Abstract
A series of indole analogs that are synthesized using the scaffold of a potent radiosensitizer, YTR107, were tested for their ability to alter the solubility of phosphorylated nucleophosmin 1 (pNPM1). NPM1 is critical for DNA double strand break (DSB) repair. In response to formation of DNA DSBs, phosphorylated T199 NPM1 binds to ubiquitinated chromatin, in a RNF8/RNF168-dependent manner, forming irradiation-induced foci (IRIF) that promote repair of DNA DSBs. A Western blot assay was developed using lead molecule, YTR107, for the purpose of screening newly synthesized molecules that target pNPM1 in irradiated cells. A colony formation assay was used to demonstrate the radiosensitization properties of the compounds. Compounds that enhanced the extractability of pNPM1 upon radiation treatment possessed radiosensitization properties.
Graphical Abstract
Novel Western blot assay was developed and tested for its applicability in identifying radiosensitizers that target nucleophosmin 1 (NPM1). Extractability of pNPM1 was used as a measurement for identification of active compounds from a library of analogs synthesized based on the lead molecule, YTR107.
Introduction
Precision medicines that target specific driver mutations have revolutionized cancer therapy. Unfortunately, not all patients will be able to take advantage of these therapies. Many will present with tumors that do not express actionable molecular driver mutations. Non-small cell lung cancer (NSCLC) represents an excellent example, as 36% of patients fall into this category1. Cytotoxic therapy continues to be a very important tool for the treatment of human cancers that do not express actionable molecular targets. Ionizing radiation is a cytotoxic agent that has a central role in cancer therapy and is used to provide local/regional control of cancer1; a requirement for preventing tumor-mediated organ failure, tumor recurrence and metastasis2–4. Recent advances in 3-D image-guided radiation therapy have significantly increased the probability of obtaining outstanding local/regional tumor control. However, a limitation to this therapy is the intrinsic radiation resistance of individual tumor cells5 due to increased DNA repair potential6–8. Thus, targeting DNA repair represents a rational strategy for overcoming radiation resistance.
The indole structure of the radiation sensitizer, indomethacin9, was used as a scaffold for the synthesis of a series of (Z)-5-((N-benzyl-1H-indol-3-yl)methylene)imidazolidine-2,4-dione and (Z)-5-((N-benzyl-1H-indol-3-yl)methylene)pyrimidine-2,4,6(1H,3H,5H)-trione derivatives that incorporated a variety of aromatic substituents in both the indole and N-benzyl moieties. Functional phenotypic screening for structure activity relationships revealed that introduction of the electron withdrawing group 4-CN into the N-benzyl moiety yielded a potent radiosensitizing compound10, capable of sensitizing six NSCLC cell lines, HT29 colorectal cells, D54 glioblastoma cells, PANC1 pancreatic cancer cells, and two breast cancer cell lines11, 12. This molecule, a substituted (Z)-5-((N-benzyl-1H-indol-3-yl)methylene)pyrimidine-2,4,6(1H,3H,5H)-trione was renamed YTR107 (Figure 1).
Figure 1.
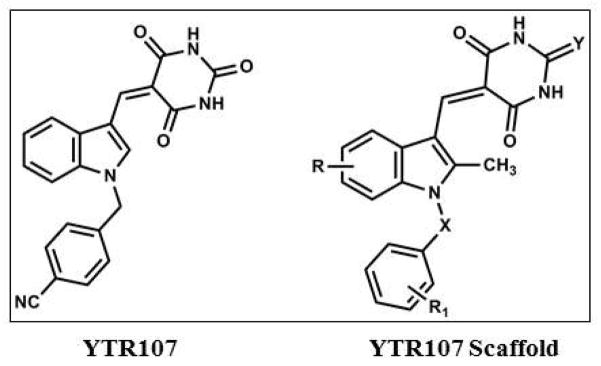
Structures of YTR107 and its scaffold
Inhibition of DNA double strand break (DSB) repair represented the mechanism responsible for YTR107-mediated radiosensitization12. YTR107 exhibited efficacious radiosensitization in 2 tumor xenografts and a syngeneic tumor model but did not produce overt normal tissue toxicity11 or normal tissue radiosensitization (unpublished results).
Use of YTR107 as a chemical probe resulted in the identification of the non-enzymatic chaperone, nucleophosmin 1 (NPM1) as a biological target that is critical for DNA DSB repair. In response to formation of DNA DSBs, phosphorylated T199 NPM1 binds to ubiquitinated chromatin, in a RNF8/RNF168-dependent manner, forming irradiation-induced foci (IRIF) that promote repair of DNA DSBs13. YTR107 binds to the amino terminus of NPM1, inhibiting IRIF formation, which in turn impairs DSB repair, and thus acts as a radiosensitizer. Genetic and cell biological approaches validated this concept by demonstrating that NPM1-deficient cells have impaired DNA DSB repair and consequently are radiosensitive. Use of NPM1-null mouse embryo fibroblasts demonstrated that the molecular basis of YTR107-mediated radiosensitization is YTR107 targeting of NPM1 and subsequent inhibition of DNA DSB repair. Although development of the YTR107 probe and discovery of its action represent a critical step for understanding a novel radiosensitizing mechanism, its use is hampered by limitations such as poor water-solubility.
In the present work, we report on a series of novel 2-methyl-N-benzyl aplysinopsin analogs, i.e. 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-oxodihydropyrimidine-4,6 (1H,5H)-triones and 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-thioxodihydro-pyrimidine-4,6(1H,5H)diones (Figure 2), which have been evaluated in a novel screening assay for their ability to modulate the extractability of phospho-nucleophosmin 1 (pNPM1 or pT199NPM1) after radiation treatment. The synthesis and anticancer properties of these analogues have recently been reported by us14. The screening assay was developed based on our novel observation that exposure of irradiated cancer cells to YTR107 increases the extractability of nuclear pNPM1 in high salt extraction buffer when compared to solvent control11. In the present report, we analyzed the solubility of pNPM1 in NP-40 and RIPA buffers and correlated the findings with radiosensitization of H460 lung cancer cells with these novel YTR107 analogs.
Figure 2.
Structures of 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-oxodihydropyri midine-4,6(1H,5H)-triones and 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-thioxodi hydropyrimidine-4,6(1H,5H)diones (1a-r).
Methods and buffer compositions
Lung cancer cells, A549, H460, and Calu1 as well as normal human lung fibroblasts, IMR-90 cells were purchased from ATCC and cultured in DMEM/F-12 (A549), RPMI-1640 (H460), and DMEM (Calu1 and IMR-90) media. Nuclei were isolated by suspending cells in nuclei isolation buffer on ice. Nuclear proteins were isolated by lysing the nuclei in high salt buffer. For total lysates, the cells were lysed either with NP-40 lysis buffer or RIPA buffer. The recipes to various buffers are given below.
NP-40 lysis buffer
50 mM Tris, pH 8.0, 150 mM NaCl, 0.01% NP-40, 0.1% SDS, 10 mM EDTA, 0.05% deoxycholic acid. Protease and phosphatase inhibitor cocktails were added before use.
RIPA buffer
10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 0.1% Triton X-100, 1% deoxycholic acid. Protease and phosphatase inhibitor cocktails were added before use.
High salt buffer
10 mM HEPES, pH 7.2, 420 mM NaCl, 0.1 mM EGTA, 1.5 mM MgCl2, 0.5 mM DTT, and 25% glycerol. Protease and phosphatase inhibitor cocktails were added before use.
Nuclei isolation buffer
50 mM Tris, pH 7.4, 50 mM NaCl, 10 mM EDTA, 0.5% NP-40. Protease and phosphatase inhibitor cocktails were added before use.
Western blot analysis
Lung cancer cells were grown to 70–80% confluency and were treated with analogs for 30 min, irradiated using Cs137 source at 2 Gy per min and incubated 90 min in 37°C cell culture incubator. The cells were washed with cold PBS and protein was extracted with various buffers described above. Protein levels were measured using BioRad protein assay reagent (Cat# 500-0006). Equal amounts of protein were resolved on 10% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was Western blotted for nucleophosmin 1 and p-nucleophosmin 1 (T199) antibodies (pT199 NPM1 from Abcam and NPM1 from LifeTechnologies). Band intensities were quantified using Adobe Photoshop software.
Colony formation assay
This assay was performed as described in our previous publications12, 15. In brief, cells were plated in T25 flasks, treated with drugs at 37°C for 30 min, irradiated and incubated for 90 min. The drugs were washed off with PBS and fresh media was added. The flasks were incubated 10–14 days. The colonies were stained with trypan blue and counted.
Extractability of pNPM1 in various buffers
In order to develop an assay to screen large number of molecules, it is necessary to demonstrate the simplicity of the assay with less processing steps. We have reported earlier on differential extractability of pNPM1 from irradiated cells with high salt buffer11. Phosphorylation of NPM1 T199 is a highly specific cell cycle event that occurs just prior to initiation of S phase. In late G1, activated CDK2/cyclin E phosphorylates NPM1 at T199, causing NPM1 to dissociate from centrosomes, an event essential for centrosome duplication 16. CDK2/cyclin A activation during S and G2 insures continued phosphorylation of T199 NPM117. During anaphase pNPM1 is dephosphorylated18, allowing it to associate with unduplicated centrosomes early in G1, and preventing centrosome duplication16. During the DNA damage response phosphorylation of T199 licenses NPM1 binding to ubiquitinated chromatin, in a RNF8/RNF168-dependent manner in response to formation of DNA DSBs13. Once bound to chromatin, pNPM1 forms irradiation-induced foci (IRIF) and promotes repair of DNA DSBs11–13.
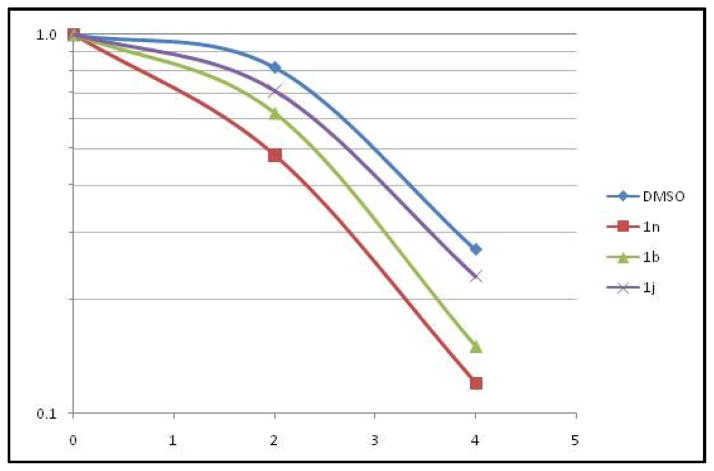
We have reported earlier that radiosensitizer, YTR107, enhances extractability of pNPM1 from nuclear extracts in high salt buffer upon radiation treatment11. Dose dependent enhancement of pNPM1 solubility was determined for YTR107 using Western blots (Fig. 3). The band intensities were quantified using Adobe photoshop and EC50 value was calculated from the graph generated from band intensities. In unirradiated A549 cells, pNPM1 can be extracted from the nuclei using a high salt buffer (data illustrating a representative immunoblot from A549 cells is shown in Figure 3, 0 Gy). However, pT199NPM1 cannot be extracted from irradiated cells by high salt buffer, as evidenced by a lack of immunoreactive protein (6 Gy, lane DMSO, Figure 3). This is a consequence of forming pNPM1 IRIF, as quantified by confocal microscopy12. The confocal analysis confirmed that the change in buffer solubility upon irradiation was not a consequence of dephosphorylation of pNPM112. YTR107 enhanced the pNPM1 solubility at 10 μM and above (Fig. 3). The EC50 was determined to be 9.7 ± 0.5 μM. This experiment demonstrates that a Western blot assay can be used to identify possible radiosensitizers that target NPM1.
Fig. 3.
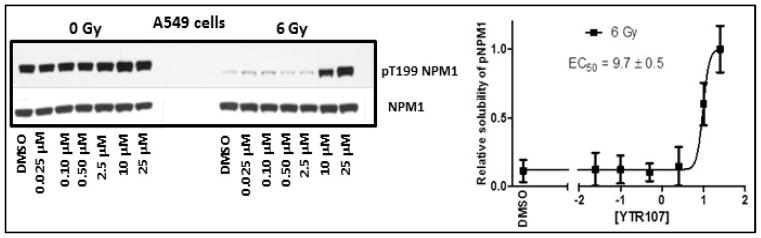
YTR107 inhibits pNPM1 recruitment to chromatin in irradiated cells.
A Representative immunoblot illustrating the extraction of non-chromatin bound pT199NPM1 in 420 mM NaCl buffer from nuclei of A549 cells. Cells were exposed to DMSO or various concentrations of YTR107 for 30 min prior to, during, and for 90 min after 0 or 6 Gy. B Quantification of buffer soluble pNPM1 (± SD) from nuclei of irradiated cells relative to non-irradiated cells, N=3 experiments.
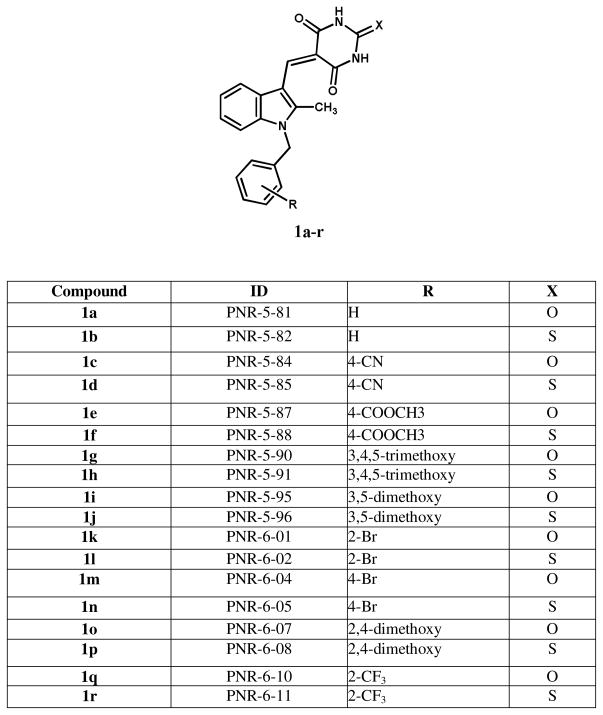
The nuclear extraction method is not suitable for the screening of large numbers of compounds, since it required isolation of nuclei, which is a laborious process and requires larger cell culture apparatus. In order to simplify the assay, we have tested two additional buffer compositions, NP-40 and RIPA buffers. This is a single step isolation of total cell lysates. Lead molecule, YTR107 was used to compare all three buffers in one assay to demonstrate the identical outcome using Western blot (Fig. 4). Both, NP-40 and RIPA, buffers yielded similar results as that of nuclear extraction with high salt buffer.
Figure 4. Effect of lysis buffers on the extractability of pNPM1 with YTR107.
A549 cells were treated with drugs (25 μM) for 30 min, irradiated, and incubated for 90 min. Nuclear extracts were made with high salt buffer (lanes 1–4). Total lysates using NP-40 buffer (lanes 5–8) and RIPA buffer (lane 9–12) were prepared and analysed by Western blot for pNPM1 and total NPM1.
Screening of New Compounds
Since we have demonstrated that the outcome from using various extraction buffers is comparable to each other, we have used NP-40 extraction method to screen newly synthesized YTR107 analogs (Western blot data not provided). NP-40 buffer is commonly used for protein isolation for Western blot analysis whereas RIPA buffer is routinely used for immunoprecipations. The band intensities were quantified using Adobe Photoshop and ratios were calculated between (Compound + 6 Gy) and (DMSO + 6 Gy). Ratios higher than 1 indicate that the molecule was capable of enhancing pNPM1 solubility upon radiation treatment (Table 1).
Table 1.
Band intensities were quantified from Western blots and ratio of band intensity from radiation + drug was divided by DMSO + radiation. Higher values indicate that the extractability pNPM1 is enhanced upon treatment with drug.
| Compound | Ratio in A549 Cells (Compound+6Gy/DMSO+6Gy) 25 μM |
|---|---|
| DMSO | 1.00 |
| 1a | 1.53 |
| 1b | 1.96 |
| 1c | 1.58 |
| 1d | 1.86 |
| 1e | 1.48 |
| 1f | 1.74 |
| 1g | 1.16 |
| 1h | 1.97 |
| 1i | 1.63 |
| 1j | 1.97 |
| 1k | 1.05 |
| 1l | 1.68 |
| 1m | 1.23 |
| 1n | 1.78 with 2.5 μM drug |
| 1o | ND# |
| 1p | (1.22) |
| 1q | 1.16 |
| 1r | 1.54 |
| YTR107 | 1.51 |
ND – Not determined
Dose response data obtained utilizing a molecule from the sub-library illustrated in Figure 2, PNR605 (1n), indicated that the EC50 value to enhance extractability of pNPM1 is below 2.5 μM (Fig. 5B). Identical extraction conditions were used for determining the EC50 values for YTR107 and 1n. Compound 1n is thus 4 times more potent than the lead compound, YTR107, in enhancing the solubility of pNPM1 upon radiation. Similar observations were made when A549 cells were treated with PNR605 and total protein lysates were prepared with NP-40 lysis buffer (Fig. 5C).
Figure 5. Extractability of pNPM1 by compound 1n.
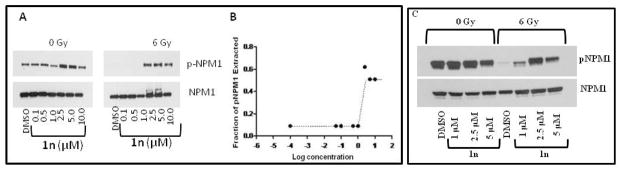
H460 and A549 cells were treated with PNR605 (1n) for 30 min, irradiated (6 Gy), and incubated for 90 min. A) Nuclei were isolated from H460 cells and proteins were extracted with high salt buffer and immunoblotted. B) Bands were quantified from 5A using Adobe Photoshop and plotted using GraphPad Prism 4 software. EC50 was calculated at which 50% of pNPM1 extracted. C) Total lysates were prepared from A549 cells with NP-40 lysis buffer and immunoblotted for pNPM1 and NPM1 antibodies.
In order to screen large number of compounds using the Western blot method, it is necessary to reduce the number of samples to be processed. So we have eliminated drug alone treatment and analysed the samples that were treated in combination with radiation. Solvent control (DMSO) was used to determine basal levels of pNPM1 present in the cancer cells (Figure 5). We have tested two concentrations of drug to evaluate the dose response of Calu1 cells. This approach also enables the determination of the dose-response of drugs in the extractability of pNPM1. For example, PNR584 (1c) at 2.5 μM concentration was unable to alter the extractability when compared to DMSO + 6 Gy control (Figure 6). However, this molecule was able to alter pNPM1 solubility at 5 μM. Similarly, PNR595 (1i) was unable to change the solubility of pNPM1 at both tested doses (Figure 6).
Figure 6. Extractability of pNPM1 in Calu1 cells after irradiation in the presence of various YTR107 analogs.
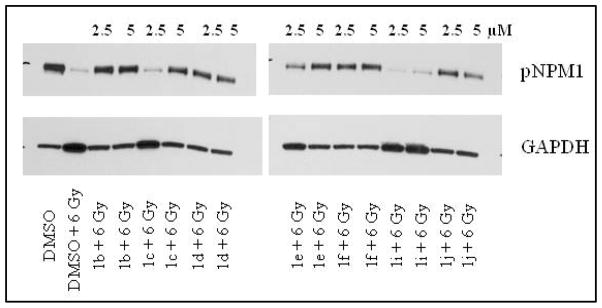
Calu1 cells were treated with DMSO/drugs for 30 min, irradiated, and incubated for 90 min. Total proteins were isolated with NP-40 lysis buffer and analyzed by Western blots.
Normal cells will not respond to YTR107-mediated solubility of pNPM1
Recruitment of pNPM1 to chromatin is essential for the repair of radiation induced DSBs. During radiation treatment, normal tissue is also exposed to the radiation field, and may be sensitized by YTR107. We have used IMR90 normal lung fibroblasts to test whether YTR107 alters the extractability of pNPM1 upon radiation treatment. Western blot analysis revealed that YTR107 was unable to enhance the extractability of pNPM1 upon irradiation (Figure 7). This observation suggests that DSB repair is not hindered even when normal tissue is exposed to both drug and radiation. Thus, the extractability of pNPM1 seems to be specific to cancer cells.
Figure 7. Effect of YTR107 on IMR90 cells.
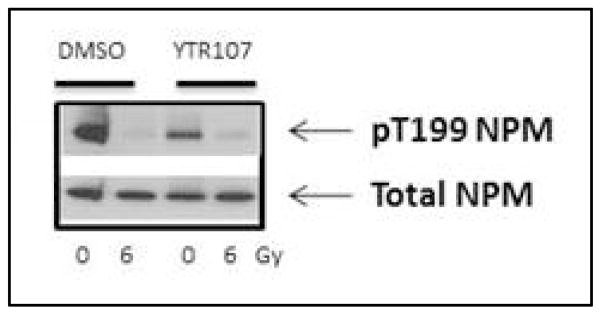
IMR90 cells were treated with YTR107 (25 μM) for 30 min, irradiated, and incubated for 90 min. Total lysate was prepared with NP-40 lysis buffer and analyzed by Western blot.
Validation of Assay
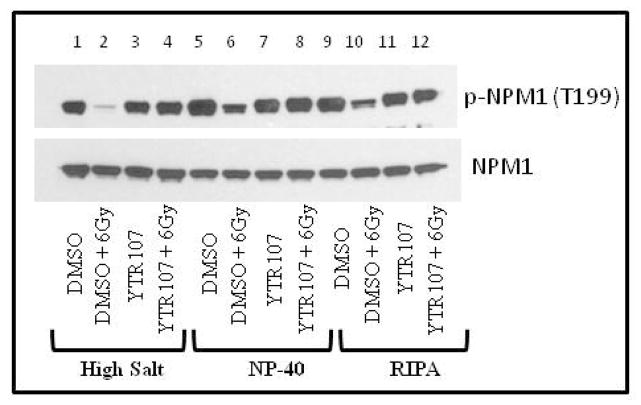
In order to demonstrate that the compounds which increased the extractability of pNPM1 upon radiation treatment are radiosensitizers, we have randomly selected three compounds [PNR582 (1b), PNR605 (1n), and PNR596 (1j)] (Figure 2) and carried out a detailed analysis of radiosensitization effects on H460 cells. H460 cells were treated with the compounds and subjected to various doses of radiation. The cells were then subjected to colony formation assays (Figure 8). All three compounds have been shown to enhance the extractability of pNPM1 (Table 1), thus the compounds are expected to be radiosensitizers. In fact, all three compounds enhanced the effect of radiation on H460 cells (Figure 8). Compound 1n exhibited superior radiosensitization when compared to other two molecules.
Figure 8. Radiosensitization of H460 cells by YTR107 analogs.
H460 cells were treated with 5 μM of 1n, 1b, or 1j for 30 min, irradiated, and incubated for 90 min. The drugs were then washed off and the flasks were incubated for 10–14 days, stained with trypan blue and colonies were counted and analyzed.
Conclusion
The molecular chaperone nucleophosmin 1 (NPM1) is frequently overexpressed in cancer, where it supports oncogenesis-driven ribosome biogenesis19. Emerging research has suggests that NPM1 may be a critical factor for homologous recombination (HR)-mediated repair of DSBs. NPM1 is recruited to sites of DSBs, binds ubiquitinated chromatin and promotes RAD51-mediated HR13. Loss of HR imposes a radiosensitization phenotype on cells subjected to fractionated irradiation20. Hence, we hypothesized that targeting pNPM1 would radiosensitize cancer cells. Since NPM1 is not an enzyme, screening of chemical libraries for NPM1 inhibitors would be challenging. We have observed that the extractability of pNPM1 is reduced when cancer cells are irradiated, and indole analog YTR107 reverses the effects of radiation on the extractability of pNPM1. Based on this observation, we have developed a Western blot assay to screen YTR107 analogs to identify molecules that could be effective radiosensitizers. The extractability of pNPM1 did not change with drug plus radiation treatment in normal lung fibroblasts, suggesting that the event is specific for cancer cells. Compounds identified through this assay radiosensitized lung cancer cells, suggesting the validity of the assay and the druggability of NPM1.
Acknowledgments
This work was supported by NIH NCI/NIEHS (CA 140409 and 1R41ES025596) grants and to the Arkansas Research Alliance (ARA) for financial support.
Footnotes
Disclosures
The compounds identified in this manuscript are covered by US patent 8,304,421 B2. The authors of this manuscript are co-inventors on this patent.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.NCCN. NCCN Guidelines Version 3.2011. 2011 [Google Scholar]
- 2.Suit HD, Sedlacek RS, Gillette EL. Radiology. 1970;95:189. doi: 10.1148/95.1.189. [DOI] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative, G. Darby S, McGale P, Correa C, Taylor C, Arriagada R, Clarke M, Cutter D, Davies C, Ewertz M, Godwin J, Gray R, Pierce L, Whelan T, Wang Y, Peto R. Lancet. 2011;378:1707. [Google Scholar]
- 4.Patel AR, Stephenson AJ. Nature reviews. Urology. 2011;8:385. doi: 10.1038/nrurol.2011.80. [DOI] [PubMed] [Google Scholar]
- 5.Gerweck LE, Vijayappa S, Kurimasa A, Ogawa K, Chen DJ. Cancer Res. 2006;66:8352. doi: 10.1158/0008-5472.CAN-06-0533. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed KM, Li JJ. Free Radic Biol Med. 2008;44:1. doi: 10.1016/j.freeradbiomed.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Nature. 2006;444:756. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 8.Chen MS, Woodward WA, Behbod F, Peddibhotla S, Alfaro MP, Buchholz TA, Rosen JM. J Cell Sci. 2007;120:468. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury CM, Markovina S, Wei SJ, Rene LM, Zoberi I, Horikoshi N, Gius D. Cancer Res. 2001;61:7689. [PubMed] [Google Scholar]
- 10.Reddy YT, Sekhar KR, Sasi N, Reddy PN, Freeman ML, Crooks PA. Bioorg Med Chem Lett. 2010;20:600. doi: 10.1016/j.bmcl.2009.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekhar KR, Reddy YT, Reddy PN, Crooks PA, Venkateswaran A, McDonald WH, Geng L, Sasi S, Van Der Waal RP, Roti JL, Salleng KJ, Rachakonda G, Freeman ML. Clin Cancer Res. 2011;17:6490. doi: 10.1158/1078-0432.CCR-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekhar KR, Benamar M, Venkateswaran A, Sasi S, Penthala NR, Crooks PA, Hann SR, Geng L, Balusu R, Abbas T, Freeman ML. International journal of radiation oncology, biology, physics. 2014;89:1106. doi: 10.1016/j.ijrobp.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike A, Nishikawa H, Wu W, Okada Y, Venkitaraman AR, Ohta T. Cancer Res. 2010;70:6746. doi: 10.1158/0008-5472.CAN-10-0382. [DOI] [PubMed] [Google Scholar]
- 14.Penthala NRKA, Sekhar KR, Freeman ML, Eoff RL, Balusu R, Crooks PA. ACS Chem Biol. 2015 doi: 10.1016/j.bmc.2015.10.019. Communicated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekhar KR, Reddy YT, Reddy PN, Crooks PA, Venkateswaran A, McDonald WH, Geng L, Sasi S, Van Der Waal RP, Roti JLR, Salleng KJ, Rachakonda G, Freeman ML. Clinical Cancer Research. 2011;17:6490. doi: 10.1158/1078-0432.CCR-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. The Journal of biological chemistry. 2001;276:21529. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- 17.Tarapore P, Shinmura K, Suzuki H, Tokuyama Y, Kim SH, Mayeda A, Fukasawa K. FEBS letters. 2006;580:399. doi: 10.1016/j.febslet.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Negi SS, Olson MO. J Cell Sci. 2006;119:3676. doi: 10.1242/jcs.03090. [DOI] [PubMed] [Google Scholar]
- 19.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Nature. 2005;437:147. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 20.Utsumi H, Elkind MM. Radiation research. 2001;155:680. doi: 10.1667/0033-7587(2001)155[0680:rfrodd]2.0.co;2. [DOI] [PubMed] [Google Scholar]