Abstract
Background
Altered miR-148/152 family expression contributes to human carcinogenesis. This study was designed to detect the potential for using miR-148/152 family as biomarkers for NSCLC patients.
Material/Methods
The relative expression levels of miR-148/152 family (miR-148a, miR-148b, and miR-152) in serum of 36 non-small-cell lung carcinoma (NSCLC) patients, 20 patients with benign pulmonary diseases (BPD), and 10 healthy individuals were assessed by real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR).
Results
The expression of all three miRNAs were significantly lower in the serum of NSCLC than that of BPD and healthy controls (all p<0.01), and their expression levels were strongly correlated with each other (r=0.781, 0.720, and 0.645, respectively). Downregulation of miR-148/152 family was found to be corrected with more aggressive tumors. The area under the receiver operating characteristic curves (AUCs) for miR-148a, miR-148b, and miR-152 discriminating NSCLC from BPD were 0.775, 0.725, and 0.774, respectively, all higher than that of CEA (0.506). Combining the three miRNAs increased the discrimination performance, yielding an AUC of 0.789 (95% confidence interval, 0.643 to 0.895), with a sensitivity of 72.2% and a specificity of 90.0%.
Conclusions
The results of present study suggest that the expression levels of circulating miR-148/152 family may serve as biomarkers for NSCLC.
Keywords: Biological Markers; Carcinoma, Non-Small-Cell Lung; MicroRNAs; Serum
Background
Non-small cell lung cancer (NSCLC) is the third most common cause of cancer-related deaths worldwide, accounting for about 80% of all lung cancer [1]. Surgical resection remains the main treatment for NSCLC. Despite advances in early detection and improvements in treatment, about 75% of patients are diagnosed at the advanced stage and less than 50% of patients among those who can undergo curative resection, with a very low 5-year overall survival rate (5%); however, the 5-year survival for early-stage NSCLC after curative resection can increase to 30~60% [2]. Thus, promising non-invasive biomarkers for identifying high-risk individuals, combined with the current main diagnostic technologies, including X-ray, computed tomography (CT) and positron emission tomography (PET), are still urgently needed.
MicroRNAs (miRNAs), a class of small, regulatory, non-coding RNAs, have emerged as important regulators involved in various biological and pathological events, such as cellular growth, differentiation, and apoptosis and metastasis in tumorigenesis and deterioration [3,4]. Due to the versatile roles played by miRNAs, it is not surprising that the expression levels of miRNAs may be indicators of the intrinsic characteristics of tumors. Because molecules function at the epigenetic level, aberrant expression of miRNAs occurs prior to phenotypic changes in the staged progression to carcinoma. In addition, miRNAs can be reliably detected and quantified in every-day diagnostic biological material such as formalin-fixed paraffin-embedded (FFPE) samples, even in minimally invasive material such as whole blood, serum, and sputum, and are amenable to assessment by standard biotechnology techniques [5]. Taken together, miRNAs have been identified as a kind of new blood-based biomarker for prognosis, diagnosis, and treatment selection for cancer patients [6,7].
MiR-148a, miR-148b, and miR-152 are the three members of the miR-148/152 family [8]. All these three miRNAs have been identified to downregulate in NSCLC specimens and cell lines [9–12]. In addition, they are all key modulators of cell biological behaviors of NSCLC: miR-148a regulates epithelial-to-mesenchymal transition (EMT) by targeting ROCK1 in NSCLC [9]; miR-148b suppresses the proliferation and migration of NSCLC cell lines by targeting CEA [10]; and miR-152 reportedly inhibits the proliferation and invasion of NSCLC by downregulating the expression of FGF2 and ADAM17 [11, 12]. However, the expression of miR-148/152 family in the serum of NSCLC patients has not yet been reported.
In the present study, we detected the expression of miR-148/152 family in serum of NSCLC patients, patients with benign pulmonary diseases (BPD), and healthy individuals. We investigated the association between their expression levels and clinicopathological characteristics and determined whether these three miRNAs could be biomarkers for NSCLC.
Material and Methods
Subjects
Serum samples were obtained from 36 patients with primary NSCLC and 20 BPD patients prior to surgery who visited our hospital between February 2010 and April 2013. Additionally, 10 healthy volunteers with no history of cancer were recruited. The detailed medical histories, including age, sex, clinical, and histopathological parameters of all participants are summarized in Table 1. The tumor stage at the time of diagnosis was determined according to guidelines of the American Joint Committee on Cancer (http://www.cancerstaging.org/). All samples were collected from the antecubital vein using 10-mL clotting tubes, centrifuged at 2,000 × g for 20 min at 4°C, and aliquoted into separate Eppendorf tubes and stored at −80°C. Written informed consent was obtained from each participant, which was reviewed and approved by the Institutional Review Board of our Hospital.
Table 1.
Patients’ characteristics and correlation of relative levels of miRNAs with clinicopathological parameters.
| Parameters | Patients | miRNA expression level (Mean±SD) | ||
|---|---|---|---|---|
| miR-148a | miR-148b | miR-152 | ||
| NSCLC patients | 36 | |||
| Age (years) | 56 years (range, 27–82 years) | 0.06±0.09 | 0.04±0.06 | 0.13±0.16 |
| ≤60 | 11 | 0.09±0.05 | 0.03±0.09 | 0.09±0.05 |
| >60 | 23 | 0.05±0.03 | 0.05±0.12 | 0.15±0.06 |
| Gender | ||||
| Male | 25 | 0.05±0.03 | 0.04±0.12 | 0.15±0.04 |
| Female | 11 | 0.07±0.04 | 0.05±0.06 | 0.12±0.02 |
| Tumor size (cm) | ||||
| ≤3 | 20 | 0.07±0.04 | 0.07±0.17b | 0.08±0.06d |
| >3 | 16 | 0.04±0.09 | 0.02±0.06b | 0.23±0.50d |
| Histology | ||||
| SCC | 16 | 0.05±0.04 | 0.06±0.09 | 0.20±0.08 |
| ADC | 20 | 0.07±0.09 | 0.04±0.03 | 0.11±0.06 |
| Differentiation | ||||
| Moderate-well | 23 | 0.07±0.04 | 0.04±0.06 | 0.11±0.08 |
| Poorly | 13 | 0.05±0.02 | 0.04±0.13 | 0.14±0.04 |
| Lymph node metastasis | ||||
| Negative | 6 | 0.09±0.04a | 0.08±0.13c | 0.21±0.16e |
| Positive | 30 | 0.03±0.09a | 0.03±0.09c | 0.08±0.09e |
| TNM stage | ||||
| I | 8 | 0.05±0.04 | 0.05±0.08 | 0.24±0.46f |
| II~IV | 28 | 0.07±0.02 | 0.04±0.12 | 0.03±0.13f |
| BPD patients | 20 | |||
| Age | 61 years (range 32–81 years) | 0.91±0.51 | 0.51±0.26 | 0.38±0.21 |
| Gender | ||||
| Male | 12 | 1.04±0.07 | 0.52±0.17 | 0.41±0.25 |
| Female | 8 | 0.92±0.03 | 0.53±0.36 | 0.33±0.14 |
| Healthy individuals | 10 | |||
| Age | 52 years (range 26–73 years) | 0.43±0.06 | 0.21±0.25 | 1.26±0.42 |
| Gender | ||||
| Male | 5 | 0.58±0.08 | 0.19±0.08 | 1.13±0.60 |
| Female | 5 | 0.77±0.06 | 0.25±0.17 | 1.42±0.91 |
p=0.027;
p=0.009;
p=0.031;
p=0.012;
p=0.045;
p=0.047.
BPD – benign pulmonary disease SCC – squamous cell carcinoma; ADC – adenocarcinoma; TNM – tumour-node-metastasis staging system.
RNA extraction and real-time reverse transcription quantitative polymerase chain reaction
Total RNA was extracted from serum using the BioTeKe miRNA extraction kit (BioTeKe Corporation, China) according to the manufacturer’s instructions. The expression levels of miRNAs in samples were measured by RT-qPCR. Isolated RNA was reverse transcribed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantification of miRNA was performed on the Eppendorf Mastercycler EP Realplex (Eppendorf, Germany) using real-Time PCR System at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. U6 snRNA served as the internal control. Relative miRNA expression levels were calculated using the 2−ΔΔCt method. All reactions were run in triplicate and Ct data were determined using default threshold settings.
Serum CEA determination
Serum CEA levels were measured using ELISA (no. ab4451; Abcam, Cambridge, UK), dilutions for excessive CEA concentration, setup, adjustments, and quality controls were performed according to the manufacturer’s instructions. The assay employed anti-human CEA antibody coated onto a 96-well plate. Standard or serum samples were pipetted into the wells, and CEA present in samples were bound to the wells by the immobilized antibody. The wells were then washed and biotinylated anti-human CEA antibodies were added. Following wash of unbound biotinylated antibody and horseradish peroxidase-conjugated streptavidin was pipetted into the wells. The wells were washed again, TMB substrate solution was added, and color developed in proportion to the amount of bound CEA. Absorbance value was measured at 450 nm. CEA concentration in serum was determined according to standard curves.
Statistical analysis
Differences between groups were assessed using the Mann-Whitney U test. The correlations between the expression levels of miR-148/152 family were evaluated by Pearson’s regression analysis. A receiver operating characteristic (ROC) curve based on the ΔCt (Ct[miRNA]-Ct[U6]) of miRNA expression in samples was plotted and the area under the curve (AUC) was calculated to evaluate the diagnostic performance of miRNAs. All statistical analyses were performed using SPSS 20.0 (SPSS Inc., Chicago, IL). The MedCalc 10.4.7.0 (MedCalc, Mariakerke, Belgium) software was used to perform the ROC analysis. All p-values were two-sided and p<0.05 was considered statistically significant.
Results
Participant characteristics
A total of 66 serum samples (36 NSCLC, 20 BPD, and 10 healthy) were analyzed by RT-qPCR. Table 1 shows the clinicopathologic characteristics of all participants. Of the 36 NSCLC cases, the median age was 56.75±8.45 years (range, 27 to 82 years), 8 were stage I, 10 were stage II, 12 were stage III, and 6 were stage IV. Histologically, 16 of the 36 tumors were squamous cell carcinomas (SCC) and 20 were adenocarcinomas (ADC). Of the 20 BPD patients, the median age was 61.75±6.76 years (range, 32 to 81 years), 9 were tuberculosis, 4 were pseudotumors, 4 were pulmonary sclerosing hemangiomas, and 3 were pulmonary granulomas (PSH). The median age of the 10 healthy volunteers was 52.36±11.47 years (range, 26–73 years). No differences were observed in age and sex distribution among the three groups (P>0.05).
miRNAs expression profile
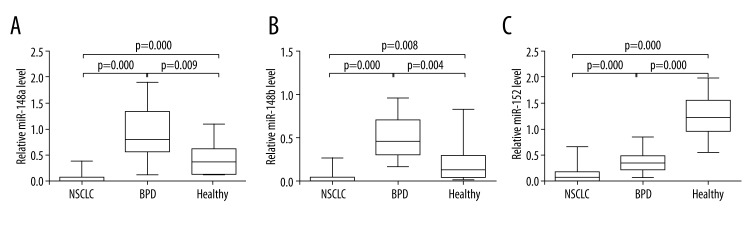
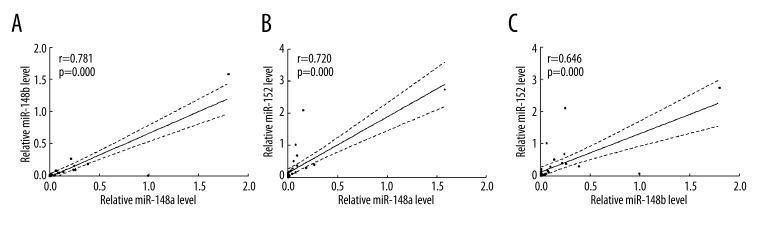
As shown in Figure 1, the expression levels of miR-148a, miR-148b, and miR-152 in NSCLC samples were significantly lower than that of BPD and healthy individuals (all p<0.01). The mean ±SD value of miR-148a, miR-148b, and miR-152 were 0.06±0.09, 0.04±0.06, and 0.13±0.16, respectively, in NSCLC samples; 0.91±0.51, 0.51±0.26, and 0.38±0.21, respectively, in BPD samples; and 0.43±0.36, 0.21±0.25, and 1.26±0.42, respectively, in samples from healthy participants. Intriguingly, while miR-152 showed continuous downregulation in the serum from healthy individuals to BPD patients and NSCLC patients, compared with healthy individuals, miR-148a and miR-148b levels were higher in BPD patients and lower in NSCLC patients. Pearson’s regression analysis showed strong correlations of the expression levels of miR-148a, miR-148b, and miR-152 with each other in NSCLC patients, and their correlation coefficients were 0.781, 0.720, and 0.645, respectively (Figure 2).
Figure 1.
The miR-148/152 family level was significantly decreased in NSCLC serum. Comparison of miR-148a (A), miR-148b (B), and miR-152 (C) expression levels in NSCLC patients, BPD patients, and healthy volunteers. Statistical significance was determined by the Mann-Whitney U test.
Figure 2.
Correlations of expression of miR-148/152 family in NSCLC patients. (A) Correlation of the expression of miR-148a and miR-148b in NSCLC patients (Pearson r=0.781, p=0.000). (B) Correlation of the expression of miR-148a and miR-152 in NSCLC patients (Pearson r=0.720, p=0.000). (C) Correlation of the expression of miR-148b and miR-152 in NSCLC patients (Pearson r=0.646, p=0.000).
The association between miR-148/152 family expression and clinicopathologic parameters in NSCLC
The correlation between miR-148/152 family expression levels and clinicopathological parameters of patients is summarized in Table 1. Low miR-148a expression was significantly correlated with presence of lymphatic metastasis (P=0.027); low miR-148b expression was significantly correlated with larger tumor size (P=0.009) and presence of lymphatic metastasis (P=0.031); and low miR-152 expression was significantly correlated with larger tumor size (P=0.012), presence of lymphatic metastasis (P=0.045), and advanced clinical stage (P=0.047). However, no correlation was observed between the expression levels of each miRNAs and other clinicopathologic factors, including age, sex, tumor differentiation, and histology (all p>0.05; Table 1).
Diagnostic performance of miR-148/152 family and CEA for NSCLC
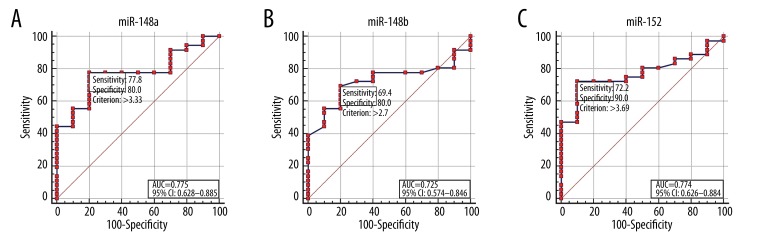
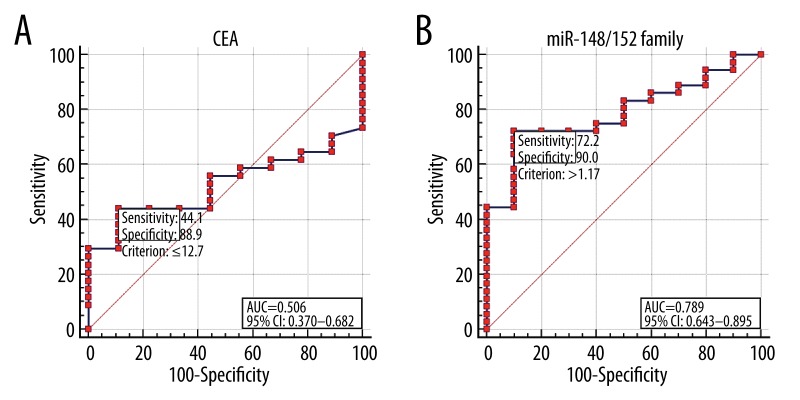
The diagnostic performance of miR-148/152 family in differentiating NSCLC from BPD was evaluated by ROC curve analysis (Figure 3). Cut-off points were determined such that they maximized the sum of sensitivity and specificity. The cut-off points for miR-148a, miR-148b, and miR-152 were 3.33, 2.70, and 3.69, respectively (Figure 3A–3C). The diagnostic accuracy of miR-148a, miR-148b, and miR-152, as measured by the AUC, were 0.775, 0.725, and 0.774, respectively (Figure 3). The diagnostic accuracy of CEA was 12.7 ng/mL and AUC for CEA was 0.506, with a sensitivity of 44.1% and a specificity of 88.9% (Figure 4A). Based on a logit model from the combination of these three miRNAs [logit=−0.745 + (0.283×expression level of miR-148a) + (−0.298×expression level of miR-148b) + (0.454×expression level of miR-152)], the predicted probability of patients being diagnosed with NSCLC was used to construct a ROC curve. The AUC for the model was 0.789 (95% confidence interval [CI], 0.643 to 0.895), with a sensitivity of 72.2% and a specificity of 90.0% (Figure 4B).
Figure 3.
Receiver operating characteristic (ROC) curve analysis according to independent miR-148/152 family level. The ROC plots for miR-148a (A), miR-148b (B), and miR-152 (C) were used to differentiate NSCLC from BPD. AUC, area under the receiver operating characteristic curve; CI, confidence interval.
Figure 4.
Receiver operating characteristic (ROC) curve analysis according to CEA and combined miR-148/152 family level. The ROC plots for carcinoembryonic antigen (CEA) (A), a combination of miR-148/152 family (B) were used to differentiate NSCLC from BPD. AUC, area under the receiver operating characteristic curve; CI, confidence interval.
Discussion
In the present study, we demonstrated that the expression level of miR-148/152 family (miR-148a, miR-148b, and miR-152) was significantly different among NSCLC patients, BPD patients (pulmonary tuberculosis, pseudotumor, pulmonary sclerosing hemangioma, and PSH), and healthy volunteers, and their expression profiles can be used to differentiate NSCLC and BPD. In addition, the diagnostic performance of the combination panel of miR-148/152 family (AUC, 0.789) was significantly higher than that of CEA (AUC, 0.506). As a widely used diagnostic biomarker for NSCLC patients, CEA is found to confer good specificity but relatively poor sensitivity for the diagnosis of NSCLC [13]. In addition to having a higher diagnostic value, the sensitivity of the miR-148/152 family panel was also higher (72.2%) than that of CEA. Our current data indicate that serum miR-148/152 family should be further evaluated as novel non-invasive biomarkers for discriminating NSCLC from benign lung tumors.
Studies have revealed that miRNAs constitute a robust regulatory network in the post-transcription regulation of coding genes [14]. By using detection analyses such as RNA sequencing and microarray, altered expression of miRNAs has been reported in varies of malignancies [3,15–18], and multiple deregulated miRNAs have been implicated in physiological and pathological processes of lung cancer [19–22]. Based on the deregulated miRNAs’ expression profiling and their association with the biological and clinical properties of lung cancer, specific miRNAs could be utilized to distinguish benign and malignant lesions [23,24]. In addition to cancer tissues, expression profiles of miRNAs are also altered in the serum or plasma from patients with lung cancers [25–27]. Given that miRNAs secreted from tumor cells into the circulatory system can resist RNase activity through being packed into highly stable complexes, miRNAs may be developed into reliable blood-based fingerprints for lung cancer diagnosis [28]. In the present study, we focused on the functional miR-148/152 family, which were reported to deregulate in lung cancer tissues, and examined their expression in serum samples obtained from NSCLC patients, BPD patients, and healthy individuals by RT-qPCR. We found that the expression levels of miR-148/152 family were lower in NSCLC than in BPD and healthy individuals. Therefore, the investigation of serum miR-148/152 family levels in NSCLC patients, as presented in this study, may be of great clinical interest.
As shown on the miRbase Website, the three members of the miR-148/152 family having the same seed sequence of approximately 6–7 nucleotides, which is well-reflected in the present study results showing strong correlations between these three miRNAs. Moreover, our results showed negative correlations between 148/152 family expression and malignant phenotypes of NSCLC, which may be good explanations for how all three miRNAs can function as tumor suppressors in NSCLC. Functional assays demonstrated that miR-148a inhibits EMT in NSCLC cells by directly targeting ROCK1, a metastasis promoter [9]. miR-148b suppresses cell proliferation and migration in NSCLC cell lines by targeting CEA [10]. Similarly, miR-152 was reported to inhibit the proliferation and invasion of NSCLC by downregulating the expression of FGF2 and ADAM17 [11,12]. In addition, low expression of miR-152 in plasma is associated with poor survival in NSCLC patients [29]. All these findings together suggest that although the three members of the miR-148/152 family have the same seed sequence, they may have different target sets and perform specific functions in different physiological and pathological processes, thereby exhibiting different expression profiles in specific disease types and stages. Therefore, it is understandable that although these three miRNAs are all downregulated in NSCLC serum, compared with healthy individuals, miR-152 showed a continuously downregulation in the serum from healthy individuals to BPD patients and NSCLC patients; while both miR-148a and miR-148b levels were higher in BPD patients and lower in NSCLC patients.
The current findings are preliminary and may have some limitations. First, this was a small study sample and the results require confirmation in further large-scale and multi-center studies before any recommendations can be generated. The second limitation of the study was that the expression of all three miRNAs were down-regulated in NSCLC, and the cut-off points that had to be detected in clinical samples for down regulated miRNAs were very low. In addition, since the function and mechanism of the miR-148/152 family in NSCLC pathogenesis have been shown thoroughly, combined with our investigation into their diagnostic value, the miR-148/152 family may be useful biomarkers for diagnosis, provide additional targets and strategies for treatment, and lead to increasingly personalized cancer therapy.
Conclusions
In summary, the present study showed downregulation of miR-148/152 family levels in serum of NSCLC patients, and their expression profiles were correlated with malignant phenotypes. Furthermore, the miR-148/152 family panel showed a diagnostic performance comparable with that of CEA, with much better sensitivity than that of CEA. Thus, we propose that miR-148/152 family could be utilized to develop invasive screening tools for NSCLC. More in-depth studies are required to confirm the diagnostic values of the miR-148/152 family in discriminating NSCLC from benign pulmonary diseases.
Footnotes
Source of support: Departmental sources
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Murray N. Reality check for pemetrexed and maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2014;32:482–83. doi: 10.1200/JCO.2013.53.3448. [DOI] [PubMed] [Google Scholar]
- 2.Ponn RB, Lo Cicero J, III, Daly BD. Surgical treatment of non-small cell lung cancer. General Thoracic Surgery. 2005;6:1548–87. [Google Scholar]
- 3.Rutnam ZJ, Yang BB. The involvement of microRNAs in malignant transformation. Histol Histopathol. 2012;27:1263–70. doi: 10.14670/HH-27.1263. [DOI] [PubMed] [Google Scholar]
- 4.Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 5.Skrzypski M, Dziadziuszko R, Jassem J. MicroRNA in lung cancer diagnostics and treatment. Mutat Res. 2011;717:25–31. doi: 10.1016/j.mrfmmm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–18. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y, Yao Q, Chen Z, et al. Genetic and molecular alterations in pancreatic cancer: implications for personalized medicine. Med Sci Monit. 2013;19:916–26. doi: 10.12659/MSM.889636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer. 2013;12:43. doi: 10.1186/1476-4598-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Song Y, Wang Y, et al. MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem. 2013;380:277–82. doi: 10.1007/s11010-013-1682-y. [DOI] [PubMed] [Google Scholar]
- 10.Liu GL, Liu X, Lv XB, et al. miR-148b functions as a tumor suppressor in non-small cell lung cancer by targeting carcinoembryonic antigen (CEA) Int J Clin Exp Med. 2014;7:1990–99. [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Z, Ma R, Tan W, Zhang L. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med. 2014;46:e112. doi: 10.1038/emm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y, Wang Y, Zhou H, et al. MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett. 2014;588:1983–88. doi: 10.1016/j.febslet.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Bekci TT, Senol T, Maden E. The efficacy of serum carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), carbohydrate antigen 19-9 (CA19-9), carbohydrate antigen 15-3 (CA15-3), alpha-fetoprotein (AFP) and human chorionic gonadotropin (hCG) levels in determining the malignancy of solitary pulmonary nodules. J Int Med Res. 2009;37:438–45. doi: 10.1177/147323000903700219. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Qi F, Cao Y, et al. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Monit. 2014;20:812–17. doi: 10.12659/MSM.890300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Cai Q, Jiang Z, et al. Prognostic role of MicroRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668–74. doi: 10.12659/MSM.892096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Zhao J, Zhang PY, et al. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012;18(9):BR299–308. doi: 10.12659/MSM.883262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Tian X, Han R, et al. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33:1181–89. doi: 10.1038/onc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J, Wu J, Zhang H, et al. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73:756–66. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- 21.Cai J, Fang L, Huang Y, et al. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73:5402–15. doi: 10.1158/0008-5472.CAN-13-0297. [DOI] [PubMed] [Google Scholar]
- 22.Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zandberga E, Kozirovskis V, Abols A, et al. Cell-free microRNAs as diagnostic, prognostic, and predictive biomarkers for lung cancer. Genes Chromosomes Cancer. 2013;52:356–69. doi: 10.1002/gcc.22032. [DOI] [PubMed] [Google Scholar]
- 24.Fanini F, Vannini I, Amadori D, Fabbri M. Clinical implications of microRNAs in lung cancer. Semin Oncol. 2011;38:776–80. doi: 10.1053/j.seminoncol.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Rani S, Gately K, Crown J, et al. Global analysis of serum microRNAs as potential biomarkers for lung adenocarcinoma. Cancer Biol Ther. 2013;14:1104–12. doi: 10.4161/cbt.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Gu J, Roth JA, et al. Pathway-based serum microRNA profiling and survival in patients with advanced stage non-small cell lung cancer. Cancer Res. 2013;73:4801–9. doi: 10.1158/0008-5472.CAN-12-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markou A, Sourvinou I, Vorkas PA, et al. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–96. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–48. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 29.Sanfiorenzo C, Ilie MI, Belaid A, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]