Abstract
The signal recognition particle is a ribonucleoprotein complex that is essential for the translocation of nascent proteins into the endoplasmic reticulum. It has been shown that the RNA component (SRP RNA) is exported from the nucleus by CRM1 in the budding yeast. However, how SRP RNA is exported in higher species has been elusive. Here, we show that SRP RNA does not use the CRM1 pathway in Xenopus oocytes. Instead, SRP RNA uses the same export pathway as pre-miRNA and tRNA as showed by cross-competition experiments. Consistently, the recombinant Exportin-5 protein specifically stimulated export of SRP RNA as well as of pre-miRNA and tRNA, whereas an antibody raised against Exportin-5 specifically inhibited export of the same RNA species. Moreover, biotinylated SRP RNA can pull down Exportin-5 but not CRM1 from HeLa cell nuclear extracts in a RanGTP-dependent manner. These results, taken together, strongly suggest that the principal export receptor for SRP RNA in vertebrates is Exportin-5 unlike in the budding yeast.
Introduction
The majority of the RNA species, following their synthesis and processing in the nucleus, are exported to the cytoplasm. It has been shown that different RNA species are exported from the nucleus via distinct export pathways, that is, by distinct sets of export factors (Jarmolowski et al. 1994; Komeili & O'Shea 2001; Cullen 2003). Among many RNA species, mRNA and spliceosomal U snRNA are initially m7G-capped and this structure plays important roles for export of corresponding RNA species (Cullen 2003; Cheng et al. 2006; Nojima et al. 2007). In contrast, other RNA species such as rRNA, tRNA and major pre-miRNA do not possess the structure and instead their RNA bodies are highly structured. The small (18S) and large (25S in yeasts and 28S in vertebrates, respectively) rRNAs are commonly exported by CRM1 (Moy & Silver 1999; Gadal et al. 2001). However, Exportin-5 and Nxf1 also export the large rRNA in vertebrates and the budding yeast, respectively (Yao et al. 2007; Wild et al. 2010). Similarly, tRNA is exported by either Exportin-t or Exportin-5 (Arts et al. 1998; Kutay et al. 1998; Bohnsack et al. 2002; Calado et al. 2002), whereas major pre-miRNA by Exportin-5 (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). Adenovirus VA1 RNA, which is also highly structured, is also exported by Exportin-5 (Gwizdek et al. 2001, 2003). Thus, it appears that Exportin-5 and CRM1 are two major export receptors for uncapped structured RNA species.
Another example of uncapped structured RNA species is the RNA component of the signal recognition particle (SRP) that is involved in the translocation of nascent polypeptides into the endoplasmic reticulum (ER) lumen (Egea et al. 2005). SRP is composed of 6 protein components, SRP9, 14, 19, 54, 68 and 72 in vertebrates, as well as the single RNA component, SRP RNA. Vertebrate SRP can be divided into 3 domains, Alu domain, S domain and the linker region that connects the other two domains. Alu domain consists of the 5′ and 3′ terminal regions of SRP RNA and the bound SRP9/14 protein heterodimer. S domain consists of the central region of SRP RNA, SRP19, SRP54 and the SRP68/72 heterodimer (see Fig. S1 in Supporting Information). The former domain is involved in the translational arrest activity of SRP, whereas the latter is involved in the recognition, as mediated by the bound SRP54 protein component, of both the signal sequence of nascent polypeptides and the SRP receptor on the ER membrane (Egea et al. 2005).
Experiments using the budding yeast, Xenopus oocytes and mammalian cells led to a proposal of an assembly model in which SRP RNA associates with all the protein components except SRP54 in the nucleolus, is then exported to the cytoplasm, and finally associates with SRP54 in the cytoplasm (He et al. 1994; Ciufo & Brown 2000; Politz et al. 2000, 2002; Grosshans et al. 2001; Sommerville et al. 2005). SRP RNA accumulates in the nucleus in the xpo1-1 ts mutant yeast strain at a nonpermissive temperature, suggesting CRM1 exports this RNA in the budding yeast (Ciufo & Brown 2000; Grosshans et al. 2001). It has been suggested that CRM1 may also export SRP RNA in vertebrates as leptomycin B (LMB) treatment of rat cells led to an increased SRP RNA signal in the nucleolus (Alavian et al. 2004). However, this could be an indirect effect of LMB as the authors discussed in the study because the LMB treatment has to be very long (as long as 20 h) to be able to see the effect, that is, shorter treatments did not lead to the SRP RNA accumulation. Moreover, association of CRM1 with SRP RNA has never been showed (neither in the case of the budding yeast). Thus, how SRP RNA is exported in higher species has been elusive. In this report, we carried out Xenopus oocyte RNA microinjection as well as biochemical experiments and gathered several lines of evidence to conclude that Exportin-5 but not CRM1 is the principal export receptor for SRP RNA in vertebrates.
Results
SRP RNA is exported via a CRM1-independent pathway in Xenopus oocytes
SRP RNA was previously suggested to use the CRM1-dependent export pathway in the budding yeast (Ciufo & Brown 2000; Grosshans et al. 2001). Whether or not vertebrate SRP RNA is also exported via the same CRM1 pathway was examined in the well-established Xenopus oocyte microinjection experiments. To this end, a well-characterized inhibitor of the CRM1-dependent export was used. This inhibitor was a conjugate of NES peptides coupled to BSA (BSA-NES), which was shown to saturate CRM1-dependent export (Fischer et al. 1995; Masuyama et al. 2004).
In the control situation, approximately 50% of the injected in vitro-transcribed 32P-labeled Xenopus SRP RNA as well as of DHFR mRNA had been exported to the cytoplasm during 1.5 h after nuclear microinjection, whereas the nonexported U6Δss RNA control stayed in the nucleus (Fig.1A, lanes 5 and 6). Approximately 30% of U1ΔSm had been exported, whereas tRNAphe had been almost completely exported. When the same RNA mixture was injected with a saturating amount of BSA-NES, the export of DHFR mRNA and tRNAphe was hardly affected as DHFR mRNA uses NXF1/p15 and tRNAphe uses Exportin-t or Exportin-5 for export (Arts et al. 1998; Kutay et al. 1998; Komeili & O'Shea 2001; Bohnsack et al. 2002; Calado et al. 2002; Katahira 2012). In contrast, the export of U1ΔSm was severely inhibited as this RNA uses the CRM1-dependent export pathway (Fig.1A, lanes 3 and 4 and Fig.1B) (Fornerod et al. 1997). Remarkably, the export of Xenopus SRP RNA was unaffected (Fig.1A, lanes 3 and 4 and Fig.1B). Very similar results were obtained when human, instead of Xenopus, SRP RNA was used (Fig.1C, D). These results suggested that vertebrate SRP RNA is exported via a CRM1-independent pathway.
Figure 1.
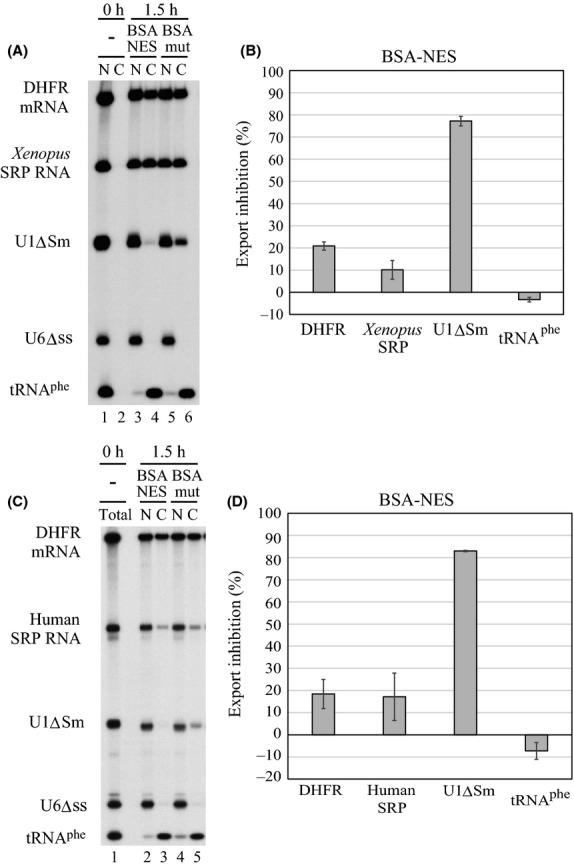
Nuclear export of SRP RNA is independent of CRM1. (A) A mixture of in vitro-transcribed 32P-labeled DHFR mRNA, Xenopus SRP RNA, U1ΔSm RNA, U6Δss RNA and tRNAphe was injected into the nucleus of Xenopus oocytes either with 0.2 μg/oocyte of BSA-NES (lanes 3 and 4) or with the same amount of BSA-mut (mutated NES, lanes 5 and 6). DHFR mRNA and U1ΔSm RNA were m7G-capped, and other RNAs were uncapped. RNA was analyzed immediately (0 h; lanes 1 and 2) or 1.5 h (1.5 h; lanes 3–6) after injection. N, nuclear; C, cytoplasmic fractions. (B) Quantitation of RNA export from four independent experiments as in A. (C) The same as A except that human SRP RNA instead of Xenopus SRP RNA was injected into the nucleus of oocytes either with 0.3 μg/oocyte of BSA-NES or with the same amount of BSA-mut. (D) Quantitation of RNA export from three independent experiments as in C.
Vertebrate SRP RNA is exported via the common pathway with pre-miRNA and tRNA
To further characterize the export pathway of vertebrate SRP RNA, we carried out cross-competition experiments (Jarmolowski et al. 1994). If SRP RNA shares the same export pathway with certain RNA specie (e.g., RNA A), export of SRP RNA should be specifically saturated and inhibited if a large amount of RNA A is co-injected. As SRP RNA is uncapped and structured, we first focused on pre-miRNA and tRNA as competitors. Co-injection of increasing amounts of human pre-miR-31 strongly inhibited export of Xenopus SRP RNA as well as of pre-miR-31 itself, whereas the nucleo-cytoplasmic distributions of U1ΔSm and U6Δss RNAs were unaffected (Fig.2A). Export inhibition of SRP RNA was quite effective. Its export was inhibited by 93.4% by even the lowest amount of competitor. Similarly, the tRNAphe competitor specifically inhibited export of Xenopus SRP RNA as well as of tRNAphe itself (Fig.2B). However, export inhibition by the tRNA competitor was not as effective as that by the pre-miRNA competitor. In contrast, the U1ΔSm competitor inhibited export of only U1ΔSm itself (Fig.2C). These results strongly suggested that the primary candidate for the export receptor for vertebrate SRP RNA is Exportin-5, considering that pre-miRNA is exported by Exportin-5 and tRNA is exported by Exportin-5 or Exportin-t.
Figure 2.
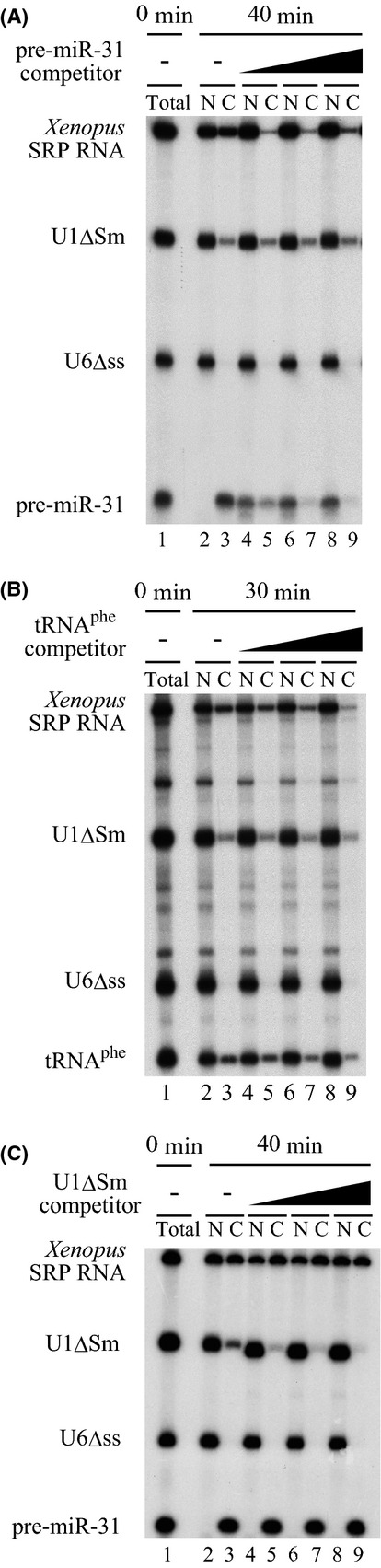
SRP RNA shares the same export pathway with pre-microRNA and tRNA. (A) A mixture of 32P-labeled Xenopus SRP RNA, U1ΔSm RNA, U6Δss RNA and human pre-miR-31 was injected into the nucleus of oocytes either alone (lanes 2 and 3) or with increasing amounts (0.2, 0.5, and 1.0 pmol/oocyte) of unlabeled pre-miR-31 (lanes 4–9). RNA was analyzed immediately (total; lane 1) or 40 min (40 min; lanes 2–9) after injection. (B) The same as A except that 32P-labeled tRNAphe and unlabeled tRNAphe (0.2, 1.0, and 5.0 pmol/oocyte) were used instead of pre-miR-31, and the incubation time was 0 or 30 min. (C) The same as A except that unlabeled U1ΔSm RNA (0.5, 1.0, and 2.0 pmol/oocyte) was used instead of unlabeled pre-miR-31.
Exportin-5 mediates export of vertebrate SRP RNAs
Consistent with the above results, microinjection of the recombinant human Exportin-5 in Xenopus oocytes stimulated export of microinjected Xenopus SRP RNA as well as of tRNAphe, but not of U1ΔSm RNA (Fig.3A, lanes 2–5 and Fig.3B), whereas microinjection of the recombinant human Exportin-t stimulated only tRNAphe export (Fig.3A, lanes 6 and 7 and Fig.3B). Very similar results were obtained when human, instead of Xenopus, SRP RNA was used (Fig.3C, D). These results suggested that vertebrate SRP RNA uses the Exportin-5-dependent export pathway.
Figure 3.
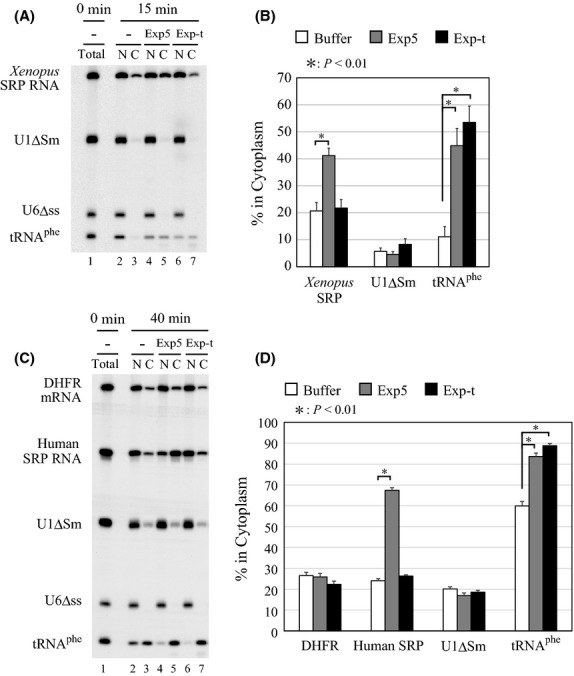
Exportin-5 mediates nuclear export of SRP RNA. (A) A mixture of 32P-labeled Xenopus SRP RNA, U1ΔSm RNA, U6Δss RNA and tRNAphe was injected into the nucleus of oocytes either alone (lanes 2 and 3) or with 150 fmol/oocyte of Exportin-5 (lanes 4 and 5) or Exportin-t (lanes 6 and 7). RNA was analyzed immediately (lane 1) or 15 min (lanes 2–7) after injection. (B) Quantitation of RNA export from the six independent experiments as in A. (C) A mixture of 32P-labeled DHFR mRNA, human SRP RNA, U1ΔSm RNA, U6Δss RNA and tRNAphe was injected either alone (lanes 2 and 3) or with 60 fmol/oocyte of Exportin-5 (lanes 4 and 5) or Exportin-t (lanes 6 and 7). RNA was analyzed immediately (lane 1) or 40 min (lanes 2–7) after injection. (D) Quantitation of RNA export from the three independent experiments in C.
To further confirm the above conclusion, a specific antibody raised against human Exportin-5 or a control antibody raised against hnRNP K was microinjected together with 32P-labeled RNAs. As expected, export of Xenopus SRP RNA as well as of tRNAphe and pre-miR-31 was inhibited only by the anti-Exportin-5 antibody, whereas export of U1ΔSm was unaffected (Fig.4A, B). Export inhibition of tRNAphe and pre-miR-31 by the antibody was much weaker than that of SRP RNA. This was because export of tRNAphe and pre-miR-31 is kinetically much faster than that of SRP RNA. The same antibody but not the control antibody also inhibited export of human SRP RNA (Fig.4C, D). In the latter experiment, we used longer incubation time (1.5 h) as export kinetics of human SRP RNA is slower than that of the Xenopus counterpart. Under these conditions, therefore, export of tRNAphe was apparently unaffected by the antibody, because of the fast export kinetics. Nevertheless, these results, taken together, strongly suggested that Exportin-5 is the export receptor for vertebrate SRP RNA.
Figure 4.
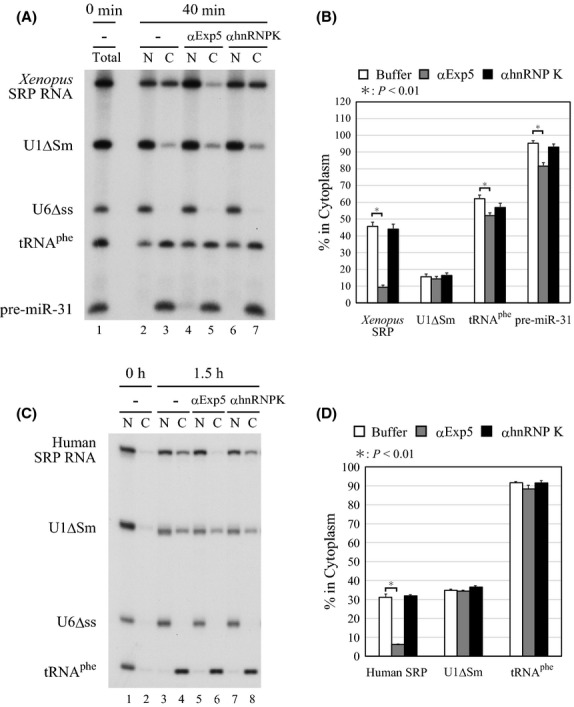
Endogenous exportin-5 mediates nuclear export of SRP RNA. (A) A mixture of 32P-labeled Xenopus SRP RNA, U1ΔSm RNA, U6Δss RNA, tRNAphe and pre-miR-31 was injected into the nucleus of oocytes either alone (lanes 2 and 3) or with 38 ng/oocyte of anti-Exportin-5 antibody (lanes 4 and 5) or control antibody (lanes 6 and 7). RNA was analyzed immediately (lane 1) or 40 min (lanes 2–7) after injection. (B) Quantitation of RNA export from the four independent experiments as in A. (C) The same as A except that human SRP RNA was used and pre-miR-31 was omitted, and the incubation was 0 or 1.5 h. (D) Quantitation of RNA export from the three independent experiments as in C.
Exportin-5 interacts with SRP RNA
The results so far prompted us to test whether Exportin-5 can interact with vertebrate SRP RNA. As this interaction could be indirect, that is, adaptor-mediated, we first tested whether Exportin-5 could be pulled down from HeLa cell nuclear extracts (HNE) together with exogenously added human SRP RNA. In vitro-transcribed biotinylated human SRP RNA was incubated with HNE in the presence or absence of recombinant RanQ69LGTP, a constitutively active mutant form of Ran loaded with GTP, the RNA was then pulled down with streptavidin beads, and the precipitated protein was analyzed by Western blotting (Fig.5). Exportin-5 was only weakly pulled down by human SRP RNA alone (Fig.5A, lane 11), but this interaction was greatly enhanced by the presence of RanQ69LGTP (lane 12). In contrast, CRM1 was not pulled down under the same conditions. To further confirm the specificity of the interaction between Exportin-5 and human SRP RNA, cross-competition experiments were carried out (Fig.5B). Addition of increasing amounts of nonbiotinylated VA1 RNA as well as nonbiotinylated human SRP RNA itself could competitively inhibit the interaction between Exportin-5 and biotinylated human SRP RNA (Fig.5B, lanes 3-9), whereas nonbiotinylated U1ΔSm had no effect. These results are consistent with the idea that human SRP RNA is one of the specific export substrates for Exportin-5.
Figure 5.
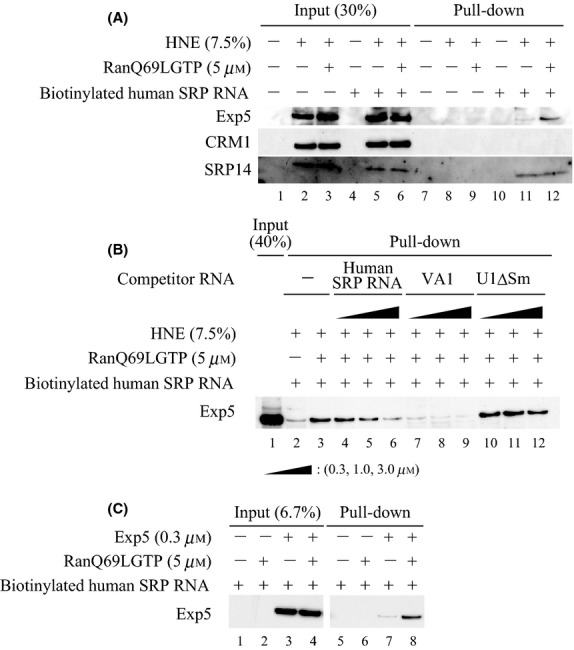
Exportin-5 interacts with SRP RNA. (A) Biotinylated human SRP RNA was incubated with HeLa cell nuclear extracts (HNE) with or without RanQ69LGTP in buffer A for 30 min at 30°C, and then biotinylated human SRP RNA was pulled down by streptavidin beads. The precipitated proteins were analyzed by Western blotting. (B) The same as A except that the nonbiotinylated RNA competitors were used. (C) The pull-down assay using biotinylated human SRP RNA, the recombinant Exportin-5 and RanQ69LGTP was similarly carried out as A. The precipitated Exportin-5 was analyzed by Western blotting using anti-His tag antibody.
As exogenously added SRP RNA could associate with SRP proteins as showed by the Western blotting with the anti-SRP14 antibody (Fig.5A), it was possible that an adaptor protein(s) in HNE bridged the interaction between Exportin-5 and SRP RNA. To test this possibility, we next examined the direct interaction between recombinant Exportin-5 and SRP RNA (Fig.5C). Biotinylated SRP RNA could pull down recombinant Exportin-5 only weakly, but this interaction was greatly stimulated by the presence of RanQ69LGTP (Fig.5C, lanes 7, 8). These results indicated that Exportin-5 can directly recognize human SRP RNA.
Discussion
In this report, we have provided several lines of evidence suggesting that Exportin-5 is the principal nuclear export receptor for SRP RNA in vertebrates, unlike in the budding yeast in which CRM1 plays an important role. Two major questions arise: How Exportin-5 recognizes vertebrate SRP RNA and why the export pathway differs between vertebrates and the budding yeast?
Recognition of vertebrate SRP RNA by Exportin-5
Exportin-5 mediates nuclear export of many uncapped structured RNA species including pre-miRNA, tRNA, large rRNA and adenovirus VA1 RNA (Gwizdek et al. 2001, 2003; Bohnsack et al. 2002, 2004; Calado et al. 2002; Yi et al. 2003; Lund et al. 2004; Wild et al. 2010). Exportin-5 directly recognizes the structure of double-stranded (ds) RNA with 3′ overhang in a Ran-GTP dependent manner. An atomic structure analysis of the Exportin-5, pre-miRNA and RanGTP complex showed that the overall structure resembles a baseball mitt and that the mitt accommodates the dsRNA stem structure of pre-miRNA (Okada et al. 2009). In addition, a tunnel-like structure at the bottom of the mitt recognizes the RNA's 3′ end protruding from the stem.
Previous structural studies suggested that the 3′-end of the vertebrate SRP RNA is located at the end of helix 5, which forms the linker region and a portion of Alu and S domains, and is protruding from the helix (Halic et al. 2004). Therefore, we propose a model in which helix 5 and the 3′ overhang are directly recognized by Exportin-5. However, it is still possible that bound protein factors influence the interaction between Exportin-5 and SRP RNA, for instance, by keeping the proper RNA secondary structure that enables more efficient Exportin-5 association or by sterically inhibiting the access of the export receptor.
Difference between yeast and vertebrate in SRP RNA export
What is the reason why yeast SRP RNA does not use the Exportin-5 (Msn5p in yeasts) export pathway? There are several differences in the structure of SRP RNA between vertebrates and the budding yeast (Zwieb et al. 2005; Andersen et al. 2006; Rosenblad et al. 2009). Most importantly, the structure near the 3′-end is quite different. In the case of the budding yeast SRP RNA, four extra helices (helix 9-12) are inserted into helix 5 (Fig. S1 in Supporting Information). Therefore, we speculate that Exportin-5 cannot recognize the 3′-end of yeast SRP RNA due to the steric hindrance. Consistent with this idea, yeast SRP RNA was hardly exported in Xenopus oocytes and became unstable upon longer incubations (Fig. S2 in Supporting Information), suggesting that Exportin-5 cannot efficiently function for yeast SRP RNA as an export receptor.
Moreover, the protein components of SRP are somewhat different between the two species. Yeast SRP similarly contains six protein components, Srp21, 14, 54, 68, 72 and Sec65, in which Srp21 and Sec65 are homologues of vertebrate SRP9 and 19, respectively (Hann & Walter 1991; Stirling & Hewitt 1992; Brown et al. 1994; Mason et al. 2000; Rosenblad et al. 2004). However, vertebrate SRP9 forms a heterodimer with SRP14, whereas yeast counterpart (Srp21) does not. Instead, yeast Srp14 forms a homodimer (Strub et al. 1999; Mason et al. 2000). These differences in the protein composition may affect the recognition by the export receptors in both negative and positive ways. There has been no evidence that CRM1 can directly recognize RNA. Therefore, it is plausible to assume that one of the yeast Srp proteins or other SRP-associating protein(s) has an NES-like sequence and serves as an adaptor between yeast CRM1 and yeast SRP RNA.
Experimental procedures
DNA constructs, templates for in vitro transcription and antibodies
For preparation of the plasmid encoding Xenopus laevis SRP RNA, a DNA fragment containing the sequence of Xenopus laevis SRP RNA and EcoRI, DraI, and SalI sites was generated by RT-PCR using RNA from Xenopus laevis oocytes and the following primers: 5′-AAAGAATTCTAATACGACTCACTATAGCCGGGCGCTGTGGCG-3′ and 5′-AAAGTCGACTTTAAAAGAACTGTGTCTCG-3′, and then the amplified fragment was inserted into the EcoRI-SalI sites of pSP65. Cloning of human SRP RNA was similarly carried out using RNA from HeLa cells and specific primers: 5′-AAAGAATTCTAATACGACTCACTATAGCCGGGCGCGGTGG-3′ and 5′-AAAGTCGACTTTAAAAGAGACGGGGTCTCG-3′. For preparation of the plasmid encoding VA1 RNA, a DNA fragment corresponding to the sequence of VA1 RNA was generated by PCR using Adeno-X System 1 Viral DNA (Clontech) and the specific primers: 5′-AAATCTAGAGGGCACTCTTCCGTGGTC-3′ and 5′-AAACTGCAGAAAAGGAGCGCTCCCCCGTTG-3′, and the fragment was inserted into the XbaI-PstI sites of pUC118. A DNA fragment containing the T7 promoter-VA1 RNA was generated by PCR using the pUC118-VA1 plasmid and specific primers: 5′-AAAGAATTCTAATACGACTCACTATAGGGCACTCTTCCGTGGTC-3′ and 5′-AAAGTCGACTTTAAAAGGAGCGCTCCCCCGTTG-3′, and then the fragment was inserted into the EcoRI-SalI sites of pSP65.
The pSP65-based plasmids for Xenopus SRP RNA and VA1 RNA were digested with DraI for in vitro transcription, whereas the template for human SRP RNA was produced by PCR using the pSP65-human SRP RNA plasmid and the following primers: 5′-TAATACGACTCACTATAGCCGGGCGCGGTGGCGCGTG-3′ and 5′-AGAGACGGGGTCTCGCTATGTTGCCCAGGCTGGAG-3′. In vitro transcription of these templates would yield the precursor form containing UCUUUU 3′-end and the mature form containing UCU 3′-end for Xenopus and human SRP RNAs, respectively. The template for in vitro transcription of pre-miR-31 was generated as previously described (Lund et al. 2004).
For preparation of the plasmid encoding Saccharomyces cerevisiae SRP RNA, a DNA fragment containing the sequence of Saccharomyces cerevisiae SRP RNA was generated by PCR using DNA from Saccharomyces cerevisiae and the following primers: 5′-AGGCTGTAATGGCTTTCTGGTGGGATGG-3′ and 5′-AAAAATATGGTTCAGGACACACTCCATCC-3′, and then the amplified fragment was inserted into pCR™4-TOPO using TOPO TA Cloning Kit for Sequencing (Life Technologies). The template for in vitro transcription of Saccharomyces cerevisiae SRP RNA was generated by PCR using the plasmid encoding Saccharomyces cerevisiae SRP RNA and the following primers: 5′-TAATACGACTCACTATAAGGCTGTAATGGCTTTCTGGTGGGATGG-3′ and 5′-AAAAATATGGTTCAGGACACACTCCATCC-3′.
The pQE60 plasmid for expressing His-tagged human Exportin-5 was kindly gifted from Dr. Jun Katahira (Shibata et al. 2006). For preparation of the plasmid for expressing His-tagged Exportin-t, a DNA fragment containing the Exportin-t coding sequence was generated by RT-PCR using RNA from HeLa cells and specific primers: 5′-AAACCATGGATGAACAGGCTCTAT-3′ and 5′-TTCATGATCATGAAGTAGGCACAGG-3′, and then PCR was carried out using the DNA fragment generated by RT-PCR and the following primers: 5′-AAACCATGGATGAACAGGCTCTAT-3′ and 5′-AAAGGATCCGGGCTTTGCTCTCTGG-3′. The amplified product was inserted into the NcoI-BamHI sites of pQE60. Polyclonal antibodies against coilin, Exportin-5, hnRNP K and SRP14 were from Santa Cruz (sc-32860), Sigma (SAB4200003), MBL (RN019P) and Abcam (ab155004), respectively. Monoclonal antibodies against CRM1 and His tag were from BD (611833) and MBL (D291-3), respectively.
Recombinant proteins
Expression and purification of His-tagged Exportin-5 and RanQ69LGTP were carried out as described previously (Ohno et al. 2000; Shibata et al. 2006). Expression and purification of His-tagged Exportin-t were carried out as His-tagged Exportin-5.
RNA-protein binding assay
Biotinylated human SRP RNA was synthesized using MEGAscript (Ambion) and Biotin-16-UTP (Roche). The competitor RNAs were also synthesized using MEGAscript. Biotinylated human SRP RNA was incubated with RanQ69LGTP and HeLa cell nuclear extracts, with or without the competitor RNAs in buffer A [20 mm Tris-HCl (pH7.5), 100 mm KCl, 2.5 mm MgCl2, 20% glycerol, 0.5 mm DTT, 0.1% NP-40 and protease inhibitor cocktail, complete (Roche)] for 30 min at 30°C, and then the mixture was incubated with streptavidin-sepharose beads (GE Healthcare) for 1 h at 4°C. After the beads were washed four times with buffer A, the bound material was recovered with 1 × SDS sample buffer and analyzed by Western blotting. The pull-down assay using the recombinant Exportin-5 was similarly carried out.
Microinjection into Xenopus oocytes
Preparation of 32P-labeled RNAs and their microinjection into Xenopus oocytes were carried out as described previously (Masuyama et al. 2004). Preparation of BSA-NES and BSA-mut was as described previously (Masuyama et al. 2004).
Acknowledgments
We thank Dr Jun Katahira for the pQE60 plasmid for expressing His-tagged human Exportin-5. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas ‘Neo-taxonomy of noncoding RNAs’ (No. 26113004 to M.O.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web site:
Figure S1 The secondary structures of human and Saccharomyces cerevisiae SRP RNAs.
Figure S2 Saccharomyces cerevisiae SRP RNA was hardly exported to the cytoplasm in Xenopus oocytes.
References
- Alavian CN, Politz JC, Lewandowski LB, Powers CM. Pederson T. Nuclear export of signal recognition particle RNA in mammalian cells. Biochem. Biophys. Res. Commun. 2004;313:351–355. doi: 10.1016/j.bbrc.2003.11.126. [DOI] [PubMed] [Google Scholar]
- Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T. Zwieb C. The tmRDB and SRPDB resources. Nucleic Acids Res. 2006;34(Database issue):D163–D168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M. Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr. Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K. Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E. Görlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Hann BC, Medzihradszky KF, Niwa M, Burlingame AL. Walter P. Subunits of the Saccharomyces cerevisiae signal recognition particle required for its functional expression. EMBO J. 1994;13:4390–4400. doi: 10.1002/j.1460-2075.1994.tb06759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Müller EC, Otto A. Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A. Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Ciufo LF. Brown JD. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr. Biol. 2000;10:1256–1264. doi: 10.1016/s0960-9822(00)00743-0. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Nuclear RNA export. J. Cell Sci. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- Egea PF, Stroud RM. Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW. Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M. Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauß D, Kessl J, Trumpower B, Tollervey D. Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Deinert K, Hurt E. Simos G. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 2001;153:745–762. doi: 10.1083/jcb.153.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Bertrand E, Dargemont C, Lefebvre JC, Blanchard JM, Singer RH. Doglio A. Terminal minihelix, a novel RNA motif that directs polymerase III transcripts to the cell cytoplasm. Terminal minihelix and RNA export. J. Biol. Chem. 2001;276:25910–25918. doi: 10.1074/jbc.M100493200. [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Ossareh-Nazari B, Brownawell AM, Doglio A, Bertrand E, Macara IG. Dargemont C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 2003;278:5505–5508. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J. Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hann BC. Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- He XP, Bataillé N. Fried HM. Nuclear export of signal recognition particle RNA is a facilitated process that involves the Alu sequence domain. J. Cell Sci. 1994;107:903–912. doi: 10.1242/jcs.107.4.903. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E. Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J. mRNA export and the TREX complex. Biochim. Biophys. Acta. 2012;1819:507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Komeili A. O'Shea EK. New perspectives on nuclear transport. Annu. Rev. Genet. 2001;35:341–364. doi: 10.1146/annurev.genet.35.102401.090720. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E. Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol. Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE. Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mason N, Ciufo LF. Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K, Taniguchi I, Kataoka N. Ohno M. RNA length defines RNA export pathway. Genes Dev. 2004;18:2074–2085. doi: 10.1101/gad.1216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy TI. Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Hirose T, Kimura H. Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M. Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y. Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- Politz JC, Lewandowski LB. Pederson T. Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J. Cell Biol. 2002;159:411–418. doi: 10.1083/jcb.200208037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C. Pederson T. Signal recognition particle components in the nucleolus. Proc. Natl Acad. Sci. USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad MA, Larsen N, Samuelsson T. Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- Rosenblad MA, Zwieb C. Samuelsson T. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genomics. 2004;5:5. doi: 10.1186/1471-2164-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J. Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerville J, Brumwell CL, Politz JC. Pederson T. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J. Cell Sci. 2005;118:1299–1307. doi: 10.1242/jcs.01726. [DOI] [PubMed] [Google Scholar]
- Stirling CJ. Hewitt EW. The S. cerevisiae SEC65 gene encodes a component of yeast signal recognition particle with homology to human SRP19. Nature. 1992;356:534–537. doi: 10.1038/356534a0. [DOI] [PubMed] [Google Scholar]
- Strub K, Fornallaz M. Bui N. The Alu domain homolog of the yeast signal recognition particle consists of an Srp14p homodimer and a yeast-specific RNA structure. RNA. 1999;5:1333–1347. doi: 10.1017/s1355838299991045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E. Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Roser D, Köhler A, Bradatsch B, Bassler J. Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG. Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, van Nues RW, Rosenblad MA, Brown JD. Samuelsson T. A nomenclature for all signal recognition particle RNAs. RNA. 2005;11:7–13. doi: 10.1261/rna.7203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The secondary structures of human and Saccharomyces cerevisiae SRP RNAs.
Figure S2 Saccharomyces cerevisiae SRP RNA was hardly exported to the cytoplasm in Xenopus oocytes.


