Abstract
We have reported previously that p115Rho guanine nucleotide exchange factor, its upstream activator Gα13, and its effector RhoA are able to inhibit HIV-1 replication. Here, we show that RhoA is able to inhibit HIV-1 gene expression through the NFAT-binding site in the HIV long-terminal repeat. Constitutively active NFAT counteracts the inhibitory activity of RhoA, and inhibition of NFAT activation also inhibits HIV-1 gene expression. We have shown further that RhoA inhibits NFAT-dependent transcription and IL-2 production in human T cells. RhoA does not inhibit nuclear localization of NFAT but rather, inhibits its transcriptional activity. In addition, RhoA decreases the level of acetylated histone H3, but not NFAT occupancy, at the IL-2 promoter. These data suggest that activation of RhoA can modulate IL-2 gene expression by inhibiting the transcriptional activity of NFAT and chromatin structure at the IL-2 promoter during T cell activation.
Keywords: GTPase, HIV-1, IL-2
INTRODUCTION
The HIV-1 virus targets the CD4+ T cell, subverting biological processes in the cell to further its own reproduction. Replication of HIV requires many of the same transcription factors used by T cells during activation, including NF-κB and NFAT [1–7]. We have reported previously that p115Rho guanine nucleotide exchange factor (GEF) interacts directly with the C terminus of HIV gp41 [8]. Furthermore, expression of p115RhoGEF, its upstream activator Gα13, or its downstream effector RhoA causes a decrease in HIV replication [9]. Given the close relationship between T cell activation and HIV replication, we were interested in looking at how RhoA inhibits HIV replication and whether it also affects T cell activation.
RhoA is a small GTPase with a well-characterized role in cytoskeletal rearrangement. Upon activation, RhoA causes the formation of stress fibers in cells [10]. In lymphocytes, this function of RhoA is critical for the proper homing of cells to areas of infection [11]. Evidence has shown that RhoA is required for the process of leukocyte rolling and diapedesis or migration beneath the endothelial layer of cells [12]. In addition, RhoA is involved in the assembly of the immunological synapse, an area on the cell surface where elements of the immune signaling apparatus congregate, allowing for proper activation of the T cell [13].
In some cells, RhoA has been shown to play a more direct role in signal transduction. Welsh et al. [14] demonstrated a dependence on RhoA for ERK activity during the G1 phase of the cell cycle. Without ERK activation, there is no cyclin D induction, leading to a block in cell cycle progression [14]. The specific role of RhoA in signaling pathways turned on during T cell activation has not been examined thoroughly, although reports have shown that the GTPase is able to activate the NFAT-binding partner AP-1 [15] as well as NF-κB [16 –18]. In addition, RhoA and its upstream activator Gα13 have been shown to increase the activity of phospholipase Cε (PLCε), leading to increased levels of intracellular calcium [19].
The NFAT family of proteins was first discovered by identification of factors involved in the up-regulation of IL-2 in response to TCR stimulation [20]. Since that time, NFAT proteins have been implicated in a wide variety of cellular processes including cardiac hypertrophy, learning and memory, and adipocyte differentiation [21, 22]. Immunologically relevant genes regulated by NFAT include IL-2, IL-4, IL-5, GM-CSF, TNF-α, CD40 ligand (CD40L), and FasL. NFAT family proteins are regulated primarily through calcium levels in the cell. Upon stimulation, an increase in intracellular calcium turns on the serine/threonine phosphatase calcineurin, which then binds to NFAT and dephosphorylates the protein, allowing NFAT nuclear translocation. The immunosuppressive drugs FK506 and Cyclosporin A work through inhibition of calcineurin—preventing NFAT-dependent transcription [23]. Studies have shown that p38MAPK, ERK, and JNK can potentially phosphorylate NFAT, inhibiting its translocation [24].
Other pathways have also been implicated in NFAT regulation at the level of nuclear translocation and DNA binding. Once the protein enters the nucleus, kinases, including glycogen synthase kinase 3, phosphorylate NFAT, preventing DNA binding and leading to nuclear export [25]. Furthermore, NFAT proteins typically have a binding partner, which stabilizes their interaction with the DNA. Although several transcription factors can act in this capacity, the predominant protein is AP-1 [26], a heterodimer comprised of c-jun and Fos [27], which are activated by MAPK pathways that can also be initiated upon TCR ligation. Fos is transcriptionally up-regulated through ERK, and JNK phosphorylates jun leading to its activation and nuclear translocation [27, 28]. Some studies have shown that NFAT plays a role in anergy induction as well as activation in T cells. Stimulation of T cells with ionomycin alone activates NFAT in the absence of factors including AP-1 and leads to the up-regulation of “anergic factors,” which block activation instead of the up-regulation of IL-2 production and proliferation [29, 30]. In addition, recent reports have shown that alternate family members can replace Fos and jun in the formation of a heterodimer, which is still able to bind DNA cooperatively with NFAT but give the complex an inhibitory function [27, 31–33].
Here, we demonstrate that activated RhoA is able to inhibit NFAT-dependent transcription from the HIV long-terminal repeat (LTR) and the IL-2 promoter. RhoA did not affect NFAT nuclear localization in response to Ca++-mediated activation. The RhoA-GTPase did, however, inhibit the transactivation potential of NFAT. In stimulated T cells, expression of activated RhoA also led to a decrease in the level of acetylated histone H3 at the IL-2 promoter but not the occupancy of NFAT at the IL-2 promoter. RhoA activation may, therefore, affect chromatin remodeling at the IL-2 promoter and the ability of NFAT to transactivate DNA.
MATERIALS AND METHODS
Luciferase assays
Jurkat T cells were transfected according to the manufacturer’s protocol using Geneporter transfection reagent from Gene Therapy Systems (San Diego, CA, USA). Approximately 1 ug DNA was transfected into 2 × 105 cells in 30 ul RPMI with 6 ul Geneporter and 130 ul unsupplemented RPMI. After 4 h, 3.6 ul Transfection Booster #1 (GTS Genlantis, San Diego, CA, USA) along with 162.4 ul complete RPMI was added to the cells. When a GFP control construct was added to DNA mixtures to monitor transfection efficiency, 20–25% of cells were transfected using this method. After 2 days, T cells were activated with 2 ug/ml PHA and 1 uM ionomycin for 5 h. T cells were then lysed in 0.5% Nonidet P-40 lysis buffer, and luciferase activity was measured using the AutoLumat luminometer from Perkin Elmer (Wellesley, MA, USA). HIV LTR luciferase deletion mutants [34] were the gift of Drs. Jianming Li and Xiao-Fan Wang (Duke University, Durham, NC, USA). The HIV LTR luciferase point mutants used were as described in ref. [1], and the NFAT-luciferase construct [35] was the gift of Dr. Gerald Crabtree (Stanford University, Stanford, CA, USA).
Transactivation assay
For the transactivation assays, 5× GAL4-luciferase was transfected along with NFAT/GAL4 or NF-κB/GAL4 (gifts of Dr. Mohamed Oukka, Harvard Medical School, Boston, MA, USA; see ref. [36]) and RhoA63L or vector as described above. Unstimulated Jurkat T cells were cultured for 2 days and then lysed using 5× lysis buffer from the Promega luciferase assay system. Luciferase activity was measured using the Fluostar Optima luminescent plate reader from BMG Labtech (Germany).
Retroviral transduction
Retroviruses were made in A293T cells by transfection using calcium phosphate as described previously [37]. Retrovirus was added to Jurkat T cells or human PBMCs in the presence of 8 ug/ml polybrene. Cells sat at room temperature for 30 min followed by 3 h of centrifugation at room temperature at 2000 g. After 2 days in culture, cells were monitored for GFP expression by FACScan analysis.
Intracellular cytokine staining
Transduced Jurkat T cells were activated with 2 ug/ml PHA and 1 uM ionomycin for 5 h. After the 1st h, 10 ug/ml Brefeldin-A was added to the cultures. After 5 h, cells were permeabilized in Becton Dickson Permeabilization 2 buffer and stained for 30 min with a monoclonal PE-conjugated anti-IL-2 (Caltag Laboratories, Burlingame, CA, USA). Cells were gated for GFP and analyzed by FACS. Transduced, primary, human PBMCs were stimulated with anti-CD3 and anti-CD28 for 24 h. During the last 4 h, Brefeldin-A was added to cultures, and cells were stained as described above.
Immunofluorescence staining
U20S cells (1×105) were plated on coverslips in 12-well plates and allowed to grow overnight. The following day, the cells were transfected with flag-tagged, wild-type NFAT [24], along with pCDNA3 or activated RhoA and serumstarved overnight. Cells were activated for 30 min with 5 mM CaCl2 and 1 uM ionomycin and then fixed with 4% paraformaldehyde for 15 min at room temperature, rinsed with PBS, permeabilized with 0.2% Trition for 5 min at 4°C, and rinsed again three times with PBS. Cells were blocked in PBS, 0.5% BSA, at room temperature and then stained in PBS, 0.5% BSA, with a mouse antiflag antibody (Sigma Chemical Co., St. Louis, MO, USA) for 1 h, followed by three washes in PBS and secondary staining with rhodamine-conjugated anti-mouse antibody for 1 h at room temperature. Cells were washed five times for 5 min in PBS. In the final wash, 1 ul 4′,6-diamidino-2-phenylindole was added to 1 ml PBS for nuclear staining. Coverslips were mounted on slides and allowed to dry overnight in the dark at room temperature. Several hundred NFAT+ cells were then scored blindly for nuclear NFAT.
Chromatin immunoprecipitation (ChIP)
For ChIP analysis, the ChIP assay kit from Upstate Cell Signaling Solutions (Lake Placid, NY, USA) was used according to protocol. Briefly, 1 × 107-transduced Jurkat T cells for each immunoprecipitation condition were left unstimulated or stimulated with 2 ug PHA and 1 uM ionomycin for 40 min followed by a 20-min incubation with 1% formaldehyde at 37°C. After incubation with formaldehyde, glycine was added to the cultures to a final concentration of 125 mM. Cells were then washed twice with cold 1× PBS plus PMSF and protease inhibitors. Cells were lysed in 300 ul SDS lysis buffer and incubated with shaking for 10 min at 4°C followed by sonication for a single 1-min burst and eight rounds of 35 s (1-s pulse, 0.8-s rest). This process produced fragments ranging between 500 and 2000 bp. Sonicates were then precleared with 60 ul salmon sperm DNA/protein A agarose slurry for 30 min with rotation at 4°C. After clearing, the lysates were divided evenly for overnight incubation with no antibody, with 10 ug α-acetylated H3 (K9 and K14) antibody (Upstate Cell Signaling Solutions) or with 25 ug α-NFATc1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). On the following day, the antibody/protein complexes were immunoprecipated with 60 ul salmon sperm/ protein A agarose slurry for 2 h, followed by one wash each at 4°C with low salt, high salt, and LiCl wash buffers and two washes with 1× tris-EDTA buffer. Complexes were eluted in elution buffer (1% SDS, 0.1 M NaHCO3) during two washes for 15 min at room temperature. NaCl (20 ul 5 M) was added to 500 ul eluate and left overnight at 65°C to reverse the protein DNA cross-linking. The following day, 10 ul 0.5 M EDTA, 20 ul Tris-HCl (pH 6.5), and 2 ul 10 mg/ml proteinase K were added and incubated for 1 h at 45°C. DNA was then recovered by phenol/chloroform extraction and resuspended in 30 ul water.
Real-time PCR
Real-time PCR was used to analyze DNA pulled down by ChIP. We designed primers for the IL-2 promoter (forward: CACCTAAGTGTGGGCTAATGTAACA; reverse: CTGATGACTCTTTGGAATTCTTTAAACC), along with the TAM/FAM-labeled probe (AGAGGGATTTCACCTACATCCATTCAGTCAGTC). DNA from final ChIP preparations was diluted 1:2, and 5 ul of this dilution was used per reaction. A standard curve was produced using a dilution series from genomic DNA from the same cell type used for ChIP. Samples were run in duplicate. The average of duplicate samples was normalized by dividing the average of the quantitation from the input DNA of the same cell type and condition.
RESULTS
RhoA specifically inhibits transcription from the HIV LTR through the NFAT-binding site
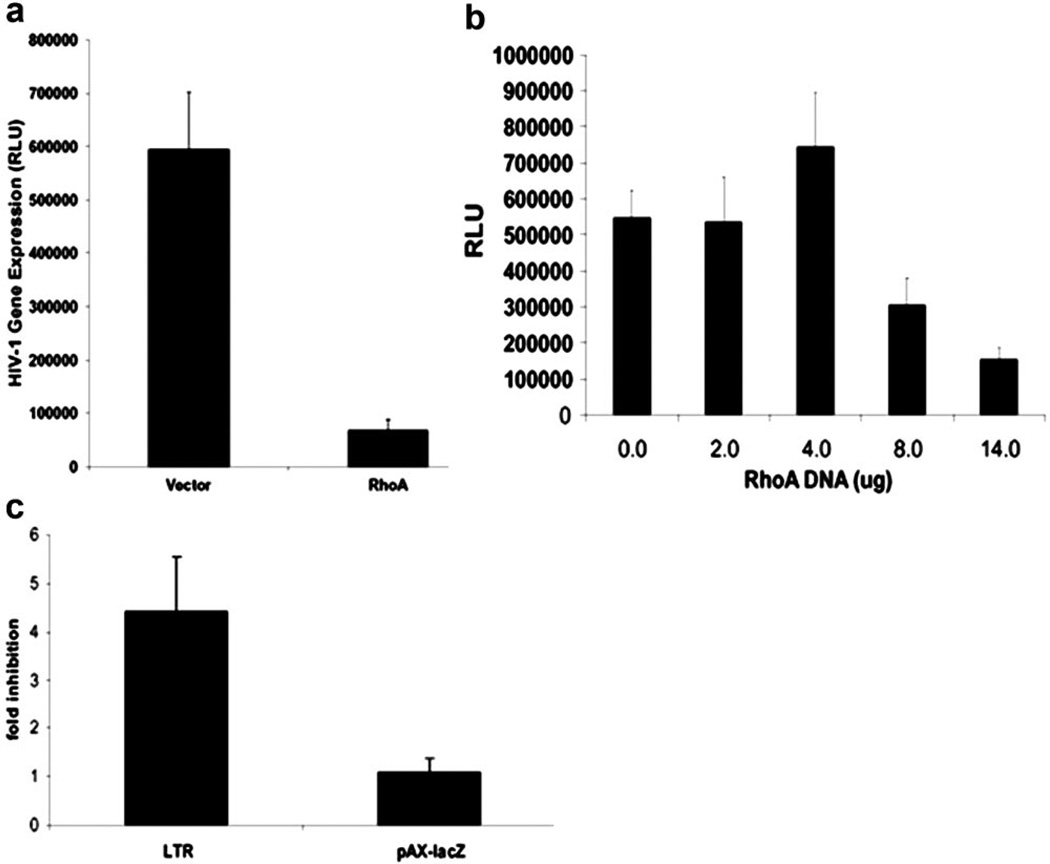
We reported previously that RhoA, its upstream activator p115RhoGEF (p115), and Gα13, a heterotrimeric G protein that activates p115, are able to inhibit HIV replication [9]. Similarly, ectopic expression of constitutively active RhoA (RhoA-63L) in Jurkat T cells inhibited HIV-1 gene expression upon coinfection with a vesicular stomatitis virus (VSV)-G-pseudotyped HIV-luciferase reporter virus (Fig. 1a). To further analyze how this pathway is able to inhibit HIV-1 infection, we transfected Jurkat T cells with the HIV-1 LTR, driving a luciferase reporter gene along with RhoA63L. We found that expression of activated RhoA leads to decreased LTR-luciferase expression in a dose-dependent manner (Fig. 1b). To show the specific inhibition of HIV LTR, we transfected pAX-lacZ, a promoter shown previously to be unaffected by Dbl family proteins including p115 [38, 39], into Jurkat cells, along with the HIV LTR, in the presence or absence of constitutively active RhoA. We showed that RhoA is able to inhibit transcription specifically from the HIV LTR but not from the pAX promoter (Fig. 1c).
Fig. 1.
RhoA specifically inhibits HIV-1 LTR activity. (a) Jurkat T cells were transduced with control or RhoA63L vectors, followed by infection with HIV-luciferase virus pseudotyped with VSV-G. After 2 days, cells were harvested, and luciferase activity was measured. RLU, Relative light units. (b) Jurkat T cells were cotransfected with HIV-1 LTR-luciferase, the HIV Tat protein, and vector (pcDNA3) or the constitutively active RhoA63L. After 2 days, cells were harvested, and luciferase activity was measured. Shown are data representative of at least 10 experiments. (c) Jurkat T cells were transfected with the HIV LTR-luciferase and pAX-lacZ in the presence or absence of RhoA63L. Post-transfection (48 h), cell lysates were measured for luciferase activity as well as lacZ activity. In the presence of RhoA63L, the LTR was inhibited by 4.5-fold, and lacZ expression from the pAX promoter was unaffected. Data from six independent experiments are summarized.
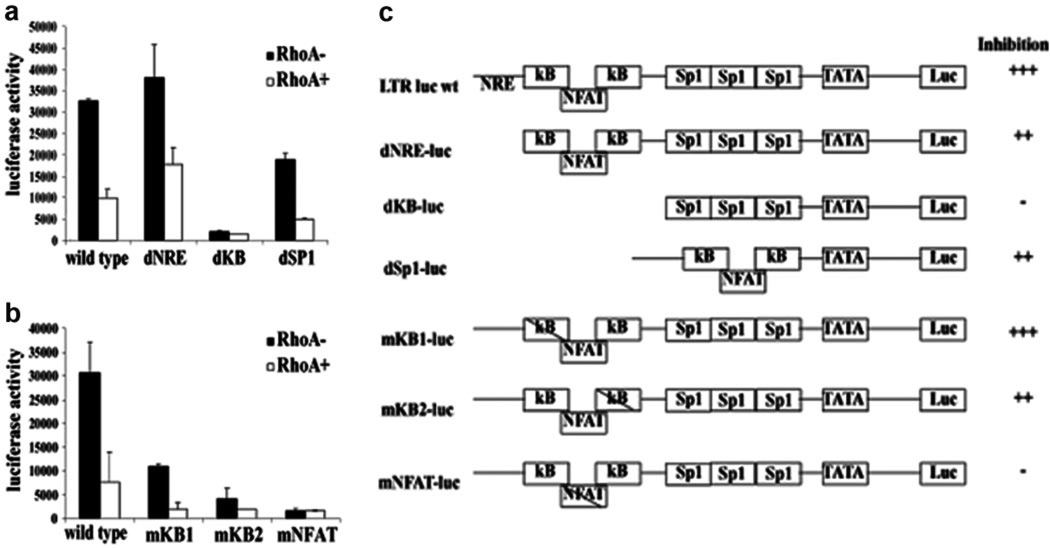
To understand the mechanism of this inhibition, we looked for the involvement of a specific cis-acting element in the HIV LTR. Wild-type or mutant LTR luciferase constructs were used to identify mutations, which become resistant to inhibition by RhoA. Analysis of a series of LTR deletion mutants showed that the core region of the promoter, containing two NF-κB sites and a NFAT-binding site, was important for the ability of RhoA to inhibit transcription (Fig. 2a). To determine which of these sites was important in RhoA inhibition of the LTR, we used a group of LTR point mutants [1]. Although mutations in the NF-κB sites of the LTR led to reduced overall activity from the promoter, only the mNFAT mutant showed consistent lack of inhibition by RhoA (Fig. 2b). A summary of the relative inhibition by RhoA from the deletion and site-specific mutants is presented in Figure 2c. These data indicate that RhoA is able to inhibit the HIV LTR through the NFAT-binding site.
Fig. 2.
RhoA inhibition of HIV LTR is dependent on the NFAT-binding site. The HIV LTR deletion mutants (a) or point mutants of the HIV LTR NF-κB/NFAT enhancer (b) were transfected into Jurkat T cells with vector or RhoA63L, and luciferase activity was measured 48 h post-transfection. (c) Summary of the HIV LTR deletion and point mutants used to define cis-acting elements involved in RhoA-mediated inhibition of the promoter. +++, Strong inhibition (>3×); ++, moderate inhibition (1.5–3×); –, no inhibition (<1.5×). Data shown are representative of four experiments. Error bars represent the standard deviation of duplicate samples in one experiment. NRE, Negative regulatory element; KB, NF-κB; SP1, specificity protein 1; mKB1, point mutation in NF-κB1; luc wt, luciferase wild-type.
RhoA inhibits NFAT-dependent transcription in T cells
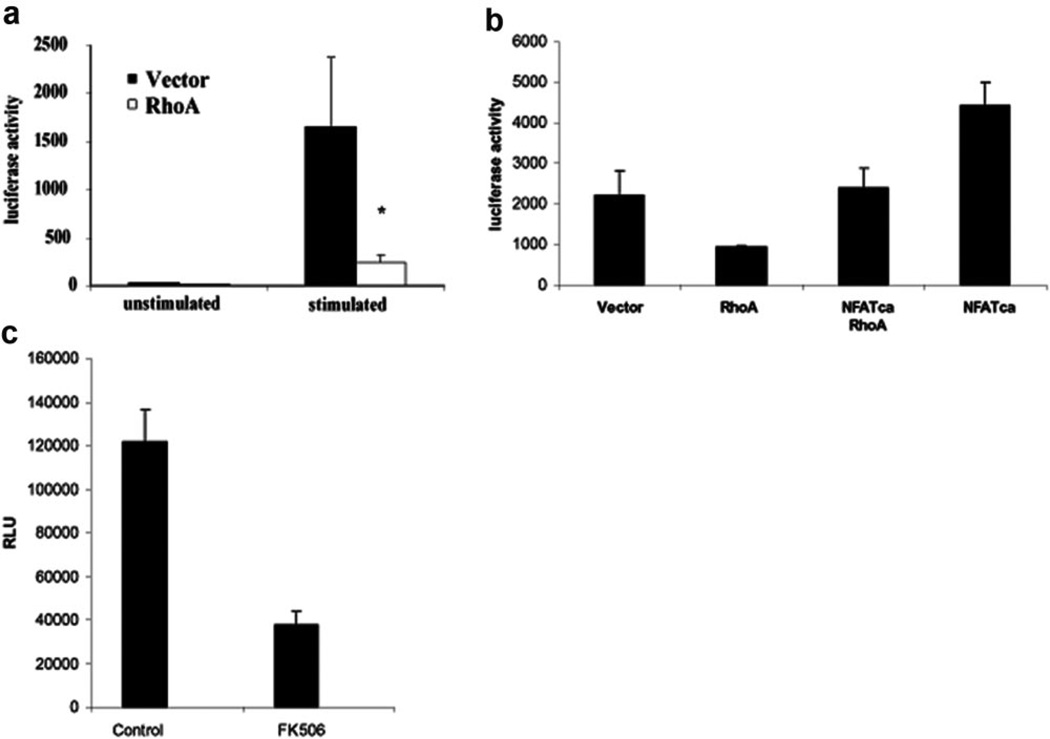
The fact that RhoA is inhibiting HIV transcription through the NFAT-binding site in the LTR implies that RhoA is acting on the NFAT pathway in the T cell. NFAT activity has been reported to enhance HIV-1 gene expression [1], and inhibition of NFAT activation with FK506 reduced HIV-1 gene expression in Jurkat T cells infected with VSV-G-pseudotyped HIV-luciferase reporter virus (Fig. 3a). To further support the finding that RhoA inhibits the LTR in a NFAT-dependent manner, we made use of a NFAT construct with serine-to-alanine mutations at serines 172 and 187 (NFATca). These mutations prevent the phosphorylation of NFAT, causing the nuclear localization of the protein and making it constitutively active [24]. The cotransfection of LTR-luciferase and RhoA63L with the NFATca mutant counteracts inhibition of the HIV promoter by RhoA (Fig. 3b). We proceeded to examine whether RhoA also inhibited other NFAT-dependent promoters. We used a NFAT luciferase construct with three copies of the NFAT-binding site from the IL-2 promoter, driving a luciferase reporter to study this question [35]. In the presence of RhoA, transcription from the 3× NFAT promoter decreased up to fivefold in activated T cells (Fig. 3c), further supporting the conclusion that RhoA inhibits HIV-1 LTR promoter activity by inhibiting NFAT-dependent transcription.
Fig. 3.
RhoA inhibits NFAT to inhibit HIV-1 LTR expression. (a) Jurkat T cells (2×105) were transfected with the 3 NFAT-luc construct in the presence or absence of RhoA. After 2 days, cells were stimulated with PHA and ionomycin for 5 h or left unstimulated. RhoA inhibited luciferase expression significantly (*, P<0.01) from the NFAT promoter in stimulated Jurkat T cells. Shown are representative data for at least six independent experiments. (b) Constitutively active NFAT counteracts RhoA inhibition. Jurkat T cells were transfected with HIV LTR-luciferase construct and the HIV Tat protein in the presence or absence of RhoA63L and/or constitutively active NFAT. The presence of constitutively active NFAT prevents HIV LTR inhibition by RhoA. NFATca,. (c) Jurkat T cells were pretreated with FK506 to inhibit NFAT activation, followed by infection with HIV-luciferase virus pseudotyped with VSV-G and activation with PHA and ionomycin. After 2 days, cells were harvested, and luciferase activity was measured. Data shown are representative of four experiments. Error bars represent the standard deviation of duplicate samples in one experiment. NFATca, constitutively active NFAT.
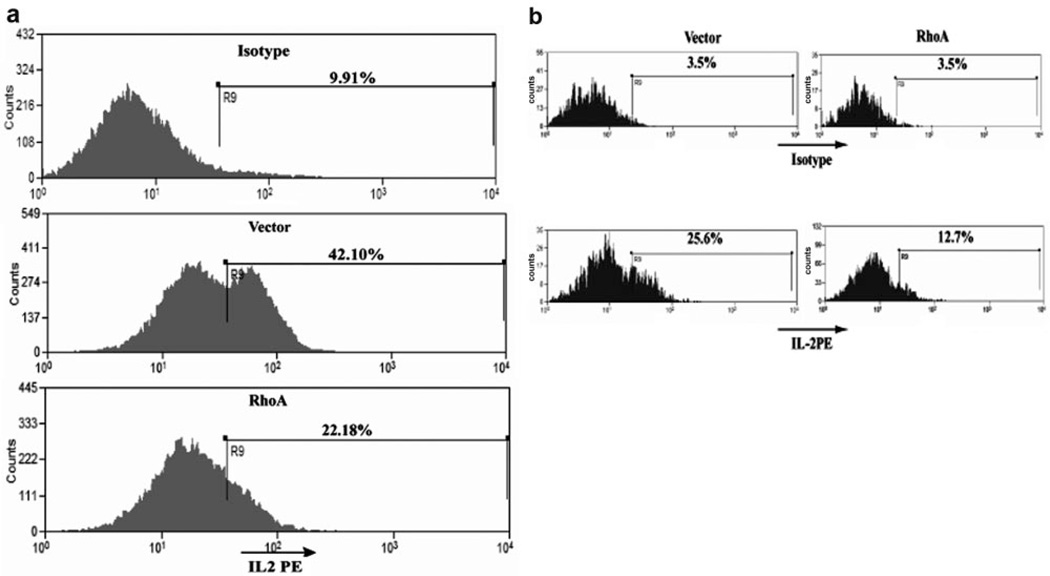
We looked at the ability of RhoA to affect an endogenous indicator of NFAT activity—production of IL-2. We transduced Jurkat T cells with a retrovirus expressing RhoA63L and GFP or with the GFP control vector. Transduction with both viruses resulted in >90% positive cells, monitored by GFP expression (data not shown). We proceeded to activate the transduced Jurkat cells. Five hours after activation, the cells were stained for intracellular IL-2. Production of IL-2 was reduced by twofold (Fig. 4a) in cells expressing RhoA63L. Primary, human PBMCs were also transduced with control or RhoA63L retrovirus vectors. IL-2 production in the GFP+ populations of these primary cells was then assessed by intracellular staining. In human primary cells expressing RhoA63L, IL-2 was again reduced by approximately twofold (Fig. 2b). Together, these results demonstrate that activation of RhoA signaling leads to inhibition of NFAT activity and IL-2 gene expression.
Fig. 4.
RhoA inhibits IL-2 expression during T cell activation. (a) Jurkat T cells transduced with vector or with RhoA63L were stimulated with PHA and ionomycin for 5 h. Cells were then stained for intracellular IL-2. The presence of active RhoA decreased IL-2 production by ~50% (n=2). (b) Primary human PBMCs were transduced retrovirally with control vector or constitutively active RhoA. After activation, intracellular IL-2 was measured. The percentage of IL-2-positive cells is shown. Data shown are representative of two independent experiments.
RhoA inhibits the transactivation activity of NFAT but not NFAT nuclear translocation
To explain the effect of RhoA on NFAT-dependent transcription and IL-2 production, we examined whether RhoA was affecting the nuclear translocation of the NFAT protein. After activation, cells transfected with RhoA or vector showed efficient NFAT nuclear localization (Supplemental Fig. 1), demonstrating that activated RhoA does not inhibit nuclear translocation of NFAT during Ca2+-mediated activation. Nuclear extracts were also prepared from untransduced or RhoA63L-transduced Jurkat T cells, and EMSA was performed using the NFAT-binding site from the IL-2 promoter. NFAT showed no decrease in binding to the probe in the presence of activated RhoA, with or without activation of the cells (Supplemental Fig. 1).
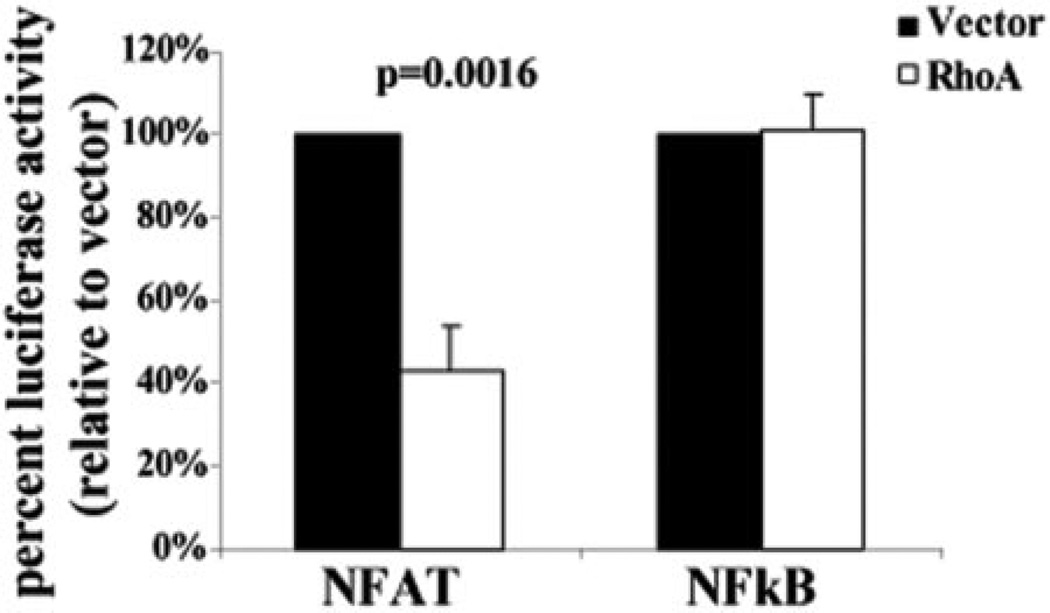
An alternative explanation for the decrease in NFAT-dependent transcription in the presence of constitutively active RhoA could be that it is a result of a change in the ability of NFAT to transactivate gene expression. To determine whether RhoA was affecting this capacity, we transfected Jurkat T cells with a 5× GAL4-luciferase construct, along with GAL4/NFAT or GAL4/NF-κB fusion constructs in the presence or absence of RhoA [36]. We found that there was an approximate twofold decrease in transcription from the 5× GAL4-luciferase promoter driven by GAL4/NFAT in the presence of RhoA in Jurkat T cells (Fig. 5). In contrast, active RhoA showed no effect of the GTPase on the transactivation activity of NF-κB fused with the GAL4 DNA-binding domain. These data suggest that RhoA inhibition of NFAT-dependent transcription is caused by an effect on the transactivation activity of NFAT.
Fig. 5.
RhoA decreases the transactivation activity of NFAT. Jurkat T cells were transfected with 5× GAL4-luciferase and plasmids encoding the GAL4-NFAT or GAL4-NF-κB fusion proteins in the presence or absence of RhoA63L. After 48 h, the cells were harvested, and luciferase activity was measured to determine transcriptional activation. Data are shown as percent activity relative to vector from four (GAL4-NFAT) or three (GAL4-NF-κB) independent experiments.
RhoA decreases levels of acetylated histone 3 at the IL-2 promoter
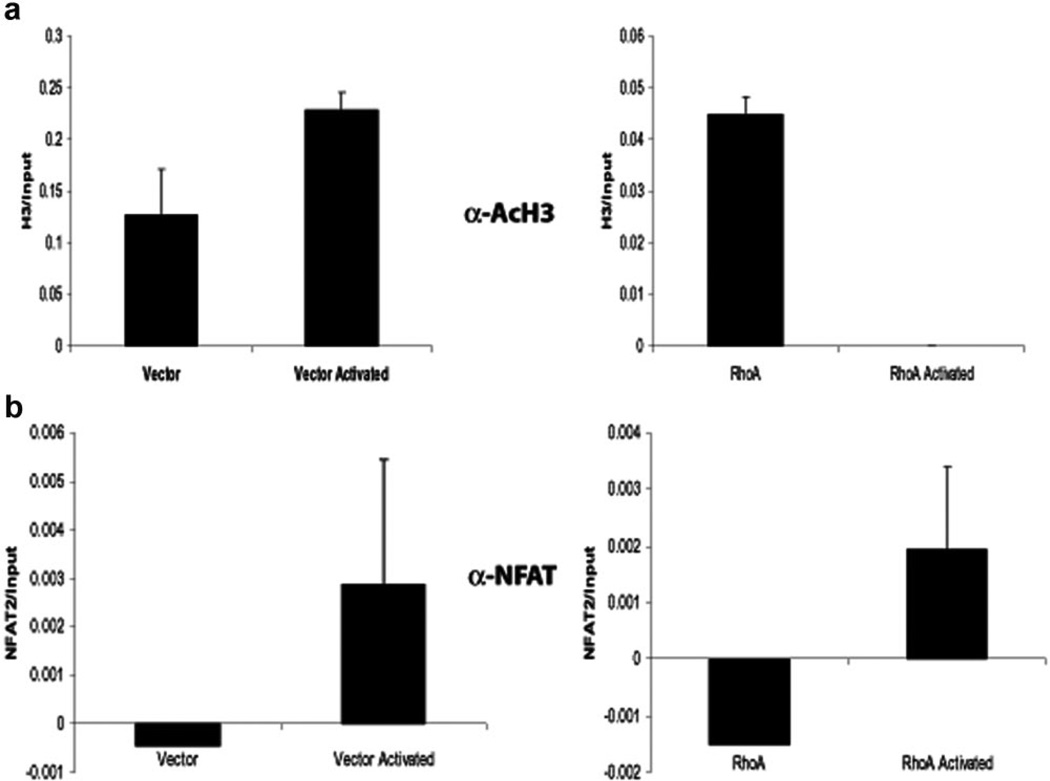
Recent work has shown that NFAT can play an important role in the production and maintenance of anergy in T cells [40]. In addition, studies have shown that NFAT is able to influence the process of chromatin remodeling, allowing for access of transcription factors to the promoter region of genes regulated by NFAT [41–43]. We therefore questioned whether expression of activated RhoA in T cells caused changes in the chromatin structure of genes involved in T cell activation. To investigate this question, we analyzed the IL-2 promoter using ChIP. Jurkat cells transduced with constitutively active RhoA or a control vector were activated for 40 min with PHA and ionomycin or left untreated, then fixed, lysed, and sonicated. The sonicated lysates were immunoprecipitated with an antibody against NFAT or with an antiacetylated histone 3 antibody as an indication of active chromatin. Immunoprecipitated DNA was analyzed by real-time PCR for the IL-2 promoter. Consistent with the NFAT nuclear localization and the EMSA-based DNA-binding assays, ChIP, with α-NFATc1, showed that there was increased binding of the protein to the IL-2 promoter upon cellular activation in the presence or absence of RhoA (Fig. 6b). When α-AcH3 ChIP was similarly analyzed, RhoA-expressing cells showed an approximately threefold decrease compared with vector-expressing cells in the level of AcH3 at the IL-2 promoter in the unstimulated condition (Fig. 6a). We also found that although the level of AcH3 at the IL-2 promoter increased with stimulation in vector-transduced cells, the amount of active chromatin in RhoA-transduced cells decreased under the same conditions (Fig. 6a). The acetylation of H3 at the actin promoter was not affected by RhoA (data not shown). These findings suggest that activated RhoA causes a change in the IL-2 promoter at the level of chromatin organization.
Fig. 6.
RhoA decreases the level of histone 3 acetylation at the IL-2 promoter in T cells. Jurkat T cells transduced with vector or RhoA63L were left unstimulated or activated with PHA and ionomycin. ChIP, using an antiacetylated H3 antibody (a) or an anti-NFATc1 antibody (b), was performed. The amount of ChIP DNA was measured by real-time, quantitative PCR of the IL-2 promoter. Input chromatin DNA (10% of total) was also measured for each sample. The relative NFAT binding is determined by the amount of anti-NFATc1 ChIP/input DNA. Data shown are representative of five independent experiments.
DISCUSSION
We have shown that RhoA decreases HIV replication by decreasing LTR-driven gene expression and that the inhibition of the HIV LTR is dependent on its NFAT-binding site. In addition, we have shown that RhoA is able to inhibit NFAT-dependent transcription as well as IL-2 production. We have gone on to demonstrate that RhoA did not inhibit NFAT nuclear translocation or NFAT DNA binding. Expression of activated RhoA did, however, decrease the transactivation activity of NFAT in T cells. Finally, we have shown by ChIP that in the presence of constitutively active RhoA, the chromatin at the IL-2 promoter is less acetylated upon activation of the T cell compared with the chromatin of vector control cells, indicating a decrease in promoter activity.
The dependence of HIV on T cell activation to further its own replication has made the virus a useful tool for furthering our understanding of factors important during this process. Earlier work from this lab showed that the Gα13, p115RhoGEF, and RhoA pathway is able to inhibit HIV replication [8, 9]. It is interesting that mice lacking G2A, a G-protein-coupled receptor (GPCR), which has been shown to activate the Gα13 pathway, develop a late onset, autoimmune disorder, implying an important role for the Gα13 signaling pathway, including RhoA, in the regulation of T cell activation [44, 45]. Although the underlying mechanism behind the development of autoimmunity in these mice is not fully understood, we show here that activation of the downstream effector of Gα13, RhoA, can lead to inhibition of transcription from the HIV LTR and the IL-2 promoter. These findings suggest that extracellular signaling through specific GPCRs may be able to modulate not only T cell activation but also HIV transcription, thus providing a potential target pathway for helping to control the virus.
T cell anergy is a state of unresponsiveness to TCR stimulation characterized in part by lack of IL-2 production [46], a phenotype similar to that seen here in the RhoA63L-expressing T cells. Recent work has indicated that NFAT plays an important role, not only in promoting T cell activation but also in preventing it. NFAT1 has been identified as a potential anergy factor, shown to be important in up-regulation of a panel of genes associated with anergy, and T cells lacking NFAT1 are resistant to anergy induction [29]. NFAT functions as a transcriptional regulator of anergy when AP-1 is not present to serve as a binding partner—in a situation where the T cell is not fully activated. In addition, work by Heissmeyer et al. [40] shows that pretreatment of cells with ionomycin leads to cyclosporine A-sensitive anergy induction. Here, we have demonstrated a reduction in T cell responsiveness in the presence of constitutively active RhoA. It is interesting that although constitutively active RhoA did not inhibit the nuclear translocation of NFAT upon Ca2+ stimulation of U2OS cells, we did observe that RhoA63L expression in these cells led to an increase in nuclear translocation of NFAT in the absence of stimulation (Supplemental Fig. 1). It is unlikely that RhoA activates NFAT family proteins directly to cause translocation. One possible candidate effector in this process is PLCε. Wing et al. [19] showed recently that RhoA was able to activate PLCε directly through a unique, ~65 amino acid insert in the protein’s Y box [19]. PLC isozymes are responsible for cleaving phosphatidylinositol 4,5, bisphosphate to produce 1,4,5-triphosphate (IP3) and diacylglycerol [47]. IP3 causes the release of intracellular calcium stores, which can then lead to the activation of calcineurin [48] and subsequently, NFAT. These findings, in combination with our observation, may provide a potential mechanism of RhoA-mediated inhibition of T cell activation. The possibility exists that early translocation of NFAT puts the protein in contact with different binding partners in the nucleus, which result in NFAT acting as a repressor of IL-2 transcription rather than as an activator. This is supported by the data presented in Figure 5, showing that RhoA decreases the ability of NFAT to transactivate DNA. RhoA may thus be able to affect which cofactors are available for NFAT-dependent transcription on several different levels, through a direct effect on NFAT-binding partners and through an indirect effect on the timing of NFAT translocation, perhaps preventing the recruitment of a transcriptional activation complex. In support of this possibility, Bodor and colleagues [49, 50] have described the interaction of NFAT with the inducible cAMP early repressor, which lacks a transactivation domain, leading its interaction with NFAT to result in the prevention of the recruitment of the CBP/p300 histone acetylase complex to the promoter and thus, to inhibition of transcription. Further analysis of which cofactors bind with NFAT at the IL-2 promoter in the presence of constitutively active RhoA may shed light on how NFAT is able to induce T cell anergy as well as activation.
RhoA is a downstream effector of many different pathways, and under some conditions, activation of the GTPase could actually lead to enhancement of IL-2 transcription. For example, RhoA has well-documented roles in the activation of NF-κB and AP-1 [15–18]. It is interesting that Jurkat T cells cotransfected with constitutively active NFAT and RhoA63L actually showed consistently higher levels of activity from the NFAT-luciferase construct than cells transfected with constitutively active NFAT alone (data not shown). Again, an effect of activated RhoA on NFAT-binding partners including AP-1 may explain the enhancement when constitutively active NFAT and RhoA63L are expressed.
Finally, although it has been shown that RhoA can cause inhibition of HIV, a mechanism for this inhibition has not been described. Here, we have shown that activation of RhoA can lead to a decrease in the ability of NFAT to activate transcription, but the overall influence of RhoA on NFAT-dependent transcription may be the result of a combination of effects. In this study, we have used the constitutively active mutant of RhoA, RhoA63L, to study the effects of the GTPase on T cell activation. Previous work from this lab, however, has shown that expression of a mutant of p115RhoGEF, which lacks GEF activity and therefore, cannot activate RhoA but is still able to inhibit Gα12 and Gα13, leads to enhancement of HIV replication and NFAT-dependent transcription (ref. [9] and data not shown). These observations suggest that the Gα13-p115RhoGEF-RhoA pathway plays a role specifically in down-modulating T cell activation and HIV replication. Further examination of ligands and GPCRs, which use Gα13, may therefore lead to novel methods of modulating T cell activation with potential uses in controlling not only HIV but also autoimmune disorders.
Supplementary Material
ACKNOWLEDGMENTS
The project was supported partially by grants from National Institutes of Health (5T32AI07273 to W. S. H. and AI5380402 and AI04840704 to L. S.). We acknowledge Dr. C. Der for the generous gift of the constitutively active RhoA construct, Dr. G. Crabtree for the NFAT-luciferase construct, Drs. J. Li and X. Wang for the LTR deletion mutants, and Dr. M. Oukka for kindly sending us the 5× GAL4-luciferase and GAL4-NFAT constructs. We thank H. Zhang and Dr. W. O’Connor for their contributions during the initial stages of the project. We also thank members of the Su lab for their input and assistance during this project as well as Dr. V. Anest for helpful discussions about the ChIP and EMSA experiments. Finally, we are grateful to Drs. J. Ting, R. Tisch, G. Matsushima, and D. Siderovski for critical reading and discussion of the manuscript.
REFERENCES
- 1.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 2.Kinoshita S, Chen BK, Kaneshima H, Nolan GP. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 3.Nabel GJ, Rice SA, Knipe DM, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Genin A, Cron RQ. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology. 2004;321:323–331. doi: 10.1016/j.virol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Pessler F, Cron RQ. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun. 2004;5:158–167. doi: 10.1038/sj.gene.6364047. [DOI] [PubMed] [Google Scholar]
- 6.Cron RQ. HIV-1, NFAT, and cyclosporin: immunosuppression for the immunosuppressed? DNA Cell Biol. 2001;20:761–767. doi: 10.1089/104454901753438570. [DOI] [PubMed] [Google Scholar]
- 7.Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ, Lewis DB. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin. Immunol. 2000;94:179–191. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Wang L, Kao S, Whitehead IP, Hart MJ, Liu B, Duus K, Burridge K, Der CJ, Su L. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr. Biol. 1999;9:1271–1274. doi: 10.1016/s0960-9822(99)80511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Zhang H, Solski PA, Hart MJ, Der CJ, Su L. Modulation of HIV-1 replication by a novel RhoA effector activity. J. Immunol. 2000;164:5369–5374. doi: 10.4049/jimmunol.164.10.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 11.Worthylake RA, Burridge K. Leukocyte transendothelial migration: orchestrating the underlying molecular machinery. Curr. Opin. Cell Biol. 2001;13:569–577. doi: 10.1016/s0955-0674(00)00253-2. [DOI] [PubMed] [Google Scholar]
- 12.Worthylake RA, Lemoine S, Watson JM, Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J. Cell Biol. 2001;154:147–160. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaguillaumie A, Lagaudriere-Gesbert C, Popoff MR, Conjeaud H. Rho GTPases link cytoskeletal rearrangements and activation processes induced via the tetraspanin CD82 in T lymphocytes. J. Cell Sci. 2002;115:433–443. doi: 10.1242/jcs.115.2.433. [DOI] [PubMed] [Google Scholar]
- 14.Welsh CF, Roovers K, Villanueva J, Liu Y, Schwartz MA, Assoian RK. Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat. Cell Biol. 2001;3:950–957. doi: 10.1038/ncb1101-950. [DOI] [PubMed] [Google Scholar]
- 15.Chang JH, Pratt JC, Sawasdikosol S, Kapeller R, Burakoff SJ. The small GTP-binding protein Rho potentiates AP-1 transcription in T cells. Mol. Cell. Biol. 1998;18:4986–4993. doi: 10.1128/mcb.18.9.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montaner S, Perona R, Saniger L, Lacal JC. Multiple signaling pathways lead to the activation of the nuclear factor κB by the Rho family of GTPases. J. Biol. Chem. 1998;273:12779–12785. doi: 10.1074/jbc.273.21.12779. [DOI] [PubMed] [Google Scholar]
- 17.Pan ZK, Ye RD, Christiansen SC, Jagels MA, Bokoch GM, Zuraw BL. Role of the Rho GTPase in bradykinin-stimulated nuclear factor-κB activation and IL-1 gene expression in cultured human epithelial cells. J. Immunol. 1998;160:3038–3045. [PubMed] [Google Scholar]
- 18.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 19.Wing MR, Snyder JT, Sondek J, Harden TK. Direct activation of phospholipase C-ε by Rho. J. Biol. Chem. 2003;278:41253– 41258. doi: 10.1074/jbc.M306904200. [DOI] [PubMed] [Google Scholar]
- 20.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 21.Graef IA, Chen F, Crabtree GR. NFAT signaling in vertebrate development. Curr. Opin. Genet. Dev. 2001;11:505–512. doi: 10.1016/s0959-437x(00)00225-2. [DOI] [PubMed] [Google Scholar]
- 22.Ichida M, Finkel T. Ras regulates NFAT3 activity in cardiac myocytes. J. Biol. Chem. 2001;276:3524–3530. doi: 10.1074/jbc.M004275200. [DOI] [PubMed] [Google Scholar]
- 23.Clipstone NA, Crabtree GR. Identification of calcineurin as a key signaling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 24.Porter CM, Havens MA, Clipstone NA. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J. Biol. Chem. 2000;275:3543–3551. doi: 10.1074/jbc.275.5.3543. [DOI] [PubMed] [Google Scholar]
- 25.Neal JW, Clipstone NA. Glycogen synthase kinase-3 inhibits the DNA binding activity of NFATc. J. Biol. Chem. 2001;276:3666–3673. doi: 10.1074/jbc.M004888200. [DOI] [PubMed] [Google Scholar]
- 26.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 27.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 28.Rincon M, Flavell RA, Davis RJ. Signal transduction by MAP kinases in T lymphocytes. Oncogene. 2001;20:2490–2497. doi: 10.1038/sj.onc.1204382. [DOI] [PubMed] [Google Scholar]
- 29.Macian F, Garcia-Cozar F, Im SH, Horton HF, Byrne MC, Rao A. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 30.Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, Skrenta H, Hollenhorst M, Fathman CG, Soares L. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity. 2003;18:535–547. doi: 10.1016/s1074-7613(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 31.Bower KE, Zeller RW, Wachsman W, Martinez T, McGuire KL. Correlation of transcriptional repression by p21(SNFT) with changes in DNA.NF-AT complex interactions. J. Biol. Chem. 2002;277:34967–34977. doi: 10.1074/jbc.M205048200. [DOI] [PubMed] [Google Scholar]
- 32.Iacobelli M, Wachsman W, McGuire KL. Repression of IL-2 promoter activity by the novel basic leucine zipper p21SNFT protein. J. Immunol. 2000;165:860–868. doi: 10.4049/jimmunol.165.2.860. [DOI] [PubMed] [Google Scholar]
- 33.Cuevas BD, Uhlik MT, Garrington TP, Johnson GL. MEKK1 regulates the AP-1 dimer repertoire via control of JunB transcription and Fra-2 protein stability. Oncogene. 2005;24:801–809. doi: 10.1038/sj.onc.1208239. [DOI] [PubMed] [Google Scholar]
- 34.Li JM, Shen X, Hu PP, Wang XF. Transforming growth factor β stimulates the human immunodeficiency virus 1 enhancer and requires NF-κB activity. Mol. Cell. Biol. 1998;18:110–121. doi: 10.1128/mcb.18.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Northrop JP, Ullman KS, Crabtree GR. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J. Biol. Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 36.Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-κB to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehead IP, Khosravi-Far R, Kirk H, Trigo-Gonzalez G, Der CJ, Kay R. Expression cloning of lsc, a novel oncogene with structural similarities to the Dbl family of guanine nucleotide exchange factors. J. Biol. Chem. 1996;271:18643–18650. doi: 10.1074/jbc.271.31.18643. [DOI] [PubMed] [Google Scholar]
- 39.Whitehead IP, Abe K, Gorski JL, Der CJ. CDC42 and FGD1 cause distinct signaling and transforming activities. Mol. Cell. Biol. 1998;18:4689–4697. doi: 10.1128/mcb.18.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 41.Johnson BV, Bert AG, Ryan GR, Condina A, Cockerill PN. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol. Cell. Biol. 2004;24:7914–7930. doi: 10.1128/MCB.24.18.7914-7930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avots A, Buttmann M, Chuvpilo S, Escher C, Smola U, Bannister AJ, Rapp UR, Kouzarides T, Serfling E. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity. 1999;10:515–524. doi: 10.1016/s1074-7613(00)80051-5. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Rodriguez C, Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/ CREB-binding protein (CBP) J. Exp. Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le LQ, Kabarowski JH, Weng Z, Satterthwaite AB, Harvill ET, Jensen ER, Miller JF, Witte ON. Mice lacking the orphan G protein-coupled receptor G2A develop a late-onset autoimmune syndrome. Immunity. 2001;14:561–571. doi: 10.1016/s1074-7613(01)00145-5. [DOI] [PubMed] [Google Scholar]
- 45.Offermanns S, Mancino V, Revel JP, Simon MI. Vascular system defects and impaired cell chemokinesis as a result of Gα13 deficiency. Science. 1997;275:533–536. doi: 10.1126/science.275.5299.533. [DOI] [PubMed] [Google Scholar]
- 46.Macian F, Im SH, Garcia-Cozar FJ, Rao A. T-cell anergy. Curr. Opin. Immunol. 2004;16:209–216. doi: 10.1016/j.coi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Berridge MJ. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu. Rev. Biochem. 1987;56:159–193. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- 48.Jayaraman T, Marks AR. Calcineurin is downstream of the inositol 1,4,5-trisphosphate receptor in the apoptotic and cell growth pathways. J. Biol. Chem. 2000;275:6417–6420. doi: 10.1074/jbc.275.9.6417. [DOI] [PubMed] [Google Scholar]
- 49.Bodor J, Habener JF. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J. Biol. Chem. 1998;273:9544–9551. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
- 50.Bodor J, Bodorova J, Gress RE. Suppression of T cell function: a potential role for transcriptional repressor ICER. J. Leukoc. Biol. 2000;67:774–779. doi: 10.1002/jlb.67.6.774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.