Abstract
Objectives:
Exclusive enteral nutrition (EEN) is commonly used to treat pediatric Crohn's disease (CD). Meta-analysis of pediatric studies that have compared the effect of EEN with other treatments have shown that EEN induces remission in up to 80–85% of patients. We aimed to gain a comprehensive understanding of the effect of EEN on the microbiota of CD patients.
Methods:
We used 16S rRNA gene and whole-genome high throughout sequencing to determine changes in the fecal microbiota of five CD children, before, during, and after EEN therapy and compared this with five healthy controls.
Results:
The microbial diversity observed in CD patients tended to be lower than that in controls (CD: 2.25±0.24, controls: 2.75±0.14, P=0.11). In all CD patients, dysbiosis was observed prior to therapy. EEN therapy had a positive effect in all patients, with 80% going into remission. In some patients, the positive effect diminished following the conclusion of EEN therapy. Significantly, the number of operational taxonomic units (OTU) decreased dramatically upon starting EEN and this corresponded with CD remission. Recurrence of CD corresponded with an increase in OTUs. Six families within the Firmicutes were found to correlate with disease activity during and following EEN therapy, a finding that was confirmed by whole-genome high throughput sequencing.
Conclusions:
Our results demonstrate that EEN leads to common and patient-specific alterations in the microbiota of CD patients, a number of which correlate with disease activity.
Introduction
Crohn's disease (CD) is a chronic relapsing idiopathic disease of the gastrointestinal tract, which is responsible for significant morbidity and mortality worldwide.1, 2 A range of therapies are employed in the treatment of CD, including the use of aminosalicylate or corticosteroid drugs, immunosuppressing drugs, biologic therapies that target tumor necrosis factor or integrin, exclusive enteral nutrition (EEN), or in those that do not respond to other therapies, surgery.
In pediatric CD, EEN is one of the more commonly used treatments. This involves the provision of a liquid diet using elemental or polymeric formulae, given exclusively over a prolonged period of up to 12 weeks. Meta-analysis of pediatric studies which have compared the effect of EEN with other CD treatments have shown that EEN has equivalent efficacy to corticosteroids,3 inducing remission in up to 80% of patients with active disease.4, 5 Reductions in disease activity based on the Pediatric Crohn's Disease Activity Index (PCDAI) scores and a decrease in inflammatory markers (e.g., C-reactive protein) have been reported to occur following 8–12 weeks of EEN therapy.5 In comparison with steroids, EEN has been shown to lead to superior mucosal healing and nutritional improvements, as well as substantially less side effects.6, 7 Moreover, EEN has been shown to have beneficial effects on markers of bone turnover and specific gut-derived pro-inflammatory proteins, with children on EEN showing mean weight gains of 4.7±3.5 kg.8, 9
Over recent years, the mechanism by which EEN induces remission has been a major topic of interest. Proposed mechanisms include direct anti-inflammatory effects, improved epithelial barrier function, and modulation of the gut microbiota.10 A recent study using a Helicobacter trogontum murine model of colitis has shown that administration of EEN leads to reductions in histological inflammatory scores, improved epithelial barrier function (via modifications in tight junction proteins) and reduced levels of pro-inflammatory cytokines as compared with untreated mice.11 Indeed, treatment with EEN, but not hydrocortisone, not only led to these changes but also a reduction in H. trogontum load.
This latter finding is consistent with a number of studies that have shown EEN therapy to have a significant effect on the intestinal microbiota.12, 13, 14, 15, 16, 17, 18 Although the findings of these studies are of great interest, to date, only the study of D'Argenio and colleagues17 has employed the highly sensitive technique of high throughput sequencing (HTS) to characterize the effect of EEN therapy on the intestinal microbiota of CD patients. Given that their study included only one CD patient and a matched control and that samples were only collected prior to and following EEN therapy, if we are to gain a comprehensive understanding of the effect of EEN on the intestinal microbiota, larger studies using HTS are required. Given this, in the current study, we used 16S rRNA gene and whole-genome HTS to determine changes in the fecal microbiota of five CD children, before, during, and after EEN therapy and compared this with five healthy controls.
Methods
Children with CD and healthy controls
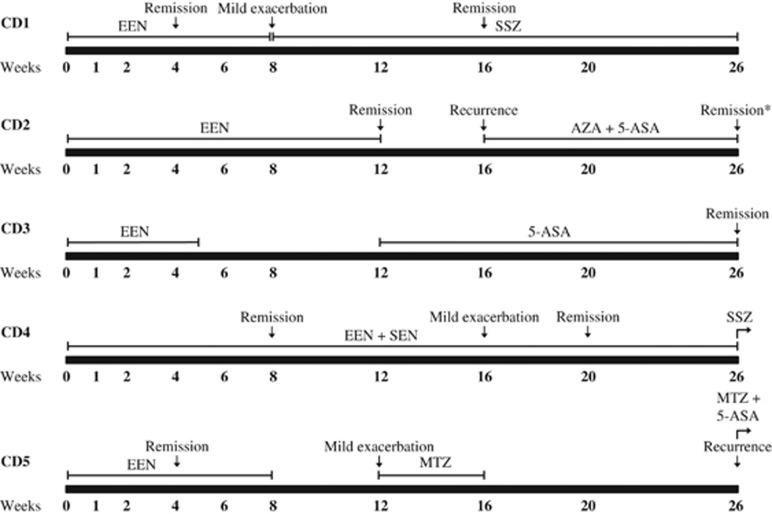
Five children attending the Sydney Children's Hospital (SCH), Randwick, Inflammatory Bowel Disease (IBD) clinic were recruited prospectively at the time of diagnosis of CD (Figure 1,Table 1). The diagnosis of CD was based upon standard histologic, endoscopic, and radiologic criteria, after performing upper gastrointestinal endoscopy, colonoscopy, and radiological assessment of the small bowel.19 Following diagnosis, all five children were managed with EEN following a standard protocol5 to induce remission. The children were prescribed Osmolite (Abbott Laboratories; Cronulla, NSW, Australia), a polymeric formula containing whole polypeptides and all nutritional requirements, to be taken as their sole nutritional source.5 EEN therapy commenced within 2 days of diagnosis and continued for 8–12 weeks in most cases. In the case of unresponsiveness or recurrence, patients were treated with antibiotics or anti-inflammatory therapy (Figure 1). Disease location was based on endoscopic and histological findings and classified as L1 (terminal ileum), L2 (colon), L3 (ileocolon), L4 (upper gastrointestinal tract), or a combination as outlined by the Montreal classification.20 The PCDAI for each patient was calculated at diagnosis (baseline, week 0) and at 2, 4, 6, 8, 16, and 26 weeks post commencement of EEN.21 Disease remission was defined as PCDAI <10.
Figure 1.
Treatment and disease activity profiles of five children with Crohn's disease over 26 weeks. *Patient was classified as in remission, however, PCDAI was calculated to be 10. 5-ASA, 5-aminosalicyclic acid; AZA, azathioprine; EEN, exclusive enteral nutrition; MTZ, metronidazole; PCDAI, Pediatric Crohn's Disease Activity Index; SEN, supplementary enteral nutrition; SSZ, sulfasalazine.
Table 1. Patient details and clinical characteristics.
| Patient | Gender | Age (years) | Disease location | Clinical values |
Weeks |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 4 | 6 | 8 | 12 | 16 | 20 | 26 | |||||
| CD1 | M | 8.6 | L3+L4 | PCDAI | 25 | — | — | 0 | — | 10 | — | 0 | — | 0 |
| Albumin (g/dl) | 34 | — | — | 38 | — | 38 | — | 48 | — | 49 | ||||
| C-reactive protein (mg/l) | 7 | — | — | 1 | — | 4 | — | 0 | — | 1 | ||||
| ESR (mm/h) | 37 | — | — | 19 | — | 16 | — | 5 | — | 8 | ||||
| Hemoglobin (g/dl) | 107 | — | — | 120 | — | 126 | — | 130 | — | 130 | ||||
| CD2 | M | 9.7 | L3 | PCDAI | 35 | — | — | — | 30 | — | 5 | 15 | — | 10 |
| Albumin (g/dl) | 40 | — | — | — | 32 | — | 39 | 39 | — | 39 | ||||
| C-reactive protein (mg/l) | 1 | — | — | — | 1 | — | 5 | 1 | — | 1 | ||||
| ESR (mm/h) | 38 | — | — | — | 38 | — | 16 | 18 | — | 19 | ||||
| Hemoglobin (g/dl) | 113 | — | — | — | 93 | — | 108 | 117 | — | 120 | ||||
| CD3 | M | 7.2 | L3+L4 | PCDAI | 42.5 | — | 17.5 | 15 | 25 | 40 | 25 | — | — | 0 |
| Albumin (g/dl) | 22 | — | 29 | 31 | 23 | 23 | — | — | — | — | ||||
| C-reactive protein (mg/l) | 80 | — | 17 | 41 | 58 | 58 | — | — | — | — | ||||
| ESR (mm/h) | 43 | — | 16 | 24 | 40 | 40 | — | — | — | — | ||||
| Hemoglobin (g/dl) | 79 | — | 94 | 97 | 93 | 93 | — | — | — | — | ||||
| CD4 | M | 13.8 | L3+L4 | PCDAI | 45 | — | 15 | — | 12.5 | 7.5 | 5 | 10 | 0 | 2.5 |
| Albumin (g/dl) | 33 | — | 34 | — | 34 | 35 | — | 33 | — | 40 | ||||
| C-reactive protein (mg/l) | 9 | — | 1 | — | 1 | 2 | — | 6 | — | 4 | ||||
| ESR (mm/h) | 25 | — | 23 | — | 25 | 23 | — | 17 | — | 24 | ||||
| Hemoglobin (g/dl) | 97 | — | 96 | — | 108 | 114 | — | 120 | — | 120 | ||||
| CD5 | M | 10.1 | L3+L4 | PCDAI | 20 | — | — | 0 | 5 | — | 10 | 10 | — | 15 |
| Albumin (g/dl) | 37 | — | — | 45 | 43 | — | — | 43 | — | 37 | ||||
| C-reactive protein (mg/l) | 1 | — | — | 1 | 1 | — | — | 1 | — | 2 | ||||
| ESR (mm/h) | 2 | — | — | 1 | 1 | — | — | 5 | — | 6 | ||||
| Hemoglobin (g/dl) | 131 | — | — | 139 | 139 | — | — | 140 | — | 135 | ||||
CD, Crohn's disease; ESR, erythrocyte sedimentation rate; M, male; PCDAI, Pediatric Crohn's Disease Activity Index.
Five healthy children from families and staff at SCH volunteered as controls. These children did not have a current diagnosis of gastrointestinal disease, did not display gastrointestinal symptoms, and did not have a significant change in diet over the study period.
Informed consent was obtained from all children (or their parent/guardian for younger children) to be included in the study. This study was approved by the Research Ethics Committees of the University of New South Wales and the South Eastern Sydney and Illawarra Area Health Service Research Ethics Committee (ethics no. 03/163, 03/165, and 06/164).
Exclusion criteria
No child involved in the study had undergone antibiotic or anti-inflammatory therapy in the 4 weeks prior to the study. Exclusion criteria for enrolment of CD patients were previous diagnosis of CD or UC and severe colitis requiring intensive medical or surgical management. Exclusion criteria for enrolment of healthy controls were any relationship to a CD patient.
Fecal sample collection and DNA extraction
Fecal samples from the five CD patients were collected at baseline (prior to endoscopy preparation, week 0) and then at 1, 2, 4, 6, 8, 12, 16, and 26 weeks following diagnosis. Owing to the lack of material, not all samples were available for analysis, however, all children had a minimum of a baseline, two time-points during EEN, 8- and 26-week samples. A major advantage of this approach is that each CD patient can also act as their own control, hence minimizing the effect of variation in the bacterial community between individuals. A baseline fecal sample was collected from each of the healthy control subjects. Samples were collected in sterile collection containers (Techno-Plas; St Marys, SA, Australia) and immediately stored at −20 °C, then transported frozen to the laboratory where they were stored at −80 °C. DNA was extracted using the method of Griffiths and colleagues.22 The concentration and quality of DNA was measured using a Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies; Wilmington, DE, USA).
Tag-encoded FLX amplicon pyrosequencing
The microbial community was assessed by high-throughput sequencing of the 16S rRNA gene. Bacterial tag-encoded FLX amplicon pyrosequencing was performed as described previously23, 24, 25, 26, 27 using the primers Gray28F (5′TTTGATCNTGGCTCAG) and Gray519r (5′ GTNTTACNGCGGCKGCTG) numbered in relation to E. coli 16S rRNA (variable regions 1–3). Generation of the sequencing library utilized a one-step PCR with a total of 30 cycles, a mixture of Hot Start and HotStar high fidelity Taq polymerases, and amplicons originated and sequencing extended from the 28F with an average read length of 400 bp. Tag-encoded FLX amplicon pyrosequencing analyses utilized a Roche 454 FLX instrument (Indianapolis, IN, USA) with Titanium reagents. This bacterial tag-encoded FLX amplicon pyrosequencing process was performed at the Research and Testing laboratory (Lubbock, TX, USA) based upon established and validated protocols.
Pyrosequencing data analysis
The sequence data derived from the high-throughput sequencing process were analyzed employing two different methods to confirm the reproducibility of our analyses. The first method utilized a pipeline developed at the Research and Testing laboratory. Sequences were first depleted of barcodes and primers, then short sequences (<200 bp), sequences with ambiguous base calls, and sequences with homopolymer runs exceeding 6 bp were removed. Sequences were then de-noised and chimeras were removed (Black Box Chimera Check software B2C2).28 Operational taxonomic units (OTU) were defined after removal of singleton sequences (sequences appearing only once in the whole dataset) with clustering set at 3% divergence (97% similarity).23, 24, 25, 26, 27 OTUs were then taxonomically classified using BLASTn against a curated GreenGenes database29 and compiled into each taxonomic level. Taxonomy was defined based on the following percentages: >97%, species; between 97 and 95%, unclassified species; between 95 and 90%, unclassified genus; between 90 and 85%, unclassified family; between 85 and 80%, unclassified order; between 80 and 77%, unclassified phylum; <77%, unclassified.
The second method analyzed the resulting raw sequence reads using the sequence analysis pipeline Mothur.30 Sequences were initially quality-filtered using the pyronoise algorithm31 followed by removing sequences that were too short (<180 bp), contained ambiguous bass calls or had >six homopolymers. Quality-filtered sequences were aligned using the Silva 16S reference alignment,32 screened to include only overlapping regions of sequence, pre-clustered (diffs=2), and chimera checked (uchime). Sequences were classified using the Ribosomal database project 9 taxonomy (RDP9).33 Sequences were then clustered into OTUs at 97% similarity with consensus taxonomy. All statistical analyses on relative abundance levels of bacterial taxa from both methods were performed using the Primer-E package.34
Shotgun metagenomic analysis
DNA extracted from the patient fecal samples was prepared using the Ion AmpliSeq Library Kit 2.0 (Life Technologies, Grand Island, NY, USA) and analyzed on an Ion Proton System (Life Technologies) to generate whole-genome sequence data. Raw unassembled whole-genome sequence reads were analyzed using the GENIUS software package (CosmosID, College Park, MD, USA) for rapid identification of bacterial species and relative abundance. GENIUS creates sample libraries from unassembled short whole-genome sequence reads using two algorithms, 5VCE and NmerCE, and utilizes GeneBook reference libraries derived from curated genomic databases to assign taxonomic membership of sample libraries, employing probabilistic matching.35 Identification is achieved at species, sub-species, and/or strain level, depending on adequate representation of relevant reference genomes in the GeneBook libraries.
Results
Fecal microbiota of CD patients and healthy controls
Over the last decade, microbial dysbiosis has been implicated in the etiology of CD, however, the exact nature of the breakdown in the balance between commensal and pathogenic intestinal bacteria remains a topic of debate.36, 37 Microbial community analyses were performed on fecal samples from five CD patients and five healthy controls through the pyrosequencing of the 16S rRNA gene and subsequent analysis of the read data using two different methods. The average number of processed reads per sample amounted to 5,960±559 and 4,357±432 for method 1 and method 2, respectively. Upon taxonomic classification, method 1 identified a total of 373 OTUs within the 39 samples whereas method 2 had higher differentiating power with 1,350 OTUs detected within the same samples.
The average number of OTUs at baseline (week 0, prior to therapy) for CD patients was 79±9 and 121±33 using method 1 and method 2, respectively. This was similar to the average number of OTUs detected in the healthy controls (method 1: 62±4, P=0.24; method 2: 117±12, P=0.92). Analysis of Shannon's microbial diversity (H′) between CD patients and controls showed that while method 1 did not identify any difference between the two groups (CD: 2.33±0.12, control: 2.30±0.11, P=0.86), a lower trend in the microbial diversity of CD patients was calculated from abundance levels generated through method 2 (CD: 2.25±0.24, control: 2.75±0.14, P=0.11).
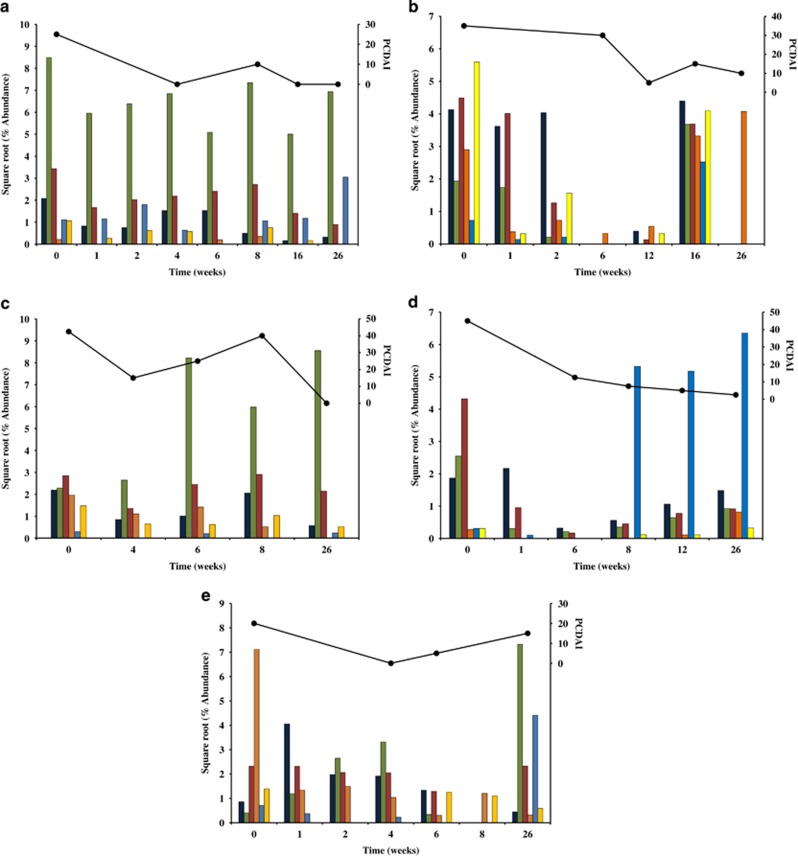
A clear dysbiosis was observed in the microbiota at the phyla level for four of the five CD patients (CD2-5) before therapy (week 0) as compared with the controls (Figures 2 and 3). Patient CD1 appeared initially to have a similar microbial profile to the healthy controls, however, a more detailed inspection found the level of Firmicutes and Fusobacteria to be elevated whereas levels of Bacteroidetes, Proteobacteria, and Actinobacteria were reduced (Figure 3).
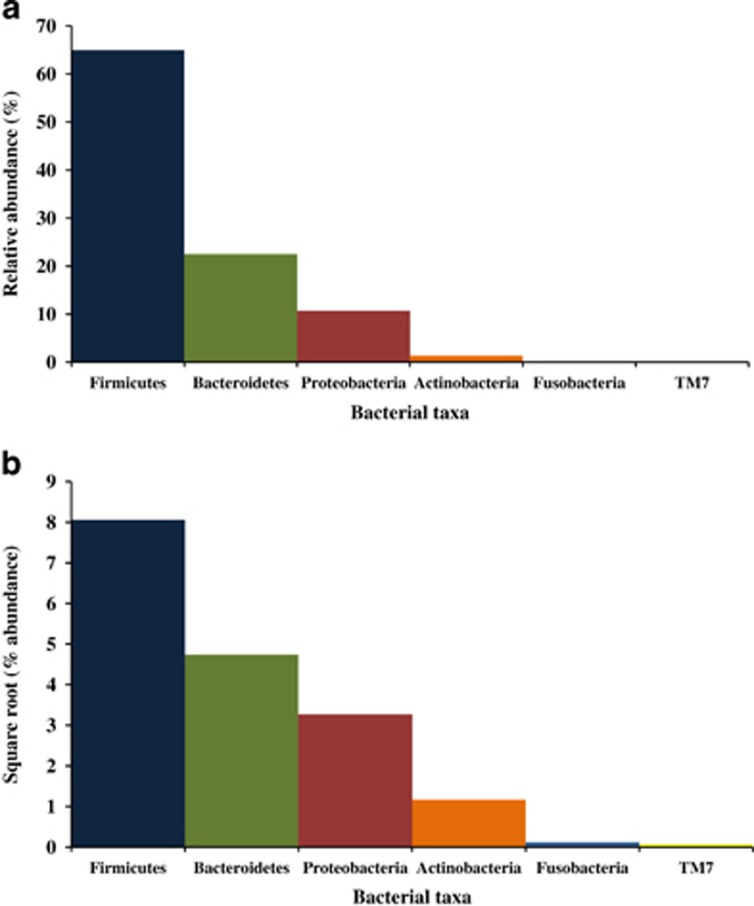
Figure 2.
Microbial profile of the healthy controls at the phylum level. (a) Average relative abundance (%) of bacterial taxa. (b) Standardized relative abundance levels (square root) of bacterial taxa. Abundance levels determined from method 2 were used.
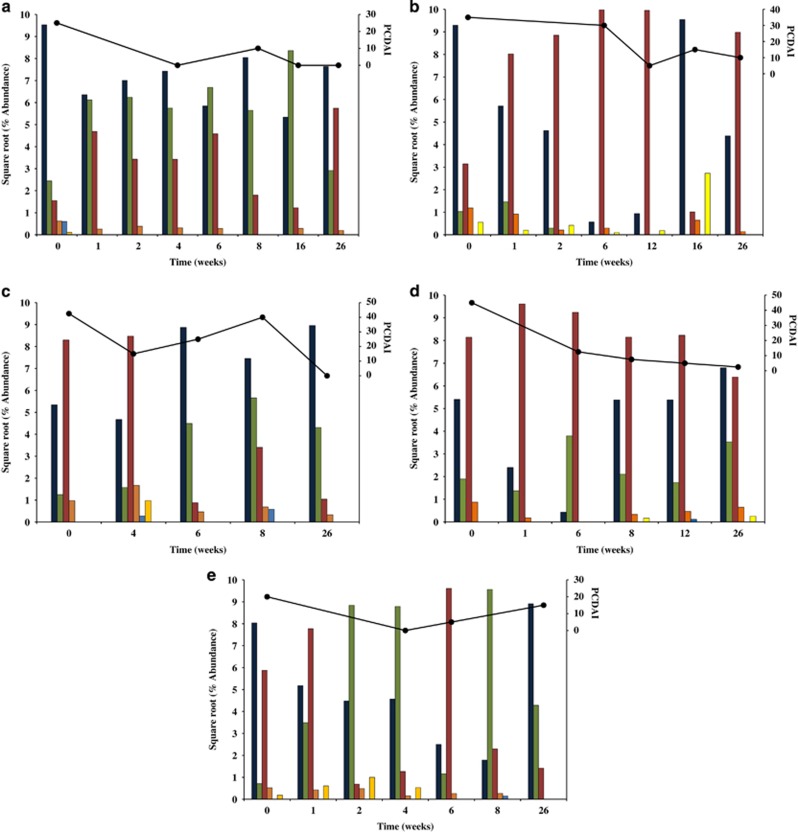
Figure 3.
Changes in the relative abundance of phyla in CD patients undergoing EEN therapy. (a) CD1, (b) CD2, (c) CD3, (d) CD4, and (e) CD5. Standardized relative abundance levels (square root) of bacterial taxa were plotted. Abundance levels determined from method 2 were used. Firmicutes (dark blue), Bacteroidetes (green), Proteobacteria (red), Actinobacteria (orange), Fusobacteria (light blue), and TM7 (yellow). CD, Crohn's disease; EEN, exclusive enteral nutrition.
Effect of EEN therapy on the fecal microbiota of CD patients
Four of the five patients (CD1, 2, 4, and 5) responded positively to EEN, with remission achieved within 4–12 weeks of therapy (Figure 1) as determined by the PCDAI scores (Table 1). Although patient CD3 responded positively to EEN, the therapy was terminated at 5 weeks prior to the disease going into remission (Figure 1, Table 1). Three of the five patients (CD1, CD2, and CD5) experienced a mild exacerbation or recurrence of the disease after the conclusion of EEN therapy and were commenced on medical therapies (Figure 1). One patient (CD4) experienced a mild exacerbation at week 16 of the 26-week period of observation, however, he was again in remission by week 20 (Figure 1).
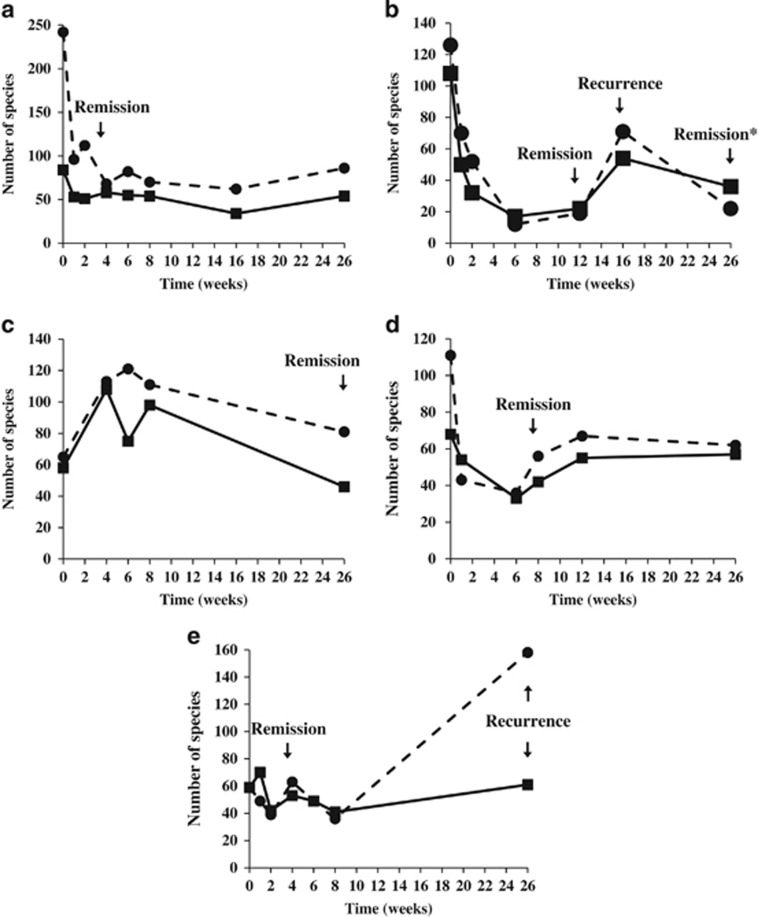
The fecal microbiota of the patients during EEN and post therapy were analyzed using two methods and compared with their microbiota prior to therapy. Changes in the number of microbial OTUs were graphed across the collection time-points and related back to the disease activity of the patient (Figure 4). Significantly, the number of OTUs decreased dramatically upon starting EEN therapy, which corresponded to disease remission (Figure 4a and d). In contrast, recurrence of disease corresponded with an increase in the number of OTUs (Figure 4b). Interestingly, patient CD3, whose therapy was ceased at 5 weeks because of the lack of remission, responded differently to EEN, with a dramatic increase in the number of OTUs occurring upon starting EEN (Figure 4c).
Figure 4.
Changes in the number of OTUs in the fecal microbiota of CD patients undergoing EEN therapy. (a) CD1, (b) CD2, (c) CD3, (d) CD4, and (e) CD5. Solid and dotted lines correspond to the number of OTUs from method 1 and method 2, respectively. CD, Crohn's disease; EEN, exclusive enteral nutrition; OTU, operational taxonomic unit.
The nature of these changes in the CD patients was investigated further by examining the effect of EEN therapy on the microbiota at the phylum level (Figure 3). As expected, the microbiota of the CD patients behaved differently given the initial variations between the baseline samples and differences in the patients' responses to therapy (Figure 3a and e). However, interesting patterns emerged from each patient. For example, the relative levels of Bacteroidetes increased with EEN in patients CD1 and CD5 (Figure 3a), suggesting that in some cases, EEN may support the growth of these bacterial taxa. In addition to these changes, EEN therapy resulted in the elimination of Fusobacteria and TM7 in patient CD1 and a decrease in Proteobacteria in patient CD5 (Figure 3a).
The microbiota of patient CD2 became dominated by Proteobacteria during EEN therapy; in particular, this was associated with an OTU corresponding to Ralstonia. This increase resulted in the abundance of other taxa falling to negligible levels, a change that was associated with improvement in PCDAI (Figure 3b). Following the conclusion of EEN therapy, the abundance of Ralstonia decreased dramatically (week 16) and other microbial taxa dominated the microbiota of this patient (Figure 3b), a change that corresponded with disease recurrence. Treatment of patient CD2 with maintenance drug therapies resulted in the re-establishment of dominance of Ralstonia (week 26) and disease activity improvement (Figure 3b). The microbiota of patients CD3 and CD4 behaved differently at the phylum level (Figure 3c), which is not unexpected given the broad nature of the phylum classification. Interestingly, patient CD3 experienced an increase in both Fusobacteria and disease activity at week 8, while at week 26 (remission) the profile of this patient's microbiota had shifted closer to the microbial profile of the controls (Figure 2).
Microbial taxa of interest in CD patients undergoing EEN therapy
Given the dominance of Firmicutes in the microbiota of humans and its variable response to EEN therapy (Figure 3), the effect of EEN on families within this phylum was investigated to gain a better understanding of the role of these microbial taxa in disease. Six families (Erysipelotrichaceae, Ruminococcaceae, Lachnospiraceae, Streptococcaceae, Veillonellaceae, and Peptostreptococcaceae) were found in some cases to correlate with disease activity during and following EEN therapy (Figure 5).
Figure 5.
Changes in the relative abundance of Firmicutes taxa in CD patients undergoing EEN therapy. (a) CD1, (b) CD2, (c) CD3, (d) CD4, and (e) CD5. Standardized relative abundance levels (square root) of bacterial taxa were plotted. Abundance levels determined from method 2 were used. Erysipelotrichaceae (dark blue), Ruminococcaceae (green), Lachnospiraceae (red), Streptococcaceae (orange), Veillonellaceae (light blue), and Peptostreptococcaceae (yellow). CD, Crohn's disease; EEN, exclusive enteral nutrition.
In patient CD1, the levels of Erysipelotrichaceae, Ruminococcaceae, Lachnospiraceae, and Peptostreptococcaceae decreased following EEN therapy, and this corresponded with an improvement in disease activity (Figure 5a). At week 8, patient CD1 had a mild exacerbation of disease which appeared to be associated with an increase in Ruminococcaceae, Lachnospiraceae, and Peptostreptococcaceae (Figure 5a). At weeks 16 and 26, the levels of Lachnospiraceae and Peptostreptococcaceae decreased, which corresponded once again with an improvement in the patient's PCDAI score (Figure 5a). The sequence data from patient CD2 indicated that the decrease in disease activity by week 12 (shortly after the completion of EEN therapy) was related to a drop in the abundance of these six families (Figure 5b), and most likely resulting from the dominance of Ralstonia (discussed earlier). The six families were found to increase in abundance dramatically at week 16 (no EEN) at which point, this patient had an exacerbation of their symptoms. Following treatment with drug therapies (week 26), this patient's disease activity improved and once again this was related to the eradication of Erysipelotrichaceae, Ruminococcaceae, Lachnospiraceae, Veillonellaceae, and Peptostreptococcaceae. However, Streptococcaceae remained high which may explain why inflammation did not completely subside in this patient (PCDAI=10; Table 1).
Strikingly similar patterns were observed for both Erysipelotrichaceae and Lachnospiraceae and the PCDAI scores of patient CD3 (Figure 5c). The levels of Streptococcaceae also behaved in a similar manner to the PCDAI scores, except at week 8 (Figure 5c). Large drops in the levels of Erysipelotrichaceae, Ruminococcaceae, and Lachnospiraceae once again correlated with both EEN therapy and improvement in disease activity in patient CD4 (Figure 5d). Although, in this patient, a large increase in the abundance of Veillonellaceae was observed between week 8 and 26 of EEN therapy, this did not correlate with disease activity (Figure 5d). In contrast, in patient CD5, the above microbial families behaved differently. In this patient, the microbiota appeared to be dominated by Streptococcaceae, which decreased considerably in abundance following EEN therapy (Figure 5e). The recurrence of disease, however, did not appear to be associated with Streptococcaceae (Figure 5e).
To determine which OTUs were associated with EEN therapy and disease activity, abundance levels at the genus level were examined in each patient and microbial taxa exhibiting relevant patterns were plotted (Supplementary Figure S1). In patient CD1, EEN therapy resulted in a decrease in levels of Faecalibacterium and an increase in Bacteroides (Supplementary Figure S1A). The levels of Bacteroides also increased in response to EEN in patients CD4 and CD5 (Supplementary Figure S1D, E). Other microbial taxa that increased with EEN regardless of disease activity included Flavonifractor (CD1, CD3, CD4, CD5) and Caulobacter (CD2, CD3, CD4) (Supplementary Figure S1).
Several microbial taxa exhibited patterns in their abundance levels that correlated with disease activity including Lachnospiraceae unclassified (all patients), Lachnospiraceae incertae sedis (CD1, CD2, CD4, CD5), Clostridium XI (CD1, CD2, CD3, CD5), Clostridium XVIII (CD2, CD4), Ruminococcaceae unclassified (CD3, CD4), Streptococcus (CD2, CD5), Dialister (CD2, CD5), Escherichia/Shigella (CD2, CD3), and Fusobacterium (CD1, CD3).
Microbial community analyses using whole-genome sequencing
To gain a more in-depth understanding of the changes in the fecal microbiota of CD patients undergoing EEN therapy and to confirm our findings using HTS on the 16S rRNA gene, microbial community analysis using metagenomics was performed on a subset of the samples. In particular, the transition of patient CD2 from remission (PCDAI=5, week 12) to active disease (PCDAI=15, week 16) and once again to remission (PCDAI=10, week 26) and the dominance of Ralstonia during therapy were investigated further (Supplementary Data 1). In addition, patient CD3, in whom EEN therapy was ceased after 5 weeks, was investigated post-EEN therapy at week 6 (post-EEN therapy, PCDAI=25), week 8 (re-establishment of moderate/severe disease, PCDAI=40), and week 26 (remission, PCDAI=0) (Supplementary Data 1).
The dominance of Ralstonia in patient CD2, specifically Ralstonia pickettii, during both EEN and drug treatments observed using the 16S rRNA gene HTS was confirmed using metagenomics (Supplementary Data 1). The lack of Ralstonia pickettii in samples from week 0 and week 16 (Supplementary Figure S1B, Supplementary Data 1) ruled out any contamination or sequencing errors. Moreover, the decrease in Ruminococcaceae observed in patient CD2, particularly in week 6 and 12 (Figure 5b), was confirmed in the metagenomics data through the depletion of Ruminococcus sp 5_1_39B_FAA and Faecalibacterium cf. prausnitzii KLE1255. Interestingly, the increase in Faecalibacterium at week 16 (Supplementary Figure S1B), 4 weeks after the completion of EEN therapy, was due to an increase in the abundance of a different OTU, namely, Faecalibacterium prausnitzii L2-6 (Supplementary Data 1). This confirmed our earlier findings that OTUs corresponding to Faecalibacterium behave differently during therapy and in relation to disease activity. Another notable finding was the confirmation of Lachnospiraceae bacterium 5_1_63FAA as the microbial taxa responsible for the pattern (depletion at week 6 and 12, return at week 16, depletion at week 26) observed for Lachnospiraceae unclassified in patient CD2 (Supplementary Figure S1B).
The fecal microbiota of patient CD3 exhibited a larger number of taxa post-EEN therapy in the metagenomic data (Supplementary Data 1), as was observed in the 16S rRNA gene results. No dominance in Ralstonia was observed in the fecal microbiota of this patient during or outside of therapy. Increases in the relative abundance levels of Bacteroides species at week 6, 8, and 26 (Supplementary Figure S1C) were confirmed by the metagenomics data (Supplementary Data 1). Moreover, as with patient CD2, microbial taxa within Erysipelotrichaceae (Figure 5c), Ruminococcaceae (Supplementary Figure S1C), and Lachnospiraceae (Figure 5c, Supplementary Figure S1C) responsible for patterns relevant to disease activity were identified. These included Erysipelotrichaceae bacterium 6_1_45 (week 6: 1.28%, week 8: 4.76%, week 26: 0.48%), Ruminococcus gnavus ATCC 29149 (week 6: 3.15%, week 8: 7.61%, week 26: 0.23%), Subdoligranulum spp. 4_3_54A2FAA (week 6: 0.64%, week 8: 4.15%, week 26: 0.23%), and Lachnospiraceae bacterium 5_1_63FAA (week 6: 1.13%, week 8: 7.69%, week 26: 0.16%). Notably, this analysis also identified substantial colonization by Bifidobacterium species at remission (week 26, total relative abundance: 6.22%), which were absent in the patient at week 6 and week 8 (Supplementary Data 1).
Discussion
EEN is now considered a common and effective therapy in pediatric CD; however, the mechanism of action by which remission is induced by EEN is currently unclear but has been suggested to involve direct anti-inflammatory effects, improved epithelial barrier function, and modulation of the gut microbiota.10 In line with this, several studies have investigated the effect of EEN on the gut microbiota of CD patients. For example, in 2004, Pryce-Miller and colleagues12 analyzed the microbiota of ileocolonic biopsy samples from six children using denaturing gradient gel electrophoresis, and showed that EEN can lead to restoration of the diversity of the microbiota in CD patients to that observed in controls. These findings were supported by Lionnetti and colleagues,13 who demonstrated that EEN therapy resulted in a significant modification of the fecal microbiota. Further, Leach and colleagues14 using denaturing gradient gel electrophoresis examined serial stool samples collected from six children with CD prior to EEN, during EEN treatment, and after EEN, and showed that EEN had a significant and sustained effect on the intestinal bacterial composition and that an association existed between the Bacteroides-Prevotella group and reduced disease activity.
More recently, Shiga and colleagues15 have reported, based on terminal restriction fragment length polymorphism analysis, that in patients with CD, species diversity was reduced by total parenteral nutrition, however, not by an elemental diet. Following the elemental diet, these authors observed a significant decrease in Bacteroides fragilis, whereas after total parenteral nutrition, Enterococcus was significantly increased. Further, Gerasimidis and colleagues16 have reported based on temporal temperature gradient gel electrophoresis, a similar decrease in bacterial diversity and the Bacteroides/Prevotella group in CD patients undergoing EEN therapy. However, when the children returned to their normal diet, microbial levels returned to pre-treatment levels. In addition, D'Argenio and colleagues17 employed 16S rRNA gene HTS to characterize the ileal mucosal microbiome of one CD patient and a matched control. While prior to EEN therapy bacterial diversity was significantly lower, Proteobacteria more abundant and Bacteroidetes less abundant in the CD patient as compared with the control, following therapy, these differences between the CD patient and the control were no longer observed. In line with these findings, Tjellstrom and colleagues18 have reported 6-weeks EEN therapy to modulate the activity of the microbiota in ileal/colonic CD patients, resulting in an anti-inflammatory short chain fatty acid pattern. In contrast, children with perianal disease showed no clinical or biochemical improvement following EEN therapy.18
In the current study, we used 16S rRNA gene and whole-genome HTS to determine changes in the fecal microbiota of five CD children, before, during, and after EEN therapy and compared this with five healthy controls in an attempt to gain a comprehensive understanding of the effect of EEN on the intestinal microbiota. We observed a reduced microbial diversity in CD patients as compared with controls, a finding that has been well documented in the literature.37, 38 Furthermore, we observed dysbiosis within the microbiota of CD patients as compared with controls, which is in line with other studies that consistently report dysbiosis to play a pivotal role in the pathogenesis of CD.37 Interestingly, the microbial profiles of the five CD patients were remarkably different as evidenced by the large variation around the mean values. The variability in the microbiota of CD patients supports the overall conclusion that several microbial species may be involved in the pathogenesis of this disease, as is evidenced by the large number of bacteria associated with this disease.37 However, these differences could also relate to the length of time that the child has had symptoms prior to having colonoscopy as well as the severity of inflammation. Given this, for a reliable evaluation of the microbial etiological agent(s) of CD, it was essential to analyze each patient independently.
Overall, EEN therapy appeared to have a positive effect on these children with 80% going into remission, similar to previous reports.4, 5 However, in some of these patients, following the conclusion of EEN therapy, the positive effect appeared to subside over time. Our findings based on a comprehensive microbial community profiling technique support the decrease in microbial diversity reported previously in patients undergoing EEN therapy,15, 16 but go further in that they associate these fluctuations in microbial OTUs with disease activity. For example, the eradication of Fusobacteria in patient CD1 is of particular interest as Fusobacterium spp. have previously been associated with the development of IBD.39, 40
More importantly, our findings in relation to Erysipelotrichaceae, Ruminococcaceae, Lachnospiraceae, Streptococcaceae, Veillonellaceae, and Peptostreptococcaceae are of particular interest given a number of recent studies. In a study by Zhu and colleagues41 using an animal model of colonic cancer, a significant increase in Erysipelotrichaceae was observed within the tumor group. Given that inflammatory bowel diseases are associated with an increased risk of developing cancers such as colorectal cancer and colitis-associated adenocarcinoma,42 the pathogenic potential of this group of microbial species should be investigated further. Further, Berry and colleagues43 have reported an increased abundance of Ruminococcaceae in mice with dextran sodium sulfate-induced colitis. In that study, the authors also identified contrasting abundance changes among taxa of the family Lachnospiraceae, where one taxon drastically decreased during inflammation in wild-type and STAT1−/− mice, whereas another drastically increased during inflammation in STAT1−/− mice.43 These findings, in addition to our own findings, strongly advocate that further investigation of the levels of Erysipelotrichaceae, Ruminococcaceae, and Lachnospiraceae in the etiology of pediatric CD are warranted.
Our findings from the whole-genome sequencing analyses not only confirm our pyrosequencing results but also identify specific OTUs that were responsible for the correlation with disease activity, suggesting that these taxa may be of major interest in the etiology of CD. Our results shed light on a recent contentious issue related to the changes in abundance of Faecalibacterium following EEN therapy. The decrease in Faecalibacterium levels following EEN therapy and its lack of correlation with improvement in disease has been previously documented.16, 44 Notably, in our study, different OTUs classified under Faecalibacterium responded differently to EEN therapy, which may explain the variations observed between patients in relation to changes in abundance of this microbial taxa during EEN therapy (Supplementary Figure S1A and E). Of particular interest was the correlation of specific Lachnospiraceae species with therapy and disease activity in all patients, given that bacteria from this family have been implicated in disease-associated immunity in CD.45, 46 Furthermore, this analysis also identified substantial colonization by Bifidobacterium species at remission, which were absent in the patient during disease, suggesting that these bacterial species may form the basis of potentially beneficial probiotic therapies that can supplement EEN therapy.
Our most consistent finding across both analyses is the decrease in number of OTUs and microbial diversity within CD patients, and the correlation of this phenomenon with disease remission (Figure 4). Furthermore, the re-colonization of patients with specific microbial taxa belonging to six Firmicutes families was also clearly correlated with disease recurrence in our patients (Figure 4). These results may be reconciled through the findings of a recent comprehensive study by Palm and colleagues,47 which found that CD patients are colonized by inflammatory commensals that illicit higher immune responses than the same commensals from healthy controls. Thus, depletion of these inflammatory commensals by EEN therapy would result in a decrease in immune reactivity towards the CD microbiota; however, upon re-establishment of dominance following cessation of EEN, these taxa will once again illicit an immune response. Furthermore, the depletion of the inflammatory commensal microbiota may also explain why patient CD2 suffered no adverse effects when their microbiota was depleted and then dominated by Ralstonia pickettii. Interestingly, Kamada and colleagues48 have previously shown that in a Citrobacter rodentium mouse model, germ-free mice harbored 10-fold more C. rodentium, yet remarkably the recruitment of neutrophils, inflammatory macrophages, and CD3+ T cells in response to infection as compared with specific pathogen-free mice was similar. Furthermore, despite high and persistent pathogen burdens in the germ-free mice, pathology scores declined in both germ-free and specific pathogen-free mice suggesting that the commensal microbiota plays a role in perpetuating inflammation.
In conclusion, all newly diagnosed pediatric CD patients in this study showed a positive clinical response to EEN, with four out of the five patients achieving remission by the end of the therapy. However, subsequently, some CD patients suffered an exacerbation of their disease. Of particular interest, the disease activity in these CD patients correlated with the number of OTUs in their fecal microbiota. Our results clearly demonstrate that EEN leads to common and patient-specific alterations in the microbiota, some of which correlate with disease activity. Specific species within the Lachnospiraceae, Ruminococcaceae, and Erysipelotrichaceae were among the key taxa identified to potentially be involved in the perpetuation of gut inflammation in CD. Given the decrease in the number of OTUs and microbial diversity secondary to EEN, and the correlation of disease recurrence with the re-colonization of eradicated taxa post-EEN, a sequential EEN-probiotic therapy may prove beneficial in the improvement of the long-term efficacy of EEN therapy in pediatric CD. However, given the relatively small sample size examined in this study, further confirmation of our results in a larger group of CD patients is clearly required. Moreover, future studies should include comparisons of the microbial composition of patients undergoing EEN with patients on other therapies to determine whether these microbial changes are specific to EEN therapy as well as in-depth metagenomic pathway analysis.
Study Highlights

Guarantor of the article: Hazel M. Mitchell, Dip. Ed, PhD.
Specific author contributions: H.M.M. and A.S.D. conceived the idea; H.M.M., A.S.D., D.A.L., and N.O.K. contributed reagents; A.S.D., S.T.L., and D.A.L. collected and organized all samples; S.T.L., A.S.D., and D.A.L. performed clinical parameter measurements, N.O.K. extracted and prepared all D.N.A. samples; N.O.K. and S.N. performed the microbial community analyses; N.O.K., S.N., and H.M.M. analyzed the results; N.O.K., H.M.M., A.S.D., S.T.L., S.N., and D.A.L. drafted the manuscript.
Financial support: N.O.K. is supported by an Early Career fellowship from the National Health and Medical Research Council, Australia. N.O.K. and H.M.M. would like to acknowledge financial support from The University of New South Wales.
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- Talley NJ, Abreu MT, Achkar JP, et al. An evidence-based systematic review on medical therapies for inflammatory bowel disease. Am J Gastroenterol. 2011;106:S2–25. doi: 10.1038/ajg.2011.58. [DOI] [PubMed] [Google Scholar]
- Heuschkel RB, Menache CC, Megerian JT, et al. Enteral nutrition and corticosteroids in the treatment of acute Crohn's disease in children. J Pediatr Gastroenterol Nutr. 2000;31:8–15. doi: 10.1097/00005176-200007000-00005. [DOI] [PubMed] [Google Scholar]
- Breese EJ, Michie CA, Nicholls SW, et al. The effect of treatment on lymphokine-secreting cells in the intestinal mucosa of children with Crohn's disease. Aliment Pharmacol Ther. 1995;9:547–552. doi: 10.1111/j.1365-2036.1995.tb00419.x. [DOI] [PubMed] [Google Scholar]
- Day AS, Whitten KE, Lemberg DA, et al. Exclusive enteral feeding as primary therapy for Crohn's disease in Australian children and adolescents: a feasible and effective approach. J Gastroenterol Hepatol. 2006;21:1609–1614. doi: 10.1111/j.1440-1746.2006.04294.x. [DOI] [PubMed] [Google Scholar]
- Berni Canani R, Terrin G, Borrelli O, et al. Short- and long-term therapeutic efficacy of nutritional therapy and corticosteroids in paediatric Crohn's disease. Dig Liver Dis. 2006;38:381–387. doi: 10.1016/j.dld.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Critch J, Day AS, Otley A, et al. Use of enteral nutrition for the control of intestinal inflammation in pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2012;54:298–305. doi: 10.1097/MPG.0b013e318235b397. [DOI] [PubMed] [Google Scholar]
- Nahidi L, Leach ST, Sidler MA, et al. Osteoprotegerin in pediatric Crohn's disease and the effects of exclusive enteral nutrition. Inflamm Bowel Dis. 2011;17:516–523. doi: 10.1002/ibd.21361. [DOI] [PubMed] [Google Scholar]
- Whitten KE, Leach ST, Bohane TD, et al. Effect of exclusive enteral nutrition on bone turnover in children with Crohn's disease. J Gastroenterol. 2010;45:399–405. doi: 10.1007/s00535-009-0165-0. [DOI] [PubMed] [Google Scholar]
- Day AS, Mitchell HM, Leach ST, et al. Comment to: Changes of faecal microflora in patients with Crohn's disease treated with an elemental diet and total parenteral nutrition. Dig Liver Dis. 2013;45:177. doi: 10.1016/j.dld.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Nahidi L, Leach ST, Mitchell HM, et al. Inflammatory bowel disease therapies and gut function in a colitis mouse model. Biomed Res Int. 2013;2013:909613. doi: 10.1155/2013/909613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce-Millar E, Murch SH, Heuschkel RB, et al. Enteral nutrition therapy in Crohn's disease changes the mucosal flora. J Pediatr Gastroenterol Nutr. 2004;39:S289. [Google Scholar]
- Lionetti P, Callegari ML, Ferrari S, et al. Enteral nutrition and microflora in pediatric Crohn's disease. JPEN J Parenter Enteral Nutr. 2005;29:S184–S188. doi: 10.1177/01486071050290S4S173. [DOI] [PubMed] [Google Scholar]
- Leach ST, Mitchell HM, Eng WR, et al. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn's disease. Aliment Pharmacol Ther. 2008;28:724–733. doi: 10.1111/j.1365-2036.2008.03796.x. [DOI] [PubMed] [Google Scholar]
- Shiga H, Kajiura T, Shinozaki J, et al. Changes of faecal microbiota in patients with Crohn's disease treated with an elemental diet and total parenteral nutrition. Dig Liver Dis. 2012;44:736–742. doi: 10.1016/j.dld.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Gerasimidis K, Bertz M, Hanske L, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis. 2014;20:861–871. doi: 10.1097/MIB.0000000000000023. [DOI] [PubMed] [Google Scholar]
- D'Argenio V, Precone V, Casaburi G, et al. An altered gut microbiome profile in a child affected by Crohn's disease normalized after nutritional therapy. Am J Gastroenterol. 2013;108:851–852. doi: 10.1038/ajg.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellstrom B, Hogberg L, Stenhammar L, et al. Effect of exclusive enteral nutrition on gut microflora function in children with Crohn's disease. Scand J Gastroenterol. 2012;47:1454–1459. doi: 10.3109/00365521.2012.703234. [DOI] [PubMed] [Google Scholar]
- IBD Working Group of the European Society for Paediatric Gastroenterology Heptagology, Nutrition Inflammatory bowel disease in children and adolescents: recommendations for diagnosis—the Porto criteria. J Pediatr Gastroenterol Nutr. 2005;41:1–7. doi: 10.1097/01.mpg.0000163736.30261.82. [DOI] [PubMed] [Google Scholar]
- Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 2005. 19:5A–36A. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- Hyams JS, Mandel F, Ferry GD, et al. Relationship of common laboratory parameters to the activity of Crohn's disease in children. J Pediatr Gastroenterol Nutr. 1992;14:216–222. doi: 10.1097/00005176-199202000-00017. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, et al. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66:5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti R, Perez de Leon AA, Dowd SE, et al. Assessment of bacterial diversity in the cattle tick RhipicephalusBoophilusmicroplus through tag-encoded pyrosequencing. BMC Microbiol. 2011;11:6. doi: 10.1186/1471-2180-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Parry NM, et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun. 2010;78:1509–1519. doi: 10.1128/IAI.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Delton Hanson J, Rees E, et al. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J Wound Care. 2011;20:40–47. doi: 10.12968/jowc.2011.20.1.40. [DOI] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Wolcott RD, et al. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis. 2008;5:459–472. doi: 10.1089/fpd.2008.0107. [DOI] [PubMed] [Google Scholar]
- Gontcharova V, Youn E, Wolcott RD, et al. Black Box Chimera Check (B2C2): a Windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J. 2010;4:47–52. doi: 10.2174/1874285801004010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Austr J Ecol. 1993;18:117–143. [Google Scholar]
- Hasan NA, Young BA, Minard-Smith AT, et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One. 2014;9:e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Day AS, Huinao KD, et al. Microbial dysbiosis in pediatric patients with Crohn's disease. J Clin Microbiol. 2012;50:3258–3266. doi: 10.1128/JCM.01396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kaakoush NO, Mitchell HM. The role of bacteria and pattern-recognition receptors in Crohn's disease. Nat Rev Gastroenterol Hepatol. 2011;8:152–168. doi: 10.1038/nrgastro.2011.3. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Borruel N, Casellas F, et al. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- Ohkusa T, Okayasu I, Ogihara T, et al. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Jin Z, Wu W, et al. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9:e90849. doi: 10.1371/journal.pone.0090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. The role of inflammation in the pathogenesis of colorectal cancer. Curr Colorectal Cancer Rep. 2009;5:69–74. doi: 10.1007/s11888-009-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Schwab C, Milinovich G, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012;6:2091–2106. doi: 10.1038/ismej.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Whitehead RN, Griffiths L, et al. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn's disease. FEMS Microbiol Lett. 2010;310:138–144. doi: 10.1111/j.1574-6968.2010.02057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duck LW, Walter MR, Novak J, et al. Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- Ye J, Lee JW, Presley LL, et al. Bacteria and bacterial rRNA genes associated with the development of colitis in IL-10(-/-) mice. Inflamm Bowel Dis. 2008;14:1041–1050. doi: 10.1002/ibd.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim Y-G, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.