Abstract
Aims
Differentiation of heart failure with reduced (HFrEF) or preserved (HFpEF) ejection fraction independent of echocardiography is challenging in the community. Diagnostic strategies based on monitoring circulating microRNA (miRNA) levels may prove to be of clinical value in the near future. The aim of this study was to identify a novel miRNA signature that could be a useful HF diagnostic tool and provide valuable clinical information on whether a patient has HFrEF or HFpEF.
Methods and results
MiRNA biomarker discovery was carried out on three patient cohorts, no heart failure (no-HF), HFrEF, and HFpEF, using Taqman miRNA arrays. The top five miRNA candidates were selected based on differential expression in HFpEF and HFrEF (miR-30c, −146a, −221, −328, and −375), and their expression levels were also different between HF and no-HF. These selected miRNAs were further verified and validated in an independent cohort consisting of 225 patients. The discriminative value of BNP as a HF diagnostic could be improved by use in combination with any of the miRNA candidates alone or in a panel. Combinations of two or more miRNA candidates with BNP had the ability to improve significantly predictive models to distinguish HFpEF from HFrEF compared with using BNP alone (area under the receiver operating characteristic curve >0.82).
Conclusion
This study has shown for the first time that various miRNA combinations are useful biomarkers for HF, and also in the differentiation of HFpEF from HFrEF. The utility of these biomarker combinations can be altered by inclusion of natriuretic peptide. MiRNA biomarkers may support diagnostic strategies in subpopulations of patients with HF.
Keywords: MicroRNA, Heart failure, Ejection fraction, Natriuretic peptide, Biomarker, Diagnosis
Translational perspective
Diagnosis of heart failure (HF) remains challenging. HF with preserved ejection fraction (HFpEF) and reduced ejection fraction HF (HFrEF) cannot be readily differentiated on clinical grounds but requires additional imaging tests, which are not frequently used in patients with a community diagnosis. We identified a simple reverse transcription–PCR (RT–PCR)-based biomarker approach improving diagnosis of HFpEF vs. HFrEF. Correct identification will be important as there is a great need for stratifying HF patient cohorts for different management and potential novel future upcoming treatment regimes.
Introduction
Heart failure (HF) has reached epidemic proportions, affecting ∼1–3% of the population within European countries, amounting to 15 million people.1 The syndrome is characterized by significant morbidity and reduced life expectancy, with 5-year mortality of newly diagnosed HF as high as 50%2 and 10-year survival of 26.7%.3 Effective management of HF depends on an accurate and rapid diagnosis. Currently, this requires assessment of symptoms and physical signs in combination with advanced and expensive imaging of cardiac structure and function. Symptom-based diagnostic challenges occur as co-morbidities of HF have similar presentations, and practical challenges are posed by the fact that the majority of HF cases are found in the community setting, where imaging equipment and expertise are not readily available.4 In addition, it has been well demonstrated that HF with preserved ejection fraction (HFpEF) and HF with reduced ejection fraction (HFrEF) cannot be readily differentiated on clinical grounds and requires assessment by these imaging tests.5 This barrier to effective diagnosis and work-up of heart failure is underscored by data from the UK indicating that more than half of patients with a community diagnosis of HF do not have echocardiography performed and therefore have a ‘questionable diagnosis’.6 Having the ability to diagnose HF more confidently within the community and to be able to differentiate between HFpEF and HFrEF would be of significant clinical value as the management of these conditions differs and patients could be more readily triaged while waiting for extensive clinical work-up within the cardiology department in hospitals.
Given that the emerging opinion is that the pathophysiology of HFpEF and HFrEF is different,7 it provides an opportunity to identify biomarker candidates that could aid in HF diagnosis and stratification between these two forms of the disease. Natriuretic peptide (NP) is an effective diagnostic biomarker used to support the diagnosis of HF in the community and is generally accepted to be the most effective biomarker for this purpose presently available.8,9 However, despite its widespread clinical use, NP is still encumbered by reduced specificity, and, furthermore, while modest differences have been shown to exist between HFrEF and HFpEF, it does not behave in a significantly distinct manner to allow accurate differentiation of these subtypes of heart failure.10,11 Therefore a novel biomarker approach is required instead of or to work in tandem with NP. The aim of this project was to identify a novel circulating biomarker signature that could be used either alone or in conjunction with established diagnostic tests such as NPs, to identify HF and help differentiate HFpEF and HFrEF. Unlike other studies, we focused on the analysis of circulating microRNAs (miRNAs) which comprise a class of small, non-coding RNAs that control expression of complementary target mRNAs. Altered miRNA expression patterns have been shown to reflect pathological processes including those which are cardiovascular related, and have a realistic potential to serve as diagnostic biomarkers in these diseases.12,13 The discovery of altered disease-specific miRNA signatures that could be used as a novel diagnostic biomarker test for HF classification have yet to be defined. These could be used either alone or in combination with current diagnostic tests, including NPs, to improve diagnostic accuracy. Disease-specific miRNA signatures may also provide insight into underlying disease mechanisms and pose as therapeutic targets.
Methods
Patient population
The total study population involved 270 patients from St. Vincent's University Hospital, Dublin. This population consisted of 90 HFpEF, 90 HFrEF, and 90 asymptomatic patients (no-HF) with cardiovascular risk factors for future heart failure development. The asymptomatic patients were recruited from the STOP-HF programme (The St Vincent's Screening to Prevent Heart Failure Study), which has recently been described in detail.14 The 180 stable HF patients were recruited from the Heart Failure Unit.
To be eligible for inclusion in the no-HF cohort, patients must have been deemed asymptomatic following assessment at the time of presentation by an experienced attending cardiologist and were required to have had an EF ≥50% at echocardiography. This population were aged ≥40 years and had at least one risk factor for LV dysfunction including type 2 diabetes mellitus (DM), hypertension, obesity, hypercholesterolaemia, orCAD.
To be eligible for inclusion in the HFpEF cohort, patients were required to have had a hospitalization for proven NYHA class IV HF (confirmed by an attending cardiologist), continued symptoms of at least NYHA class II HF, and an LVEF ≥50% with Doppler abnormalities of diastolic dysfunction but no Doppler echocardiographic evidence of significant valvular heart disease. The HFrEF cohort also had to have a prior hospitalization for HF with an LVEF <50% and continued to have at least NYHA class II symptoms.
All subjects gave written informed consent to participate in the study. The study protocol was approved by the ethics committee of St. Vincent's University Hospital, which conformed to the principles of the Declaration of Helsinki.
Clinical assessment
All study patients underwent a full history and physical examination, NYHA functional class assessment, and phlebotomy. Physical examination included assessment of waist circumference, calculation of body mass index (BMI), and heart rate and blood pressure measurement. Peripheral venous blood samples were tested for levels of creatinine, glucose, and lipid profile. Point of care BNP was measured using a Triage meter (Biosite). Assay sensitivity for BNP was 5 ng/mL. Serum samples were obtained from venous blood following clot formation and centrifugation at 2500 g for 10 min. Samples were aliquoted and stored at −80 °C until required. All patients had Doppler echocardiography performed.
Doppler echocardiography
Doppler echocardiographic assessment was performed by one of two blinded operators in accordance with the European Society of Echocardiography recommendations. All data represent the mean of three measurements on sequential cardiac cycles. The LV mass was calculated using the Devereux method and was indexed to body surface area. Left atrial volume was calculated using the biplane area length method and was also indexed to body surface area. The LV filling pressures were non-invasively assessed with tissue Doppler measurements taken at the lateral mitral annulus. The LVEF was calculated by the Teichholz method.
Classification of LV diastolic dysfunction was based on having either a left atrial volume index (LAVI) ≥34 mL/m2 or a tissue lateral e' value ≤10 cm/s.15
MicroRNA analysis
Biomarker discovery and independent validation
Total RNA was extracted from 100 µL of serum collected from each of the 270 study patients (three patient cohorts representing n = 90 per group of no-HF, HFrEF, and HFpEF) using a Qiagen miRNeasy 96 kit as per the manufacturer's instructions.
A miRNA expression analysis was initially carried out on the three patient cohorts with samples pooled from n = 15 per group using Taqman array Human MicroRNA card set v3 (Applied Biosystems, Life Technologies #4444913) which has a 745 microRNA Taqman probe on a 384-well microfluidic card (Applied Biosystems). MiRNAs with fold change ≥2 compared with no-HF and those which could differentiate HFpEF and HfrEF were selected, and among them top candidates with higher fold change and expression were selected for validation. Array data were analysed using Data Assist software. Five miRNA candidates that showed potential as a HF diagnostic that could differentiate HFpEF and HFrEF were selected and further verified in the same patient population using Taqman miRNA assays (Applied Biosystems). The diagnostic utility of the five selected miRNAs was then further validated in the remaining 225 patients (n = 75 per group).
Statistical analysis
All analyses were carried out using R statistical software version 2.7.2. The four outcomes tested were (i) HF present/absent; (ii) HFpEF/HFrEF (within an HF-present subsample); (iii) EF; and (iv) LAVI. The first two (binary) outcomes were analysed using Generalized Linear Modelling with a binomial outcome; the latter two outcomes were analysed using ordinary linear regression with transformations.
For each miRNA, the initial process of analysis was as follows: (i) run the single variable linear model; (ii) run the same model adjusted for BNP (log-transformed); (iii) run the same model adjusted for age and gender; and (iv) run the same model adjusted for age, gender, and BNP. For the binary outcomes, the area under the curve (AUC) is reported as a measure of fit, and for the continuous outcomes, the R2 is reported. Each of the models tested in steps (ii), (iii), and (iv) above were then compared with the unadjusted model using a χ2 test on gain in residual sum of squares per additional degree of freedom.
All miRNA values were log-transformed prior to analysis and missing values were handled using listwise deletion—only patients with zero non-missing data are included in the models.
Results
Patient population
The patient population is described in Table 1. All three populations have a mean age >70 years old and had an equivalent gender balance. The average systolic blood pressure (SBP) and diastolic blood pressure (DBP) were higher in the no-HF control group compared with both HF cohorts. This is possibly reflective of different medication usage between the three populations. The HFpEF cohort demonstrates many of the typical characteristics of this HF subtype, being marginally older than the other two cohorts, having a greater BMI, and a high prevalence of hypertension and AF. The HFrEF cohort also demonstrates typical characteristics with dominant ischaemic aetiology. The NP levels are elevated in both HF cohorts as one would anticipate. The lower level of NP in the HFrEF group compared with the HFpEF group might be explained by the impact of effective use of disease-modifying therapy in the HFrEF cohort. Echocardiographic findings are again typical of these groups with normal EF and markedly increased left atrial size in the HFpEF cohort, though this measurement is probably influenced by AF in many of this group. Both HF groups exhibit an increase in LV mass compared with the control group.
Table 1.
Patient characteristics per study group
| Variables | No-HF (n = 75) | HFpEF (n = 75) | HFrEF (n = 75) | P-values |
|---|---|---|---|---|
| Age, years | 72 ± 9 | 75 ± 7 | 70 ± 11 | 0.008‡ |
| Male, % | 44 (59%) | 46 (61%) | 55 (73%) | 0.15 |
| SBP, mmHg | 136 ± 20 | 124 ± 21 | 116 ± 21 | <0.001*† |
| DBP, mmHg | 81 ± 13 | 74 ± 12 | 70 ± 10 | <0.01* <0.001† |
| Heart rate, b.p.m. | 67 ± 10 | 70 ± 13 | 67 ± 13 | 0.45 |
| Body mass index, kg/m2 | 28 ± 4 | 31 ± 6 | 28 ± 4 | <0.001*‡ |
| Hypertension, % | 60 (80%) | 63 (84%) | 25 (33%) | <0.001†‡ |
| Diabetes mellitus, % | 14 (19%) | 23 (31%) | 11 (15%) | 0.02‡ |
| Dyslipidaemia, % | 47 (63%) | 47 (63%) | 56 (75%) | 0.20 |
| Ischaemic heart disease, % | 19 (25%) | 35 (47%) | 48 (64%) | 0.007* <0.001† 0.03‡ |
| Atrial fibrillation, % | 9 (12%) | 60 (80%) | 43 (57%) | <0.001*† 0.003‡ |
| BNP, pg/mL | 32 (15–70) | 215 (126–353) | 139 (71–254) | <0.001*† <0.05‡ |
| Medications | ||||
| ACE inhibitor or ARB, % | 47 (63%) | 52 (70%) | 68 (91%) | <0.001†‡ |
| Beta-blocker, % | 28 (37%) | 65 (87%) | 60 (80%) | <0.001*† |
| Calcium channel blocker, % | 24 (32%) | 24 (32%) | 4 (5%) | <0.001†‡ |
| Diuretic, % | 32 (43%) | 70 (93%) | 55 (73%) | <0.001*† 0.002‡ |
| Statin, % | 51 (68%) | 46 (61%) | 51 (68%) | 0.61 |
| Antiplatelet, % | 43 (57%) | 38 (51%) | 46 (61%) | 0.41 |
| Warfarin, % | 5 (7%) | 23 (31%) | 29 (39%) | <0.001*† |
| Echocardiography | ||||
| Ejection fraction, % | 66 ± 9 | 62 ± 7 | 36 ± 12a | <0.05* <0.001†‡ |
| Interventricular septum, mm | 11.5 ± 1.9 | 12.6 ± 2.7 | 11.5 ± 3.0 | 0.06 |
| Posterior wall, mm | 9.9 ± 1.5 | 11.0 ± 2.2 | 9.7 ± 2.2 | <0.01*‡ |
| LVMI, g/m2 | 95 ± 24 | 114 ± 36 | 126 ± 38 | <0.01* <0.001† |
| LAVI, mL/m2 | 30 ± 11 | 52 ± 19 | 46 ± 14 | <0.001*† |
| Deceleration time, ms | 193 ± 46 | 215 ± 63 | 235 ± 84 | 0.004† |
| e′ (lat), cm/s | 8.3 ± 2.7 | 9.6 ± 2.2 | 8.1 ± 3.7 | <0.05*‡ |
| Peak E, cm/s | 70 ± 18 | 99 ± 24 | 66 ± 23 | <0.001*‡ |
| E/e′ ratio | 9.0 ± 3.1 | 11.1 ± 3.6 | 9.6 ± 4.9 | 0.008* |
Values are mean ± SD, n (%), or median (interquartile range).
P-values for numerical variables are obtained from one-way analysis of variance with Bonferroni post-hoc analysis.
P-values for categorical variables are obtained from contingency χ2 or Fisher's test of the three study groups.
DBP, diastolic blood pressure; E/e', ratio of mitral early diastolic flow velocity over tissue Doppler lateral mitral annular lengthening velocity; HF, heart failure; HFpEF, heart failure with preserved ejection fraction, HFrEF, heart failure with reduced ejection fraction; LAVI, left atrial volume index; LVMI, left ventricular mass index; SBP systolic blood pressure.
*, †, and ‡ indicate significant differences between no-HF and HFpEF, no-HF and HFrEF, and HFpEF and HFrEF groups, respectively.
Ejection fraction values recorded at the time of diagnosis of HFrEF.
Biomarker discovery for heart failure diagnostics using a global microRNA profiling approach
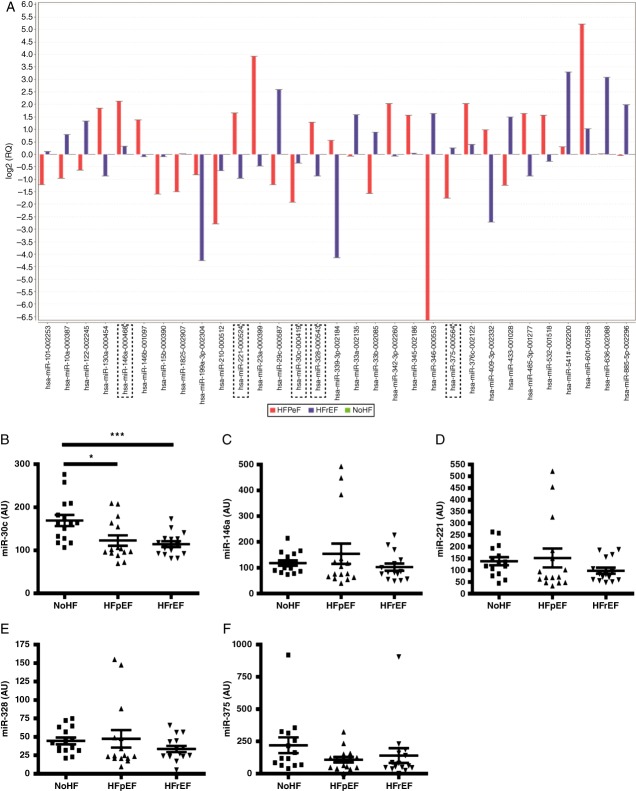
We performed miRNA profiling from the plasma RNA pool of the discovery cohort and found several miRNAs which can differentiate between the three groups (Figure 1). Five miRNA candidates, miR-30c, miR-146a, miR-221, miR-328, and miR-375, were selected from the miRNA screen carried out using the discovery cohort based on high expression levels and high discriminatory capacity. These selected miRNA candidates showed potential utility as biomarkers that could differentiate between HFpEF and HFrEF populations (Figure 1).
Figure 1.
Identification of five microRNA (miRNA) biomarker candidates for heart failure diagnostics. miRNA biomarker discovery was carried out using pooled serum from three cohorts: no heart failure (no-HF); heart failure with preserved ejection fraction (HFpEF); and heart failure with reduced ejection fraction (HFrEF) (A). Five miRNA candidates, miR-30c, miR-146a, miR-221, miR-328, and miR-375, were selected for further analysis and were subsequently quantified in the individual patients from the same discovery cohort (n = 45), by miRNA reverse transcription–PC (RT–PCR (B–E). Data are presented in arbitrary units (AU). Bars represent fold expression changes; data represent the mean ± SEM. **P < 0.01, ***P < 0.001.
Independent validation of the five selected microRNA candidates as heart failure diagnostics
An independent cohort of an additional 225 patients consisting of 75 no-HF, 75 HFpEF, and 75 HFrEF (clinical characteristics described in Table 1) was used to validate the differential expression patterns of the five selected miRNA candidates and to determine whether they had any potential value as HF diagnostics, including the ability to differentiate between HFpEF and HFrEF. Graphical representation of the expression levels of the five miRNA candidates measured within the independent validation cohort is illustrated in Figure 2. In all cases, the average level of the miRNAs was significantly reduced in HF compared with the no-HF control group.
Figure 2.
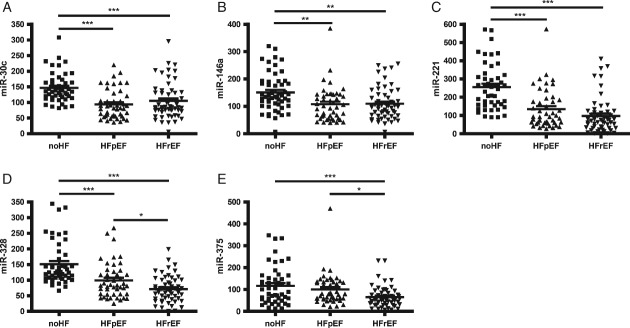
Comparative expression levels of the five microRNA (miRNA) biomarker candidates within the independent validation cohort. The circulating levels of five miRNAs (miR-30c, miR-146a, miR-221, miR-328, and miR-375) were quantified in the three study cohorts;: no heart failure (no-HF); heart failure with preserved ejection fraction (HFpEF); and heart failure with reduced ejection fraction (HFrEF), n = 75 patients per group. Analysis was carried out using reverse transcription–PC (RT–PCR). Data are presented in arbitrary units (AU). Data represent mean the ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
The diagnostic utility of individual miRNA biomarker candidates was initially assessed for HF discrimination followed by differentiation of HFpEF and HFrEF subtypes within the HF population (Table 2).
Table 2.
Statistical modelling to determine the utility of individual microRNA biomarker candidates for heart failure diagnosis and differentiation of heart failure with preserved ejection fraction and heart failure with reduced ejection fraction subtypes
| miRNA | Models: AUC (95% CI)a (χ2
P-value)b |
||||
|---|---|---|---|---|---|
| No covariates | BNP | Age and gender | BNP, age, and gender | ||
| HF or no-HFc | None | – | 0.88 (0.82–0.93) (P < 0.001) | 0.54 (0.46–0.63) (P = 0.45) | 0.91 (0.86–0.95) (P < 0.001) |
| miR-146a | 0.68 (0.60–0.76) | 0.88 (0.83–0.93) (P < 0.001) | 0.67 (0.59–0.75) (P = 0.41) | 0.91 (0.87–0.96) (P < 0.001) | |
| miR-221 | 0.80 (0.73–0.87) | 0.90 (0.85–0.95) (P < 0.001) | 0.80 (0.73–0.87) (P = 0.29) | 0.93 (0.88–0.97) (P < 0.001) | |
| miR-328 | 0.75 (0.68–0.82) | 0.88 (0.82–0.93) (P < 0.001) | 0.74 (0.66–0.81) (P = 0.26) | 0.91 (0.86–0.96) (P < 0.001) | |
| miR-30c | 0.71 (0.63–0.79) | 0.88 (0.82–0.93) (P < 0.001) | 0.68 (0.60–0.76) (P = 0.69) | 0.90 (0.86–0.95) (P < 0.001) | |
| miR-375 | 0.56 (0.47–0.64) | 0.88 (0.82–0.93) (P < 0.001) | 0.55 (0.47,0.64) (P = 0.41) | 0.91 (0.86–0.96) (P < 0.001) | |
| HFpEF or HFrEFd | None | – | 0.65 (0.57–0.74) (P < 0.001) | 0.64 (0.55–0.72) (P = 0.003) | 0.68 (0.60–0.77) (P < 0.001) |
| miR-146a | 0.57 (0.48–0.66) | 0.69 (0.61–0.78) (P < 0.001) | 0.69 (0.60–0.77) (P < 0.001) | 0.71 (0.63–0.80) (P < 0.001) | |
| miR-221 | 0.65 (0.56–0.74) | 0.74 (0.66–0.82) (P < 0.001) | 0.74 (0.66–0.82) (P < 0.001) | 0.77 (0.69–0.85) (P < 0.001) | |
| miR-328 | 0.67 (0.58–0.76) | 0.78 (0.70–0.85) (P < 0.001) | 0.77 (0.69–0.85) (P < 0.001) | 0.80 (0.73–0.88) (P < 0.001) | |
| miR-30c | 0.51 (0.41–0.60) | 0.68 (0.60,0.77) (P < 0.001) | 0.66 (0.57,0.74) (P = 0.001) | 0.69 (0.61,0.78) (P < 0.001) | |
| miR-375 | 0.75 (0.67–0.83) | 0.80 (0.73–0.87) (P < 0.001) | 0.80 (0.72–0.87) (P = 0.001) | 0.82 (0.75–0.89) (P < 0.001) | |
AUC, area under the curve; CI, confidence interval; HFpEF, heart failure with preserved ejection fraction, HFrEF, heart failure with reduced ejection fraction; miRNA, microRNA.
95% CI = AUC ± Z(0.975)SE. Method for estimating SE taken from Hanley and McNeil (1982).16
χ2 test for difference on residual sum of squares compared with no-covariate model.
Logistic model with full sample.
Logistic model with heart failure only subsample.
Two miRNAs (miR-221 and miR-328) showed a reasonable ability to distinguish HF from no-HF, with AUCs of 0.80 and 0.75, respectively. Age and gender did not improve these models, but the addition of BNP significantly improved all to AUC ∼0.90 in all cases. Of note, using BNP alone to predict HF yielded an AUC of 0.875 (Table 3).
Table 3.
Comparisons of single use and combinations of microRNAs as predictors of heart failure and assessing their value by comparing with the clinically accepted biomarker brain natriuretic peptide
| Model description | MicroRNA(s) | AUC (95% CI)a | BNP-only model comparisons |
|
|---|---|---|---|---|
| LR test P-value | IDI (95% CI)b | |||
| Single miRNA | 375 | 0.60 (0.51–0.68) | – | – |
| 146a | 0.72 (0.64–0.79) | – | – | |
| 30c | 0.74 (0.67–0.82) | – | – | |
| 328 | 0.79 (0.72–0.86) | – | – | |
| 221 | 0.81 (0.74–0.88) | – | – | |
| Two miRNA combinationc | 221, 328 | 0.81 (0.75–0.88) | – | – |
| Three miRNA combinationc | 146a, 221, 328 | 0.83 (0.77–0.90) | – | – |
| Four miRNA combinationc | 146a, 221, 328, 30c | 0.83 (0.77–0.90) | – | – |
| All miRNAs | 146a, 221, 328, 375, 30c | 0.84 (0.77–0.90) | – | – |
| Log (BNP) | – | 0.88 (0.82–0.93) | – | – |
| Log (BNP) + single miRNA | 375 | 0.89 (0.83–0.94) | 0.004 | 0.030 (0.003–0.057) |
| 146a | 0.88 (0.82–0.94) | 0.020 | 0.018 (0.000–0.035) | |
| 30c | 0.89 (0.83–0.94) | 0.005 | 0.030 (0.008–0.053) | |
| 328 | 0.90 (0.84–0.95) | <0.001 | 0.053 (0.026–0.080) | |
| 221 | 0.90 (0.85–0.95) | <0.001 | 0.061 (0.028–0.091) | |
| Log (BNP) + two miRNA combinationd | 221, 375 | 0.90 (0.85–0.95) | <0.001 | 0.071 (0.037–0.103) |
| Log (BNP) + three miRNA combinationd | 146a, 221, 375 | 0.91 (0.86–0.96) | <0.001 | 0.081 (0.045–0.122) |
| Log (BNP) + four miRNA combinationd | 146a, 221, 328, 375 | 0.91 (0.86–0.96) | <0.001 | 0.089 (0.054–0.124) |
| Log (BNP) + all miRNAs | 146a, 221, 328, 375, 30c | 0.91 (0.86–0.96) | <0.001 | 0.089 (0.054–0.125) |
AUC, area under the curve; CI, confidence interval; IDI, integrated discrimination improvement; LR, likelihood ratio; miRNA, microRNA.
95% CI = AUC ± Z(.975)SE. Method for estimating SE taken from Hanley and McNeil 1982.16
Bootstrapped confidence interval with 1000 repetitions.
Best model combination according to AUC.
Best model combination according to observed IDI (see Kerr et al. 2011).17
For distinguishing HFpEF from HFrEF, all miRNAs had an AUC >0.7, except for miR-375 (Table 2). Both age/gender and BNP, separately and in combination, significantly improved these models in all cases. MiR-328 and miR-375 had the greatest estimated AUCs for their fully adjusted models at 0.80 and 0.82, respectively.
Following these analyses, we investigated the impact of miRNA biomarker combinations in predicting HF, and their ability to differentiate HFpEF from HFrEF. In addition, with the added value of BNP in the previous models, we also compared the diagnostic models of miRNA combinations with and without BNP, and also compared whether a diagnostic model using miRNAs was superior to BNP models alone. These analyses are detailed in Table 3. In brief, the data from Table 3 indicate that BNP is better than using any miRNA combination as a diagnostic test for HF in general. However, the predictive power of BNP is significantly improved by adding any single miRNA to the analysis, with miR-221 and miR-328 having the greatest effect, as indicated when analysed using integrated discrimination improvement (IDI) and likelihood ratio (LR) tests (bootstrapping and parametric methods of analysis) to determine if one model is better at predictive probability over another model. Using the same analytical approach, a further improvement to the model occurs when examining the addition of miRNA combinations with BNP. However, it is of note that when applying a separate analysis, the 95% confidence interval (CI) for the AUCs would indicate that either miRNAs alone (miR-328 and miR-221) or the miRNA combinations presented are as good as BNP in identifying HF but do not improve the model.
The utility of miRNA biomarker combinations to differentiate HFpEF from HFrEF was investigated, and the findings are summarized in Table 4 and Figure 3. According to AUC analysis, using a single miRNA to differentiate HFpEF from HFrEF was just as good as BNP alone. The use of a single miRNA (miR-375, −146a, −328, or −221) with BNP compared with BNP alone can significantly improve the model, as indicated by IDI and LR tests. The addition of miRNA combinations of two or more to BNP significantly improves the model of BNP alone at differentiating HFpEF from HFrEF, as evident with all forms of statistical analysis (AUC, IDI, and LR tests).
Table 4.
Comparisons of single use and combinations of microRNAs as predictors of heart failure with preserved ejection fraction from heart failure with preserved ejection fraction and assessing their diagnostic value by comparing with the heart failure biomarker brain natriuretic peptide
| Model description | MicroRNA(s) | AUC (95% CI)a | BNP-only model comparisons |
|
|---|---|---|---|---|
| LR test P-value | IDI (95% CI)b | |||
| Single miRNA | 375 | 0.75 (0.67–0.83) | – | – |
| 146a | 0.57 (0.48–0.66) | – | – | |
| 30c | 0.52 (0.42–0.61) | – | – | |
| 328 | 0.66 (0.58–0.75) | – | – | |
| 221 | 0.64 (0.55–0.73) | – | – | |
| Two miRNA combinationc | 30c, 375 | 0.76 (0.68–0.84) | – | – |
| Three miRNA combinationc | 221, 328, 30c | 0.79 (0.72–0.86) | – | – |
| Four miRNA combinationc | 221, 328, 30c, 375 | 0.79 (0.72–0.87) | – | – |
| All miRNAs | 146a, 221, 328, 375, 30c | 0.80 (0.72–0.87) | – | – |
| Log (BNP) | – | 0.66 (0.58–0.75) | – | – |
| Log (BNP) + single miRNA | 375 | 0.78 (0.71–0.85) | <0.001 | 0.101 (0.055–0.146) |
| 146a | 0.69 (0.61–0.78) | 0.034 | 0.028 (0.002–0.052) | |
| 30c | 0.67 (0.58–0.75) | 0.529 | 0.003 (−0.006 to 0.011) | |
| 328 | 0.77 (0.70–0.85) | <.001 | 0.118 (0.066– 0.165) | |
| 221 | 0.72 (0.63–0.80) | 0.005 | 0.055 (0.016– 0.091) | |
| Log (BNP) + two miRNA combinationd | 328, 30c | 0.82 (0.75–0.89) | <0.001 | 0.205 (0.143– 0.268) |
| Log (BNP) + three miRNA combinationd | 221, 328, 30c | 0.84 (0.78–0.91) | <0.001 | 0.255 (0.187–0.329) |
| Log (BNP) + four miRNA combinationd | 221, 328, 30c, 375 | 0.85 (0.79–0.92) | <0.001 | 0.272 (0.202–0.345) |
| Log (BNP) + all miRNAs | 146a, 221, 328, 375, 30c | 0.86 (0.79–0.92) | <0.001 | 0.275 (0.202–0.341) |
AUC, area under the curve; CI, confidence interval; IDI, integrated discrimination improvement; LR, likelihood ratio; miRNA, microRNA.
95% CI = AUC ± Z(.975)SE. Method for estimating SE taken from Hanley and McNeil 1982.16
Bootstrapped confidence interval with 1000 repetitions.
Best model combination according to AUC.
Best model combination according to observed IDI (see Kerr et al. 2011).17
Figure 3.
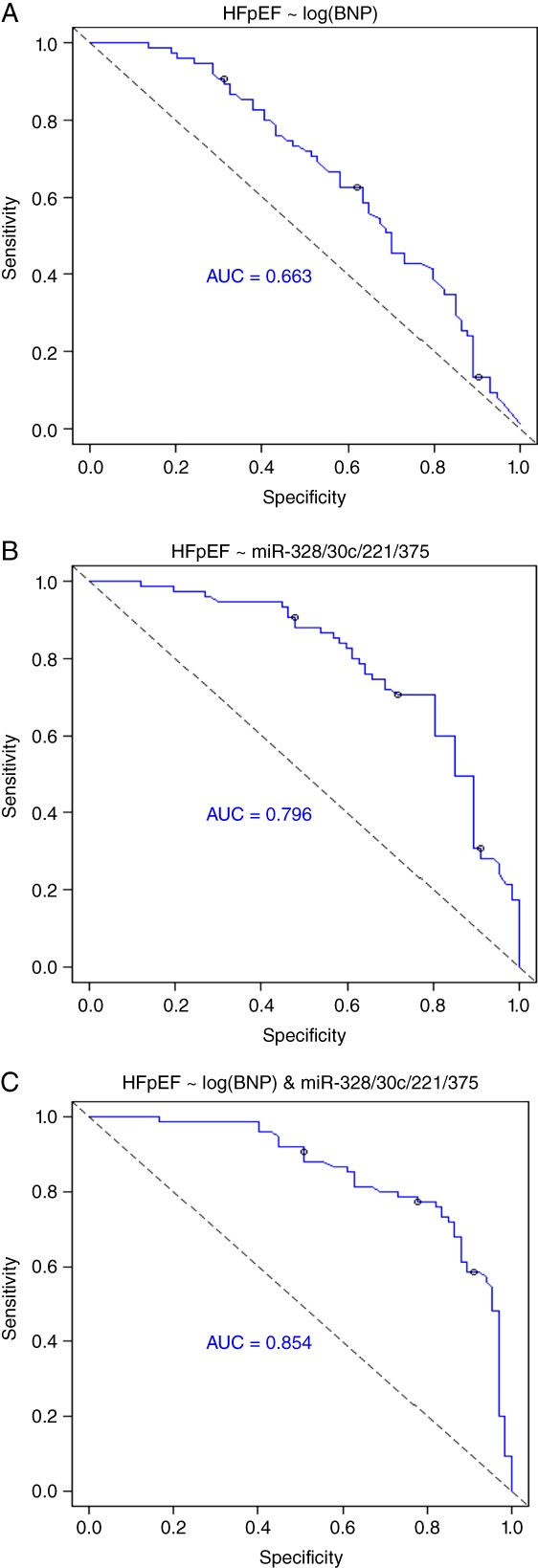
Receiver operating characteristic (ROC) analysis for the use of microRNAs (miRNAs) to improve the diagnostic utility of BNP in the differentiation of heart failure with preserved ejection fraction (HFpEF) from heart failure with reduced ejection fraction (HFrEF). ROC analysis highlights that the diagnostic value of using miRNA combinations to differentiate HFpEF from HFrEF is superior to using BNP on its own. The best diagnostic test includes BNP with a combination of four miRNAs, namely miR-328, miR-30c, miR-221, and miR-375, with an area under the curve (AUC) of 0.854.
Graphical representations of some of these examples are highlighted in Figure 3, with the strongest predictor consisting of a combination of BNP with miR-30c, miR-221, miR-328, and miR-375, generating an AUC of 0.854 for distinguishing HFpEF from HFrEF.
Predictive models to determine the associations of microRNAs with ejection fraction and left atrial volume index
Using statistical modelling, the five miRNA biomarker candidates were investigated to determine their relationship with the echocardiographic parameters EF and LAVI (Table 5). Analysis revealed that none of the miRNAs show good R2 values for predicting EF or LAVI. For EF, the full model including age, gender, and BNP was the best, with 25% (combined with miR-146a or miR-221). However, BNP alone captured almost all the explained variation in LAVI, with age and gender making no contribution once BNP was included, and the relationship of the miRNAs with LAVI was effectively zero. The finding that there was no relationship between LAVI or EF and any of the miRNAs was surprising, but this may suggest that whatever is altering the circulating level of miRNA is independent of the classical disease mechanisms that affect EF and LAVI.
Table 5.
Micr RNA model results investigating the relationship between the echocardiographic parameters ejection fraction and left atrial volume index
| miRNA | Models: R2 (95% CI)a (χ2
P-value)b |
||||
|---|---|---|---|---|---|
| No covariates | BNP | Age and gender | BNP, age, and gender | ||
| EFc | None | – | 0.10 (0.02–0.18) (P < 0.001) | 0.08 (0.01–0.15) (P < 0.001) | 0.21 (0.11–0.30) (P < 0.001) |
| miR-146a | 0.05 (−0.01 to 0.11) | 0.13 (0.04–0.21) (P < 0.001) | 0.15 (0.06–0.24) (P < 0.001) | 0.25 (0.15–0.35) (P < 0.001) | |
| miR-221 | 0.04 (−0.01 to 0.10) | 0.11 (0.03–0.19) (P < 0.001) | 0.17 (0.08–0.27) (P < 0.001) | 0.25 (0.15–0.35) (P < 0.001) | |
| miR-328 | 0.04 (−0.01 to 0.09) | 0.11 (0.03–0.18) (P < 0.001) | 0.16 (0.07–0.26) (P < 0.001) | 0.24 (0.14–0.34) (P < 0.001) | |
| miR-30c | 0.01 (−0.02 to 0.04) | 0.10 (0.02–0.18) (P < 0.001) | 0.10 (0.02–0.17) (P < 0.001) | 0.22 (0.12–0.31) (P < 0.001) | |
| miR-375 | 0.01 (−0.01 to 0.03) | 0.11 (0.03–0.19) (P < 0.001) | 0.09 (0.02–0.17) (P < 0.001) | 0.22 (0.12–0.32) (P < 0.001) | |
| LAVId | None | – | 0.41 (0.30–0.52) (P < 0.001) | 0.14 (0.05–0.23) (P < 0.001) | 0.42 (0.31– 0.53) (P < 0.001) |
| miR-146a | 0.01 (−0.02–0.05) | 0.41 (0.30–0.52) (P < 0.001) | 0.15 (0.05–0.24) (P < 0.001) | 0.42 (0.31–0.53) (P < 0.001) | |
| miR-221 | 0.04 (−0.02 to 0.10) | 0.41 (0.30–0.53) (P < 0.001) | 0.18 (0.08–0.28) (P < 0.001) | 0.42 (0.31–0.53) (P < 0.001) | |
| miR-328 | 0.01 (−0.02 to 0.04) | 0.41 (0.30–0.52) (P < 0.001) | 0.16 (0.06–0.25) (P < 0.001) | 0.42 (0.31–0.53) (P < 0.001) | |
| miR-30c | 0.00 (−) | 0.41 (0.30–0.52) (P < 0.001) | 0.13 (0.04–0.22) (P < 0.001) | 0.41 (0.30–0.52) (P < 0.001) | |
| miR-375 | 0.00 (−) | 0.42 (0.30–0.53) (P < 0.001) | 0.14 (0.04–0.23) (P < 0.001) | 0.42 (0.31–0.53) (P < 0.001) | |
CI, confidence interval LAVI, left atrial volume index; miRNA, microRNA.
95% CI = R2d ± t(0.975, n-k-1)SE. n = size of sample, k = number of predictors, Olkin–Finn method for estimating the SE.
χ2 test for difference on residual sum of squares compared with no-covariate model.
Linear model with full sample, outcome = EF2.
Linear model with full sample, outcome = log-transformed LAVI.
Discussion
This work provides original data on the potential role of miRNAs as a diagnostic aid in HF and also as a tool to differentiate HFrEF and HFpEF. The data demonstrate that the studied miRNAs are of diagnostic utility in differentiating patients with heart failure from asymptomatic individuals with cardiovascular risk factors. On their own, their performance is no better than BNP but combined with BNP they may provide better diagnostic certainty than BNP alone. In addition, the ability of miRNAs to differentiate between HFpEF and HFrEF is described for the first time in this study.
Five miRNA candidates were selected (miR-30c, −146a, −221, −328, and −375) from a miRNA biomarker discovery study following the profiling of pooled serum samples from a cohort of no-HF patients, HFpEF patients, and HFrEF patients. When investigating the discriminating power of these individual miRNAs to identify HF, none of them achieved a greater AUC than BNP, which was 0.875. However the value of the miRNAs was evident in HF diagnosis when these were used in conjunction with BNP, and was further improved with the addition of age and gender, generating an AUC of ≥0.90 for all individual miRNA candidates. Similarly, the data in Table 3 highlight that the use of miRNAs either individually or in combinations along with BNP had a significantly better diagnostic accuracy for predicting HF when compared with a BNP alone model, with similar AUCs being generated when using miRNA combinations of between two and five biomarkers (all >0.90). When assessing the clinical and practical added value of increasing the number of miRNAs within the combinations with the small gain in AUC (range 0.904–0.910), a balance between diagnostic assay costs and gain in discrimination by increasing the number of miRNAs in the assay needs to be considered. In this case it may be more clinically viable to use a two-miRNA combination (miR-221 and miR-375) with BNP, generating an AUC of 0.904 in this study, rather than generating a diagnostic test consisting of five miRNA components to generate an additional AUC gain of only 0.006.
When applying the five miRNA biomarker candidates individually to a model for differentiating HFpEF from HFrEF within a symptomatic population, miR-375 yielded the highest AUC of 0.75. All AUCs improved with the addition of BNP and age and gender, both individually and combined, with miR-375 remaining the most accurate diagnostically with an AUC of 0.82. When investigating the use of miRNA biomarker panels to differentiate HFpEF from HFrEF, combinations of two or more miRNAs was an improvement on BNP alone. The best miRNA combination was found to be miR-221, miR-328, miR-30c, and miR-375 (AUC 0.796) which generated an AUC of 0.662. Combinations of miRNAs of two or more along with BNP yields the best AUC values and are significantly better than using BNP on its own. The strongest predictor consisted of a combination of BNP with miR-30c, miR-221, miR-328, and miR-75, generating an AUC of 0.854 for distinguishing HFpEF from HFrEF. MiRNA combinations with BNP were superior to using NPs alone to differentiate HFpEF from HFrEF effectively.
Collectively, the data highlight that the use of miRNAs may have the potential to be more specific than NPs for differentiating this syndrome.
Although it was not possible to identify the cellular source of circulating miRNAs, it is noteworthy that the five miRNA biomarker candidates investigated herein have been described to various degrees in other studies relating to cardiovascular disease. In this study, all five miRNAs were reduced in HF; however miR-375 was only reduced in HFrEF. MiR-328 and miR-375 levels were significantly different between HFpEF and HFrEF when comparing average circulating levels.
Previous work has implicated miR-30c to have a beneficial regulatory function in myocardial matrix remodelling by negatively regulating connective tissue growth factor, a key profibrotic protein that contributes to heart disease including pathological LV hypertrophy.18 Increased miR-146a expression was shown to protect the myocardium from ischaemia/reperfusion injury via attenuation of proinflammatory pathways.19 Reduced miR-146a levels have previously been reported in type 2 diabetes patients and were associated with insulin resistance, poor glycaemic control, and increased proinflammatory cytokine genes including tumour necrosis factor α (TNFα) and interleukin 6 (IL6),20 and restoring expression of miR-146 reduced extracellular matrix protein fibronectin production and had beneficial effects in diabetes.21 An interesting study quantified miR-146a levels within circulating angiogenic cells (CACs) and plasma in an old chronic HF population (median age 81) and showed that this miRNA was increased 1000-fold in CACs and 2-fold in plasma compared with an age-matched control group, and was linked to increased cellular senescence within this cell type.22 In a recent study, Roldan et al. have identified polymorphisms in the miR-146a gene which is linked to increased adverse cardiovascular risk in anticoagulated patients with AF.23 Cardiac tissue levels of miR-221 have been reported to be deregulated during the development of hypertrophy in vivo, with an initial decrease in expression followed by up-regulation later in the pathogenesis.24 Detailed studies have described the impact of increased miR-221 within the myocardium and the mechanisms that drive the cardiomyocyte hypertrophic response.25 MiR-328 has also been implicated in the development of cardiac hypertrophy by mechanisms involving targeting SERCA2a,26 and has been shown to be a predictor for acute myocardial infarction, with increased levels associated with increased risk of mortality and development of HF.27 Finally, miR-375, which was the only down-regulated miRNA in HFpEF patients, has been linked to diabetes, with a study highlighting its expression in pancreatic islet cells and its requirement for normal pancreatic development, cell growth and proliferation, and insulin secretion.28 Circulating miR-375 levels have also been shown to be a valuable marker to detect β-cell death and predict diabetes in disease models.29
In summary, diagnosis of HF remains challenging, as is the ability to differentiate the subclasses which exhibit reduced or preserved EFs independent of echocardiography. Clinical application of biomarkers to aid accurate and specific diagnosis appears to be an attractive companion tool within HF care, particularly in the community setting. Diagnostic strategies based on quantifying and monitoring circulating miRNA levels may prove to be of clinical value in the near future. In particular, as new therapies emerge that are specific to HFpEF, a need for stratifying patient cohorts for different management and treatment regimes will arise. Having a biomarker-based diagnostic strategy may prove beneficial in this regard.
Future direction
Although we highlight the potential diagnostic utility in the ability of miRNAs to discriminate between symptomatic HF and asymptomatic no-HF patients, it would be of significant clinical value to determine if these miRNA biomarkers could be used to discriminate between patients with symptomatic HF and those with respiratory disease such as COPD, as well as for examining the influence of other cardiovascular factors including the impact of medications. Patients with COPD can present with similar clinical symptoms to HF patients, and work recently reported by Ellis and colleagues has identified a miRNA panel combined with NT-proBNP that can differentiate these two conditions.30
Limitations
It would be important to investigate the circulating levels of the five miRNA candidates in a control no-HF cohort who are free from any evidence of ventricular dysfunction in order to gain a better appreciation of expression differences across a wider spectrum of health and disease. In addition, expanding validation studies into larger geographically independent cohorts is necessary to test fully the utility of these miRNA biomarker candidates. From a technical perspective, caution needs to be exercised when undertaking miRNA biomarker candidate discovery studies to ensure adequate patient numbers are present in each cohort for sample pooling prior to miRNA array analysis as this will maximize the likelihood that findings will be maintained between data sets from discovery arrays and individual miRNA quantification assays during validation stages. In addition, sample pooling may result in missing potentially useful miRNA candidates.
Funding
This work was supported by the European Commission FP7 project FIBRO-TARGETS, the Integrated Research and Treatment Center Transplantation (Hannover), and the Health Research Board of Ireland [grant nos 602904; CSA-2012-36, respectively].
Conflict of interest: none declared.
References
- 1.Seferovic PM, Stoerk S, Filippatos G, Mareev V, Kavoliuniene A, Ristic AD, Ponikowski P, McMurray J, Maggioni A, Ruschitzka F, van Veldhuisen DJ, Coats A, Piepoli M, McDonagh T, Riley J, Hoes A, Pieske B, Dobric M, Papp Z, Mebazaa A, Parissis J, Ben Gal T, Vinereanu D, Brito D, Altenberger J, Gatzov P, Milinkovic I, Hradec J, Trochu JN, Amir O, Moura B, Lainscak M, Comin J, Wikstrom G, Anker S. Organization of heart failure management in European Society of Cardiology member countries: survey of the Heart Failure Association of the European Society of Cardiology in collaboration with the Heart Failure National Societies/Working Groups. Eur J Heart Fail. 2013;15:947–959. doi: 10.1093/eurjhf/hft092. [DOI] [PubMed] [Google Scholar]
- 2.Levy D, Kenchaiah S, Larson MG, Benjamin EJ, Kupka MJ, Ho KK, Murabito JM, Vasan RS. Long-term trends in the incidence of and survival with heart failure. N Engl J Med. 2002;347:1397–1402. doi: 10.1056/NEJMoa020265. [DOI] [PubMed] [Google Scholar]
- 3.Taylor CJ, Roalfe AK, Iles R, Hobbs FD. Ten-year prognosis of heart failure in the community: follow-up data from the Echocardiographic Heart of England Screening (ECHOES) study. Eur J Heart Fail. 2012;14:176–184. doi: 10.1093/eurjhf/hfr170. [DOI] [PubMed] [Google Scholar]
- 4.Fox KF, Cowie MR, Wood DA, Coats AJ, Poole-Wilson PA, Sutton GC. A Rapid Access Heart Failure Clinic provides a prompt diagnosis and appropriate management of new heart failure presenting in the community. Eur J Heart Fail. 2000;2:423–429. doi: 10.1016/s1388-9842(00)00108-2. [DOI] [PubMed] [Google Scholar]
- 5.McMurray JJ, Adamopoulos S, Anker SD, Auricchio A, Bohm M, Dickstein K, Falk V, Filippatos G, Fonseca C, Gomez-Sanchez MA, Jaarsma T, Kober L, Lip GY, Maggioni AP, Parkhomenko A, Pieske BM, Popescu BA, Ronnevik PK, Rutten FH, Schwitter J, Seferovic P, Stepinska J, Trindade PT, Voors AA, Zannad F, Zeiher A, Bax JJ, Baumgartner H, Ceconi C, Dean V, Deaton C, Fagard R, Funck-Brentano C, Hasdai D, Hoes A, Kirchhof P, Knuuti J, Kolh P, McDonagh T, Moulin C, Reiner Z, Sechtem U, Sirnes PA, Tendera M, Torbicki A, Vahanian A, Windecker S, Bonet LA, Avraamides P, Ben Lamin HA, Brignole M, Coca A, Cowburn P, Dargie H, Elliott P, Flachskampf FA, Guida GF, Hardman S, Iung B, Merkely B, Mueller C, Nanas JN, Nielsen OW, Orn S, Parissis JT, Ponikowski P. ESC guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: the Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2012;14:803–869. doi: 10.1093/eurjhf/hfs105. [DOI] [PubMed] [Google Scholar]
- 6.Hobbs FD, Roalfe AK, Davis RC, Davies MK, Hare R. Prognosis of all-cause heart failure and borderline left ventricular systolic dysfunction: 5 year mortality follow-up of the Echocardiographic Heart of England Screening Study (ECHOES) Eur Heart J. 2007;28:1128–1134. doi: 10.1093/eurheartj/ehm102. [DOI] [PubMed] [Google Scholar]
- 7.Paulus WJ, Tschope C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J Am Coll Cardiol. 2013;62:263–271. doi: 10.1016/j.jacc.2013.02.092. [DOI] [PubMed] [Google Scholar]
- 8.van Veldhuisen DJ, Linssen GC, Jaarsma T, van Gilst WH, Hoes AW, Tijssen JG, Paulus WJ, Voors AA, Hillege HL. B-type natriuretic peptide and prognosis in heart failure patients with preserved and reduced ejection fraction. J Am Coll Cardiol. 2013;61:1498–1506. doi: 10.1016/j.jacc.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 9.Maisel A, Mueller C, Adams K, Jr, Anker SD, Aspromonte N, Cleland JG, Cohen-Solal A, Dahlstrom U, DeMaria A, Di Somma S, Filippatos GS, Fonarow GC, Jourdain P, Komajda M, Liu PP, McDonagh T, McDonald K, Mebazaa A, Nieminen MS, Peacock WF, Tubaro M, Valle R, Vanderhyden M, Yancy CW, Zannad F, Braunwald E. State of the art: using natriuretic peptide levels in clinical practice. Eur J Heart Fail. 2008;10:824–839. doi: 10.1016/j.ejheart.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Bishu K, Deswal A, Chen HH, LeWinter MM, Lewis GD, Semigran MJ, Borlaug BA, McNulty S, Hernandez AF, Braunwald E, Redfield MM. Biomarkers in acutely decompensated heart failure with preserved or reduced ejection fraction. Am Heart J. 2012;164:763–770. doi: 10.1016/j.ahj.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitzman DW, Little WC, Brubaker PH, Anderson RT, Hundley WG, Marburger CT, Brosnihan B, Morgan TM, Stewart KP. Pathophysiological characterization of isolated diastolic heart failure in comparison to systolic heart failure. JAMA. 2002;288:2144–2150. doi: 10.1001/jama.288.17.2144. [DOI] [PubMed] [Google Scholar]
- 12.Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013;113:676–689. doi: 10.1161/CIRCRESAHA.113.300226. [DOI] [PubMed] [Google Scholar]
- 13.Mayr M, Zampetaki A, Willeit P, Willeit J, Kiechl S. MicroRNAs within the continuum of postgenomics biomarker discovery. Arterioscler Thromb Vasc Biol. 2013;33:206–214. doi: 10.1161/ATVBAHA.112.300141. [DOI] [PubMed] [Google Scholar]
- 14.Ledwidge M, Gallagher J, Conlon C, Tallon E, O'Connell E, Dawkins I, Watson C, O'Hanlon R, Bermingham M, Patle A, Badabhagni MR, Murtagh G, Voon V, Tilson L, Barry M, McDonald L, Maurer B, McDonald K. Natriuretic peptide-based screening and collaborative care for heart failure: the STOP-HF randomized trial. JAMA. 2013;310:66–74. doi: 10.1001/jama.2013.7588. [DOI] [PubMed] [Google Scholar]
- 15.Nagueh SF, Appleton CP, Gillebert TC, Marino PN, Oh JK, Smiseth OA, Waggoner AD, Flachskampf FA, Pellikka PA, Evangelista A. Recommendations for the evaluation of left ventricular diastolic function by echocardiography. J Am Soc Echocardiogr. 2009;22:107–133. doi: 10.1016/j.echo.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 16.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;43:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 17.Kerr KF, McClelland RL, Brown ER, Lumley T. Evaluating the incremental value of new biomarkers with integrated discrimination improvement. Am J Epidemiol. 2011;174:364–374. doi: 10.1093/aje/kwr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. 6p following 178. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Ha T, Liu L, Zou J, Zhang X, Kalbfleisch J, Gao X, Williams D, Li C. Increased expression of microRNA-146a decreases myocardial ischaemia/reperfusion injury. Cardiovasc Res. 2013;97:432–442. doi: 10.1093/cvr/cvs356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasubramanyam M, Aravind S, Gokulakrishnan K, Prabu P, Sathishkumar C, Ranjani H, Mohan V. Impaired miR-146a expression links subclinical inflammation and insulin resistance in Type 2 diabetes. Mol Cell Biochem. 2011;351:197–205. doi: 10.1007/s11010-011-0727-3. [DOI] [PubMed] [Google Scholar]
- 21.Feng B, Chen S, McArthur K, Wu Y, Sen S, Ding Q, Feldman RD, Chakrabarti S. miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes. 2011;60:2975–2984. doi: 10.2337/db11-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivieri F, Lazzarini R, Recchioni R, Marcheselli F, Rippo MR, Di Nuzzo S, Albertini MC, Graciotti L, Babini L, Mariotti S, Spada G, Abbatecola AM, Antonicelli R, Franceschi C, Procopio AD. MiR-146a as marker of senescence-associated pro-inflammatory status in cells involved in vascular remodelling. Age (Dordr) 2013;35:1157–1172. doi: 10.1007/s11357-012-9440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roldan V, Arroyo AB, Salloum-Asfar S, Manzano-Fernandez S, Garcia-Barbera N, Marin F, Vicente V, Gonzalez-Conejero R, Martinez C. Prognostic role of MIR146A polymorphisms for cardiovascular events in atrial fibrillation. Thromb Haemost. 2014;112:781–788. doi: 10.1160/TH14-01-0092. [DOI] [PubMed] [Google Scholar]
- 24.Feng HJ, Ouyang W, Liu JH, Sun YG, Hu R, Huang LH, Xian JL, Jing CF, Zhou MJ. Global microRNA profiles and signaling pathways in the development of cardiac hypertrophy. Braz J Med Biol Res. 2014;47:361–368. doi: 10.1590/1414-431X20142937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Wang S, Zhao P, Wang X, Wang J, Wang Y, Song L, Zou Y, Hui R. MiR-221 promotes cardiac hypertrophy in vitro through the modulation of p27 expression. J Cell Biochem. 2012;113:2040–2046. doi: 10.1002/jcb.24075. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Li X, Gao X, Zhang R, Zhang Y, Liang H, Xu C, Du W, Liu X, Ma N, Xu Z, Wang L, Chen X, Lu Y, Ju J, Yang B, Shan H. MicroRNA-328 as a regulator of cardiac hypertrophy. Int J Cardiol. 2014;173:268–276. doi: 10.1016/j.ijcard.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 27.He F, Lv P, Zhao X, Wang X, Ma X, Meng W, Meng X, Dong S. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol Cell Biochem. 2014;394:137–144. doi: 10.1007/s11010-014-2089-0. [DOI] [PubMed] [Google Scholar]
- 28.Li X. MiR-375, a microRNA related to diabetes. Gene. 2014;533:1–4. doi: 10.1016/j.gene.2013.09.105. [DOI] [PubMed] [Google Scholar]
- 29.Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology. 2013;154:603–608. doi: 10.1210/en.2012-1744. [DOI] [PubMed] [Google Scholar]
- 30.Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138–1147. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]