5 (primarily Golgi-localized ARFs) are recruited to inclusions in an InaC-dependent manner, while ARF6 (plasma membrane and endosome localized ARF) is not (D’Souza-Schorey and Chavrier., 2006; Donaldson and Jackson., 2011) (Figure 4C). Strikingly, the localization of ARF1, 4, 5 and 14-3-3 to inclusions is markedly increased in strains over-expressing InaC (Figure 4C). These data suggest that InaC is necessary for and mediates the recruitment of 14-3-3s and Golgi-specific ARFs to the inclusion.
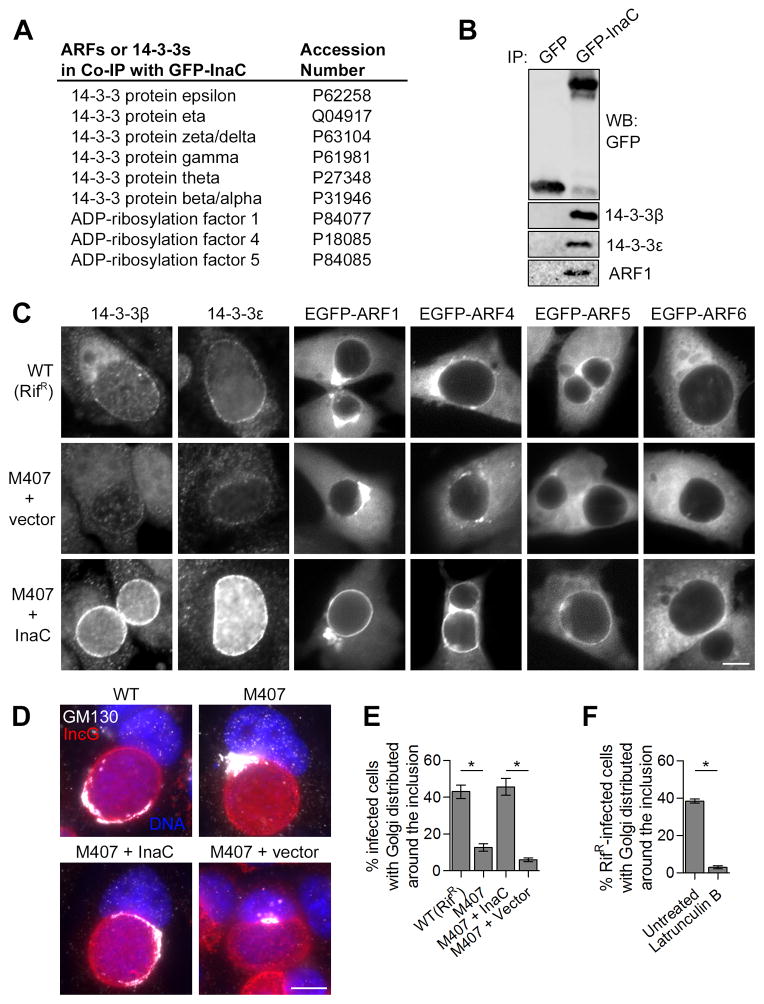
Figure 4. The recruitment of ARF and 14-3-3 proteins and Golgi redistribution to the inclusion requires InaC.
(A and B) ARF and 14-3-3 proteins interact with InaC. Lysates from HEK 293T cells expressing either GFP or a GFP-InaC fusion protein were incubated with GFP-Trap resin. Bound proteins were identified by LC-MS/MS (Table S5). Proteins of ARF and 14-3-3 families are shown with corresponding accession numbers (A). The specificity of these interactions was confirmed by SDS-PAGE followed by western blot with specific antibodies (B). (C) InaC mediates the recruitment of 14-3-3β, -ε, and Golgi-specific ARF GTPases to the inclusion. HeLa cells were infected with the indicated C. t strains and either processed at 30 hpi for immunofluorescence using anti-14-3-3 antibodies (left) or transfected with EGFP-ARF isoforms for 24 hours before processing at 30 hpi for microscopy (right). Note that the intensity of 14-3-3 and ARF1, 4 and 5 recruitment to inclusions correlates with InaC expression. The divergent ARF6 was not recruited to inclusions or bound to GFP-InaC. (D and E) Golgi redistribution around the inclusion requires InaC. HeLa cells were infected as in (C) and processed at 30 hpi for immunofluorescence using anti-GM130 (grayscale) anti-IncG (red) antibodies and Hoechst (blue), and the frequency of Golgi distribution was quantified. Golgi distribution at least halfway around inclusions was scored as a positive (E), as detailed in Supplemental Experimental Procedures and Figure S3A. (F) F-actin assembly is required for Golgi redistribution at inclusions. HeLa cells were infected with a wild type LGV-L2 strain and treated with 500nM Latrunculin B for 30 minutes before processing as in (E) at 30 hpi. Mean ± SEM for three independent experiments is shown. At least 40 (E) or 90 (F) inclusions were enumerated in each triplicate for each experiment. * indicates P < 0.05 by one-way ANOVA and Newman-Keuls post hoc (E) or Student’s t test (F). Scale bars represent 10μm.
Golgi redistribution around the inclusion requires InaC and F-actin
The Golgi apparatus is a dynamic membrane system in which organelle architecture and membrane traffic are fundamentally linked (Lippincott-Schwartz et al., 1998). ARFs are mediators of vesicular traffic within this system and critical for maintaining Golgi structure and identity (Donaldson and Jackson., 2011; Lippincott-Schwartz et al., 1998). The actin cytoskeleton supports membrane traffic within the secretory system and is directly involved in the maintenance of Golgi architecture (Egea et al., 2006). In C. trachomatis infected cells, the Golgi fragments into ministacks that envelop the inclusion (Heuer et al., 2009). Since InaC recruits ARFs and is required for F-actin assembly at the inclusion, we tested if InaC plays a role in modulating Golgi architecture during C. trachomatis infection. The Golgi appeared distributed at least halfway around 40% of wild type inclusions (Figure 4D and 4E, Figure S3A). In contrast, the Golgi retained a compact morphology at a perinuclear region adjacent to inclusions lacking InaC. Given the importance of F-actin in maintaining the shape of the Golgi in mammalian cells (Egea et al., 2006), we tested whether Golgi redistribution around the inclusion is dependent on F-actin. The Golgi was no longer distributed around wild type inclusions treated with the F-actin polymerization inhibitor Latrunculin B (Figure 4F). These results indicate that Golgi redistribution around C. trachomatis inclusions requires InaC and intact actin filaments.
InaC-mediated Golgi redistribution is dispensable for Chlamydia sphingolipid acquisition and growth
Since vesicular traffic through the Golgi and via ARF regulated pathways deliver sphingolipids to Chlamydia (Elwell et al., 2011; Hackstadt et al., 1996) and Golgi redistribution around inclusions has been proposed to facilitate sphingolipid acquisition by Chlamydia (Heuer et al., 2009; Rejaman Lipinski et al., 2009), we assessed whether InaC contributes to Chlamydia growth and its ability to acquire sphingolipids (Figure S3). While we found a 3-fold decrease in the recovery of infectious progeny due to background mutations in M407 compared to its wild type RifR parent (data not shown), we detected no significant difference in either the recovery of infectious progeny from infected cells (Figure S3B), or the average area of inclusions (Figure S3C) upon restoring InaC expression in M407. When we incubated wild type, InaC-deficient, and InaC-restored infected cells with a fluorescent ceramide analog, and assessed the fluorescence intensity of inclusions in live cells, we observed no significant differences. As a control, we treated infected cells with Brefeldin A, which inhibits sphingolipid acquisition by Chlamydia (Elwell et al., 2011; Hackstadt et al., 1996) and observed a significant decrease in the fluorescent ceramide acquisition (60%) in all strains tested (Figure S3D and S3E). Collectively, these data demonstrate that InaC itself, including InaC-mediated ARF recruitment and Golgi redistribution around the inclusion is not required for C. trachomatis replication, inclusion expansion, and efficient sphingolipid trafficking to the inclusion in cultured epithelial cells.
DISCUSSION
The first report of stable transformation of plasmid DNA into Chlamydia (Wang et al., 2011) was a landmark event for Chlamydia genetics. Recently, a report describing targeted insertional mutagenesis with Type II Introns (Johnson and Fisher., 2013) further suggests that modern molecular genetic approaches to disrupt genes can be implemented in C. trachomatis. However, these tools are not yet suitable to generate large collections of mutant strains. With these limitations in mind, we developed a platform for forward and reverse genetic analysis in C. trachomatis by generating and sequencing a collection of 934 chemically mutagenized strains. As the costs of genome sequencing continue to plummet there is a renewed interest in the application of chemical and radiation-induced mutagenesis as tools in microbial genetics. From the identification of suppressors, adaptive mutations, and conditional alleles, point mutations provide a richer source of information for genetic analysis than gene disruption strategies. It is in this context that the resource we generated offers unique advantages such as strains with hypo and hypermorphic mutations, including mutations in genes that are likely essential for infection. Furthermore, point mutations are unlikely to exhibit polar effects on the expression of other genes within an operon as is often observed during insertional mutagenesis.
Mapping of all SNVs in the collection facilitated the identification of strains bearing mutant alleles of interest. Indeed, mutants from this collection have been recently characterized (Chen et al., 2014; Snavely and Kokes et al., 2014; Brown et al., 2014), underscoring its utility in reverse genetic applications. Mutant strains were sequenced in pools of 20 to lower the overall DNA sequencing costs (~$15/genome). To identify which strain within a pool harbors a mutant allele of interest, each strain (which are individually arrayed) can be readily genotyped in a cost-effective manner by standard PCR based techniques such as TILLING (Colbert et al., 2001; Kari et al., 2011; Supplemental Experimental Procedures), or direct sequencing of amplicons spanning the mutated region. Although the genome of the strain of interest should be re-sequenced to identify accompanying mutations, there is no need to sequence the genomes of additional members of the original pool. Genetic linkage between a mutant allele and phenotype of interest can be established either by linkage analysis (Nguyen and Valdivia., 2012) or by trans-complementation through plasmid transformation (Wang et al., 2011).
The mutant collection is well suited for forward genetic screens. We described a screen for mutants impaired in F-actin assembly at the inclusion and identified InaC, an inclusion membrane protein that mediates F-actin reorganization, ARF and 14-3-3 recruitment, and Golgi redistribution. InaC is highly conserved in all serovars of C. trachomatis, including ocular, genital and LGV strains and orthologs can be found in other Chlamydiales. We determined that InaC recruits Golgi-specific ARF GTPases and 14-3-3 proteins to the inclusion, both of which have been found to influence F-actin processes. For instance, ARF1 at the Golgi stimulates actin assembly via recruitment and regulation of Cdc42 (reviewed in (Myers and Casanova., 2008)) and 14-3-3 proteins can modulate T-cell adhesion by binding to an intracellular region of integrins which are membrane receptors that bridge the intracellular F-actin network and the extracellular matrix (Takala et al., 2008). The precise mechanism by which InaC, presumably through ARF and/or 14-3-3s interactions, regulates F-actin assembly and Golgi redistribution at the inclusion remains to be determined.
Cellular F-actin contributes to the stability of the inclusion (Kumar and Valdivia., 2008) and to the extrusion of inclusions at late stages of infection upon bacterial cell exit (Chin et al., 2012; Hybiske and Stephens., 2007). Our findings indicate that cellular F-actin and InaC-mediated F-actin structures at the inclusion are also required for Golgi redistribution to surround the inclusion. The Golgi plays a central role in the secretory pathway, which Chlamydia manipulates to intercept ER-synthesized sphingolipids destined for the plasma membrane both by engaging the ceramide transporter CERT at ER-inclusion membrane contact sites (Derré et al., 2011; Elwell et al., 2011) and by hijacking Golgi vesicular traffic (Elwell et al., 2011; Hackstadt et al., 1995; Hackstadt et al., 1996). Disruption of Golgi vesicular traffic and interference with ARF regulated pathways using Brefeldin A or by depletion of the ARF nucleotide exchange factor GBF1 disrupts sphingolipid acquisition by bacteria (Elwell et al., 2011; Hackstadt et al., 1996). In addition to vesicular traffic, Golgi morphology and localization may affect sphingolipid trafficking to the inclusion since inhibition of Golgi dispersal during infection using a caspase inhibitor or by depletion of Rab6 or Rab11 impairs sphingolipid acquisition by Chlamydia (Heuer et al., 2009; Rejman Lipinski et al., 2009). Surprisingly, we find that InaC-mediated Golgi redistribution or ARF recruitment does not appear to have any discernable effect on the delivery of sphingolipids to the inclusion.
Based on the analysis of nonsense mutations tolerated in the mutagenized Chlamydia strains, we gained insights into this pathogen’s metabolic requirements for completion of its infectious cycle, including a limited role for a functional TCA cycle in the LGV L2 biovar as previously suggested (Thomson et al., 2008). In addition to the loss of fumC (Fumerase) (Thomson et al., 2008), the C. trachomatis lymphogranuloma biovars can tolerate further degradation of the TCA cycle through loss of SucA. These observations suggests that oxidative phosphorylation (respiration) in these biovars is not required for ATP production. Instead, C. trachomatis L2 likely generates most of its ATP by substrate level phosphorylation (Glycolysis) by taking advantage of its efficient glycolytic (EMP) pathway that avoids expenditure of two ATP molecules by importing glucose-6-phosphate from the host through the UhpC transporter and by utilizing inorganic pyrophosphate (PPi) as a phosphate donor by the Chlamydia atypical phosphofructose kinase (PfkAB) (Island et al., 1992; McClarty., 1999). Further evidence for a bias against employing oxidative phosphorylation by LGV C. trachomatis biovars, comes from observations that the succinate dehydrogenase cytochrome B subunit (SdhC), a central component of the electron transport chain, is a pseudogene (Thomson et al., 2008).
Loss of oxidative phosphorylation would result in lower levels of reactive oxygen species (ROS), a major byproduct of respiration. This might be a desirable trait since C. trachomatis lacks genes encoding the oxidative damage repair proteins MutT and MutM (Stephens et al., 1998; Thomson et al., 2008). Low levels of endogenous ROS might also increase tolerance to oxidative bursts generated by immune cells allowing for LGV biovar strains to establish infections in human mononuclear phagocytes (MNPs) (Yong et al., 1987; Beagley et al., 2009).
In summary, we described a robust system for generating and mapping mutations in the absence of classic molecular genetic tools which we have applied for genetic dissection of a complex trait in Chlamydia. The methodology can be readily applied to any microbe even those that lack systems for genetic exchange or DNA transformation, including emerging pathogens, outbreak strains, and other experimentally intractable microbes.
EXPERIMENTAL PROCEDURES
Full description of Experimental Procedures are available in Supplemental Information.
Growth condition, cell lines, and Chlamydia strains
Vero cells (ATCC CCL-81), HeLa cells, and HEK293T cells were grown in high glucose DMEM supplemented with L-glutamine, sodium pyruvate (Gibco, Life Technologies) and 10% FBS (Mediatech, CellGro), at 37 °C in a 5% CO2 humidified incubator. C. trachomatis LGV biovar L2 434/Bu infections were performed with EBs and synchronized by centrifugation (2,500 × g for 30 min at 10 °C) onto Vero or HeLa cells, and incubated for indicated times. EB titers were determined by assessing the number of inclusion forming units (IFUs) with a Cellomics ArrayScan automated fluorescence imaging system (Thermo Scientific).
Generation of a C. trachomatis LGV-L2 mutant library
Vero cells infected with RifR LGV-L2 (Nguyen and Valdivia., 2012) were mutagenized with ethyl methyl sulfonate (EMS) and N-ethyl-N-nitrosourea (ENU) (described in (Nguyen and Valdivia., 2012)). Following mutagenesis, Vero cells seeded in 6-well plates were infected with 100 IFUs/well from independent pools of mutagenized EBs and at 2 hpi, overlaid with DMEM/agarose (0.5 % SeaKem LE agarose (Lonza), 1X DMEM, 10% FBS, 25 μg/mL gentamycin (Gibco), 1X nonessential amino acids (Gibco), 200 ng/mL cycloheximide (Sigma-Aldrich)). At 7 to 10 dpi bacteria from small plaques were harvested and expanded in Vero cells. Cell lysates containing EBs were arrayed into 96 well plates to establish an ordered collection of mutant strains and stored at −80°C.
Genome sequencing of C. trachomatis LGV-L2 mutants
Vero cells in T25 flasks were infected with each mutant strain. At time of harvest, EBs were concentrated from cell lysates by centrifugation (21,000 × g, 15 minutes, 4 °C), and resuspended in DNAse I reaction buffer (NEB Labs). Residual host DNA was digested with 8 Units of DNAse I (NEB) for 1 hour at 37 °C and EBs washed with PBS, pelleted, and resuspended in 180 μl of ATL buffer (Qiagen). Total DNA was isolated with a Qiagen DNeasy Blood and Tissue Kit (Qiagen). 150 ng of total DNA isolated from each of 20 individual strains were pooled with 150 ng of control DNA. One μg of pooled DNA was sheared with an Adaptive Focused Acoustics S220 instrument (Covaris). Sequencing libraries were prepared and barcoded with a TrueSeq DNA sample preparation kit (Illumina Inc.) and sequenced on a HiSeq2000 sequencing platform (Illumina Inc.).
Screen for mutants defective for F-actin assembly and identification of InaC
HeLa cells were grown to 50% confluence in 96-well plates and infected with 10 μl of crude cell lysates derived from cells infected with each mutant in the library or controlled amounts of wild type C. trachomatis. At 30 hpi, cells were fixed and imaged as described below to assess F-actin at the periphery of inclusions. M407 was amplified from the library, subjected to whole genome sequencing, and crossed with a wild type SpcR strain to generate recombinants as previously described (Nguyen and Valdivia., 2012). Recombinants were assessed for F-actin assembly at inclusions and genotyped for all M407 encoded SNVs (Colbert et al., 2001; Kari et al., 2011) to map the causal mutation to CTL0184. M407 was transformed (Wang et al., 2011) with either an empty shuttle vector p2TK2-SW2 (Agaisse and Derré., 2013) or a vector containing wild type CTL0184 and 250 bp of upstream sequence.
Identification of host proteins that interact with InaC
HEK 293T cells were grown to 50% confluence in 10 cm dishes and transfected with either a GFP-only or a plasmid encoding an N-terminally GFP-tagged full-length InaC. At 24 hr post transfection, cells were lysed in 25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40 supplemented with a protease inhibitor cocktail (Roche Diagnostics) and a phosphatase inhibitor cocktail (Pierce), combined with GFP-Trap (ChromoTek) resin and incubated overnight at 4°C. Bound proteins were analyzed by LC-MS/MS at the Duke Proteomics Core facility or eluted in 0.2 M glycine [pH 2.5], resolved by SDS PAGE, and subjected to immunoblot analysis.
Immunofluorescence and quantitative analysis
For high-magnification imaging, HeLa cells grown to 50% confluence on glass coverslips were infected with the indicated C. trachomatis strains at an MOI of 3. For ARF localization, cells were transfected with various EGFP-ARF plasmids at the time of infection. At 30 hpi, cells were fixed with 4% paraformaldehyde and permeabilized with cold 0.2% Triton-X 100. Samples were stained as indicated and images were captured on an Axio Observer Z1 fluorescence microscope with a 63X objective (Zeiss). For quantification, HeLa cells grown to 50% confluence in 96-well plates were treated as above. As indicated, F-actin was depolymerized with a 30 min treatment of 500 nM Latrunculin B (Life Technologies) prior to fixation. Cells were imaged with a Cellomics HTC Arrayscan (Thermo Scientific) using a 20x objective and images were viewed with Cellomics vHCS: View Software v1.6 (Thermo Scientific) to quantify the number of inclusions surrounded by F-actin, or the number of cells with Golgi elements enveloping inclusions. Alternatively, images were analyzed with a custom-designed algorithm in Cellomics Scan Software v5.6 (Thermo Scientific) to quantify the average intensity of F-actin at inclusions. Values from three technical triplicate wells were combined in each of three independent experiments. Statistically significant differences were assessed by Student’s t test or a one-way ANOVA followed by Newman-Keuls Multiple Comparison post hoc analysis with a p-value < 0.05 considered significant.
Supplementary Material
Acknowledgments
We thank the Duke Center for Genomic and Computational Biology (DCGCB) Genome Sequencing and Analysis Core facility, the Experimental Genomics Shared Resource center which provided access and use of the Cellomics HTC Arrayscan, and the Duke Proteomics and Metabolomics Shared Resource Core. We thank Dora Posfai for constructing the GFP-InaC vectors and members of the Valdivia lab for valuable discussions. This work was supported by National Institute of Health awards AI085238, AI100759, AI081694, an NRSA fellowship award AI102500-01 (to R.J.B.) and by the American Heart Association Mid-Atlantic Affiliate Predoctoral Fellowship #11PRE7360030 (to M.K.).
Footnotes
AUTHOR CONTRIBUTIONS
J.D.D, B.D.N, and J.R.B generated reagents. J.A.G analyzed data and contributed to manuscript preparation. R.H.V contributed to data analysis and manuscript preparation. M.K and R.J.B generated data, analyzed data, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agaisse H, Derré I. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PloS One. 2013;8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. Chlamydial intracellular survival strategies. Cold Spring Harb Perspec Med. 2013;3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagley KW, Huston WM, Hansbro PM, Timms P. Chlamydial infection of immune cells: altered function and implications for disease. Crit Rev Immunol. 2009;29:275–305. doi: 10.1615/critrevimmunol.v29.i4.10. [DOI] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HM, Knowlton AE, Snavely E, Nguyen BD, Richards TS, Grieshaber SS. Multinucleation during C. trachomatis infections is caused by the contribution of two effector pathways. PloS One. 2014;9:e100763. doi: 10.1371/journal.pone.0100763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen D, Sharma J, Cheng W, Zhong Y, Liu K, Jensen J, Shain R, Arulanandam B, Zhong G. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect Immun. 2006;74:4826–4840. doi: 10.1128/IAI.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M. CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Path. 2012;8:e1002842. doi: 10.1371/journal.ppat.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Path. 2014;10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin E, Kirker K, Zuck M, James G, Hybiske K. Actin recruitment to the Chlamydia inclusion is spatiotemporally regulated by a mechanism that requires host and bacterial factors. PloS One. 2012;7:e46949. doi: 10.1371/journal.pone.0046949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Bio. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A, Subtil A, Hackstadt T. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 2005;7:1714–1722. doi: 10.1111/j.1462-5822.2005.00627.x. [DOI] [PubMed] [Google Scholar]
- Derré I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Path. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Bio. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G, Lazaro-Dieguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U S A. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 2010;2:134–155. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- Hatch TP. In: Developmental biologyChlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens RS, editor. Washington, DC: American Society for Microbiology Press; 1999. pp. 69–100. [Google Scholar]
- Heuer D, Kneip C, Maurer AP, Meyer TF. Tackling the intractable - approaching the genetics of Chlamydiales. Int J Med Microbiol. 2007;297:569–576. doi: 10.1016/j.ijmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, Brinkmann V, Meyer TF. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U S A. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Island MD, Wei BY, Kadner RJ. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella Typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PloS One. 2013;8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, et al. Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. U S A. 2011;108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokes M, Valdivia RH. Cell Biology of the Chlamydial Inclusion. In: Tan M, editor. Intracellular Pathogens I: Chlamydiales. Washington, DC: ASM Press; 2012. pp. 170–191. [Google Scholar]
- Kumar Y, Cocchiaro J, Valdivia RH. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol. 2006;16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Valdivia RH. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe. 2008;4:159–169. doi: 10.1016/j.chom.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Larrea AA, Lujan SA, Kunkel TA. SnapShot: DNA mismatch repair. Cell. 2010;141:730. doi: 10.1016/j.cell.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Donaldson JG. Building a secretory apparatus: role of ARF1/COPI in Golgi biogenesis and maintenance. Histochem Cell Biol. 1998;109:449–462. doi: 10.1007/s004180050247. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:115–122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Izutsu H, Miyashita N, Ohuchi M. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol. 1998;36:3013–3019. doi: 10.1128/jcm.36.10.3013-3019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarty G. Chlamydia metabolism as inferred from the complete genome sequence. In: Stephens RS, editor. Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Washington, DC: American Society for Microbiology Press; 1999. pp. 69–100. [Google Scholar]
- Moorhead AM, Jung JY, Smirnov A, Kaufer S, Scidmore MA. Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect Immun. 2010;78:1990–2007. doi: 10.1128/IAI.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Nakane S, Shimada A, Inoue M, Iino H, Wakamatsu T, Fukui K, Nakagawa N, Masui R, Kuramitsu S. Molecular mechanisms of the whole DNA repair system: a comparison of bacterial and eukaryotic systems. J Nucleic Acids. 2010;2010:179594. doi: 10.4061/2010/179594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–192. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc. Natl. Acad. Sci. U S A. 2012;109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Path. 2010;6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, et al. Structure of the endonuclease domain of MutL: unlicensed to cut. Mol Cell. 2010;39:145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman Lipinski A, Heymann J, Meissner C, Karlas A, Brinkmann V, Meyer TF, Heuer D. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Path. 2009;5:e1000615. doi: 10.1371/journal.ppat.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Scidmore MA, Hackstadt T. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol Microbiol. 2001;39:1638–1650. doi: 10.1046/j.1365-2958.2001.02355.x. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis. 2014;71:336–351. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nature Rev Mol Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- Takala H, Nurminen E, Nurmi SM, Aatonen M, Strandin T, Takatalo M, Kiema T, Gahmberg CG, Ylänne J, Fagerholm SC. β integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood. 2008;112:1853–1862. doi: 10.1182/blood-2007-12-127795. [DOI] [PubMed] [Google Scholar]
- Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O’Connor CD, Goodhead I, Norbertzcak H, et al. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 2008;18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Menzlaff E, Arbinger B, Gutensohn M, Eckerskorn C, Flugge UI. The 2-oxoglutarate/malate translocator of chloroplast envelope membranes: molecular cloning of a transporter containing a 12-helix motif and expression of the functional protein in yeast cells. Biochemistry. 1995;34:2621–2627. doi: 10.1021/bi00008a028. [DOI] [PubMed] [Google Scholar]
- Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39:1–13. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. Transaminase activity and other enzymatic reactions involving pyruvate and glutamate in Chlamydia (psittacosis-trachoma group) J Bacteriol. 1967;93:177–184. doi: 10.1128/jb.93.1.177-184.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong EC, Chi EY, Kuo CC. Differential antimicrobial activity of human mononuclear phagocytes against the human biovars of Chlamydia trachomatis. J Immunol. 1987;139:1297–1302. [PubMed] [Google Scholar]
- Zhong G. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Micrbiol. 2009;17:467–474. doi: 10.1016/j.tim.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G. Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Front Microbiol. 2011;2:14. doi: 10.3389/fmicb.2011.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.