Abstract
Sonic Hedgehog (Shh) signaling is critical in development and oncogenesis, but the mechanisms regulating this pathway remain unclear. While protein phosphorylation clearly affects Shh signaling, little is known about phosphatases governing the pathway. Here we conducted an shRNA screen of the phosphatome and identified Eya1 as a positive regulator of Shh signaling. We find that the catalytically active phosphatase Eya1 co-operates with the DNA-binding protein Six1 to promote gene induction in response to Shh, and that Eya1/Six1 together regulate Gli transcriptional activators. We show that Eya1, which is mutated in a human deafness disorder, branchio-oto-renal syndrome, is critical for Shh-dependent hindbrain growth and development. Moreover Eya1 drives the growth of medulloblastoma, a Shh-dependent hindbrain tumor. Together, these results identify Eya1 and Six1 as key components of the Shh transcriptional network in normal development and in oncogenesis.
Graphical Abstract
Introduction
Shh is a key regulator of mammalian development, functioning as both a mitogen and morphogen (Ingham et al., 2011). Dysregulated Shh signaling results in a wide variety of devastating birth defects and cancers (Cohen, 2012; Nieuwenhuis and Hui, 2005; Traiffort et al., 2010), and so understanding the mechanisms of Shh signaling has been a major goal in developmental and cancer biology. Shh initiates signaling by binding to its receptor Patched (Ptch1). In the absence of ligand, Ptch1 inhibits Smoothened (Smo), a potent pathway activator. Upon Shh binding, Ptch1 no longer represses Smo. Once de-repressed, Smo enhances Gli transcriptional activators and inhibits Gli transcriptional repressors and so alters programs of gene expression.
The mechanisms whereby activated Smo signals to Gli transcription factors are not yet understood. Protein phosphorylation and de-phosphorylation regulate multiple physiological functions, and several screens have identified kinases involved in the Shh pathway (Evangelista et al., 2008; Hillman et al., 2011; Jacob et al., 2011; Varjosalo et al., 2008). However, the role of phosphatases in Shh signaling is largely unexplored. We conducted an shRNA screen to discover phosphatases in the Shh signaling pathway, and identified Eya1 as a positive regulator of this pathway. Eya1 is a phosphotyrosine phosphatase that is mutated in branchio-oto-renal syndrome (Abdelhak et al., 1997; Li et al., 2003; Rayapureddi et al., 2003; Tootle et al., 2003; Xu et al., 1999). Eya1 dephosphorylates histone variant H2AX and thereby impacts DNA repair and cell survival (Cook et al., 2009). In addition, catalytically active Eya1 interacts with Six family transcription factors to regulate gene expression during development (Rebay et al., 2005; Tadjuidje and Hegde, 2013). Both Eya and Six family members have been implicated in tumor proliferation and progression (Christensen et al., 2008; Pandey et al., 2010; Patrick et al., 2013).
Here we demonstrate that catalytically active Eya1, and its binding partner Six1, function as transcriptional regulators in Shh signaling pathways. Eya1 and Six1 alter the equipoise between Gli activators and repressors following Shh stimulation, and so determine the ensuing biological response. Therefore Eya1 is required for Shh-regulated proliferation and morphogenesis during hindbrain development. Constitutive activation of the Shh pathway in neural precursors of the hindbrain causes medulloblastoma, a cerebellar cancer that is the most common malignant brain tumor in children (Goodrich et al., 1997). We show that Eya1 is preferentially expressed in Shh-subtype medulloblastomas and fosters tumor growth. Together these findings identify a critical role for Eya1 and its partner Six1 in promoting Shh-dependent transcription and suggest that Eya1 represents a propitious therapeutic target in medulloblastoma.
Results
Identification of Eya1 as a regulator of Shh signal transduction by RNAi
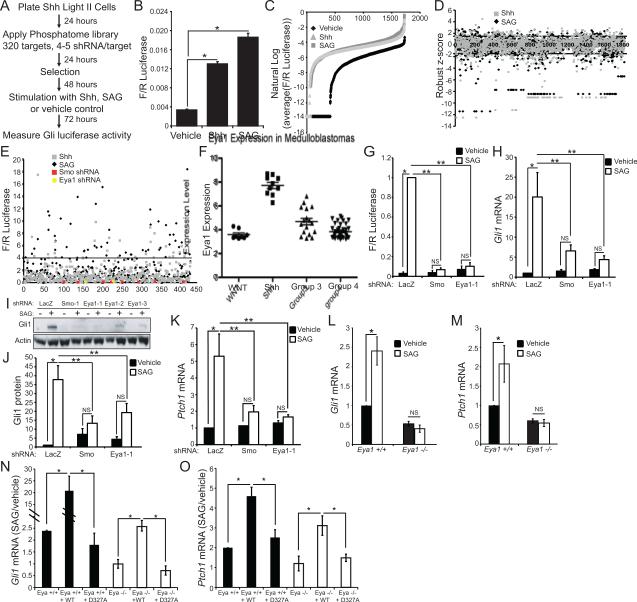
To identify phosphatases regulating the Shh signaling pathway, we conducted an shRNA screen that encompassed 320 gene targets (Table S1). The screen was conducted in ShhLightII fibroblasts, NIH3T3 cells stably expressing a Gli-dependent firefly luciferase reporter gene and a constitutive Renilla luciferase gene (Taipale et al., 2000). To distinguish phosphatases that function in the pathway between Ptch1 and Smo from those that function downstream of Smo, cells were stimulated with full-length Shh ligand, which binds Ptch1, with a direct Smo agonist (SAG)(Chen et al., 2002), or with vehicle control (Figure 1A). Data from the primary screen indicate that both Shh and SAG successfully stimulate the pathway (Figure 1B,C). We identified as potential hits those genes for which multiple targeting shRNAs achieved a robust Z-score greater than 1.5 (Figure 1D, Table S2), and we carried out a secondary screen to validate these hits (Figure 1E). The secondary, more rigorous screen identified genes for which two or more targeting sequences altered Shh responses by more than four-fold.
Figure 1.
shRNA screen to identify phosphatases in Shh signaling. (A) ShhLightII cells were infected with virus encoding a single shRNA targeting one of 320 genes; infected cells were selected with puromycin, stimulated with Shh-conditioned media, SAG, or vehicle control then Gli-responsive luciferase activity was measured. (B) Primary screen average Firefly/Renilla luciferase values confirm Shh pathway activation by Shh and SAG (N=1720 wells, *P<0.01, Error bars = SEM). (C) Replicate Firefly/Renilla luciferase values were averaged for each shRNA and converted to their natural log value. (D) Primary screen robust z-score results. Genes with two or more hairpins giving results less than −1.5 or greater than 1.5 qualified as hits. (E) A subset of hits from the primary screen were re-screened. Firefly/Renilla luciferase values were normalized to median Firefly/Renilla luciferase value of negative control shRNAs. Values greater than four-fold of the median response (above the black line) or less than 25% of the median response qualified as hits. Red squares indicate values corresponding to an shRNA targeting Smo and yellow circles indicate values corresponding to an shRNA targeting Eya1. (F) Affymetrix array of medulloblastomas reveal a significant increase in Eya1 expression in the Shh subtype compared to WNT, Group 3 or Group 4 (P<0.0001 by ANOVA). (G) An shRNA targeting Eya1 blocks SAG-mediated induction of Firefly/Renilla luciferase (N=4,*P<0.05, **P<0.01, Error bars = SEM; NS = not significant, P>0.05). (H) Eya1 shRNA blocks SAG-mediated induction of Gli1 mRNA (N=5, *P<0.05, **P<0.01, Error bars = SEM; NS = not significant, P>0.05). (I) Induction of Gli1 protein is blocked by shRNAs targeting Eya1. Actin = loading control. (J) Induction of Gli1 protein is blocked by an Eya1 shRNA (N=8,*P<0.05, **P<0.01, Error bars = SEM; NS = not significant, P>0.05). (K) An shRNA targeting Eya1 blocks induction of Ptch1 mRNA (N=5, *P<0.05, **P<0.01, Error bars = SEM; NS = not significant, P>0.05). Gli1 (L) and Ptch1 (M) mRNA induction are reduced in Eya1−/− MEFs compared to Eya1+/+ littermates (N=3, *P<0.05, Error bars = SEM, NS = not significant, P>0.05). (N) Gli1 mRNA and (O) Ptch1 mRNA in Eya1−/− and Eya1+/+ MEFs, following reintroduction of wild type or D273A Eya1 (N=3, *P<0.05, Error bars = SEM).
Several phosphatases previously implicated in Shh signaling, including catalytic and regulatory subunits of Pp2a, were identified in our screens (Hillman et al., 2011; Nybakken et al., 2005) (Table S3). None of the phosphatases tested differentially affected signaling initiated by Shh and signaling initiated by SAG, suggesting that the identified phosphatases function downstream of Smo activation (Table S3). Among the genes recognized in our screen (Table S4), three phosphatases (Ppm1a, Eya1, Eya2) are reported to be differentially expressed in Shh-subtype medulloblastoma compared with other medulloblastoma subtypes (Kool et al., 2008; Northcott et al., 2011; Thompson et al., 2006), suggesting that they are likely to be biologically important for Shh signaling. Analysis of gene expression databases encompassing more than 70 medulloblastomas tumors (Robinson et al., 2012) confirmed that Eya1 expression is consistently higher in Shh subtype medulloblastomas than in other medulloblastoma subtypes (Figure 1F). We verified that shRNAs targeting Eya1 and Smo efficiently knock-down the target mRNA and that shRNAs targeting Eya1 reduce Eya1 protein levels (Figure S1A-C), while shRNAs intended to target Eya2 do not affect Eya2 levels and instead reduce Eya1 mRNA expression (Figure S1D, S1E). We therefore focused our studies on Eya1.
Multiple, validated shRNAs targeting Eya1 clearly inhibit Shh signaling in ShhLightII cells using several readouts of pathway activation, including SAG-mediated induction of Gli-responsive firefly luciferase (Figure 1G, Figure S1F), and induction of the target genes Gli1 and Ptch1 as measured by quantitative PCR and by western blot (Figure 1H-K, Figure S1G-I). Strikingly, Eya1 knock-down inhibits Shh pathway to a similar degree as does Smo knock-down. This is a selective effect on Shh responses, as Eya1 knock-down in ShhLightII cells does not alter PDGF-induced c-fos expression (Figure S1J). Furthermore, under the conditions tested, Eya1 knock-down did not affect cell number overall, alter apoptosis or proliferation as measured by TUNEL staining and by phospho-histone H3 (pH3) staining respectively (Figure S1K, S1L). Together these data indicate that Eya1 shRNAs exert a selective effect on Shh-responsive gene induction.
Studies in mouse embryonic fibroblasts (MEFs) from wild type and Eya1−/− littermates confirm that Eya1 is needed for Shh dependent signaling, as demonstrated by SAG-induced expression of Gli1 and Ptch1 (Figure 1L, M, respectively). Re-expression of wild type Eya1 in mutant MEFs restores SAG-dependent Gli1 and Ptch1 regulation, while a mutant form of Eya1 without phosphatase activity (D327A) does not do so, indicating that catalytically active Eya1 is required for Shh signaling (Figure 1N, O and Figure S1M, S1N). We verified that endogenous Eya1 is catalytically active in the cells tested, as Eya1−/− MEFs exhibit increased tyrosine phosphorylation of the substrate H2AX (Cook et al., 2009), and knock-down of Eya1 increases H2AX phosphorylation in ShhLightII cells (Figure S1O). However, stimulation with a Smo agonist does not alter H2AX expression or phosphorylation status, suggesting that phosphatase activity of Eya1, and not changes in H2AX phosphorylation per se, are required for Eya-mediated Shh signaling (Figure S1P).
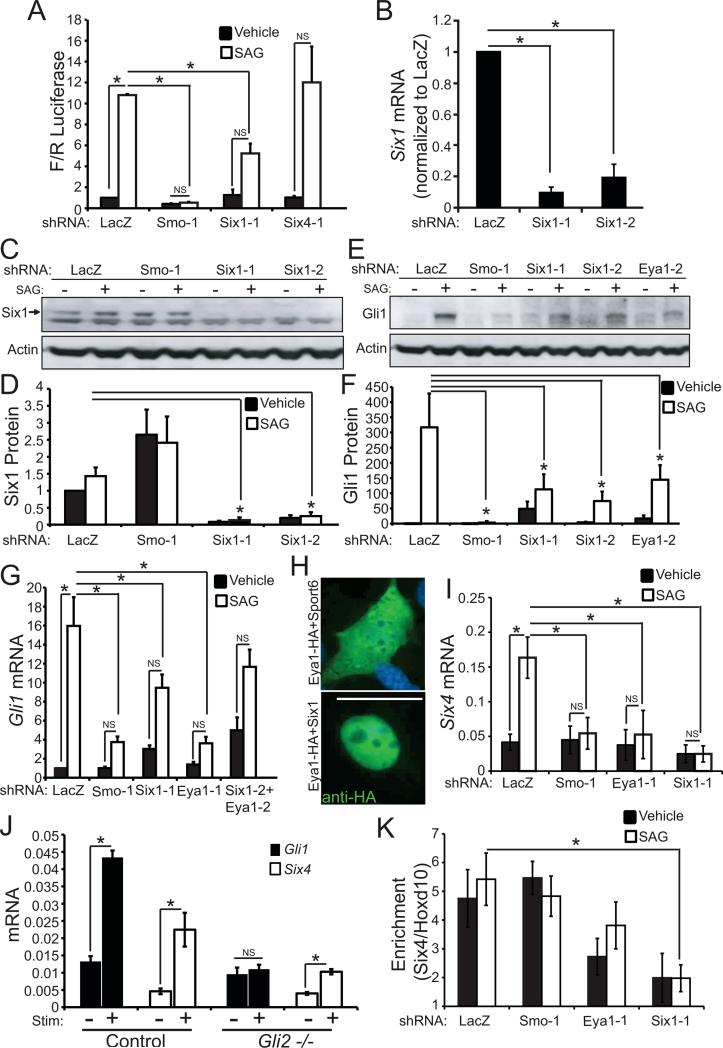
The catalytically active phosphatase Eya1 interacts with two DNA-binding proteins, Six and Dach, to regulate multiple developmental processes (Li et al., 2003). We screened shRNAs targeting diverse members of the Six and Dach protein families in ShhLightII cells, and found that Six1 is both expressed in these cells and required for Shh pathway activation (Figure 2A, Figure S2A). Two distinct shRNAs that target Six1 (Figure 2B-D) reduce Gli1 induction in response to SAG to the same extent as Eya1 shRNAs (Figure 2E-G, Figure S2B); furthermore, simultaneously knocking-down Eya1 and Six1 does not additionally impinge on the response to SAG (Figure 2G). When introduced into ShhLightII cells, HA-Tagged Eya1 is localized throughout the cell but becomes localized to the nucleus when co-expressed with Six1 (Figure 2H). Together, these data are consistent with a model wherein Eya1 and Six1 work together in the nucleus to regulate Shh signal transduction.
Figure 2.
Six1 promotes Shh pathway activation. (A) An shRNA targeting Six1, but not Six4, blocks induction of Firefly/Renilla luciferase (N=3); no puromycin selection in these studies (N=4, *P<0.05, Error bars = SEM; NS = not significant, P>0.05) . (B) Six1 shRNAs (Six1-1 and Six1-2) effectively knock-down Six1 mRNA (N=4-5, *P<0.05, Error bars = SEM) and (C) reduce Six1 protein levels (Actin = loading control). (D)Quantification of Six1 protein normalized to actin (N=6, *P<0.05, Error bars = SEM). (E) Six1 knock-down blocks Gli1 protein induction by SAG (Actin = loading control). (F) Quantification of Gli1 induction (N=6, *P<0.05, Error bars = SEM). (G) Six1 knock-down blocks Gli1 mRNA induction (N=4-5, *P<0.01, Error bars = SEM; NS = not significant, P>0.05). (H) Eya1-HA is located throughout the cell; co-transfection of Eya1-HA and Six1 results in nuclear localization of Eya1 in ShhLightII cells, anti-HA (green); scale bar = 50μm. (I) Six4 mRNA is induced by SAG in ShhLightII cells; induction requires Smo, Eya1 and Six1 as shown by knockdown of LacZ-control, Smo, Eya1 or Six1 (N=3, *P<0.05, Error bars = SEM; NS = not significant, P>0.05). (J) Gli1 and Six4 mRNA in control and Gli2−/− MEFS (N=3, *P<0.05, Error bars = SEM; NS = not significant, P>0.05). Gli2 is required for induction of Gli1 but not Six4. (K) ChIP of SAG-stimulated or control ShhLightII cells (following knockdown of LacZ-control, Smo, Eya1 or Six1) was done using antibody to Six1, followed by QPCR for Six4 or Hoxd10 promoter. Six1 interacts with Six4 promoter, not with negative control Hoxd10. Six4 promoter interaction is reduced by Six1 or Eya1 knockdown. Enrichment for Six4 relative to Hoxd10: *P<0.05 by Z-test with Bonferroni correction.
Six1 contains a DNA binding domain and binds directly to transcription promoter sites, including the Six4 promoter (Liu et al., 2010a). However, Six1 does not contain an activation domain, and so catalytically active Eya1 or another co-factor is required to alter gene transcription (Wu et al., 2013). We analyzed expression of Six4, a known Six1-target gene (Liu et al., 2010b) in ShhLightII cells and in mouse embryonic fibroblasts. Stimulation with Smo agonist (SAG) increases mRNA levels of Six4, and this response depends on Six1, Eya1, and the Shh signaling receptor Smo (Figure 2I), but does not require Gli2, the canonical Shh-pathway transcription factor (Figure 2J, Figure S2C). Chromatin immunoprecipitation studies verified that Six1 interacts with the Six4 promoter in ShhLightII cells (Figure 2K). Taken together these data indicate that Eya1 cooperates with Six1 to initiate transcription of Shh-dependent genes including Six4.
Eya1 and Six1 Act in the Shh Signaling Pathway Between Smo and Sufu
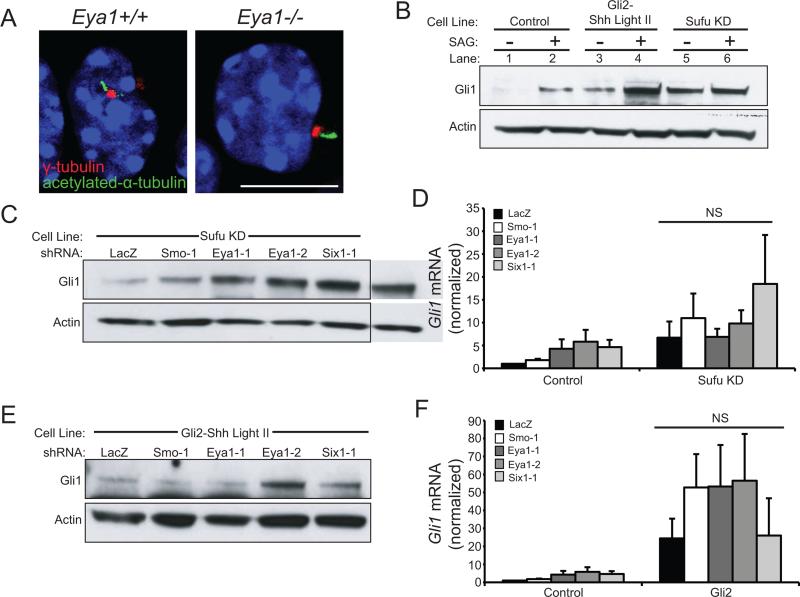
Primary cilia are essential for Shh signaling and mutants lacking cilia do not respond to pathway stimulation (Goetz and Anderson, 2010). Eya1−/− cells are ciliated, indicating that Eya1 is not necessary for ciliogenesis (Figure 3A). As both Eya1 and Six1 are required for activation of the pathway by SAG, a Smo agonist, these components must act in the pathway downstream of Smo activation.
Figure 3.
Eya1 and Six1 function in Shh transduction between Smo and Sufu. (A) Eya1−/− granule cell precursors are ciliated. γ-tubulin (red) marks basal bodies and acetylated-α-tubulin (green) marks the ciliary axoneme; nuclei are visualized by DAPI (blue); scale bar = 10 μm. (B) Stable cell lines with Sufu knock-down (Sufu KD) or Gli2 overexpression (Gli2-ShhLightII) have constitutively elevated levels of Gli1 protein (see lanes 1, 3, 5), Actin = loading control. (C) Eya1 and Six1 shRNAs do not reduce Gli1 protein levels in Sufu KD cells (Actin = loading control). (D) Eya1 and Six1 shRNAs do not reduce Gli1 mRNA in Sufu KD cells (N=5; N=3 Six1-1). (E) Eya1 and Six1 shRNA do not alter Gli1 protein in Gli2-ShhLightII cells (Actin = loading control). (F) Eya1 or Six1 shRNA do not reduce Gli1 mRNA in Gli2-ShhLightII cells (N=5, N=2 Six1-1, Error bars = SEM; NS = not significant, P>0.05).
In the absence of Shh stimulation, Gli transcription factors interact with Sufu. Following Smo activation, Gli transcription factors are released by Sufu and traffic to the nucleus where they activate or repress gene transcription. To investigate whether Eya1 and Six1 function upstream of Sufu and of Gli transcription factors we expressed shRNA against Sufu in ShhLightII cells (Figure S3A) or expressed a truncated constitutively active Gli2 and thereby activated the Shh pathway (Figure 3B). Eya1 and Six1 shRNAs do not affect the constitutive activation of the pathway induced by Sufu loss or by Gli2 expression (Figure 3B-F, Figure S3A, S3B). Together these data demonstrate that Eya1 and Six1 function upstream of Sufu and of Gli2 to regulate Shh signaling activity.
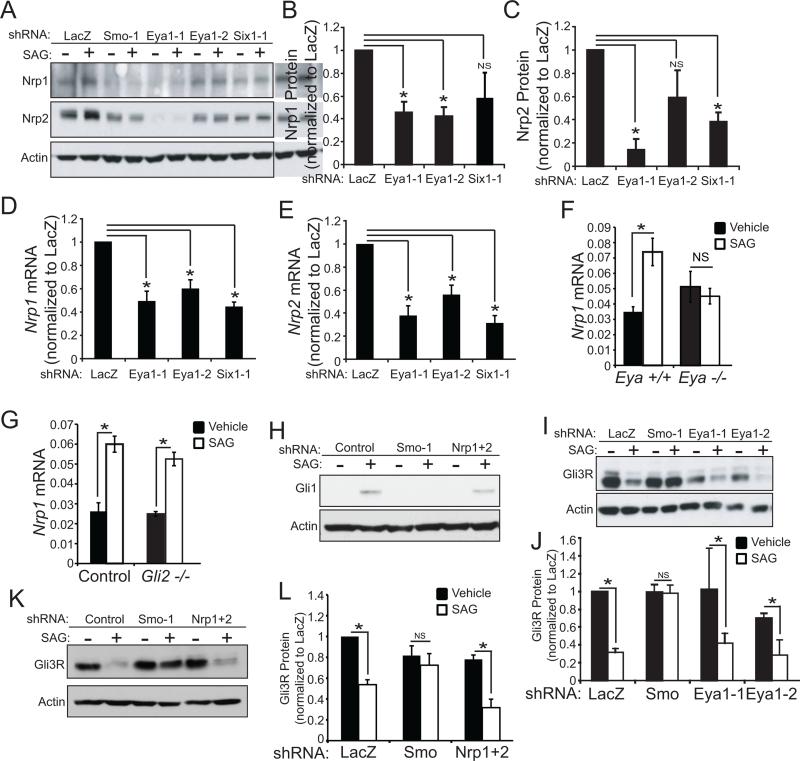
Like Eya1 and Six1, Nrp1 and Nrp2 are positive regulators of Shh signaling that mediate signal transduction between Smo and Sufu (Hillman et al., 2011). Shh pathway activation both relies on, and increases, Nrp1 expression; however Nrp1 is not a direct target of Gli transcription factors suggesting that Shh might regulate transcription by additional mechanisms other than Gli transcription factors (Hillman et al., 2011; Hochman et al., 2006).
Knock-down of Eya1 or Six1 reduce Nrp1 and Nrp2 (Figure 4A-E) in both unstimulated and SAG-stimulated cells, indicating Eya1 and Six1 are required for both basal and Shh-induced Nrp gene expression. Similarly expression of Nrp1 mRNA is reduced in Eya1−/− MEFs, (Figure 4F), and catalytically active Eya1, but not inactive D273A Eya1, increases Nrp1 expression following SAG stimulation (Figure S4). In contrast, the transcription factor Gli2 is not required for SAG-dependent Nrp gene induction (Figure 4G). Thus transcription factor Six1 and catalytically active co-activator Eya1 rather than Gli transcriptional activators control Shh-regulated Nrp1 expression, thereby conveying signal transduction between Smo and Sufu.
Figure 4.
Eya1 and Six1 regulate Nrp expression. (A) shRNAs targeting Eya1 and Six1 reduce Nrp1 and Nrp2 protein levels in unstimulated and SAG-stimulated ShhLightII cells (Actin = loading control). (B) Quantification of Nrp1 (N=3-4, *P<0.05, Error bars = SEM; NS = not significant, P>0.05). (C) Quantification of Nrp2 (N=3, *P<0.05, Error bars = SEM; NS = not significant, P>0.05). (D,E) shRNAs targeting Eya1 and Six1 reduce Nrp1 mRNA (N=5-6, *P<0.05, Error bars = SEM) and Nrp2 mRNA (N=4-5, *P<0.05, Error bars = SEM). (F,G) SAG-induced expression of Nrp1 is prevented in Eya1−/− MEFs (F), but not in Gli2−/− MEFs (G) (N=3, *P<0.05, Error bars = SEM; NS = not significant, P>0.05). (H) shRNA targeting Nrp1 plus shRNA targeting Nrp2 reduce Gli1 protein following Shh stimulation (Actin = loading control). (I) Eya1 knock-down does not alter SAG-induced changes in Gli3R formation; Smo knock-down does do so. (J) Western blot quantification of Gli3R (N=3, *P<0.05, Error bars = SEM; NS = not significant, P>0.05). (J) shRNA targeting Nrp1 plus shRNA targeting Nrp2 does not alter Gli3R formation following Shh stimulation (Actin = loading control). (K) Western blot quantification of Gli3R (N=3, *P<0.05, Error bars = SEM; NS = not significant, P>0.05).
Eya1, Six1, and Nrps do not affect Formation of Gli3 Repressors (Gli3R)
Shh pathway activity enhances expression and function of Gli-transcriptional activators, and concurrently inhibits formation of Gli transcriptional repressors (Blaess et al., 2006; Bok et al., 2007; Wang et al., 2000). Eya1 and Six1 are required for transcription mediated by Gli-activator species, as assessed by Gli1 gene induction. Similarly, Nrp1 and Nrp2 regulate Gli1 induction in response to pathway stimulation (Figure 4H). However, we find that Eya1 and Nrps are not necessary for Shh-dependent inhibition of Gli3R. Eya1 knock down, or knock-down of Nrp1 and Nrp2 do not alter SAG-induced processing of Gli3 to form the 83kD amino-terminal repressor fragment, Gli3R (Figure 4I-L). In contrast, knock-down of Smo prevents both Gli3R inhibition and SAG-stimulated Gli1 induction. These data indicate that Eya1 and Nrps are required selectively for Smo-mediated Gli activator functions and not for regulation of Gli3R, and so link Eya1 and Nrps in a pathway that differentially modulates Shh signal transduction.
Eya1 promotes Shh Signaling in hindbrain development and in tumorigenesis
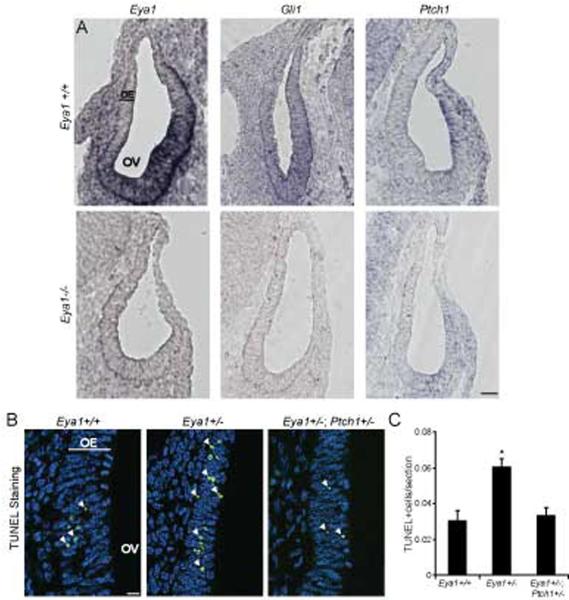
Eya1 and Six1 are expressed in Shh-responsive cells within the otic vesicle, and Eya1−/−, Six1−/−, and Shh−/− mutants exhibit similar otic vesicle phenotypes (Ozaki et al., 2004; Xu et al., 1999; Zheng et al., 2003; Zou et al., 2006). To investigate a role for Eya1 in Shh signaling within the auditory system, we analyzed expression of the Shh target genes Gli1, and Ptch1, in Eya1−/− otic vesicles (Xu et al., 1999). Eya1, Gli1 and Ptch1 are all expressed in the ventral portion of the otic vesicle in wild type animals, and expression of these genes is decreased in Eya1−/− otic vesicles, demonstrating that loss of Eya1 results in reduced Shh signaling in vivo (Figure 5A, Figure S5).
Figure 5.
Eya1 and Shh signaling in developing otic vesicle. (A) Eya1−/− otic vesicles at E10.5 have reduced Eya1, Gli1, and Ptch1 expression by in situ hybridization (OV = otic vesicle, OE = otic epithelium, scale bar = 50 μm), however expression of Gata3 is evident in ventral cells of Eya1−/− otic vesicles (Zou et al., 2006). (B) E10.5 Eya1+/− otic vesicles have increased apoptosis by TUNEL staining (green); nuclei stained by DAPI (blue); scale bar = 50 μm. White arrows indicate TUNEL-positive cells. Comparison of Eya1+/− and Eya1+/−/Ptch1+/− mice. (C) Percentage of otic vesicle cells that are TUNEL-positive (N=19 sections from 2 mutant and 2 wild type littermate mice, *P<0.01, Error bars = SEM).
Cell death is increased in Eya1−/− otic vesicles (Xu et al., 1999; Zou et al., 2006), and this phenotype is also observed in Eya1+/− otic vesicles (Figure 5B center panel, C). We find that gain of function in the Shh pathway reverses the apoptotic phenotype of Eya1+/− mice, as demonstrated by comparing TUNEL-positive cells in Eya1+/− and Eya1+/−; Ptch1+/− otic vesicles (Goodrich et al., 1997) (Figure 5B right panel, Figure 5C). These data demonstrate a genetic interaction between Eya1 and Shh signaling in vivo, and indicate that Eya1 plays a critical role in Shh-dependent hindbrain development.
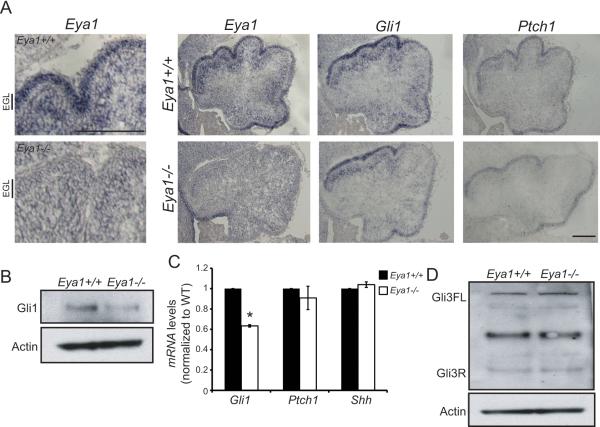
Shh has a well-recognized role as a mitogen for granule cell precursor proliferation in the developing external granule cell layer (EGL) of the cerebellum (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999). Eya1 is expressed in the developing cerebellum, and levels decrease during postnatal development (http://www.cdtdb.neuroinf.jp). In the early cerebellum, Eya1 mRNA is evident both in Purkinje cells and in granule cell precursors in the EGL, an expression pattern similar to that observed for Gli1 (Figure 6A,B,C, Figure S6). We find that Gli1 levels are reduced in Eya1−/− cerebella while expression of Shh does not change (Figure 6A,B,C). Consistent with our data in fibroblasts, Eya1 is required for Gli activators but is not required for Gli3 Repressor formation in developing cerebellum (Figure 6D). Importantly, the cerebellar phenotype of Eya1−/− embryos resembles the phenotype observed at this age with mutations in Gli2 or other mutations that cause loss of Shh activity (Corrales et al., 2004), with striking reduction in granule cell precursor proliferation as assessed by phospho-histone H3 (PH3) and Ki67 staining (Figure 7A-C), without increased cell death (Figure 7D, E). Eya1 fosters Shh-responses during embryonic and postnatal development of the cerebellum, as granule cell precursor proliferation and Gli1, Nrp1 and Ptch1 expression are also dramatically decreased in Eya1+/− mice at post-natal day 3 (Figure 7F, G, H, I). Eya1 acts cell autonomously within granule cell precursors to promote Shh responses, as purified, cultured granule cell precursors that lack Eya1 do not respond to exogenous Shh pathway agonists measured by induction of Gli1, Ptch1, Six4 and Nrp1 (Figure 7J, K, L, M). Together these data demonstrate that Eya1 promotes Shh-dependent signaling and proliferation in precursors of the developing cerebellum.
Figure 6.
Eya1 promotes Shh signaling in cerebellum. (A) Eya1 is expressed in Purkinje cells and granule cell precursors in the external granule cell layer (EGL). In situ hybridization of Eya1, Gli1 and Ptch1 in E18.5 Eya1+/+ and Eya1−/− cerebella; scale bar = 25 μm. (B) Western blot of Gli1 in Eya1+/+ and Eya1−/− E18.5 cerebellum. (C) Gli1, Ptch1 and Shh mRNA levels in Eya1+/+ and Eya1−/− cerebellum. (D) Gli1 and Gli3 immunoblots of Eya1+/+ and Eya1−/− E18.5 cerebellar lysates. Eya1−/− cerebella have decreased Gli1 protein with no change in Gli3R levels.
Figure 7.
Eya1 promotes proliferation in cerebellar granule cell precursors and in medulloblastoma. (A) Phospho-Histone H3 immunohistochemistry (red); nuclei stained with DAPI (blue) in Eya1+/+ and Eya1−/− E18.5 cerebellum. (Arrows indicate PH3-positive cells; scale bar = 50 μm). (B) Quantification of PH3-positive cells per cerebellum (N=3 per genotype, *P<0.01, Error bars = SEM). (C) Ki67-positive cells are reduced in Eya1−/−E18.5 cerebella (N=3 per genotype, *P<0.01, Error bars = SEM). (D) TUNEL-positive cells (green) in Eya1+/+ and Eya1−/− E18.5 cerebella. Nuclei stained with DAPI (blue), (Arrows indicate TUNEL-positive cells; scale bar = 50 μm). (E) Quantification of TUNEL-positive cells (N=3 per genotype, Error bars = SEM). (F) Quantification of PH3-positive cells in Eya1+/+ and Eya1+/− postnatal day 3 (P3) cerebellum (N=3 per genotype, *P<0.01, Error bars = SEM). (G) Western blot and (H) quantification of Gli1 in P3 Eya1+/+ and Eya1+/− cerebellum (N=3 per genotype, *P<0.01, Error bars = SEM). (I) qRT-PCR of Gli1, Nrp1 and Ptch1 in Eya1+/+ and Eya1+/− P3 cerebellum (N=3 per genotype, *P<0.01, Error bars = SEM). In culture, purified Eya1−/− cerebellar granule cell precursors (from E18.5) do not respond to Shh-pathway stimulation with exogenous SAG as assessed by qRT-PCR for (J) Gli1, (K) Ptch1, (L) Six4 and (M) Nrp1 (N=3 per genotype, *P<0.01, Error bars = SEM, NS = not significant, P>0.05). (N) Kaplan-Meier analysis of deaths due to medulloblastoma in Eya1+/− (N=12), Ptch1 +/− (N=64) and Eya1+/−/Ptch1+/− (N=87) (P=0.0062). (O) Smo and Eya1 shRNA reduce Gli1 mRNA in cultured mouse medulloblastoma cells (N=4-5). (P) Smo and Eya1 shRNA reduces the viability of cultured mouse medulloblastoma cells (N=3, *P<0.01, Error bars = SEM).
Constitutive activation of Shh signaling in granule cell precursors results in the cerebellar tumor, medulloblastoma (Goodrich et al., 1997; Hallahan et al., 2004; Han et al., 2009). Heterozygous mutations in Ptch1 in mice and in people confer a high risk for developing medulloblastoma (Goodrich et al. 1997). Strikingly, the incidence of medulloblastoma is greatly decreased in mice that are heterozygous for both Ptch1 and Eya1 compared to Ptch1+/− mice from the same colony (Figure 7N), providing in vivo evidence that Eya1 promotes growth of these Shh-pathway-dependent cerebellar tumors. Our lab recently generated cell lines derived from medulloblastoma of Ptch1+/− mice that are tumorigenic and retain Shh dependence in vitro (Shh-medulloblastoma or SMB cells; Zhao et al. unpublished observations). Both Smo and Eya1 knock-down reduce Gli1 mRNA levels and decrease viability of these SMB cells (Figure 7O, P), indicating that Eya1, like Smo, functions cell autonomously to promote tumor cell growth. Thus Eya1 provides a potential therapeutic target for Shh-subtype medulloblastoma.
Discussion
An unbiased shRNA screen of the phosphatome implicated Eya1, a tyrosine phosphatase and transcriptional co-activator, in Shh signaling. We find that Eya1 acts in concert with a collaborating transcription factor, Six1, to change gene expression. Eya1, Six1, and a target gene Nrp1, all function in the Shh pathway between Smo and Sufu, and preferentially regulate Gli activators rather than Gli repressors. In vivo, Eya1 is critical for Shh-dependent development of the cerebellum and inner ear, and Eya1 fosters growth of medulloblastoma, a Shh-pathway dependent tumor. Thus Eya1/Six1 function as transcriptional regulators within the Shh pathway and alter the equipoise between Gli activators and repressors during development and oncogenesis.
Eya1 and Six1 Function Together to Regulate Shh Signal Transduction
Previous studies demonstrate that Eya family members cooperate with DNA-binding proteins of the Six family to regulate transcription (Li et al., 2003). In humans, heterozygous mutations in Eya1 and Six1 cause Branchio-Oto-Renal syndrome, in which development of the inner ear, face, and kidneys are perturbed, resulting in deafness and impaired renal function (Kochhar et al., 2007). Taken together, data that Eya1 facilitates Six1 binding to the Six4 promoter and that Shh-pathway stimulation regulates Six4 expression, indicate that Shh fosters Eya1/Six1 dependent transcription and thereby instigates signal transduction. Eya and a Drosophila homolog of Six, So, are part of the Retinal Determination Gene network, a program critical for eye development in Drosophila (Kumar, 2009). Interestingly, So was identified in a genome-wide RNAi screen for components of the Hh signaling pathway, suggesting that Eya and Six family members have an evolutionarily conserved role in Hh functions (Nybakken et al., 2005).
Eya proteins contain a highly conserved C-terminal domain called the Eya domain, which is necessary for Eya to bind Six. The Eya domain contains a haloacid dehalogenase (HAD) sequence motif that is necessary for tyrosine phosphatase activity (Li et al., 2003; Rayapureddi et al., 2003; Tootle et al., 2003). Missense Eya mutations that disrupt protein phosphatase activity also impair the function of an Eya-So/Six complex (Mutsuddi et al., 2005; Rayapureddi et al., 2003; Tootle et al., 2003), however Eya1 dephosphorylation of H2AX does not depend on Six (Cook et al., 2009; Krishnan et al., 2009). Thus Eya1 phosphatase can operate either independently or in concert with Six family members. The data presented here indicate that Eya1 acts together with Six1 to mediate Shh signal transduction, and demonstrate that Eya1 phosphatase activity is essential for this function.
Eya1 and Six1 Regulate Nrp Gene Expression
A model for the roles of Eya1, Six1 and Nrp1/2 in Shh signal transduction is shown as the graphical abstract. Genetic epistasis experiments indicate that Eya1, Six1, and Nrp1/2 are all positive regulators of Shh transduction, and function in the signaling pathway between activated Smo and Sufu/Gli2. We suggest that a major mechanism by which Eya1 and Six1 regulate Shh signaling is by controlling transcription of Nrp1 and other target genes (Ahmed et al., 2012). Consistent with a paradigm in which Eya1/Six1 collaborate to directly regulate Nrp expression, Six1 binds to the Nrp1 promoter region by ChIP analysis (Liu et al., 2010b). Intriguingly, Eya1 and Nrp1/2 regulate Gli activator species but are not involved in generating the Gli3 repressor. These data suggest that the mechanisms regulating Gli activators and repressors diverge from one another subsequent to Smo activation, and that Nrp1/2 function selectively in the branch crucial for Gli activators. Recent studies indicating that expression of Gli activators and repressors exhibit distinct dynamics during development (Junker et al., 2014) accord well with the proposed model.
An uncoupling of Gli activator and Gli3R regulation has been reported in mice with a null allele of Arl13b, a small GTPase of the Arf/Arl family (Caspary et al., 2007). Mouse embryonic fibroblasts lacking Arl13b have shorter cilia and abnormal localization of Smo, Gli2, and Gli3 following pathway activation (Caspary et al., 2007; Larkins et al., 2011). In contrast, absence of Nrp1/2 or of Eya1 do not alter ciliary morphology (Figure 3) and (Hillman et al., 2011). While Arl13b preferentially affects Gli activators due to its role in ciliogenesis, it appears that Nrp1/2 functions more directly to liberate Gli activator from sequestration and promote trafficking of Gli activator to nuclear DNA binding sites.
Significance of Eya1 in Shh Signaling in Hindbrain Development and Cancer
We demonstrate that Eya1 is required for Shh signaling in NIH3T3-derived cells and in MEFs in vitro, and show that Eya1 promotes Shh signaling in the developing brainstem and hindbrain in vivo. In Eya1 mutants, Gli1 expression is decreased in developing otic vesicle and cerebellum. The genetic interaction between Eya1 and Ptch1 mutations in the otic vesicle, and the phenotypic similarity of the Eya1 mutant cerebellum to mutants with loss-of-Shh signaling, indicate that Eya1 is critical for multiple biological responses to Shh in the developing hindbrain. However, Eya1 is not uniformly involved in Shh-dependent responses. An early function for Shh signaling is patterning of the developing spinal cord, but Eya1−/− embryos do not exhibit defects in spinal cord patterning (Figure S7). Outside of the nervous system, Eya1 and Six1 function within Shh-producing cells rather than Shh-responsive cells, and so Eya1−/− lung tissue exhibits higher levels of Shh and of Gli1 compared to wild type counterparts (El-Hashash et al., 2011a; El-Hashash et al., 2011b; Lu et al., 2013). Thus, unlike Ptch1 or Smo, Eya1 is not a core component of the Shh signal transduction pathway but is preferentially involved in a subset of Shh-dependent functions in neural tissues.
Overexpression of Eya family members has been reported in many human cancers, often accompanied by misregulation of Six family members (Auvergne et al., 2013; Tadjuidje and Hegde, 2013). Neuropilins have also been implicated in growth of multiple tumors, including medulloblastoma (Hayden Gephart et al., 2013; Snuderl et al., 2013). Eya1 is highly expressed in human Shh-subtype medulloblastoma (Figure 2)(Kool et al., 2008; Northcott et al., 2011; Thompson et al., 2006), and Eya1 promotes medulloblastoma growth both in vitro and in vivo. These data raise the intriguing possibility that targeting Eya1 may provide an effective strategy for treating these cancers. Unlike many transcriptional regulators, Eya1 has enzymatic activity and is therefore a potentially druggable target. In addition, Eya proteins belong to the small HAD family of phosphatases that possess an uncommon catalytic domain. Targeting this infrequent catalytic domain has allowed the development of specific Eya2 phosphatase inhibitors, suggesting that selectively targeting Eya1 may be feasible (Krueger et al., 2013). While several promising small molecule inhibitors of Smo can treat basal cell carcinoma and medulloblastoma, not all patients respond consistently to Smo inhibitors (Kool et al., 2014; Metcalfe and de Sauvage, 2011; Rodon et al., 2014). Selective Eya1 inhibitors may provide an effective alternative approach for blocking Shh signaling and thereby treating these cancers.
Methods
Animal studies
All experimental procedures were done in accordance with National Institutes of Health guidelines and were approved by the Dana-Farber Cancer Institutional Animal Care and Use Committee. Eya1−/− mice were obtained from Pin-Xian Xu (Xu et al., 1999), Ptch1+/− mice from Jackson Laboratory (Goodrich et al. 1997). The morning of the day a vaginal plug was detected was designated E0.5.
Cell Culture and Constructs
ShhLightII (SL2) cells were obtained from American Type Culture Collection, and cultured according to their recommendations. After introduction of pLKO.1 lentiviral shRNAs, cells were selected for infection using puromycin (4μg/ml), unless otherwise noted. SMB cells were prepared from medulloblastoma tumors of Ptch1+/− mice. These cells express Shh components and cell viability is dependent on Shh pathway activity (Zhao et al, unpublished data).
Primary granule cell precursors were cultured from E18.5 cerebella as described (Zhou et al., 2007). SMB cell viability was assayed using CellTiter96 Aqueous One Solution Cell Proliferation Assay (Promega). SL2 cells were transfected using Fugene6 with mouse PiggyBac transposase and a transposon encoding V5-Gli2 or GFP. The coding sequence from full-length Eya1 (Thermo Scientific Clone ID 6848408) was cloned into pcDNA3.1(+)-KozakHAHA., and full-length Eya1 was cloned into pCMV-Sport6. Six1-pCMV-Sport6 was obtained from Open Biosystems (Clone ID 4188451).
Mouse embryonic fibroblasts (MEFs) were cultured from E12 Eya1+/+ and Eya1−/− littermates, or from Gli2−/− mice as described (Jozefczuk et al. 2012).
Shh Ligand preparation and cell stimulation
293FT cells were transfected with full-length Shh in pcDNA3 using Lipofectamine 2000; and Shh was prepared as described previously (Chan et al., 2009; Witt et al., 2013). Confluent SL2 cells were stimulated with Shh (300ng/ml) or SAG (300nM; Enzo Life Sciences) in DMEM with 0.5% calf serum for 48-72 hrs. Cells were stimulated with purified PDGF-BB (100ng/ml for 30 mins) in the same media.
shRNA Screen and Analysis
pLKO.1 lentiviral shRNAs were from the Broad Institute RNAi Consortium (TRC). SL2 cells were plated in 96-well plates, then each well was infected with a single shRNA lentivirus. shRNAs targeting RFP, GFP, LacZ and two shRNAs targeting Smo were included on each plate as negative and positive controls, respectively; duplicate plates were tested. Infected cells were selected with 4ug/ml puromycin, then cells were stimulated with Shh, SAG, or vehicle for 72 hrs, then lysed and tested. Luciferase assays were conducted using a Dual Luciferase Reagent (DLR; Promega #E1960) kit.
shRNAs with a Renilla luciferase value equal to zero were eliminated from analysis. Firefly/Renilla luciferase ratios from duplicate wells were averaged and average Firefly/Renilla luciferase ratios equal to zero were assigned a value of 1×10−6. We calculated the robust z score for each shRNA [(x-median)/(median absolute deviation); median absolute deviation = median (abs(x- median))*1.4826] using the natural log of all values. For the primary screen, genes were considered hits if two or more targeting shRNAs generated a robust z score less than −1.5 or greater than 1.5 in pathway stimulation conditions. In the secondary screen genes were considered hits if multiple shRNAs resulted in Firefly/Renilla luciferase values less than 25%, or greater than 400% of the median value of control shRNAs. Screen analysis was conducted in Microsoft Excel and in Matlab.
Lentiviral Production
Virus containing media was collected from 293T packaging cells transfected using Fugene6 reagent (Promega) in complete DMEM supplemented with 10% fetal bovine serum or in DMEM/F12 supplemented with B27 (Gibco). 293T and SL2 cells were transfected using Fugene6 using the protocol from The RNAi Consortium (TRC) at the RNAi Platform of the Broad Institute of MIT and Harvard at http://www.broadinstitute.org/rnai/public/.
shRNA Knock-Down
Lentivirus was generated from pLKO.1 or pCDF (for Eya1 WT and Eya1 D237A (Wu et al., 2013)), lentiviral shRNAs obtained from the Broad Institute RNAi Consortium (TRC). SL2 cells were plated in 12-well or 6-well plates, then infected with shRNA lentivirus. Infected cells were selected with 4μg/ml puromycin, then cells were stimulated with SAG or vehicle (300ng) for 48 or 72 hrs before RNA or protein collection.
Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted using Trizol (Invitrogen) according to manufacturer's protocol. Reverse Transcription was performed using the cDNA archive kit (Applied Biosystems) according to the manufacturer's specifications. Quantitative real-time RT-PCR was performed using Taqman gene expression assays to assess the expression of: Eya1 Mm00438796_m1, Eya2 Mm00802561_m1, Smo Mm01162710_m1, Gli1 Mm00494645_m1, Ptch1 Mm00436026_m1, Gli2 Mm01293117_m1, Dach1 Mm00473899_m1, Dach2 Mm00473899_m1, Six1 Mm00808212_m1, Six2 Mm00807058_m1, Six4 Mm00803396_m1, Six5 Mm01305439_g1, Nrp1 Mm00435371_m1, Nrp2 Mm00803099_m1. Values were normalized to gapdh levels.
Western Blotting
Cells were lysed in modified RIPA buffer and protein lysates were separated by 4-12% SDS-PAGE and blotted with primary antibodies. Bands were visualized with secondary antibodies conjugated to HRP (1:10,000: Bio-Rad) and SuperSignal chemiluminescent substrate kit. For western blot quantification, film was scanned using Epson perfection V750 pro scanner and Epson scan software. Background-subtracted band density was measured in ImageJ, and normalized to actin as a loading control.
Immunohistochemistry and in situ hybridization
Immunohistochemistry and in situ hybridization were performed as described (Chan et al., 2009). For visualization of cilia in dissociated granule cell precursors, dissociated cells were stained with antibodies to acetylated alpha tubulin (Invitrogen #322700) and to gamma tubulin (Sigma #T5192) and imaged on a Leica confocal microscope at 63x with optical zoom (Leica Microsystems, Buffalo Grove, IL). Images were acquired with the Leica Microsystems Application Suite (24.1 build 6384), then processed and analyzed using ImageJ software (NIH) and Adobe Photoshop 7.0. For Phospho-Histone H3 (PH3) detection, sections were dried at room temperature for 30-60 mins, rehydrated with PBS 2X for 5 mins and then underwent antigen retrieval in Tris-EDTA antigen retrieval solution (0.005M Tris, 0.001M EDTA), blocked and permeabilized (3% NGS, 5% BSA, 0.3% Triton-X100) for 1 hr. Tissue was incubated overnight at 4°C with antibody in (3% NGS, 0.3% Triton). Cells in M-phase of the cell cycle were detected using mouse anti-Phospho-Histone H3. Primary antibody was washed, secondary antibody was applied, and cells mounted. Phospho-Histone H3-positive cells were detected by immunofluorescence and analyzed using NIS Elements imaging software. Positive cells were manually counted. Sections were collected from three pairs of mutant and wild type animals, each pair was collected from a unique litter. The detection of apoptotic cells was conducted using DeadEnd™ Fluorometic TUNEL system staining (Progema, G3250). The percent of TUNEL-positive cells was calculated using NIS elements software to manually count TUNEL-positive cells the number DAPI-positive cells. Sections were collected from three pairs of mutant and wild type animals, each pair was collected from a unique litter.
Antibodies
Eya1 (Aviva Systems Biology #ARP32434), HA (Millipore #05-904), Actin (Cell Signaling #4968), Gli1 (Cell Signaling #2534), Six1 (Abcam #ab84329, #ab86028), Nrp1 (R&D Systems #AF566), Nrp2 (Cell Signaling #3366), Gli2 (Aviva Systems Biology #ARP31885), Gli3 (R&D Systems #AF3690), Gamma-tubulin (Sigma #T5192) and Acetylated-alpha-tubulin (Invitrogen #322700).
Gene Expression analysis
Results from Affymetrix arrays of pediatric medulloblastomas (GSE37418) were analyzed. Levels of Eya1 in samples from each of the four tumor subtypes as defined in that study (8 Wnt, 10 Shh, 17 group 3, 39 group 4) were plotted and analyzed in Prism by ANOVA, P<0.0001.
Methods for Six1 ChIP-qPCr
Approximately 20-80 million ShhLightII cells were used for a single ChIP experiment. Cells were stimulated with SAG for 3 days, incubated with crosslinking solution (1% formaldehyde, 100mM NaCl, 1mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 50 mM HEPES pH 7.9) for 10 mins at room temperature. Cross-linking was quenched with 125 mM glycine for 5 mins at room temperature, cells were rinsed with cold PBS, resuspended in PBS with protease inhibitors. To isolate nuclei, cell pellets were lysed in 2mL of Buffer I (50 mM Hepes KOH, pH 7.5, 140 mM NaCl, 1mM EDTA pH 8.0, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 10 mM B-Glycerophosphate, 10mM Sodium Fluoride, 1x protease inhibitor cocktail, 100 nM okadaic acid) and incubated 10 mins at 4°C. Nuclei were then pelleted for 10 mins at 4°C at 3000 RPM. Isolated nuclei were washed in Buffer II (200mM NaCl, 1mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10mM Tris pH 8.0, 10mM B-Glycerophosphate, 10mM Sodium Fluoride, 1x protease inhibitor cocktail, 100 nM okadaic acid), suspended in Buffer III (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-laurylsarcosine, 10mM NaF, 1x protease inhibitor cocktail, 100 nM Okadaic Acid). Nuclear lysates were sonicated using a Bioruptor sonicator, for a total of 32 cycles. After sonication, samples were centrifuged. After Triton-X 100 and SDS were added, samples were incubated for 1 hr at 95°C, then incubated with Proteinase K for 30 mins at 55°C, purified with the Qiagen PCR purification kit.
Lysates were pre-cleared with 15ul pre-rinsed Protein A Dynabeads in 200ul TBSTPb (0.01% BSA and 0.2mM PMSF in 1x TBST). Pre-cleared lysates were incubated with Six1 Abcam antibody coupled to Protein A Dynabeads overnight at 4°C. Beads were then bound to immune-complexes, collected and washed twice each with low salt buffer and high salt buffer. Immunoprecipitated materials were eluted twice with elution buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0, 1 % SDS) at 65°C for 30 mins. All eluates were reverse-crosslinked at 65°C overnight for 12-16 hrs. RNAse A was added for 1 hr at 37°C, then Proteinase K was added for 2-3 hrs at 55°C, and DNA was isolated from each sample. qPCR of ChIP samples was performed for Hoxd10 as the negative region and Six4 as the positive region. Primer sequences used were: Six4_F: ATCTGGCCGATCAGGTTTC, Six4_R: ACCGGAGGAGTCACGTTG, Hoxd10_F: GAGAAATCGGACTCACCTTCC, Hoxd10_R: CACATACCCAGGCAGAACG. For each primer set, we analyzed standard curves and melting curves to assess the quality of the primers. Sequential dilutions of input DNA were used for the qPCR standard curve.
Screen Data
Primary data from the phosphatome screen are available on figshare: http://dx.doi.org/10.6084/m9.figshare.1287817
Statistics
Within each experiment, mRNA or protein values for each condition were normalized to an internal standard (gapdh or actin). To average results across multiple independent experiments, values were normalized to the results obtained with a control virus in that experiment (LacZ). Statistical significance was determined using a z-test with Bonferroni correction for multiple comparisons, or by Student's t-test with Bonferroni correction in Microsoft Office Excel, or by ANOVA with Tukey's post-test analysis in Prism as indicated. All experiments were done with at least three independent biological replicates.
Supplementary Material
Acknowledgements
We thank William Hahn, So Young Kim and the Broad Institute RNAi Consortium for help with the shRNA screen, and Alexandra Smolyanskaya for help with analyzing the screen. We thank Lisa Goodrich, Clifford Tabin for providing probes for in situ hybridization studies, and we thank Richard Pestell for the Eya1 WT and Eya1 D237A constructs in pCDF. We thank members of the Segal Lab for help and advice. These studies were supported by grants from NIH: T32CA009361 (AE, RAS), Director's Pioneer Award DP1000839 (RAS), CA142536 (RAS) and support from the PLGA foundation; NIH R01 DK64640 (PXX).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvergne RM, Sim FJ, Wang S, Chandler-Militello D, Burch J, Al Fanek Y, Davis D, Benraiss A, Walter K, Achanta P, et al. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell Rep. 2013;3:2127–2141. doi: 10.1016/j.celrep.2013.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Bok J, Dolson DK, Hill P, Ruther U, Epstein DJ, Wu DK. Opposing gradients of Gli repressor and activators mediate Shh signaling along the dorsoventral axis of the inner ear. Development. 2007;134:1713–1722. doi: 10.1242/dev.000760. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chan JA, Balasubramanian S, Witt RM, Nazemi KJ, Choi Y, Pazyra-Murphy MF, Walsh CO, Thompson M, Segal RA. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat Neurosci. 2009;12:409–417. doi: 10.1038/nn.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Cohen DJ. Targeting the hedgehog pathway: role in cancer and clinical implications of its inhibition. Hematol Oncol Clin North Am. 2012;26:565–588. viii. doi: 10.1016/j.hoc.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004;131:5581–5590. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- El-Hashash AH, Al Alam D, Turcatel G, Bellusci S, Warburton D. Eyes absent 1 (Eya1) is a critical coordinator of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev Biol. 2011a;350:112–126. doi: 10.1016/j.ydbio.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hashash AH, Turcatel G, Al Alam D, Buckley S, Tokumitsu H, Bellusci S, Warburton D. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development. 2011b;138:1395–1407. doi: 10.1242/dev.058479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Evangelista M, Lim TY, Lee J, Parker L, Ashique A, Peterson AS, Ye W, Davis DP, de Sauvage FJ. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci Signal. 2008;1:ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden Gephart MG, Su YS, Bandara S, Tsai FC, Hong J, Conley N, Rayburn H, Milenkovic L, Meyer T, Scott MP. Neuropilin-2 contributes to tumorigenicity in a mouse model of Hedgehog pathway medulloblastoma. J Neurooncol. 2013 doi: 10.1007/s11060-013-1216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman RT, Feng BY, Ni J, Woo WM, Milenkovic L, Hayden Gephart MG, Teruel MN, Oro AE, Chen JK, Scott MP. Neuropilins are positive regulators of Hedgehog signal transduction. Genes Dev. 2011;25:2333–2346. doi: 10.1101/gad.173054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman E, Castiel A, Jacob-Hirsch J, Amariglio N, Izraeli S. Molecular pathways regulating pro-migratory effects of Hedgehog signaling. J Biol Chem. 2006;281:33860–33870. doi: 10.1074/jbc.M605905200. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Jacob LS, Wu X, Dodge ME, Fan CW, Kulak O, Chen B, Tang W, Wang B, Amatruda JF, Lum L. Genome-wide RNAi screen reveals disease-associated genes that are common to Hedgehog and Wnt signaling. Sci Signal. 2011;4:ra4. doi: 10.1126/scisignal.2001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker JP, Peterson KA, Nishi Y, Mao J, McMahon AP, van Oudenaarden A. A Predictive Model of Bifunctional Transcription Factor Signaling during Embryonic Tissue Patterning. Dev Cell. 2014;31:448–460. doi: 10.1016/j.devcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Kochhar A, Fischer SM, Kimberling WJ, Smith RJ. Branchio-oto-renal syndrome. Am J Med Genet A. 2007;143A:1671–1678. doi: 10.1002/ajmg.a.31561. [DOI] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, Kim SJ, Tonks NK. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284:16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger AB, Dehdashti SJ, Southall N, Marugan JJ, Ferrer M, Li X, Ford HL, Zheng W, Zhao R. Identification of a selective small-molecule inhibitor series targeting the eyes absent 2 (Eya2) phosphatase activity. J Biomol Screen. 2013;18:85–96. doi: 10.1177/1087057112453936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. The molecular circuitry governing retinal determination. Biochim Biophys Acta. 2009;1789:306–314. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Liu X, Ding L, Liu J, Zhang Y, Huang Z, Wang L, Chen B. [Simultaneous determination of six synthetic sweeteners in food by high performance liquid chromatography- tandem mass spectrometry]. Se Pu. 2010a;28:1020–1025. [PubMed] [Google Scholar]
- Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 2010b;38:6857–6871. doi: 10.1093/nar/gkq585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Reddy R, Berika M, Warburton D, El-Hashash AH. Abrogation of Eya1/Six1 disrupts the saccular phase of lung morphogenesis and causes remodeling. Dev Biol. 2013 doi: 10.1016/j.ydbio.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71:5057–5061. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- Mutsuddi M, Chaffee B, Cassidy J, Silver SJ, Tootle TL, Rebay I. Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics. 2005;170:687–695. doi: 10.1534/genetics.104.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis E, Hui CC. Hedgehog signaling and congenital malformations. Clin Genet. 2005;67:193–208. doi: 10.1111/j.1399-0004.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome- wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, Okamura HO, Kitamura K, Muto S, Kotaki H, et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, Hegde RS. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure- function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nat Struct Mol Biol. 2013;20:447–453. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodon J, Tawbi HA, Thomas AL, Stoller RG, Turtschi CP, Baselga J, Sarantopoulos J, Mahalingam D, Shou Y, Moles MA, et al. A phase I, multicenter, open-label, first- in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin Cancer Res. 2014;20:1900–1909. doi: 10.1158/1078-0432.CCR-13-1710. [DOI] [PubMed] [Google Scholar]
- Snuderl M, Batista A, Kirkpatrick ND, Ruiz de Almodovar C, Riedemann L, Walsh EC, Anolik R, Huang Y, Martin JD, Kamoun W, et al. Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell. 2013;152:1065–1076. doi: 10.1016/j.cell.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cell Mol Life Sci. 2013;70:1897–1913. doi: 10.1007/s00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Traiffort E, Angot E, Ruat M. Sonic Hedgehog signaling in the mammalian brain. J Neurochem. 2010;113:576–590. doi: 10.1111/j.1471-4159.2010.06642.x. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Bjorklund M, Cheng F, Syvanen H, Kivioja T, Kilpinen S, Sun Z, Kallioniemi O, Stunnenberg HG, He WW, et al. Application of active and kinase- deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Witt RM, Hecht ML, Pazyra-Murphy MF, Cohen SM, Noti C, van Kuppevelt TH, Fuller M, Chan JA, Hopwood JJ, Seeberger PH, et al. Heparan Sulfate Proteoglycans Containing a Glypican 5 Core and 2-O-Sulfo-iduronic Acid Function as Sonic Hedgehog Co-receptors to Promote Proliferation. J Biol Chem. 2013;288:26275–26288. doi: 10.1074/jbc.M112.438937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Li Z, Cai S, Tian L, Chen K, Wang J, Hu J, Sun Y, Li X, Ertel A, et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013;73:4488–4499. doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Porcionatto M, Pilapil M, Chen Y, Choi Y, Tolias KF, Bikoff JB, Hong EJ, Greenberg ME, Segal RA. Polarized signaling endosomes coordinate BDNF- induced chemotaxis of cerebellar precursors. Neuron. 2007;55:53–68. doi: 10.1016/j.neuron.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev Biol. 2006;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.