Abstract
Rationale: Genome-wide association studies (GWAS) of chronic obstructive pulmonary disease (COPD) have identified disease-susceptibility loci, mostly in subjects of European descent.
Objectives: We hypothesized that by studying Hispanic populations we would be able to identify unique loci that contribute to COPD pathogenesis in Hispanics but remain undetected in GWAS of non-Hispanic populations.
Methods: We conducted a metaanalysis of two GWAS of COPD in independent cohorts of Hispanics in Costa Rica and the United States (Multi-Ethnic Study of Atherosclerosis [MESA]). We performed a replication study of the top single-nucleotide polymorphisms in an independent Hispanic cohort in New Mexico (the Lovelace Smokers Cohort). We also attempted to replicate prior findings from genome-wide studies in non-Hispanic populations in Hispanic cohorts.
Measurements and Main Results: We found no genome-wide significant association with COPD in our metaanalysis of Costa Rica and MESA. After combining the top results from this metaanalysis with those from our replication study in the Lovelace Smokers Cohort, we identified two single-nucleotide polymorphisms approaching genome-wide significance for an association with COPD. The first (rs858249, combined P value = 6.1 × 10−8) is near the genes KLHL7 and NUPL2 on chromosome 7. The second (rs286499, combined P value = 8.4 × 10−8) is located in an intron of DLG2. The two most significant single-nucleotide polymorphisms in FAM13A from a previous genome-wide study in non-Hispanics were associated with COPD in Hispanics.
Conclusions: We have identified two novel loci (in or near the genes KLHL7/NUPL2 and DLG2) that may play a role in COPD pathogenesis in Hispanic populations.
Keywords: COPD, genome-wide association studies, Hispanics
Chronic obstructive pulmonary disease (COPD) is a common respiratory disease and a leading cause of morbidity and mortality worldwide (1). Inhalation of noxious particles (primarily cigarette smoke) is the major risk factor for COPD. However, only a minority of individuals who smoke develop COPD, suggesting that genetic factors influence disease pathogenesis (2, 3).
Genome-wide association studies (GWAS) have identified several susceptibility genes for COPD, including CHRNA3, FAM13A, and HHIP (4, 5). Although these loci were identified in GWAS conducted in populations of European and African descent, variants in these genes have also been implicated in COPD risk in Asians (6–8). To date, there has been no GWAS of COPD conducted exclusively in Hispanic populations (9). One recent study that did include a diverse group of Hispanics, along with members of other ethnic groups, identified a novel variant in MAN2B1 in Hispanics that was associated with lower values of the percentage of emphysema-like lung on computed tomography and higher plasma levels of α1-antitrypsin (10).
COPD is a major public health problem in Hispanic America, with adjusted prevalence ranging from ∼12% in Mexico City (Mexico) to ∼19% in Montevideo (Uruguay) (11). Given that the average Native American ancestry of Mexicans living in Mexico City (∼56% [12]) is markedly higher than that of Uruguayans living in Montevideo (∼1% [13]), these findings could be partly explained by protective effects of Native American ancestry against nicotine addiction (e.g., reduced intensity of smoking) or the detrimental effects of cigarette smoking on lung function or COPD progression. We hypothesized that Hispanic subgroups with a significant proportion of Native American ancestry have protective alleles against COPD, which may be undetected in GWAS of non-Hispanic populations.
In this manuscript, we report our findings from a metaanalysis of GWAS of COPD affection status in two Hispanic cohorts (living in Costa Rica and in the United States). We attempted to replicate our findings in a third cohort of Hispanics living in New Mexico. To assess the generalizability of our findings to cohorts of European and African descent, we examined our top findings for COPD in Hispanics in a U.S.-based cohort including non-Hispanic whites and African Americans (Genetic epidemiology of COPD [COPDGene]). In addition, we tested whether single-nucleotide polymorphisms (SNPs) identified in genome-wide studies of COPD in non-Hispanic populations are associated with COPD risk in two Hispanic cohorts.
Methods
Discovery Cohorts
Costa Rica
We recruited 579 subjects between April of 2003 and November of 2010, including: (1) 316 members of 13 families of probands (index cases; see Table E1 in the online supplement) with COPD, (2) 68 unrelated subjects with COPD (cases), and (3) 159 unrelated control subjects. An additional 36 individuals could not be classified as control subjects based on spirometry and were excluded from this analysis. Probands and cases were recruited from four major public hospitals in San José (Costa Rica) and through newspaper ads. Control subjects were recruited from a smoking-cessation clinic in San José (which was affiliated with the same public hospitals from where cases were recruited) and through newspaper ads. All study participants had to have at least six great-grandparents born in the Central Valley of Costa Rica (to ensure descent from the founder population, composed primarily of Europeans and Native Americans). All probands, cases, and control subjects also had to be aged 21 years or older and have a history of at least 10 pack-years of smoking. Members of the families of probands had to be 12 years and older. Other inclusion criteria for cases or probands were physician-diagnosed COPD, an FEV1 less than or equal to 65% predicted, and an FEV1/FVC ratio less than 70%. Control subjects had to have no physician-diagnosed COPD and normal spirometry.
All study participants completed a protocol including a validated questionnaire (containing questions on demographics, smoking history, and respiratory health) (14), pulmonary function testing, and collection of blood samples for DNA extraction. Spirometry was conducted with a Collins Survey Tach spirometer. All subjects had to be free of respiratory illnesses for 4 or more weeks before spirometry, and they were also instructed (when possible) to avoid use of inhaled short-acting bronchodilators (BDs) for 4 or more hours before testing. Forced expiratory maneuvers were judged to be acceptable if they met or exceeded American Thoracic Society criteria (15). Written informed consent was obtained from all participants. The study was approved by the Institutional Review Boards of the Hospital Nacional de Niños (San José, Costa Rica), Brigham and Women’s Hospital (Boston, MA), and the University of Pittsburgh (Pittsburgh, PA).
Multi-Ethnic Study of Atherosclerosis
The Multi-Ethnic Study of Atherosclerosis (MESA) is a population-based longitudinal study of subclinical cardiovascular disease (16). Between 2000 and 2002, MESA recruited 6,814 men and women 45 to 84 years of age from Forsyth County, North Carolina; New York City, New York; Baltimore, Maryland; St. Paul, Minnesota; Chicago, Illinois; and Los Angeles, California. Exclusion criteria were clinical cardiovascular disease, weight exceeding 136 kg (300 lb), pregnancy, and impediment to long-term participation. The MESA Family Study recruited 1,595 African American and Hispanic participants, generally siblings of MESA participants, using the same inclusion and exclusion criteria as MESA, except that clinical cardiovascular disease was permitted. The MESA Air Pollution Study recruited an additional 257 participants from Los Angeles and Riverside County, California, and Rockland County, New York, using the same criteria as MESA, except that participants were aged 50 to 89 years, lived in the area 50% or more of the year, and had no plans to move in the next 5 years (17). A total of 2,174 Hispanic participants consented to genetic analyses, as follows: 1,449 from MESA, 674 from MESA Family, and 51 from the MESA Air Pollution Study.
The MESA Lung Study administered a standardized respiratory questionnaire (14) and performed pre-BD spirometry in accordance with American Thoracic Society/European Respiratory Society guidelines (18) for a subset of MESA participants (19), MESA Family participants at one site, and all MESA Air Pollution Study participants. All participants attempted at least three acceptable maneuvers on the same dry rolling seal spirometers (Occupational Marketing Inc., Houston, TX) following the MESA Lung protocol; all examinations were reviewed by one investigator (20). For the current genetic analysis of Hispanics in MESA, there were 63 cases and 619 control subjects. Written informed consent was obtained from all participants. MESA and all associated studies were approved by the Institutional Review Boards of the National Heart, Lung, and Blood Institute and all participating institutions.
Follow-up Cohorts
Lovelace Smokers Cohort
The top SNPs from the metaanalysis of the GWAS in Costa Rica and MESA were replicated in a cohort of 337 unrelated smoking Hispanic adults in New Mexico, 47 (13.9%) of whom were classified as having COPD based on an FEV1 less than 80% predicted and an FEV1/FVC ratio less than 0.70. The protocols for subject recruitment and data collection for the Lovelace Smokers Cohort have been previously described in detail (21). Study participants were drawn from the Albuquerque (NM) metropolitan area. Inclusion criteria for entry into the study were age 40 to 75 years, current or former cigarette smoking (with a minimum of 10 pack-years), and the ability to understand English or Spanish. Information related to demographics and smoking history was obtained by self-report. The study was approved by the Western Institutional Review Board (Olympia, WA).
COPDGene
The protocols for subject recruitment and data collection for the COPDGene study have been previously published (22). In brief, the COPDGene Study enrolled non-Hispanic white and African American subjects with COPD (defined as Global Initiative for Chronic Obstructive Lung Disease stage II or greater) and smoking control subjects with normal spirometry. Genome-wide SNP genotyping data that met quality control standards were available on 2,812 COPD cases and 2,534 control subjects in the non-Hispanic white group and 821 COPD cases and 1,749 control subjects in the African American group. Subjects were aged 45 to 80 years with at least 10 pack-years of lifetime smoking history, and they were recruited at 21 U.S. centers. Each study site obtained local institutional review board approval to enroll participants in this project, and all subjects provided informed consent to participate in the study.
Primary Outcome
For this analysis, COPD was defined as having COPD stage II or higher according to the Global Initiative for Chronic Obstructive Lung Disease: a post-BD FEV1 lower than 80% predicted and a post-BD FEV1/FVC ratio less than 0.70. Control subjects had to have a post-BD FEV1 greater than or equal to 80% predicted and a post-BD FEV1/FVC ratio greater than or equal to 0.70. Because there are no reference FEV1 or FVC values for Costa Ricans, those published for Mexican Americans were used instead (23).
Genotyping in Discovery Cohorts
Costa Rica
Whereas the first 115 participants were genotyped using the Illumina Human610-Quad platform (containing ∼610,000 SNPs), the remaining participants (n = 464) were genotyped at a later date using the Illumina HumanOmniExpress platform (containing ∼730,000 SNPs). We applied stringent filters for quality control of each dataset by excluding SNPs: with minor allele frequency less than 1% or a completion rate less than 90%, or out of Hardy-Weinberg equilibrium. We then merged the two SNP datasets and implemented quality control measures similar to those previously conducted for each separate dataset. To assess for Mendelian inconsistencies in members of families of probands, the KING program (24) was used to determine relationships based on the genome scan marker data.
MESA
Participants in the original MESA cohort, the MESA Family Study, and the MESA Air Pollution Study who consented to genetic analyses were genotyped in 2009 using the Affymetrix Human SNP array 6.0 (Affymetrix Inc., Santa Clara, CA), with 897,981 SNPs passing study-specific quality control (10).
Genotyping in other Cohorts
In the Lovelace Smokers’ Cohort, genotyping was conducted using the Illumina GoldenGate assay with VeraCode technology on the BeadXpress platform. Genotyping accuracy was confirmed by examining genotypes between intra- and interplate duplicates and by visual checking of the genotype clustering using GenomeStudio. Details of the COPDGene genotyping, imputation, and quality control have been previously described (25). Briefly, genotyping was performed using the Illumina HumanOmniExpress, with imputation based on the 1,000 Genomes Phase I v3 reference panel.
Expression Analysis
To explore the possible functional consequences of identified SNPs, we tested whether they were associated with expression of nearby genes in adult human lung tissue using a previously published dataset of 1,111 tissue samples with both genotyping and gene expression data (26). We also examined a publically available eQTL database with information on brain and lung expression (27).
Statistical Analysis
Stage 1: metaanalysis (Costa Rica and MESA)
MaCH-Admix (28) was used in Costa Rican samples to perform imputation of an additional approximately 2 million SNPs in each race/ethnic group for inclusion in GWAS analyses, using HapMap Phase I + II reference panels with individuals of European (CEU), West African (YRI), Chinese (CHB), and Japanese (JPT) descent. Similarly, IMPUTE v2 (29, 30) was used in the imputation of MESA samples. R2 was used to assess the imputation quality. SNPs with R2 less than 0.5 were filtered out in each individual study before metaanalysis.
We accounted for within-family correlations for members of COPD pedigrees in Costa Rica using a kinship coefficient matrix, and generalized estimation equation (GEE) models were used for the analysis of COPD (a binary outcome) using the imputed genotype probabilities (dosage) generated by MaCH-Admix. All multivariate analyses were adjusted for age, sex, principal components of ancestry (to account for population stratification), smoking status (ever vs. never), and intensity of cigarette smoking (in pack-years).
For MESA samples, the KING program (24) was used to perform relationship inference and construct an unrelated subset of individuals for analysis. Analysis of these unrelated individuals was performed under an additive genetic model, using a score test accounting for imputed genotype probabilities as implemented in SNPTEST v2.2.0 (31). Regression analyses were adjusted for study site and the same covariates as described in the analysis of Costa Rican samples.
Results from Costa Rica and MESA were combined in an inverse variance weighted metaanalysis using METAL (32). All other analyses were conducted using the R package (www.r-project.org, version 2.15.0) with R package GWAF (33) and in-house R functions.
Stage 2: follow-up studies in Hispanic and non-Hispanic populations
Of the top 10 SNPs from the stage 1 metaanalysis, nine had an minor allele frequency greater than or equal to 5% and were carried forward for replication in the Lovelace Smokers Cohort study. Logistic regression was used to assess the association between SNPs and risk for COPD in the Lovelace Smokers Cohort, with adjustment for age, sex, smoking status, intensity of cigarette smoking, and principal components. Results were combined with the results from the two cohorts in stage 1 using inverse variance weighting using METAL to create a joint P value (32). After completing our analysis in Hispanic subjects only, we attempted to replicate our results in a cohort of non-Hispanic white subjects and African American subjects from the COPDGene study (22).
Results
Top Associations
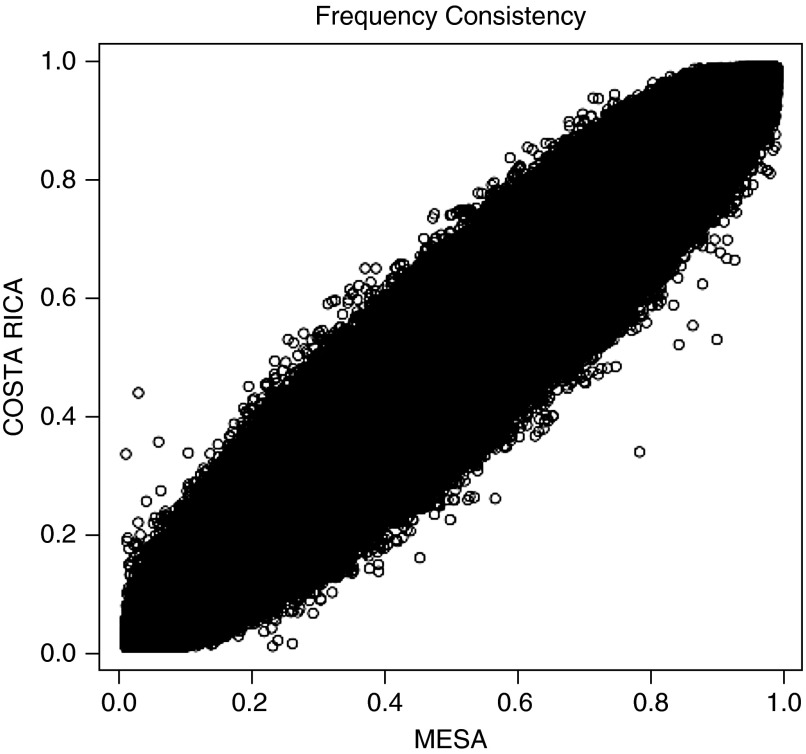
The characteristics of Hispanic cases and control subjects in the two primary cohorts (Costa Rica and MESA) are presented in Table 1. In addition to expected differences in lung function and smoking rates, subjects with COPD were more likely to be older, to be men, and to have a greater intensity of cigarette smoking but lower FEV1 and FEV1/FVC than control subjects. Figure 1 shows the overall consistency of allelic frequencies among Hispanics in the Costa Rican and MESA cohorts. The mean proportions of estimated ancestries for the Costa Rican cohort were: 62% European, 3% African, and 35% Native American. Among Hispanics in MESA, the mean proportions of estimated ancestries were: 52% European, 16% African, and 32% Native American. The plots of first two principal components are shown in Figure E1 in the online supplement.
Table 1.
Characteristics of Hispanic cases and control subjects in the Costa Rican and Multi-Ethnic Study of Atherosclerosis cohorts
| Covariate | Costa Rica |
MESA Hispanics |
||
|---|---|---|---|---|
| COPD |
Control Subjects |
COPD |
Control Subjects |
|
| (N = 94) | (N = 412) | (N = 63) | (N = 619) | |
| Age, yr | 60.1 (9.2)* | 42.6 (14.8) | 67.7 (9.4)* | 62.8 (9.5) |
| Sex, male | 58 (61.7%)* | 192 (46.6%) | 40 (63.5%)* | 268 (43.3%) |
| Height, cm | 161.8 (9.8) | 163.2 (8.8) | 165.3 (11.4) | 161.3 (8.9) |
| BMI, kg/m2 | 24.5 (5.2)* | 26.4 (4.8) | 29.4 (5.6) | 29.3 (4.9) |
| Smoking, ever | 94 (100%)* | 253 (61.9%) | 42 (66.7%)* | 242 (39.1%) |
| Smoking, current | 39 (41.5%) | 153 (37.4%) | 8 (12.7%) | 44 (7.1%) |
| Pack-years of smoking | 56.6 (35.0)* | 18.8 (25.6) | 22.5 (30.2)* | 6.2 (13.7) |
| Pre-BD FEV1, L | 1.18 (0.52)* | 3.20 (0.83) | 1.79 (0.67)* | 2.59 (0.69) |
| Post-BD FEV1, L | 1.27 (0.52)* | 3.32 (0.89) | NA | NA |
| Pre-BD FVC, L | 2.18 (0.81)* | 3.91 (0.97) | 2.93 (0.97)* | 3.26 (0.88) |
| Post-BD FVC, L | 2.34 (0.78)* | 3.97 (1.02) | NA | NA |
| Pre-BD FEV1/FVC | 0.54 (0.11)* | 0.82 (0.06) | 0.61 (0.09)* | 0.79 (0.04) |
| Post-BD FEV1/FVC | 0.54 (0.11)* | 0.84 (0.05) | NA | NA |
Definition of abbreviations: BD = bronchodilator; BMI = body mass index; COPD = chronic obstructive pulmonary disease; MESA = Multi-Ethnic Study of Atherosclerosis; NA = not applicable.
Mean (SD) and count (%) are shown for continuous and binary variables, respectively.
P < 0.05 for comparison between cases and control subjects.
Figure 1.
Comparison of allelic frequencies among Hispanics in the Costa Rica and Multi-Ethnic Study of Atherosclerosis (MESA) cohorts. Frequency of each single-nucleotide polymorphism (SNP) was flipped with the same reference allele. For each SNP, the frequency from Costa Rica is plotted against that from MESA.
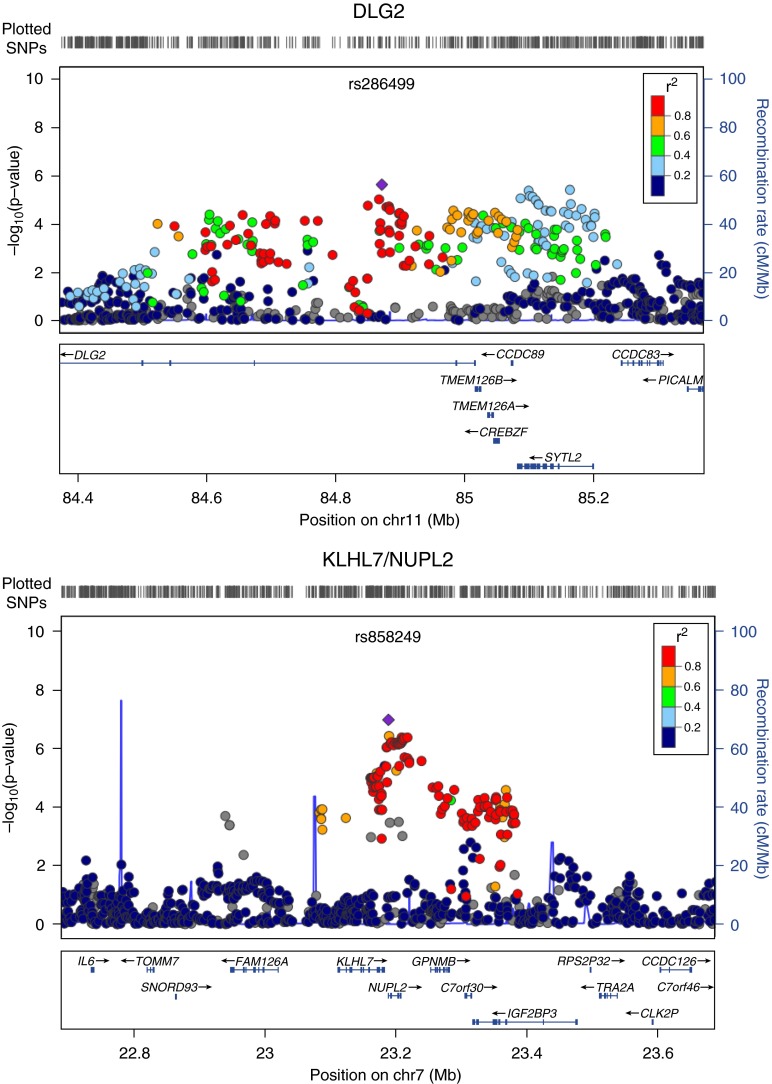
Figure 2 shows the genome-wide association signals from the metaanalysis of the GWAS in the Costa Rican and MESA cohorts (stage 1). Nine SNPs were significant in this discovery stage at P < 5 × 10−6 (Table 2), and all but one were nominally significant with the same direction of effect in both cohorts. Figure 3 shows the regional association plots for the two most significant results from this metaanalysis. The SNP most significantly associated with COPD in the stage 1 metaanalysis was rs858249, which lies between the genes nucleoporin like 2 (NUPL2) and kelch-like family member 7 (KLHL7) (P = 1.1 × 10−7). This SNP had a similar magnitude of effect in both primary cohorts. When combined with the results from the analysis among Hispanics in the Lovelace Smokers Cohorts in the stage 2 replication analysis (in which the effect was in the same direction but not statistically significant), the metaanalysis P value of all three Hispanic cohorts approached genome-wide significance (Stage 2 P value = 6.1 × 10−8). We then tested whether this SNP was associated with expression of KLHL7 in an independent dataset of adult lung tissue samples. We found that the minor allele was associated with increased expression of KLHL7 in current smokers and decreased expression of KLHL7 in never smokers (P = 0.009 for both groups).
Figure 2.
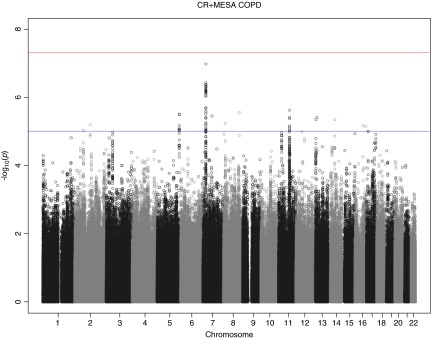
Manhattan plot for results from metaanalysis. A genome-wide plot of association significance for all single-nucleotide polymorphisms with minor allele frequency > 5%. The red and blue lines are corresponding to the threshold P = 5 × 10−8 and P = 1 × 10−5. COPD = chronic obstructive pulmonary disease; CR = Costa Rica; MESA = Multi-Ethnic Study of Atherosclerosis.
Table 2.
Top results of metaanalysis of chronic obstructive pulmonary disease in Hispanics in the Costa Rica, Multi-Ethnic Study of Atherosclerosis, and Lovelace cohorts
| SNP | Chr. | Position (Hg18) | Reference/Alternate Alleles | Reference Allele Frequency | Primary Cohorts |
Replication cohort | Combined Metaanalysis P Value‡ | Genes within 20kb§ | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Costa Rica β (P Value)* (N = 94/412) | MESA β (P Value)* (N = 63/619) | Primary Cohorts Metaanalysis P Value† | Lovelace β (P Value)* (N = 47/290) | |||||||
| rs858249 | 7 | 23187164 | A/G | 0.91 | −1.2 (1.6 × 10−5)|| | −1.1(0.0019)a | 1.1 × 10−7 | −0.84 (0.26)|| | 6.1 × 10−8 | KLHL7(+5.601kb)|NUPL2(-0.8kb) |
| rs286499 | 11 | 84871517 | G/T | 0.86 | −0.7 (0.024)¶ | −1.2 (1.5 × 10−5)¶ | 2.4 × 10−6 | −1.2 (0.0096)|| | 8.4 × 10−8 | DLG2(0) |
| rs11625461 | 14 | 51473434 | C/T | 0.89 | −1.1 (0.0087)¶ | −1.5 (1.2 × 10−4)|| | 4.6 × 10−6 | −0.63 (0.15)|| | 3.6 × 10−6 | GNG2(0) |
| rs17504027 | 13 | 29485544 | C/T | 0.94 | −1.2 (6.7 × 10−4)|| | −1.5 (0.0015)|| | 4.0 × 10−6 | −0.23 (0.67)|| | 2.0 × 10−5 | . |
| rs4638420 | 13 | 20770348 | G/A | 0.86 | −0.76 (0.0013)|| | −0.85 (0.0011)¶ | 4.6 × 10−6 | −0.13 (0.67)|| | 2.5 × 10−5 | . |
| rs652615 | 11 | 85163098 | C/T | 0.59 | −0.62 (0.0068)|| | −0.84 (1.4 × 10−4)¶ | 3.9 × 10−6 | −0.15 (0.54)|| | 2.5 × 10−5 | SYTL2(+16.41kb) |
| rs9313521 | 5 | 170083744 | C/A | 0.93 | −1 (0.011)¶ | −1.4 (7.8 × 10−5)¶ | 3.2 × 10−6 | 0.2 (0.61)|| | 0.00035 | KCNIP1(0) |
| rs7791363 | 7 | 69763300 | A/G | 0.68 | −1 (2.8 × 10−6)|| | −0.41 (0.1)¶ | 3.6 × 10−6 | 0.17 (0.57)|| | 0.00016 | AUTS2(0) |
| rs2515033 | 8 | 119564326 | T/C | 0.95 | −1.2 (2.2 × 10−4)|| | −1.1 (0.0039)¶ | 2.8 × 10−6 | 0.46 (0.34)|| | 0.0002 | SAMD12(0) |
Definition of abbreviations: Chr = chromosome; MESA = Multi-Ethnic Study of Atherosclerosis; SNP = single-nucleotide polymorphism.
Beta estimate (P value) presented for each cohort. N is number of cases/control subjects in each cohort.
Metaanalysis P value for Costa Rica and MESA only.
Metaanalysis P value for Costa Rica, MESA, and Lovelace.
Gene name (distance from the gene in kb); “.” indicates that there is no gene within 20 kb of the SNP.
Genotyping method: directly genotyped.
Genotyping method: imputed.
Figure 3.
Regional plots for the top two loci on chromosome 11 near gene DLG2 and on chromosome 7 near genes KLHL7 and NULPL2. The plots were generated by LocusZoom Software using HapMap II JPT + CHB as the reference panel for linkage disequilibrium information. SNP = single-nucleotide polymorphism.
The second most significant SNP in the stage 1 metaanalysis (rs286499) was also nominally significant in both cohorts in the same direction. In the metaanalysis combining the results in all three Hispanic cohorts it also approached genome-wide significance (Stage 2 P value = 8.4 × 10−8). This SNP is located in an intron of the gene Discs, Large Homolog 2 (DLG2) and was not significantly associated with expression of this gene in adult lung tissue samples of current or never smokers. Because DLG2 may play a role in stabilizing nicotinic acetylcholine receptors at neuronal synapses (34), we tested whether rs286499 is associated with smoking behavior. Although we found that the G allele that was associated with decreased COPD risk was associated with increased pack-years of smoking in both MESA and Cost Rica, this association became markedly attenuated and nonstatistically significant in a model restricted to subjects with at least 10 pack-years of smoking (with P values of 0.23 and 0.68 in Costa Rica and MESA, respectively). Neither rs286499 nor rs858249 was associated with COPD in non-Hispanic white subjects or African Americans in the COPDGene Study (Table E2). Notably, both of these SNPs are rare in CEU in the HapMap Project, but common in YRI and JPT + HCB in HapMap.
Previously Published Associations
Next, we examined whether SNPs in previously identified susceptibility genes for COPD (e.g., in FAM13A, HHIP, and CHRNA3) were associated with COPD among Hispanics in the Costa Rican and MESA cohorts. We downloaded previously reported SNPs in COPD studies (4, 5, 25, 35–37) from National Institutes of Health GWAS Catalog (as of November 12, 2014; Table E3) (38). Although our sample size is much smaller than those studies, we found consistent results in several genes. A full list of comparisons can be found in Table E4. The two most significant SNPs in the first published report of FAM13A (rs1903003, rs7671167) (5) were also significantly associated with COPD in the metaanalysis of GWAS from Costa Rica and MESA (P = 0.02 and 0.04, respectively, in the same direction of association as that previously reported). In contrast, we did not find significant replication of the top previously reported SNPs in HHIP and CHRNA3, although they are in the same direction of association. A recent report showed that multiple SNPs are associated with the levels of two circulatory biomarkers of COPD: Clara cell secretory protein (CC16) and surfactant protein D (SP-D) (35). One of the top SNPs in that report (rs3923564) was associated with SP-D in patients with COPD. SNP rs3923564 was associated with COPD in the metaanalysis of GWAS from the Costa Rican and MESA cohorts (P = 1.7 × 10−4, in the same direction of association as previously reported).
Discussion
In this metaanalysis of GWAS of COPD in Hispanics living in Costa Rica and the United States, we identified two novel variants associated with COPD affection status. We also show that an SNP associated with the COPD biomarker SP-D in a non-Hispanic population is associated with COPD in Hispanic cohorts. To our knowledge, this is the first GWAS of COPD conducted exclusively in Hispanic subjects.
The SNP most significantly associated with COPD in this analysis (rs858249) lies within the promoter region of NUPL2. Although not previously associated with COPD, NUPL2 is one of the top genes overexpressed in macrophage-like cells treated with nicotine (39). This gene is involved in mRNA transport out of the nucleus and is ubiquitously expressed (40). Although we did not find an association between rs858249 and expression of NUPL2 in the lung, this SNP was associated with KLHL7 expression (P = 0.08) in the nucleus accumbens (a region of the brain that may play a role in nicotine addiction) (41) and in lung tissue (from current and never smokers, albeit in opposite directions, P < 0.01 in both instances). This suggests that this gene may be differentially expressed based on smoking behavior. KLHL7 is ubiquitously expressed, with highest expression in the brain, heart, and testis (27). It appears to play a role in the proteasome-degradation pathway (42).
The second most significant SNP in our analysis (rs286499) is in an intron of DLG2, a gene that may stabilize nicotinic acetylcholine receptors at neuronal synapses (34). These are the same types of synapses that are encoded by the COPD susceptibility gene CHRNA3 (4), which may also play a role in nicotine addiction. In support of this hypothesis, the G allele of rs286499 is associated with expression of DLG2 in the substantia nigra and frontal cortex (27) and with decreased risk of COPD in MESA and Costa Rica. Although our finding for COPD may be due to effects on smoking behavior, we had limited statistical power to test for association between SNP rs286499 and pack-years of smoking in Costa Rica or MESA, given the relatively small number of subjects with significant smoking history (≥10 pack-years) in either cohort. Whether SNP rs286499 influences smoking behavior should thus be examined in future studies.
Our results highlight the importance of studying diverse populations. The minor alleles for both our top SNPs in the current study are associated with increased COPD risk. Because these alleles are rare in Europeans, they may reflect unique risk factors for COPD in Hispanic populations. Other genome-wide studies in non-European populations have revealed novel loci even in well-studied diseases such as type 2 diabetes mellitus (43). Conversely, associations identified in European-ancestry populations may be less apparent in non-European populations due to differences in linkage disequilibrium and allele frequencies (44). Both of these effects are apparent in this study, in which we identified novel associations with strong effects in Hispanic populations but found that the replication results in non-Hispanics were only nominally significant. Studies of diverse populations are necessary to comprehensively identify the genetic variability that contributes to COPD susceptibility.
Our study has several limitations. First, our top two results approached genome-wide significance after inclusion of the Lovelace replication cohort, but only one SNP showed nominal significance in Lovelace. Further replication to confirm these findings is needed. Second, we cannot draw negative inferences about nonsignificant findings due to small sample size. Third, our results are cross-sectional, so we may have misclassified some subjects who are currently healthy but will go on to develop COPD. We may have also misclassified a small subgroup of subjects who have nonreversible airflow obstruction due to asthma, who would be expected to have different genetic determinants than subjects without asthma. These types of misclassification would reduce our statistical power to detect true associations but should not bias the results. Fourth, we do not have the statistical power to explore interactions with tobacco smoke exposure, a potentially important source of “missing heritability.” Finally, “Hispanic” is a broad term that encompasses individuals with complex and varied racial admixture (45). In the current study, members of the Costa Rican and Lovelace cohorts had lower proportions of African ancestry but higher proportions of European ancestry than Hispanics in MESA, which may have reduced our statistical power. In addition, participants in all three Hispanic cohorts had more than 30% Native American ancestry, and thus our findings may not be generalizable to Hispanic populations with a lower proportion of Native American ancestry (e.g., Puerto Ricans).
In summary, we have identified two novel COPD susceptibility loci in Hispanics, which are plausibly associated with nicotine addiction. We have also shown that SNPs in a known susceptibility gene for COPD (FAM13A) and a gene encoding a COPD biomarker (SP-D) in non-Hispanics are associated with COPD per se in Hispanics. Further studies are needed to determine how these variants influence gene expression and translation in relevant tissues.
Acknowledgments
Acknowledgment
The authors thank the participants, families, investigators and study staff from Costa Rica, MESA, Lovelace, and COPDGene studies. Analyses were conducted at Children’s Hospital of Pittsburgh of UPMC under local guidelines and policies.
Footnotes
The Genetics of COPD in Costa Rica Study was supported by grant HL073373 from the U.S. National Institutes of Health (NIH). Dr. Brehm’s contribution was supported by grant K08HL111201 from the NIH. The Multi-Ethnic Study of Atherosclerosis (MESA) Lung/SHARe Study was funded by NIH grant RC1HL100543 and R01-HL077612. MESA and the MESA SHARe project are conducted and supported by contracts N01-HC-95159 through N01-HC-95169 and RR-024156 from the National Heart, Lung, and Blood Institute (NHLBI) and RR024156 and ES09089. MESA Air is conducted and supported by the United States Environmental Protection Agency in collaboration with MESA Air investigators, with support provided by grant RD83169701. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-6–4278, the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Lovelace Smokers Cohort Study was supported by grant HL068111 from the NIH. The Genetic epidemiology of COPD (COPDGene) project was supported by award number R01HL089897 and award number R01HL089856 from the NHLBI (see the Appendix in the online supplement for this manuscript). The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board composed of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, GlaxoSmithKline, and Sunovion.
Author Contributions: Conception and design: Y.T., R.G.B., and J.C.C; data analysis and interpretation: W.C., J.M.B., A.M., M.H.C., E.K.S., Q.Y., N.B., K.M.B., H.P., S.L., M.O., Y.B., K.H., R.P., S.S.R., R.G.B., and J.C.C; drafting the manuscript for intellectual content: W.C., J.M.B., A.M., P.L.E., J.I.R., C.-A.B., L.A., M.S.-Q., Y.T., R.G.B., and J.C.C. All authors participated in the review of the manuscript and approved its final version.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lopez AD, Shibuya K, Rao C, Mathers CD, Hansell AL, Held LS, Schmid V, Buist S. Chronic obstructive pulmonary disease: current burden and future projections. Eur Respir J. 2006;27:397–412. doi: 10.1183/09031936.06.00025805. [DOI] [PubMed] [Google Scholar]
- 2.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am J Respir Crit Care Med. 2013;188:941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman EK. Perspective: how can genetics help? Nature. 2012;489:S7. doi: 10.1038/489S7a. [DOI] [PubMed] [Google Scholar]
- 4.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. ICGN Investigators. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B, Zhou H, Yang J, Xiao J, Liang B, Li D, Zhou H, Zeng Q, Fang C, Rao Z, et al. Association of HHIP polymorphisms with COPD and COPD-related phenotypes in a Chinese Han population. Gene. 2013;531:101–105. doi: 10.1016/j.gene.2013.08.069. [DOI] [PubMed] [Google Scholar]
- 7.Wang B, Liang B, Yang J, Xiao J, Ma C, Xu S, Lei J, Xu X, Liao Z, Liu H, et al. Association of FAM13A polymorphisms with COPD and COPD-related phenotypes in Han Chinese. Clin Biochem. 2013;46:1683–1688. doi: 10.1016/j.clinbiochem.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Qiu F, Lu X, Huang D, Ma G, Guo Y, Hu M, Zhou Y, Pan M, Tan Y, et al. Functional polymorphisms of CHRNA3 predict risks of chronic obstructive pulmonary disease and lung cancer in Chinese. PLoS ONE. 2012;7:e46071. doi: 10.1371/journal.pone.0046071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossé Y. Updates on the COPD gene list. Int J Chron Obstruct Pulmon Dis. 2012;7:607–631. doi: 10.2147/COPD.S35294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, Budoff M, Austin JH, Washko GR, Carr JJ, et al. Genome-wide study of percent emphysema on computed tomography in the general population: The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menezes AM, Perez-Padilla R, Jardim JR, Muiño A, Lopez MV, Valdivia G, Montes de Oca M, Talamo C, Hallal PC, Victora CG PLATINO Team. Chronic obstructive pulmonary disease in five Latin American cities (the PLATINO study): a prevalence study. Lancet. 2005;366:1875–1881. doi: 10.1016/S0140-6736(05)67632-5. [DOI] [PubMed] [Google Scholar]
- 12.Lisker R, Babinsky V. Admixture estimates in nine Mexican Indian groups and five East Coast localities. Rev Invest Clin. 1986;38:145–149. [PubMed] [Google Scholar]
- 13.Sans M, Salzano FM, Chakraborty R. Historical genetics in Uruguay: estimates of biological origins and their problems. Hum Biol. 1997;69:161–170. [PubMed] [Google Scholar]
- 14.Ferris B. G. Epidemiology standardization project (American Thoracic Society) Am Rev Respir Dis. 1978;118:1–88. [PubMed] [Google Scholar]
- 15.American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 16.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman JD, Adar SD, Allen RW, Barr RG, Budoff MJ, Burke GL, Casillas AM, Cohen MA, Curl CL, Daviglus ML, et al. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air) Am J Epidemiol. 2012;176:825–837. doi: 10.1093/aje/kws169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Crapo R, Enright P, van der Grinten CP, Gustafsson P, et al. ATS/ERS Task Force. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez J, Jiang R, Johnson WC, MacKenzie BA, Smith LJ, Barr RG. The association of pipe and cigar use with cotinine levels, lung function, and airflow obstruction: a cross-sectional study. Ann Intern Med. 2010;152:201–210. doi: 10.1059/0003-4819-152-4-201002160-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hankinson JL, Kawut SM, Shahar E, Smith LJ, Stukovsky KH, Barr RG. Performance of American Thoracic Society-recommended spirometry reference values in a multiethnic sample of adults: the multi-ethnic study of atherosclerosis (MESA) lung study. Chest. 2010;137:138–145. doi: 10.1378/chest.09-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood A, Stidley CA, Picchi MA, Celedón JC, Gilliland F, Crowell RE, Belinsky SA, Tesfaigzi Y. Difference in airflow obstruction between Hispanic and non-Hispanic White female smokers. COPD. 2008;5:274–281. doi: 10.1080/15412550802363345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 24.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium GT GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu EY, Li M, Wang W, Li Y. MaCH-admix: genotype imputation for admixed populations. Genet Epidemiol. 2013;37:25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker MJ, Zhao S, Bredt DS, Sanes JR, Feng G. PSD93 regulates synaptic stability at neuronal cholinergic synapses. J Neurosci. 2004;24:378–388. doi: 10.1523/JNEUROSCI.3865-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DK, Cho MH, Hersh CP, Lomas DA, Miller BE, Kong X, Bakke P, Gulsvik A, Agustí A, Wouters E, et al. ECLIPSE, ICGN, and COPDGene Investigators. Genome-wide association analysis of blood biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186:1238–1247. doi: 10.1164/rccm.201206-1013OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansel NN, Ruczinski I, Rafaels N, Sin DD, Daley D, Malinina A, Huang L, Sandford A, Murray T, Kim Y, et al. Genome-wide study identifies two loci associated with lung function decline in mild to moderate COPD. Hum Genet. 2013;132:79–90. doi: 10.1007/s00439-012-1219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siedlinski M, Cho MH, Bakke P, Gulsvik A, Lomas DA, Anderson W, Kong X, Rennard SI, Beaty TH, Hokanson JE, et al. COPDGene Investigators; ECLIPSE Investigators. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66:894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koshi R, Sugano N, Orii H, Fukuda T, Ito K. Microarray analysis of nicotine-induced changes in gene expression in a macrophage-like human cell line. J Periodontal Res. 2007;42:518–526. doi: 10.1111/j.1600-0765.2007.00976.x. [DOI] [PubMed] [Google Scholar]
- 40.Farjot G, Sergeant A, Mikaélian I. A new nucleoporin-like protein interacts with both HIV-1 Rev nuclear export signal and CRM-1. J Biol Chem. 1999;274:17309–17317. doi: 10.1074/jbc.274.24.17309. [DOI] [PubMed] [Google Scholar]
- 41.David SP, Munafò MR, Johansen-Berg H, Smith SM, Rogers RD, Matthews PM, Walton RT. Ventral striatum/nucleus accumbens activation to smoking-related pictorial cues in smokers and nonsmokers: a functional magnetic resonance imaging study. Biol Psychiatry. 2005;58:488–494. doi: 10.1016/j.biopsych.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kigoshi Y, Tsuruta F, Chiba T. Ubiquitin ligase activity of Cul3-KLHL7 protein is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J Biol Chem. 2011;286:33613–33621. doi: 10.1074/jbc.M111.245126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, Takeuchi F, Wu Y, Go MJ, Yamauchi T, et al. DIAGRAM Consortium; MuTHER Consortium. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, Schumacher FR, Peters U, Franceschini N, Ritchie MD, et al. PAGE Consortium. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11:e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brehm JM, Celedón JC. Chronic obstructive pulmonary disease in Hispanics. Am J Respir Crit Care Med. 2008;177:473–478. doi: 10.1164/rccm.200708-1274PP. [DOI] [PMC free article] [PubMed] [Google Scholar]