Abstract
The response of a murine macrophage cell line exposed to a library of seven metal and metal oxide nanoparticles was evaluated via High Throughput Screening (HTS) assay employing luciferase-reporters for ten independent toxicity-related signaling pathways. Similarities of toxicity response among the nanoparticles were identified via Self-Organizing Map (SOM) analysis. This analysis, applied to the HTS data, quantified the significance of the signaling pathway responses (SPRs) of the cell population exposed to nanomaterials relative to a population of untreated cells, using the Strictly Standardized Mean Difference (SSMD). Given the high dimensionality of the data and relatively small dataset the validity of the SOM clusters was established via a consensus clustering technique. Analysis of the SPR signatures revealed two cluster groups corresponding to (i) sub-lethal pro-inflammatory responses to Al2O3, Au, Ag, SiO2 nanoparticles possibly related to ROS generation, and (ii) lethal genotoxic responses due to exposure to ZnO and Pt nanoparticles at a concentration range of 25 μg/mL-100 μg/mL at 12 h exposure. In addition to identifying and visualizing clusters and quantifying similarity measures, the SOM approach can aid in developing predictive quantitative-structure relations; however, this would require significantly larger datasets generated from combinatorial libraries of engineered nanoparticles.
Keywords: Nanotoxicity, Self-organizing map, clustering, HTS, cell signaling pathway
Introduction
There have been rising concerns that unintended exposures of humans and other ecological receptors to engineered nanomaterials (eNMs) may result in adverse effects that differs from those known for their bulk counterpart (1, 2). Environmental protection plans associated with the manufacture and use of eNMs requires understanding nano-bio interface interactions that govern the biological activity and potential toxicity of nanomaterials. In this regard, the rapid generation of complex toxicity datasets, integrating in-vitro information at both, the molecular and cellular levels, with in-vivo whole-organism data, has in tandem accelerated the emergence of a new multilevel paradigm for toxicity testing. An important goal of toxicity testing is to identify critical biological pathways that, when perturbed, can lead to adverse effects. Accordingly, high-throughput toxicity-pathway assays are emerging as central elements of toxicity testing (3). Specifically, high-throughput screening (HTS) aims to screen the toxicity of nanoparticle libraries in a multivariate context that usually includes multiple cell lines, exposure times and nanoparticle concentrations (4). HTS data analysis requires normalization to remove systematic errors and for comparison and combination of data acquired from different plates (5). Such data can then be used to identify similarity patterns to construct eNM categories of common mechanisms of action and thus support the development of structure-activity nanotoxicity relationships.
Statistical techniques such as cluster analysis have proven useful for “mining” the relationships hidden in multidimensional cellular activity datasets (6). Hierarchical clustering and its application to heat maps (i.e., mapping display of cell activity data) are commonly used in bioinformatics for the analysis of HTS datasets. This clustering approach does not preserve the intrinsic topology of the data (e.g., nanoparticles that are placed in consecutive leaves in the hierarchical tree structure may in fact be far apart in the original data space). Self-Organizing Map (7) analysis is an alternative approach that provides an ordered two-dimensional visualization of multidimensional HTS data where similar nanoparticles assigned to nearby SOM units are also closer in the HTS data space (i.e., preserves the original distance relationships). SOM provides more accurate and robust clustering specifically for “noisy” datasets (8). SOM analysis has been shown to be useful for the development of quantitative structure-activity (QSAR) (9) and structure-property relationships (QSPR) (10). SOM has also been effectively used for the exploratory analysis of microarray data since, in contrast to the rigid structure of hierarchical clustering (i.e., based on pairwise similarity that doesn't preserve the distances between all the elements in the data set) it allows organization of data clusters such that cluster similarity can be visually identified based on the proximity of SOM units relative to each other in the map (11). However, gene expression arrays and HTS datasets of eNM toxicity differ markedly in the sample size (e.g., thousands to tens of thousands genes), the latter usually being a smaller dataset (typically ∼10-100 nanoparticles in a specific concentration range) but of higher dimensionality (e.g., combination of different cell lines, toxicity-pathways and exposure times). Smaller HTS datasets of higher dimensionality present a fundamental challenge of determining which clusters are truly representative of the actual physical domain (12). Consensus clustering can be utilized to overcome the above difficulty by providing a quantitative measure of cluster validity as demonstrated in a recent work on nano-SAR development (13, 14).
In the current work, a strategy of SOM analysis, along with consensus clustering (15) and multi-scale bootstrap sampling (16), was demonstrated for data mining of a small eNM library (seven metal and metal oxide nanoparticles). Toxicity screening data were obtained via measurements of the activity of ten toxicity-related cell signaling pathways (hereinafter termed “signaling pathways”) for macrophage cells. Grouping of similar cell signaling pathway responses was accomplished via a consensus SOM clustering approach that provided both a quantitative and visual representation of pathway similarity and possible relationships.
Materials and Methods
Knowledge extraction
The present approach for knowledge extraction from nanoparticle (NP) HTS signaling-pathway data is summarized in Figure S1 in Supporting Information (SI). In vitro HTS data measuring signaling pathway responses (hereinafter termed “SPR”) (SI, Table S1) of macrophage cells exposed to eNM were generated as described in the supporting information. Briefly, data acquired from different HTS plates were first normalized to account for systematic experimental errors. The significance of the biological responses (i.e., data labeling), triggered in macrophage cells exposed to eNMs, was quantified with respect to responses observed in untreated cells using the approach described later in this section (17). SPR similarities were then identified via SOM cluster analysis. Given the relatively small HTS dataset, consensus clustering was required for cluster validation (12, 15). Subsequently, clusters of SOM units were identified along with their associated dominant signaling pathways. Finally, an averaged cluster pathway signature (i.e., profile with respect to the pathway and time) was determined for each cluster in order to evaluate the similarity among clusters.
Experimental toxicity data
Seven metal and metal-oxide nanoparticles (Ag, Au, Pt, Al2O3, Fe3O4, SiO2, ZnO with respective primary diameters of 13, 12, 13, 12, 8, 19, and 20 nm) were acquired from commercial sources (Table S1; SI) to test for induction of toxicity-associated cell signaling pathways using a set of RAW 264.7 luciferase reporter cell lines. Macrophages were selected in this study since they constitute one of the principal immune-system sentinels whose phagocytic activity makes them likely to interact with eNMs upon exposure (18).
In order to obtain stable luciferase reporter cell lines, RAW 264.7 (ATCC #TIB-71) murine macrophage-like cells were transduced (see SI) with Cignal Lenti Reporters (SABiosciences Corp., US). Ten cell lines containing a unique transcriptional response element (TRE) were stably transduced with each of the ten pathway-specific reporters (SI, Table 1). An additional control cell line was generated in which the TRE is absent (TRE-), leaving only a basal promoter element upstream of the luciferase gene. The TRE- data served as control samples (i.e., indicators of cytotoxic effects of nanomaterials and other non-specific events affecting cell viability) and for data normalization. Details of the experimental protocol are provided in the Supporting Information. Briefly, each individual reporter cell line was seeded at 2×104 cells per well in white 384 well plates (Thermo Fisher Sci., NH, US, Cat#4334) with nanoparticle concentrations spanning the range of 0.375 - 100 μg/mL (at two-fold increases). At specific times (i.e., t=3, 6, 12, 24 hours), luciferase levels produced, in response to nanoparticle exposure, were determined via Bright-Glo Luficerase Assay (Promega Corp., WI, US).
Table 1.
Metal and Metal-oxide nanoparticles.
| Type | Name | Formula | Primary Size (nm) | Purity (%) | Specific Surface Area (m2/g) | Supplier |
|---|---|---|---|---|---|---|
| Metal (Me) | Silver | Ag | 13 | 99.9 | 22.99 | Meliorum Tech., Inc.(1) |
| Gold | Au | 12 | 95 | 12.95 | Meliorum Tech., Inc. (1) | |
| Platinum | Pt | 13 | 99 | 10.76 | Meliorum Tech., Inc. (1) | |
|
| ||||||
| Metal Oxides (MeO) | α-Alumina | Al2O3 | 12 | 99 | 62.97 | Meliorum Tech., Inc. (1) |
| Magnetite | Fe3O4 | 8 | 99 | 71.54 | Meliorum Tech., Inc. (1) | |
| Silica amorphous | SiO2 | 19 | 99 | 60.73 | NanoAmor, Inc. (2) | |
| Zinc Oxide | ZnO | 20 | 99.3 | 25.5 | Meliorum Tech., Inc. (1) | |
Rochester, NY, USA.
Houston, TX, USA
The HTS plate layout is provided in Figure 1. Cells in the wells located in the two outer columns of the plate were left untreated (i.e., served as a control indicative of the natural signaling pathway responses). The remaining wells contained cells exposed to the library of eNMs (Table 1). Well quadruplicates in each plate, served to account for natural (i.e., biological) and systematic (i.e., experimental) variabilities (5). Luminescence data were acquired from a set of 44 × 384-well plates (i.e., one plate for each studied signaling pathway plus TRE- control at each of the four exposure times) producing a total of 16,896 luminescence data readings.
Figure 1.
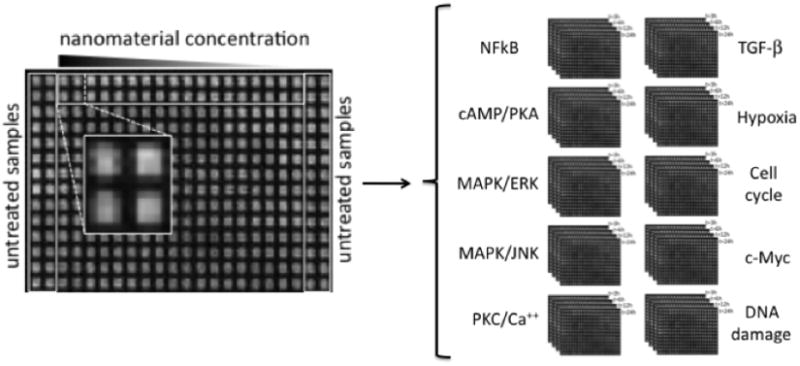
Depiction of the experimental set-up for the HTS assay and the data generation. Layout of a 384-well HTS plate (left) showing the location of untreated samples (i.e., cell not exposed to eNMs) and the four replicate wells for each nanoparticle concentration. Four different plates were employed to measure the activity of each of the 10 pathways (right) at different exposure times (3, 6, 12 and 24 h).
Data preprocessing
Data normalization was performed in two steps. First, in order to account for nonspecific luciferase expression or alterations in cell viability, HTS data were normalized by dividing the luminescence values in each well by the luminescence values of the corresponding TRE− control cells exposed to the identical experimental conditions (i.e., exposed to the same concentration of nanoparticles). Second, for each plate, the strictly standardized mean difference (17) was used to determine the statistical significance of the differences between each set of quadruplicate measurements and the untreated (i.e., not exposed to eNMs) cell population (Figure S2, SI). The strictly standardized mean difference (SSMD) for two independent populations, 1 and 2 (e.g., quadruplicate responses and untreated samples) is estimated as:
| (1) |
where X̄1 and X̄2 are the averages of each population, n1 and n2 the corresponding population sizes (i.e., n1=4 and n2=64) and s1 and s2 are estimates of the standard deviation of each population. The SSMD, similar to Student's t-test, measures the significance of differences between the two populations while accounting for their intrinsic variability. However, unlike the SSMD, the student t-test under the assumption of unequal variances tends to underestimate of the likelihood of populations similarity (i.e., lower p-values) with increasing sample size (17). For example, SSMD ≥ 3 indicates that the probability that a value from the first population being greater than a value from the second population is > 99.9% when the difference is normally distributed, or > 95% when the difference has a unimodal distribution with finite variance (17). Following the above scheme, the SPR of the macrophage cells, exposed to a nanoparticle n at a given concentration C, was described by an SPR vector , whose forty components are the SSMD (Eq. 1) values of the normalized luminescence corresponding to the ten studied pathways at each of the four exposure time.
Similarity analysis
SOM representation (as two-dimensional projection) of the multidimensional HTS SPR data was accomplished as depicted in Figure 2. SOM identification of similar SPR vectors (i.e., SPR vectors that are close to each other in the native topology of the data), corresponding to similar patterns of signaling pathway activity, can aid in the identification of common mechanisms of action for specific types of nanoparticles. Details of the implementation and properties of the SOM analysis can be found elsewhere (7). Briefly, SOM provides an ordered projection of the SPR vectors while preserving the topology of the original HTS dataset (i.e., relative distances among the processed SPR vectors) and facilitates the identification and visualization of groups of eNMs that trigger similar biological responses. The SOM was developed utilizing an optimal (20, SI) two-dimensional hexagonal 8×5 grid (i.e., 40 SOM units, Figure 2). Each SOM unit k was characterized by a weighted average of its clustered SPR vectors yielding a prototype vector pk for the unit (Figure 2). During the map construction process prototype vectors adapts such that similar map units (i.e., in terms of prototype vectors) are organized closer to each other. The SOM structure can be conveniently visualized as a set of 40 consecutive layers (i.e., component planes; (20), each representing the responses (in terms of the prototype vector components) from one of the specific ten signaling pathway at one of the four different exposure times (Fig. 2). Accordingly, the component planes c-planes are displayed by a color code corresponding to each individual prototype vector component over the SOM.
Figure 2.
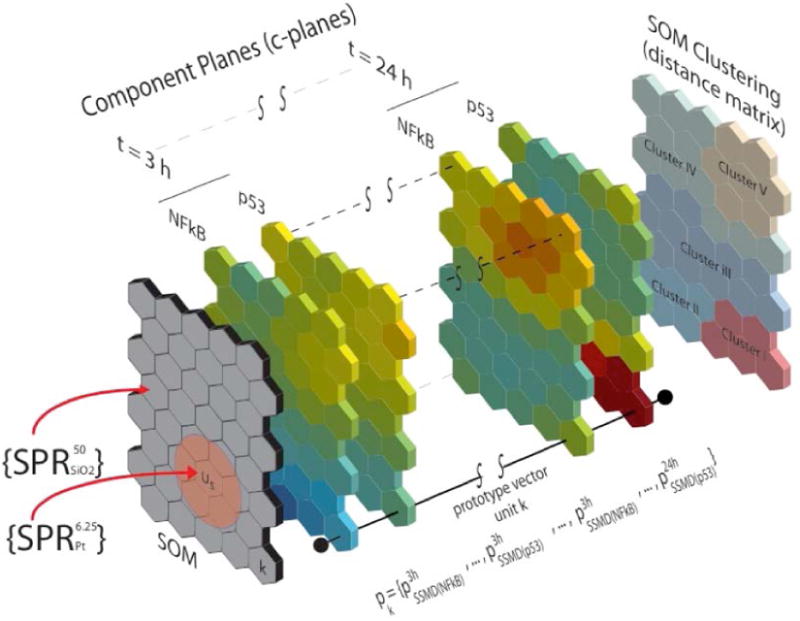
Structure and principal elements of the SOM used the current work. The gray plane (left) represents the SOM grid composed by 40 units arranged in a hexagonal configuration of 8×5 units. Subsequent color slices correspond to the visualization of each of the 40 component planes (c-planes) corresponding to each signaling pathway at each exposure time (3 h – 24 h). The plane on the right side depicts the clustering of similar SOM units based on the distance matrix.
Clustering of HTS data
Identification of clusters of SOM units (Figure 2) can be accomplished by determining the similarity among the prototype vectors (19, 20). However, cluster validation is challenging for HTS data since: (a) lack of prior knowledge about the expected cluster structure, and (b) clustering of a dataset of high dimensionality is sensitive to data quality (e.g., both in terms of dataset size and data uncertainty). Accordingly, in the present work identified HTS dataset clusters were validated using a consensus-clustering method (15). The approach consisted of randomly sampling the post-processed HTS dataset with replacement (i.e., allowing the selection of the same SPR vector multiple times) to generate replicate datasets of the same order of the original HTS dataset. Subsequently, the SOM algorithm was applied to cluster the SPR vectors in each replicate dataset. The cumulative number of clustering runs in which two response vectors were grouped together in the same SOM unit formed the elements of a symmetric consensus matrix M (dimension= n×n, where n=63, the total number of SPR vectors in the HTS dataset). This matrix was then normalized (forming a co-clustering index matrix) by dividing each element (i.e., representative of an SPR pair) by the total number of times that the corresponding SPR pair is found in the total number of replicate datasets.
The validity of a given SOM cluster (either SOM unit or cluster), k, was quantified in terms of a consensus index CI(k), from the elements of the normalized consensus matrix M as,
| (2) |
where Ik is the set of consensus matrix indices corresponding to the SPR vectors in cluster k and Nk is the number of elements (SPR vectors) in the same cluster (16). The consensus index is in the range of [0,1] where values near unity are indicative of increasing cluster validity (i.e., the SPR vectors cluster together with increased validity irrespective of the perturbations introduced in the HTS dataset).
Results and Discussion
SOM clustering of signaling-pathway response data
Validation of SOM units (i.e., grouping of SPR vectors) was assessed via the consensus clustering approach (Eq. (2)). As shown in Fig. 3a, grouping of SPR vectors within SOM units was highly robust and reproducible as verified by the consensus clustering analysis. Two SOM units had the low consensus index values of 0.83 and 0.85. The first, contained nanoparticles of Ag (12.5 μg/mL and 50 μg/mL), Al2O3 (25 μg/mL) and SiO2 (3.12 μg/mL) and the second contained Ag (0.375 μg/mL and 1.6 μg/mL), Al2O3 (0.375 μg/mL) and ZnO at (25 μg/mL). The above results and the fact that the remaining SOM units had consensus index values above about 0.9 indicate that the SOM developed from the HTS data reflects valid SOM grouping (i.e., units) of SPR vectors.
Figure 3.
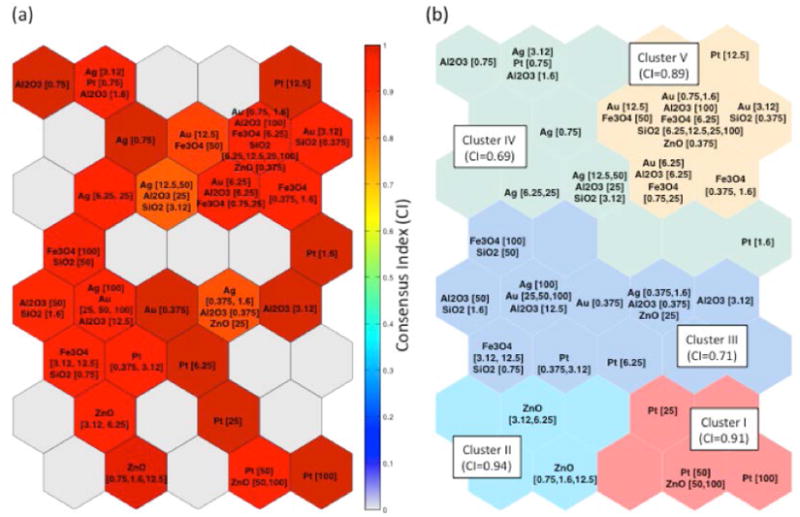
Clustering of nanoparticles according to their signaling pathway response profile using the SOM. (a) Visualization of the consensus index for each SOM unit. (b) Clusters obtained from the SOM distance matrix and its corresponding consensus index.
Five SOM clusters were identified, each representing a group of similar SOM units, based on the similarities (distances) between neighboring SOM units and validated via a consensus approach (see Consensus Matrix, SI Figure S4). The consensus index values for Clusters I and II (Figure 4b) were of 0.91 and 0.94, respectively. Cluster I included SPR vectors corresponding to ZnO (50 and 100 μg/mL) and Pt (25, 50 and 100 μg/mL) at high concentrations while Cluster II consisted of SPR vectors corresponding to ZnO nanoparticles at concentration of 0.75 – 12.5 μg/mL. Cluster V (Figure 3b) had a consensus index of 0.89 and included the SPR vectors corresponding to SiO2, Al2O3 and Fe3O4 nanoparticles at high concentrations (≥ 25 μg/mL), as well as those of SiO2, Al2O3, Fe3O4, Au, Pt, ZnO nanoparticles at moderate or low concentrations (<12.5 μg/mL). Finally, it is noted that although clusters III and IV (Figure 3b) had slightly lower consensus indices of 0.71 and 0.69, respectively, these values can be viewed as indicative of a clear clustering tendency. Finally, it is noted that for comparison, a cluster heat map of the preprocessed HTS data was also developed and details are provided in the Supporting Information (Figure S3).
Figure 4.
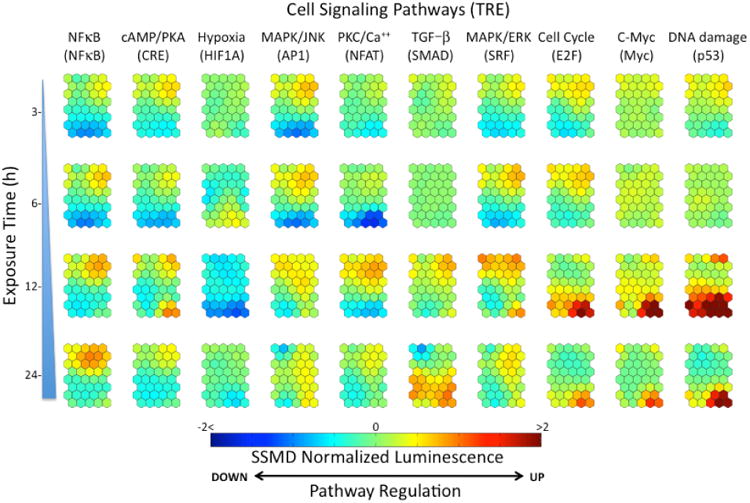
Component planes corresponding to the biological response (up-regulation/down-regulation) of the 10 cell signaling pathways for the RAW 264.7 macrophage cells at 3 h, 6 h, 12 h and 24 h of exposure to the seven nanoparticles (Table 1).
Relationships between signaling-pathway response (SPR) vectors
A color-coded SOM representation (i.e., c-planes) of the signaling-pathway responses (SI, Table 1) is shown in Figure 4. In this representation, each c-plane, displays the SOM average of the components of the SPR vectors for each unit. The content of each of the SOM units in the forty c-planes (Figure 4) is identified with the specific nanoparticles (at a given concentration) through the unit labeling shown in Figure 3.
As is revealed in Figure 4, after short exposure time 3 h to 6 h, there is an incipient up-regulation tendency of the NF-κB, MAPK/JNK and MAPK/ERK stress and inflammatory signaling pathways attributed to nanoparticles identified as members of cluster V (Figure 3b). Macrophages are known to be stimulated to secrete inflammatory mediators, such as cytokines and chemokines, when exposed to foreign substances to enhance an immune response (22). Interestingly, significant down-regulation (blue colors in Figure 4) of the inflammatory NF-κB, MAPK/JNK and PKC/Ca++ signaling pathways is observed in clusters I and II (Figure 3b) that are specifically linked to the responses associated with exposure to ZnO nanoparticles (Figure 3b).
Inspection of the c-planes (Figure 4) reveals that after 12 h of exposure there is an additional significant up-regulation of p53, c-Myc and cell cycle signaling-pathways (areas in red color in Figure 4). Specifically, nanoparticles in clusters I and II (Figure 3b) induced a significant up-regulation of the p53/DNA damage pathway. The nanoparticles responsible for these signaling pathway responses were ZnO and Pt at concentration ranges of 0.75 μg/mL - 100 μg/mL and 25 μg/mL - 100 μg/mL, respectively. A highly correlated pathway response is also observed in the c-plane (Figure 4) corresponding to the cell cycle/E2F pathway, which is responsible for delaying or halting cell cycle progression in response to DNA damage through the regulation of the G1/S cell cycle checkpoint (23).
Interestingly, the c-Myc pathway c-plane shows significant up-regulation (Figure 4) induced by ZnO at high concentrations (50 - 100 μg/mL) and Pt (25 - 100 μg/mL) nanoparticles in cluster I (Figure 3b). Deregulation of c-Myc may have a severe effect on normal cell functions, including proliferation, differentiation, and apoptosis (24). It is plausible that the stimulation of apoptosis by c-Myc may not always be directly linked to cell cycling; it can also arise through indirect actions that culminate in DNA damage (25). The combined response observed in these pathways is consistent with the known fact that damaged DNA in mammalian cells triggers the production of proteins that initiate cell-cycle arrest, DNA repair and ultimately may induce apoptosis (26). The possible induction of cell death by nanoparticles in cluster I (Figure 3b), is supported by the down-regulation of the PKC/Ca++ pathway observed after 6 h. Indeed, there is considerable evidence that a number of toxicants alter Ca2+ signaling processes and may induce cell death by apoptosis (27). For instance, the exposure of human cells to Pt nanoparticles results in genotoxic stress that increases DNA damage, accumulation of cells at the S-phase of cell cycle and apoptosis (28). Genotoxic effects were also observed in human epidermal cells after 6 h of exposure to low concentrations of ZnO nanoparticles (29). This genotoxic potential may be mediated through lipid peroxidation and oxidative stress (30).
The down-regulation of the hypoxia pathway (Figure 4) induced by the same group of nanoparticles (ZnO and Pt) could be attributed to the fact that high expression levels of p53 lead to a decrease in HIF-1α protein levels and thereby eliminate the activity of the HIF-1 reporter (31). Finally, there was significant decrease in the response of the cell cycle, c-Myc and DNA damage signaling pathways after prolonged exposure to nanoparticles (24 h). It is possible that these results may have been affected by cytotoxicity of ZnO and Pt nanoparticles (Cluster I in Figure 3b) as indicated by a significant reduction in the luminescence signal associated with TRE- control cells (not shown).
Extraction of signaling-pathway response signatures
Similarity in terms of the biological response of the five SOM clusters was evaluated by comparing their averaged pathways response. Accordingly, the prototype vectors components (i.e., SSMD values) of the SOM units in each cluster (Figure 3b) were averaged to obtain distinct pathway response (PR) signatures (Figure 5 and 6). In this representation |SSMD| ≈ 0.5 indicates a weak effect (17) with a probability less than about 70% of a response that is significantly different from the untreated cells. Conversely, for |SSMD| > 0.5 the effects are stronger (SI) with higher probability of significant up or down-regulation of the signaling pathway as observed in the PR signatures for clusters I, II and V (Figures 5 and 6). The signaling PR signatures for these clusters suggest that the significant responses (i.e., pathways with the highest SSMD values) for the above clusters are associated with sub-lethal pro-inflammatory response (Cluster V) and lethal response including DNA damage (Clusters I and II). In contrast with the above, reliable biological interpretation could not be derived from the PR signatures for clusters III and IV (not shown) since these clusters had low consensus clustering indices (Figure 3b) and low |SSMD| values (e.g. mostly |SSMD| <0.5).
Figure 5.
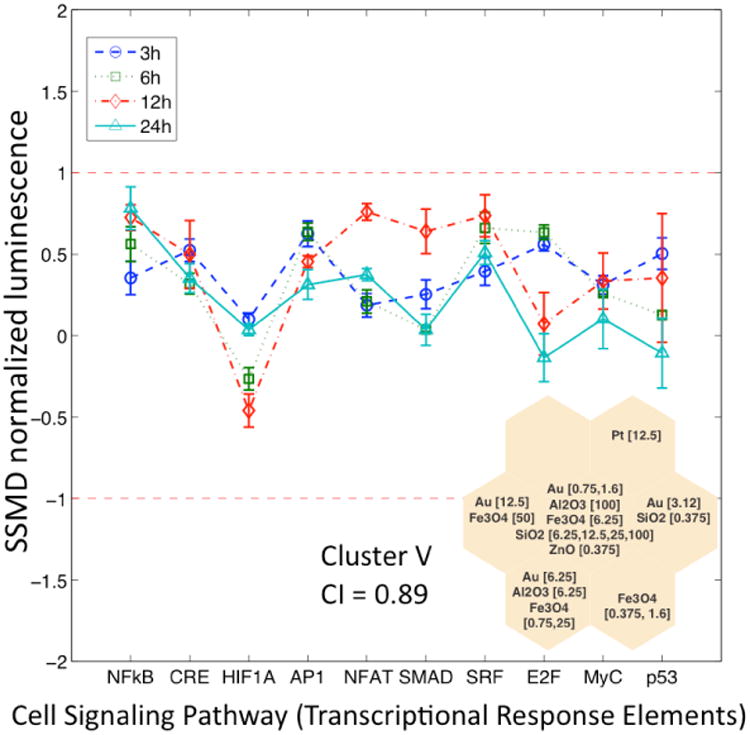
Sub-lethal pro-inflammatory signatures at 3h, 6h, 12h and 24h of exposure corresponding to SPR profiles in Cluster V. The error bars indicate the within-cluster variability of each signaling pathway.
Figure 6.
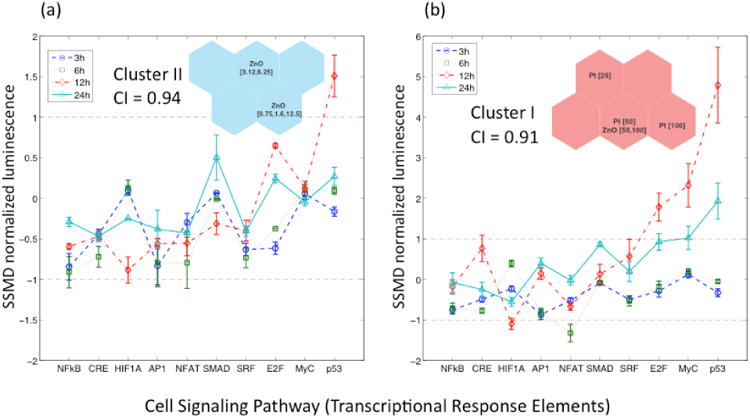
Lethal genotoxic signatures of ZnO and Pt. The error bars indicate the within-cluster variability of each signaling pathway. (a) Response signatures at 3h, 6h, 12h and 24h of exposure corresponding to SPR profiles in Cluster II, (b) Response signatures at 3h, 6h, 12h and 24h of exposure corresponding to SPR profiles in Cluster I.
The sub-lethal response inferred in Cluster V (Figure 5) can only be considered as a moderate biological effect with a probability in the range 70% - 80% of being significantly different from the untreated cells. On the other hand, the PR signatures in Cluster V, which includes silica and alumina have its more significant components in pathways related to ROS generation and inflammatory responses (e.g., NF-κB, MAPK/JNK, and MAPK/ERK). It has been reported that exposure to SiO2 nanoparticles could lead to cellular morphological modifications, mitochondrial dysfunction, and oxidative stress by generation of intracellular ROS (32). It is interesting to note that ROS generation has been reported to trigger pro-inflammatory responses, both in vivo and in vitro, by changes in the expression levels of distinct genes and pathways related to inflammatory responses and apoptosis including MAPK/ERK kinase, NF-κB and AP-1 (33, 34). Further, alumina nanoparticles have been shown to initiate inflammatory events in macrophages, including secretion of pro-inflammatory cytokines and interaction with neighboring cells (35). Activator protein-1 (AP-1) has also been demonstrated to be redox sensitive and its activation has been linked to both exogenous oxidants (36) and ligand-induced ROS (37). ROS has also been reported to activate serine/threonine phosphorylation processes (e.g., regulation of serine/threonine kinases of the MAPK family including ERKs and JNKs) (38).
Significant PR signatures are observed for clusters I and II (Figure 3b). In cluster II (ZnO nanoparticles at 0.75 – 12.5 μg/mL) the most significant signaling pathways are related to TGF-β (SMAD), cell cycle (E2F) and DNA damage (p53) (Figures 4 and 6). The activity in the genotoxicity-related pathway (p53/DNA damage) is more evident after a 12 h exposure period with SSMD > 1.5 which corresponds to a significant difference (probability > 80%) between the response of cells exposed to low ZnO concentrations (0.75 μg/mL – 12.5 μg/mL) relative to the response of untreated cells. The correlation between the c-planes (Fig. 4) corresponding to E2F and p53 (i.e., both showing significant response in the same SOM units) is consistent with the suggestion that significant DNA damage induced by the ZnO nanoparticles triggers cell cycle arrest. The above two pathways also appear to be significant for nanoparticles in cluster I (Figure 3b). However, at the higher ZnO and Pt nanoparticles (> 25 μg/mL) more significant genotoxic effects (i.e., larger SSMD values) were observed relative to the untreated cells. After 12 h of exposure, DNA damage (p53), cell cycle (E2F) and c-Myc (Myc) pathways display significant response levels (i.e., SSMD ≅ 5 for DNA damage with a >99.9% probability of being different relative to the response of the untreated cells). The above response decreased significantly after a 24 h exposure, possibly due to the cytotoxicity of the nanomaterials and the corresponding decline in the measured luminescence.
In summary, SOM enabled detailed analysis of the clustering structure of the SPR induced by a set of seven Me and MeO nanoparticles to which RAW 264.7 macrophage cells were exposed. The SOM analysis identified two groups of SPR signatures corresponding to (i) sub-lethal pro-inflammatory responses that are possibly related to ROS generation, and (ii) lethal genotoxic responses after exposure to ZnO and Pt nanoparticles at high concentrations. Comparison of the SOM c-planes relevant relationships between the cell signaling pathways. The current study demonstrates that the Self-Organizing Map is an effective tool for the analysis of HTS nanoparticle data that complements and extends the data mining capabilities of more traditional techniques such as cluster heat maps. Finally, it is important to recognize that notwithstanding the power of SOM or other methods for knowledge extraction from multidimensional nanoparticle HTS toxicity datasets, definitive generalizations and conclusions regarding clustering and activity patterns of eNM toxicity will require significantly expanded datasets including eNM structural and physicochemical properties than presently available.
Supplementary Material
Brief.
Application of Self-Organizing Maps to the analysis of cell signaling pathway responses induced by nanoparticles
Acknowledgments
This material is based upon work supported by the National Science Foundation and the Environmental Protection Agency under Cooperative Agreement Number DBI-0830117 and U19 ES019528. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the Environmental Protection Agency. This work has not been subjected to EPA review and no official endorsement should be inferred. Robert Rallo and Francesc Giralt also acknowledge support provided by CICYT (Project CTQ2009-14627), Generalitat de Catalunya (2009SGR-01529) and the EU Commission (OSIRIS, Contract No. 037017).
Footnotes
Supporting Information (SI). The supporting information provides additional details of the experimental protocol, table of the signaling pathways, data analysis, cluster validation results, and comparison with heat map hierarchical clustering. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Auffman M, Rose J, Bottero J, Lowry G, Jolivet J, Wiesner M. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat Nanotechnol. 2009;4:634–640. doi: 10.1038/nnano.2009.242. [DOI] [PubMed] [Google Scholar]
- 2.Xia T, Li N, Nel AE. Potential Health Impact of Nanoparticles. Annu Rev Public Health. 2009;30:137–150. doi: 10.1146/annurev.publhealth.031308.100155. [DOI] [PubMed] [Google Scholar]
- 3.Krewski D, Acosta D, Jr, Andersen M, Anderson H, Bailar JC, III, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S, Jr, Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L Staff of Committee on Toxicity Testing and Assessment of Environmental Agents. Toxicity Testing in the 21st Century: A Vision and a Strategy. J Toxicol Environ Health, Part B. 2010;13(2):51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George S, Pokhrel S, Xia T, Gilbert B, Ji Z, Schowalter M, Rosenauer A, Damoiseaux R, Bradley KA, Maedler L, Nel EA. Use of a Rapid Cytotoxicity Screening Approach To Engineer a Safer Zinc Oxide Nanoparticle through Iron Doping. ACS Nano. 2010;4(1):15–29. doi: 10.1021/nn901503q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birmingham A, Selfors L, Forster T, Wrobel D, Kennedy C, Shanks E, Santoyo-Lopez J, Dunican D, Long A, Kelleher D, Smith Q, Beijersbergen R, Ghazal P, Shamu C. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper G, Pickett SD. Methods for Mining HTS data. Drug Discovery Today. 2006;11(15-16):694–699. doi: 10.1016/j.drudis.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Kohonen T. Self-Organizing Maps. 2dn. Extend. Springer-Verlag; Berlin, Heidelberg: 1997. [Google Scholar]
- 8.Mangiameli P, Chen SK, West D. A comparison of SOM neural network and hierarchical clustering methods. European Journal of Operational Research. 1996;93:402–417. [Google Scholar]
- 9.Rallo R, Espinosa G, Giralt F. Using an ensemble of neural based QSARs for the prediction of toxicological properties of chemical contaminants. Trans IChemE Part B. 2005;83(B4):387–392. [Google Scholar]
- 10.Giralt F, Espinosa G, Arenas A, Amat L, Girones X, Carbo-Dorca R, Cohen Y. Prediction of activity coefficients of diverse aromatic compounds in water at infinite dilution with an integrated SOM-fuzzy ARTMAP neural system. AIChE J. 2004;50(6):1315–1343. [Google Scholar]
- 11.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E, Golub T. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Hayes DN, Nobel A, Marron JS. Statistical Significance of Clustering for High-Dimensional, Low-Sample Size Data. J Am Stat Assoc. 2008;103(483):1281–1293. [Google Scholar]
- 13.Shaw SY, Westly EC, Pittet MJ, Subramanian A, Schereiber SL, Weissleder R. Perturbational profiling of nanomaterial biological activity. Proc Natl Acad Sci USA. 2008;105(21):7387–7392. doi: 10.1073/pnas.0802878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fourches D, Pu D, Tassa C, Weissleder R, Shaw SY, Mumper RJ, Tropsha A. Quantitative Nanostructure- Activity Relationship Modeling. ACS Nano. 2010;410:5703–5712. doi: 10.1021/nn1013484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monti S, Tamayo P, Mesirov J, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 16.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89(4):552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Jones DW, Grainger DW. In vitro assessments of nanomaterial toxicity. Adv Drug Delivery Rev. 2009;61:438–456. doi: 10.1016/j.addr.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Z, Jin X, George S, Xia T, Meng H, Wang X, Suarez E, Zhang H, Hoek EMV, Godwin H, Nel EA, Zink J. Dispersion and Stability of TiO2 Nanoparticles in Cell Culture Media. Environ Sci Technol. 2010 doi: 10.1021/es100417s. online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vesanto J, Alhoniemi E, Himberg J, Kiviluoto K, Parviainen J. Self-organizing map for data mining in MATLAB: the SOM toolbox. Simulation News Europe. 1999:25–54. [Google Scholar]
- 21.Vesanto J, Hollmen J. An Automated Report Generation Tool for the Data Understanding Phase. In: Abraham A, Koppen M, editors. Hybrid Information Systems. Physica Verlag; Heidelberg: 2002. p. 611. [Google Scholar]
- 22.Gwinn MR, Vallyathan V. Respiratory burst: Role in signal transduction in alveolar macrophages. J Toxicol Environ Health, Part B. 2006;9(1):27–39. doi: 10.1080/15287390500196081. [DOI] [PubMed] [Google Scholar]
- 23.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27(50):6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 25.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 26.Fulda S, Gorman AM, Hori O, Samali A. Cellular Stress Responses: Cell Survival and Cell Death. Int J Cell Biol. 2010 doi: 10.1155/2010/214074. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajnoczky G, Davies E, Madesh M. Calcium signaling and apoptosis. Biochem Biophys Res Commun. 2003;304(3):445–454. doi: 10.1016/s0006-291x(03)00616-8. [DOI] [PubMed] [Google Scholar]
- 28.Asharani PV, Xinyi Ng, Hande MP, Valiyaveettil S. DNA damage and p53-mediated growth arrest in human cells treated with platinum nanoparticles. Nanomedicine. 2010;5(1):51–64. doi: 10.2217/nnm.09.85. [DOI] [PubMed] [Google Scholar]
- 29.Sharma V, Shukla RK, Saxena N, Parmar D, Das M, Dhawan A. DNA damaging potential of zinc oxide nanoparticles in human epidermal cells. Toxicol Lett. 2009;185:211–218. doi: 10.1016/j.toxlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Xia T, Kovochich M, Liong M, Madler L, Gilbert B, Shi H, Yeh JI, Zink JI, Nel AE. Comparison of the Mechanism of Toxicity of Zinc Oxide and Cerium Oxide Nanoparticles Based on Dissolution and Oxidative Stress Properties. ACS Nano. 2008;2(10):2121–2134. doi: 10.1021/nn800511k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou JJ, Chua YL, Chew EH, Gao J, Bushell M, Hagen T. Inhibition of hypoxia-inducible factor-1α (HIF-1α) protein synthesis by DNA damage inducing agents. PLoS One. 2010;5(5):e10522. doi: 10.1371/journal.pone.0010522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu S, Zhao H, Al-Humadi NH, Yin XJ, Joseph KH. Silica-induced apoptosis in alveolar macrophages: evidence of in vivo thiol depletion and the activation of mitochondrial pathway. J Toxicol Environ Health, Part A. 2006;69(13):1261–1284. doi: 10.1080/15287390500361875. [DOI] [PubMed] [Google Scholar]
- 33.Nel AE, Wooten MW, Galbraith RM. Molecular signaling mechanisms in T-lymphocyte activation pathways: A review and future prospects. Clin Immunol Immunopathol. 1987;44(2):167–186. doi: 10.1016/0090-1229(87)90064-x. [DOI] [PubMed] [Google Scholar]
- 34.Park E, Park K. Oxidative stress and pro-inflammatory responses induced by silica nanoparticles in vivo and in vitro. Toxicol Lett. 2009;184(1):18–25. doi: 10.1016/j.toxlet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigo A, Valles G, Saldana L, Rodriguez M, Martinez ME, Munuera L, Vilaboa N. Alumina particles influence the interactions of cocultured osteoblasts and macrophages. J Orthop Res. 2006;24:46–54. doi: 10.1002/jor.20007. [DOI] [PubMed] [Google Scholar]
- 36.Nose K, Shibanuma M, Kikuchi K, Kageyama H, Sakiyama S, Kuroki T. Transcriptional activation of early-response genes by hydrogen peroxide in a mouse osteoblastic cell line. Eur J Biochem. 1991;201:99–106. doi: 10.1111/j.1432-1033.1991.tb16261.x. [DOI] [PubMed] [Google Scholar]
- 37.Puri PL, Avantaggiati ML, Burgio VL, Chirillo P, Collepardo D, Natoli G, Balsano C, Levrero M. Reactive Oxygen Intermediates Mediate Angiotensin II-induced c-Jun•c-Fos Heterodimer DNA Binding Activity and Proliferative Hypertrophic Responses in Myogenic Cells. J Biol Chem. 1995;270(38):22129–22134. doi: 10.1074/jbc.270.38.22129. [DOI] [PubMed] [Google Scholar]
- 38.Poli G, Leonarduzzi G, Biasi F, Chiarpotto E. Oxidative stress and cell signalling. Curr Med Chem. 2004;11(9):1163–1182. doi: 10.2174/0929867043365323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


