Summary
Transcription-coupled repair (TCR) is a phenomenon that exists in a wide variety of organisms from bacteria to humans. This mechanism allows cells to repair the actively transcribed DNA strand much faster than the non-transcribed one. At the sites of bulky DNA damage RNA polymerase stalls, initiating recruitment of the repair machinery. It is a commonly accepted paradigm that bacterial cells utilize a sole coupling factor, called Mfd to initiate TCR. According to that model, Mfd removes transcription complexes stalled at the lesion site and simultaneously recruits repair machinery. However, this model was recently put in doubt by various discrepancies between the proposed universal role of Mfd in the TCR and its biochemical and phenotypical properties. Here we present a second pathway of bacterial TCR recently discovered in our laboratory, which does not involve Mfd but implicates a common repair factor, UvrD, in a central position in the process.
Keywords: DNA repair, transcription coupled repair, UvrD helicase
Introduction
A multitude of DNA damaging agents bombard the genome on a daily basis, causing multiple lesions in the genetic material. To remove damage from DNA, cells rely upon different repair pathways to maintain genomic integrity and stability. One of the major pathways in bacterial cells is nucleotide excision repair (NER), which is able to locate and remove various bulky DNA lesions such as cyclo-butane dimers, inter- and intra-strand cross-links, and a variety of massive nucleo-base modifications [1-3]. NER starts with a UvrA2B2 hetero-dimer scanning DNA for significant distortions in DNA helical structure that serve as hallmarks for DNA damage. If the distortion is indeed caused by some bulky lesion, UvrB locally melts underwound DNA by inserting a hairpin inside the helix [4, 5]. Next, the endonuclease UvrC is recruited to the repair site and makes two incisions: 8 nucleotides upstream and 4-5 nucleotides downstream from the lesion site on the damaged DNA strand. To finish the repair, UvrD helicase removes UvrC and the damage-containing DNA oligo, and DNA re-synthesis by DNA polymerase I ensues. Finally the phosphoester bonds to the neighbouring nucleotides are re-made by DNA ligase.
DNA repair is not a trivial task and must overcome two major challenges. First, DNA repair enzymes have to locate damage sites among millions of non-damaged nucleotides. In the case of NER this task is complicated by the fact that UvrABCD enzymes have to deal with a variety of non-similar lesions relying only on DNA distortion as their signal. Second, repair enzymes need to have unobstructed access to the DNA to carry out their work. This is complicated by the fact that DNA is often bound to proteins, or contains superstructures that prevent access by repair machinery. With this as an evolutionary pressure, living organisms have developed a sub-pathway of NER called transcription coupled DNA repair (TCR) [6]. The origin of TCR can be traced to the discovery that the actively transcribed DNA strand is repaired much faster than the non-transcribed one, even inside of the same gene [7]. During TCR, RNA polymerase (RNAP) serves as a surveillance vehicle that scans DNA for damage. Bulky DNA lesions stall transcription complexes if they are located on the template strand, but are mostly bypassed if they reside in the non-template strand [8]. Stalled RNA polymerase then acts as a signal for transcription repair coupling Factor (TRCF) to remove RNAP together with nascent RNA [9, 10]. TCRF then recruits UvrABCD machinery to the damaged region [11] and the repair continues via the common NER pathway. As a result, actively transcribing RNAPs not only improve the chances of locating lesions but also assist in removing obstacles from DNA, hence preparing lesion sites for repair. Moreover, speedy repair of the transcribed sequences helps cells to avoid potentially disastrous disruption of essential gene expression. Until recently the only identified TRCF in bacterial cells was Mfd protein. This protein is able to remove stalled RNAP until it is either dislodged from DNA or resumes RNA elongation [12]. About 20 years ago, Sancar and co-workers, in their elegant work showed that apparently Mfd is the only coupling factor in E. coli cells because Mfd deletion is sufficient to render TCR pathway completely inactive [9]. However, since then, many questions as to whether Mfd is the only source of transcription coupling to the repair machinery have arisen, placing this paradigm under close scrutiny.
Mfd: More questions than answers
Mfd is a single-subunit protein belonging to super-family 2 of nucleic acid translocases. In E. coli it is a 130kDa protein containing eight structural domains with canonical ATPase and RecA motifs common for helicases and translocases [13]. In addition to its translocase domain, Mfd harbors several additional elements serving various functions during TCR. The RNAP-interaction domain (RID, domain 4) plays a central role in targeting stalled elongation complexes (ECs) for Mfd binding. RID makes specific contacts with the β-subunit of RNAP near the upstream portion of the EC and allows Mfd to dislodge stalled RNAP from DNA templates [12, 14]. However, in solution, Mfd's repressor domains (Domain 2 and domain 7) block its ATPase/translocase functionality, preventing non-specific translocase activity [15].
In addition to dismantling stalled ECs -- which, by itself, inhibits DNA repair by blocking access of the UvrABC machinery to the lesion site [8] -- Mfd also actively recruits UvrA to the site of DNA damage. The UvrB-homology module in Mfd -- consisting of the D1a, D1b, and D2 domains -- is required for direct interactions between UvrA and Mfd. This module allows Mfd to bind UvrA, presumably after removing RNAP from DNA. Consequentially, Mfd is displaced by UvrB, allowing UvrA2B2 complex to initiate DNA repair [10, 11, 16, 17]. In its inactive state (not bound to stalled RNAP), the UvrB-homology module is masked by the auto-inhibitory domain D7, which prevents non-productive interactions between Mfd and the rest of the UvrABC repair system [15, 18, 19].
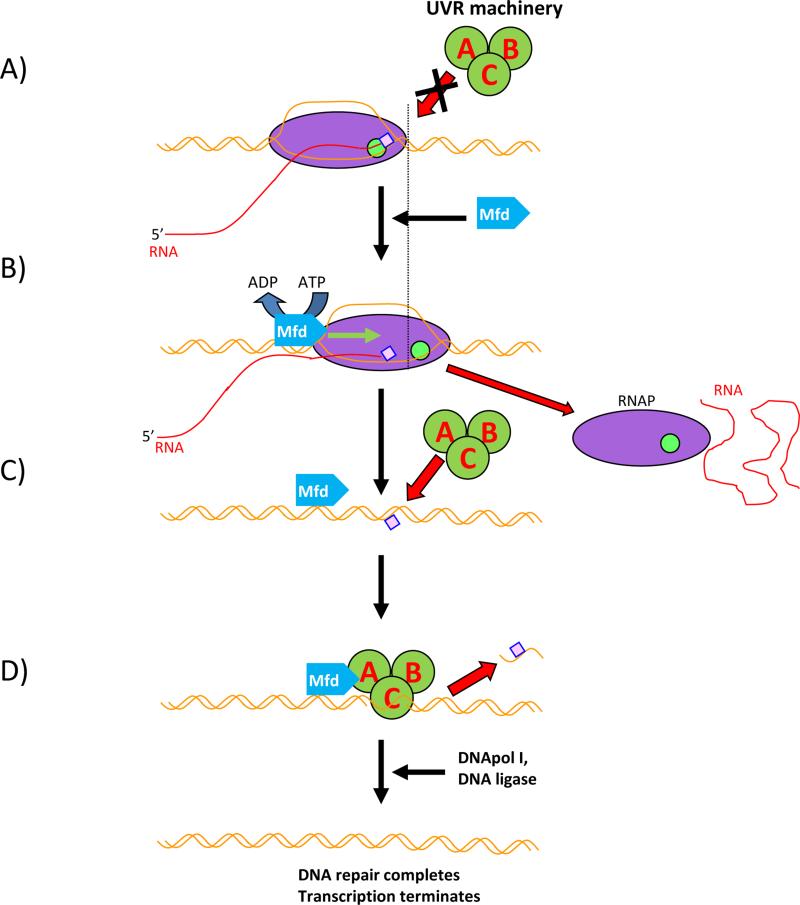
The mechanics of EC removal from lesion sites by Mfd is now well established (Fig. 1). Mfd first binds to double-stranded DNA (dsDNA) directly upstream from the stalled RNAP. Mfd then pushes RNAP forward via its ATP-dependent translocase activity, until RNAP either resumes elongation or is removed from the template, releasing nascent RNA in the process [10, 12, 13, 20, 21]. Mfd does not distinguish between various stalled ECs, be it irreversible arrest, DNA lesion, or protein bound to DNA in front of RNAP [22]. The only requirement for Mfd-dependent displacement of the stalled ECs is relatively long dwell time of ECs in the inactive state. It seems that Mfd discriminates between transiently paused ECs and inactivated ones via kinetic control. Whereas a majority of regulatory pauses are relatively short lived, ECs stopped by DNA lesions are there to stay until removed [22].
Figure 1.
Scheme of Mfd-depended TCR in bacteria. RNAP – magenta oval, active site – green circle, Mfd – blue pentagon. UvrABC system – dark green circles, bulky lesion - pink square. DNA – brown line, RNA – red line. A: RNAP encounters a bulky lesion on the template strand and stalls, unable to elongate. Elongation Complex blocks UvrABC from loading onto the DNA. B: Mfd recognises stalled Elongation Complex, loads upstream and pushes RNAP forward by using its helicase/translocase activity. Eventually RNAP is removed, RNA synthesis is terminated and the lesion becomes exposed. C: The UvrABC system is actively recruited at the DNA damage site by Mfd-UvrA interactions and UvrABCD machinery removes the lesion. D: UvrABCD leave the complex; DNApol and DNA ligase finish DNA repair.
Despite the very well established paradigm of Mfd action, some questions have arisen over the years as to whether Mfd plays a unique role in TCR coupling or whether there are alternative pathways with yet unknown players. There are several unsolved problems that have to be addressed. First, the Mfd deletion mutant is not especially UV-sensitive [9, 10, 23] unlike members of uvrABCD system which were selected precisely on account of their ability to confer UV sensitivity upon deletion. This fact seems to be counterintuitive, taking into account the presumably central role of Mfd in TCR that helps to remove thymine dimers (the major DNA lesion caused by UV exposure). Second, Mfd has a very low processivity [12, 22]. Indeed, its low rate serves as a discriminator between regulatory pauses and irreversible arrests, but even then it takes more than 3 minutes to fully disassemble stalled ECs, as shown directly by single-molecular observations [22]. During massive DNA damage, such a long reaction time should make TCR quite ineffective. Third, the mechanism of Mfd action makes it especially ill-suited for repairing actively transcribed genes where ECs are queued one after another on the same DNA molecule [24, 25]. Such arrangement means that Mfd has to repeatedly remove stalled ECs until it reaches the leading RNAP stalled by the lesion. If removal of a single EC takes at least 3 minutes, removal of several ECs would take tens of minutes. This poses a problem not only for cell survivability if the damaged gene is essential, but it could also lead to replication fork collapse and double-stranded breaks. Indeed, Kunala and Brush have previously reported [26] compromised ability of Mfd to direct TCR in actively transcribed genes. Fourth, in recent years new data have emerged that indicate the existence of pathways that do not depend upon Mfd, but somehow utilize the transcription factor NusA. However, the authors could not elucidate the mechanism of its involvement in TCR [27, 28].
Though it seems that Mfd cannot be the only TCR player in the bacterial cell, compelling data from Sancar's lab [9] indicate that that is indeed the case. Furthermore, genetic analysis could not identify new candidates for the elusive “second TCR” player. So, what is going on in the repair machinery of the bacterial cell? At this point we turn to UvrD – an old player with a new role.
UvrD to the rescue!
Our recent work identified the UvrD helicase as the “missing link” that solves the majority of discrepancies from earlier work, and establishes a new TCR paradigm where Mfd plays an important but secondary role in coupling transcription to DNA repair during global genotoxic stress [29]. UvrD is a member of UvrABCD NER machinery, and serves an important role in later steps of DNA repair by removing the oligonucleotide containing DNA lesion and UvrC endonuclease (for a review see [6]). Due to this fact, UvrD could not be identified as a TCR coupling factor via conventional genetic analysis because its deletion would compromise NER in general. The helicase is also involved directly in mismatch repair [30, 31], and works as an antagonist to RecA-directed strand exchange [32-34], playing multiple roles in DNA maintenance and homeostasis. UvrD belongs to SF1 of DNA helicases-tranclocases, harboring the typical Walker-motif common to all members of that protein family, and it utilizes ATP for powering its 3’ to 5’ movement [35-37]. UvrD can work as both a translocase and a helicase, dependent on different oligomeric states of the protein. Whereas a UvrD monomer is enough for translocase activity, for DNA unwinding UvrD must be in a dimeric state [38-41]. Those two reactions also differ in their processivity. UvrD translocation on the ssDNA strand can run for hundreds of nucleotides with no interruption, but as a helicase UvrD rarely moves beyond a few dozen of base pairs before dissociating.
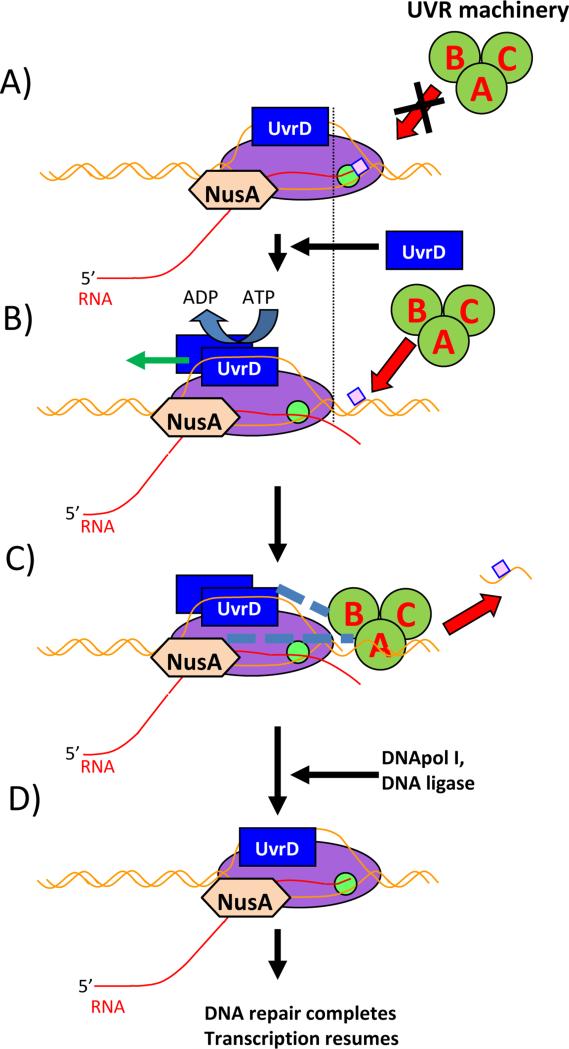
In our work [29], we found that UvrD not only binds to RNAP tightly at an abundancy level comparable to common transcription factors such as NusA or NusG, but that it can also actively push RNAP backward in the case of EC stalling. This finding allowed us to propose a novel TCR pathway that does not involve Mfd, but relies only on core UvrABCD machinery (Fig. 2). Essentially UvrD is pre-loaded at actively transcribing ECs and moves as a bona fide transient subunit during processive transcription. Upon encountering a DNA lesion, forward movement of the RNAP would be arrested, and UvrD can move the EC backwards, exposing the damaged DNA segment for repair. It was also shown that UvrD makes direct contacts with UvrB [42, 43] and thus can possibly serve as a recruitment factor to speed up DNA damage recognition. Another transcription factor, NusA, was earlier implicated in Mfd-independent TCR pathway [27, 28] and according to our data it indeed improves the pro-back-tracking activity of UvrD. Interestingly enough, NusA makes direct contacts with UvrA [28] and can also contribute to UvrA binding and subsequent UvrA2B2 assembly at the lesion site. Though we haven’t demonstrated direct recruitment of the core NER machinery to sites of DNA damage via UvrD/NusA pathway, interactions between UvrA2B2 and NusA/UvrD (Fig. 2 dashed lines) tempt us to speculate that UvrD-driven pathway speeds up repair not only via clearing lesion sites from obstructing RNAPs but also directly by rising local concentration of the repair factors at the DNA damage sites. However this hypothesys has to be tested directly to prove the point.
Figure 2.
Principal scheme of proposed UvrD assisted TCR in bacteria. RNAP – magenta oval, active site – green circle, UvrD – blue rectangle, UvrABC system – dark green circles, bulky lesion - pink square, NusA – light brown hexagon. DNA – brown line, RNA – red line. Proposed interactions are shown as dashed lines. A: RNAP encounters a bulky lesion on the template strand and stalls, unable to elongate. Elongation Complex blocks UvrABC from loading onto the DNA. B: UvrD forms a dimer and moves the Elongation Complex backward by using helicase/translocase activity. NusA assists UvrD, and the lesion becomes exposed. C: The UvrABC system is recruited at the DNA damage site and removes the lesion. D: UvrABC and one of the UvrD monomers leave the complex; DNApol and DNA ligase repair DNA. RNAP resumes elongation.
According to our cross-linking data UvrD interacts with the EC near the upstream portion of the transcription bubble. UvrD binds with high affinity to as little as 4 nucleotides in the 3’->5’ portion of ssDNA adjusted to dsDNA of DNA fork junctions [38]. The non-transcribed strand in the transcription bubble is at least 10 nucleotides long, fully exposed to solution [44], and serves as an excellent substrate for loading UvrD. In our model we have proposed that UvrD binds at the upstream junction of the transcription bubble and moves the EC backward by either melting upstream dsDNA or by actively pushing RNAP backward. In the first case, unwinding the upstream part of the bubble leads to net loss of the EC's stored energy, which must be compensated for by simultaneous rewinding of the front part of the transcription bubble, forcing RNAP to back-track. In the second case, energy of ATP hydrolysis directly transforms into motility force to push RNAP backward. We cannot exclude also some kind of combination between two mechanisms, though we are inclined to believe that the former mechanism -- unwinding the upstream part of the bubble -- plays the predominant role. Our titration experiments have shown that the extent of UvrD-directed back-tracking depends upon UvrD concentration and that dependence perfectly fits into a sigmoid curve – a signature of protein dimerization. Also pro-backtracking activity of UvrD can be washed out from stalled ECs that were immobilized on beads and yet in the same time UvrD can pull down non-tagged RNAP from solution [29]. This fact points out at dimeric state of pro-back-tracking UvrD complex if active dimer forms transiently. Bearing in mind that DNA unwinding is conducted exclusively by the UvrD dimers [38], helicase activity seems the more attractive option.
Our model allows us to address several inherent problems that plagued the Mfd-centric TCR pathway. Unlike Mfd, UvrD acts fast. In fact, under saturation conditions, it can back-track a majority of paused ECs in just seconds. Repair can commence immediately after encountering the lesion site without prolonged delays. Also, UvrD is an abundant protein; the amount of active monomer [45] is comparable to RNAP in the bacterial cell65, allowing the helicase to be available at the first appearance of DNA damage to initiate TCR. The mechanism of UvrD loading and action also resolves the long standing conundrum of how Mfd helps to repair long arrays of stalled ECs. While Mfd has to disassemble them one at a time, in UvrD-directed TCR each stalled EC can be pushed back by its own UvrD dimer. Together the entire array will be moved away from the lesion site quickly and efficiently. In addition, UvrD-directed back-tracking does not involve termination of the RNA synthesis, as opposed to the Mfd-centric model, thus allowing cells to resume elongation as soon as the DNA damage is repaired. Retention of the nascent transcript is beneficial for cells for several reasons. First, it could be crucial for some weakly expressed genes where RNA synthesis has to start anew if the transcript is terminated; second, it helps to conserve resources by preventing futile transcription; third, lack of premature termination prevents appearance of aberrant small RNAs, which can hinder gene expression.
When too much help is bad
Despite many advantages of UvrD-directed TCR over the Mfd pathway it has its own drawbacks. Cells have thus developed several ways to deal with it. Back-tracking on its own is harmful to bacteria: it delays transcription, makes RNA synthesis less efficient, and in some extreme cases, leads to irreversible transcription arrests. It was also reported that backtracking could lead to double strand DNA breaks during replication and cause genome instability [46]. UvrD's pro-backtracking activity in vitro rapidly produces transcription arrest and at saturation concentration prevents almost all ECs from reaching the end of their transcription units. How do cells cope with such indiscriminate behaviour of the UvrD helicase? We discovered that anti-back-tracking activities of the bacteria help to solve that problem. GreA and GreB proteins stimulate intrinsic cleavage activity of the RNAP [47, 48] and thus allow the EC to cleave out the 3’-end of nascent RNA in backtracked complexes, restoring RNA's 3’-end in the catalytic site in active conformation. Indeed, our in vitro experiments confirmed that GreB protein can rapidly restore activity of the UvrD-back-tracked complexes and complete transcription. Interestingly, though the UvrD null-mutant is very sensitive to UV and other DNA damaging agents, the triple UvrD/GreA/GreB deletion strain has much better resistance [29] to those factors. Obviously in living cells the pro-back-tracking activity of UvrD is balanced out by GreA/B anti-back-tracking action. In the case of UvrD deletion, lack of Gre proteins would allow spontaneous RNAP backtracking at the DNA-damage site and would preclude ECs from blocking repair proteins’ access to the lesions. At the same time, GreA/B-assisted RNA cleavage would restore activity to transcription complexes that were accidentally inactivated by UvrD. This phenotype also supports leading role of UvrD in TCR during genotoxic stress. Deletion of UvrD much less deleterious when anti-back-tracking machinery is disabled as well. There is a report that DNA PolI plays a significant role in displacing lesion-containing oligos during NER [49], making UvrD's role in general NER at least partly dispensable.
Another anti-back-tracking mechanism that keeps UvrD-directed pro-back-tracking activity in check relies upon coupling transcription and translation in the bacterial cells. Bacterial protein synthesis starts as soon as the ribosome-binding motif emerges from transcribing RNAP. The rate of transcription is tightly coupled to the rate of translation [50, 51]. This not only prevents premature termination by Rho factor, but also helps to keep RNA synthesis pause-free, supposedly by preventing spontaneous backtracking by trailing ribosomes. Indeed, slowing down translation by treatment with sub-lethal concentrations of antibiotics has the same phenotypical effect as triple UvrD/GreA/GreB deletion [29].
We cannot rule out the possibility that other anti-back-tracking pathways are also involved in controlling how many stalled ECs UvrD are allowed to move backward, or to what extent. For example, the co-operative nature of transcription works in a similar manner to transcription-coupled translation. In the case of actively transcribed genes, leading RNAP is pushed forward by trailing RNAP molecules that synthesize RNA on the same gene, preventing leading RNAP from back-tracking [52]. Another possible control mechanism may utilize the termination factor Rho [53], which would disassemble irreversibly back-tracked transcription complexes and free DNA from blockades.
When the cells yell “SOS”!
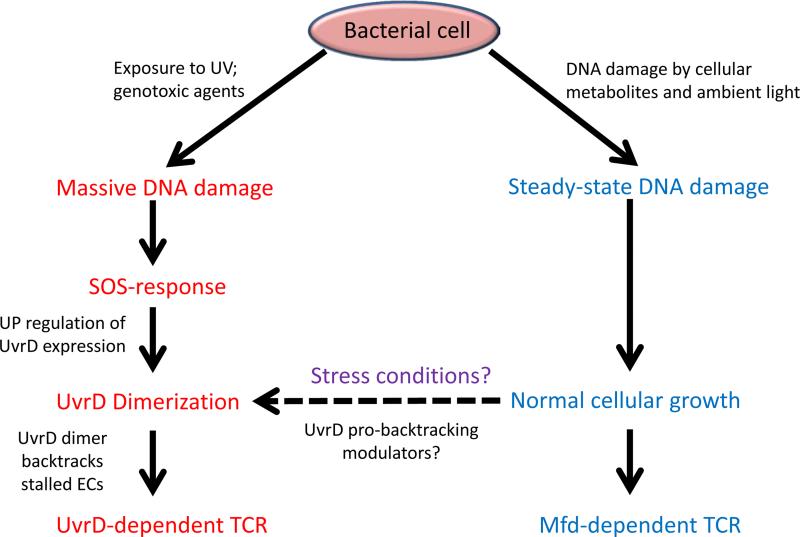
Both RNAP and UvrD are abundant proteins, with UvrD numbering about 3,000 copies per exponentially growing cell [45] whereas RNAP numbers lie between 1,600 and 8,000 copies [65], depending on the rate of growth. This ratio leads to mostly monomeric UvrD bound to transcribing RNAPs. However, the sigmoid form of UvrD's titration curve and washing experiments (see above) leads us to believe that UvrD must dimerize to have pro-back-tracking activity. This finding, together with the fact that UvrD is an SOS-inducible protein, suggests a very elegant mechanism of TCR regulation that minimizes harmful backtracking during normal growth, while maximizing coupling effects during massive DNA damage. According to this model (Fig. 3), in non-stressed cells, monomeric UvrD is bound to transcribing RNAPs and does not interfere with transcription except at extremely long-lived pauses/arrests, where spontaneous dimerization is possible, hence keeping harmful back-tracking to a minimum. However, after suffering massive DNA damage and inducing the SOS-response, cellular levels of UvrD are increased at least three-fold via Lex-dependent up-regulation [54-56]. This increase in UvrD concentration leads to dimerization of the helicase, and forces RNAP to back-track during delays in transcription elongation, which is presumably caused by DNA damage and requires repair. Short-lived ubiquitous pauses would be poor substrates for UvrD-driven back-tracking because of the existence of multiple anti-back-tracking mechanisms;, while any significant delay in transcription elongation would serve as a trigger for UvrD-directed TCR. At the same time, it was reported that repair of cyclo-butane pyrimidine dimers does not depend upon transcription during SOS-response [66], which seemingly contradicts our model. However, the timing of the SOS-dependent induction could play a crucial role and explains the contradiction. The authors calculated that before irradiation cells carried 200 molecules of UvrA and 400 molecules of UvrB per chromosome whereas after exposure to UV in 40 minutes the molecule count increased to 1,200 and 2,000 molecules correspondingly. Constitutive expression of the SOS proteins leads even to higher concentration of UvrA/B [66]. At such amounts the NER machinery probably can locate and repair DNA lesions without TCR assistance but in 10 minutes after UV irradiation amount of the UvrA/B increases only twofold [66], which could be not high enough for efficient repair without TCR. In the same time UvrD is a plentiful protein even without SOS induction and slight increase in UvrD concentration following SOS-activation at earlier stages can play important role for cell survival via dimerization mechanism described above. Though this model seems quite attractive it needs a direct verification, for example constructing UvrD mutant with diminished dimerization activity and testing it in vitro and in vivo.
Figure 3.
Two pathways of cellular TCR. UvrD-dependent TCR (left). Under conditions of massive DNA damage, the SOS response increases UvrD output, which leads to UvrD dimerization, RNAP backtracking, and TCR.Mfd-dependent TCR (right). Cells with a steady-state level of DNA damage have low concentrations of UvrD, thus the UvrD-dependent pathway is repressed. DNA damage is located and removed via Mfd-dependent TCR. Proposed switch from Mfd to UvrD pathway under non-SOS stress conditions is shown by the dashed line.
This type of regulation also explains the discrepancy between the proposed UvrD-directed model of bacterial TCR and complementation experiments performed by Sancar and co-workers [9], where Mfd deletion led to the abolition of TCR in vitro and coupling could be restored only by lysate from Mfd+ cells but not from Mfd- cells. In those experiments the authors used lysates containing final UvrD concentrations too low to initiate TCR in Mfd's absence. Though there are no doubts that Mfd indeed works as a TCR coupling factor, it is interesting to see if complementation experiments would provide a different results under different conditions such as different level of transcription, level of UV dose, SOS induction etc.
So, where does the Mfd-pathway fit in the picture of UvrD-centric coupling between transcription and repair during acute genotoxic stress? We propose that Mfd in fact helps to clean up irreversibly back tracked ECs after cells start to recover from DNA damage. In the outgrowth phase, the cellular level of UvrD goes down, but a multitude of irreversibly arrested complexes are left behind. Though some of these complexes would be reactivated via anti-back-tracking mechanisms discussed above, other ECs would be irreparable, and would have to be removed. It is easy to imagine that some of the improperly back-tracked complexes would be covering hitherto unrepaired DNA lesions. The indiscriminate nature of Mfd-directed transcription termination would be ideal for dealing with such “transcription wreckages”, followed by the recruitment of the NER proteins to clean up the few unrepaired lesion sites left behind. Our model predicts that a lack of Mfd would compromise recovery from stress, but would otherwise not influence UV sensitivity or vulnerability to DNA-damaging chemicals in the bacterial cells. Indeed, it is reported that Mfd is required for rapid recovery of transcription after UV exposure [57]. We propose that Mfd also works in non-stressed conditions when levels of UvrD are low and UvrD-directed TCR is not efficient. In this view, Mfd-directed pathway helps to protect cells from DNA damage during normal growth condition but cells rely mostly upon UvrD-directed pathway in case of an acute genotoxic stress (Fig.3). According to this model two pathways work together to make DNA repair most efficient under different physiological conditions. UvrD dimerization is dependent upon cellular concentration of the helicase, but efficiency of that process can be regulated. For example, NusA improves UvrD's ability to push RNAP backward [58] and binds to transcriptional complexes in the same general vicinity as UvrD [29, 59]. At the present time we cannot say whether NusA stabilizes UvrD dimer, increases its pro-back-tracking activity or prevents RNAP from returning to the active state. NusA does, however, allow UvrD to act at lower concentration. It is easy to imagine that bacterial cells would develop specialized systems that regulate the extent of UvrD-driven back-tracking depending on the physiological state of the cell, its damage status, and the nature of the stress. In cases of serious DNA damage, the SOS response would activate UvrD-directed TCR and take care of lesions even at the cost of compromising transcriptional efficiency. However when DNA damage is significant but not severe enough to trigger the SOS response, we propose that other modulators can possibly activate UvrD-directed TCR as well. This approach allows cells better flexibility in their response to DNA damage: they can trade off improved DNA-damage recognition for the cost of less efficient transcription and fine tune that ratio to cellular needs.
A second job for transcribing RNAP: Sensing DNA damage on genome wide scale
Efficiency of UvrD-dependent TCR sheds a new light on the role of RNAP in DNA damage recognition. In our view, RNAP serves as a surveillance device, constantly scanning template DNA for damage. This housekeeping role explains why transcription is so ubiquitous and covers the entire genome even in non-coding regions in both pro- and eukaryotes [60, 61]. Some small RNA transcripts do play a role in cellular metabolism, but in our opinion, the majority of those RNA species are by-products of DNA scanning. Cells do trade off spending some resources at non-productive RNA synthesis, but in return they get better damage recognition, which increases chances of survival. We also suggest that eukaryotic cells could utilize UvrD-like mechanisms for their own TCR, taking into account how conservative the core RNAP structure is. Eukaryotes do not have repair factors similar to Mfd, but their own multiprotein coupling factor TFIIH does have DNA-helicases among its subunits [62, 63]. It was suggested that eukaryotic helicase CSB work via mechanism similar to UvrD by pushing stalled RNAPs backward (reviewed in [64]). We propose that due to the complexity of the transcriptional apparatus eukaryotic cells could have redundant helicases that back each other up and are selectively activated in various tissues under different conditions. It remains to be seen whether UvrD-like TCR works in higher eukaryotes, but its discovery would drastically change our understanding of how cells maintain integrity of their DNA.
Conclusion
In the view of our recent findings it has become clear that bacterial cells developed several TCR pathways each uniquely fit for different physiological conditions and those pathways are tightly regulated to minimize harmful effects of premature termination (Mfd pathway) and extensive backtracking (UvrD pathway). It remains to be seen if similar multiple pathways exist in eukaryotic organisms and whether additional factors participate in the bacterial TCR alongside with known players. Another interesting question is: is this the end of the story or more hidden TCR-pathways exist in prokaryotes and if they do, what kind of conditions triggers them?
Acknowledgements
The author would like to thank Evgeny Nudler, Venu Kamarthapu, Katelyn McGary, and Vladimir Svetlov for their invaluable contributions in establishing discussed model and Bradley Benjamin for critical reading of the manuscript. This work was supported by NIH grant GM058750-13S1
Abbreviations
- EC
elongation complex
- NER
nucleotide excision repair
- RNAP
RNA polymerase
- TCR
transcription coupled DNA repair
References
- 1.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 2.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–52. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 3.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moolenaar GF, Höglund L, Goosen N. Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. EMBO J. 2001;20:6140–9. doi: 10.1093/emboj/20.21.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malta E, Verhagen CP, Moolenaar GF, Filippov DV, et al. Functions of base flipping in E. coli nucleotide excision repair. DNA Repair (Amst) 2008;7:1647–58. doi: 10.1016/j.dnarep.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog Mol Biol Transl Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 7.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–8. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265:21330–6. [PubMed] [Google Scholar]
- 9.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci USA. 1991;88:11574–8. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–8. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 11.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–5. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 12.Park JS, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–67. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 13.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–20. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 14.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–64. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–11. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–9. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 17.Manelyte L, Kim YI, Smith AJ, Smith RM, et al. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40:714–24. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy MN, Gong P, Ralto K, Manelyte L, et al. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–53. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava DB, Darst SA. Derepression of bacterial transcription-repair coupling factor is associated with a profound conformational change. J Mol Biol. 2011;406:275–84. doi: 10.1016/j.jmb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103:4870–5. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–69. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howan K, Monnet J, Fan J, Strick TR. Stopped in its tracks: The RNA polymerase molecular motor as a robust sensor of DNA damage. DNA Repair (Amst) 2014;20:49–57. doi: 10.1016/j.dnarep.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Witkin EM. Radiation-induced mutations and their repair. Science. 1966;152:1345–53. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 24.Foe VE. Modulation of ribosomal RNA synthesis in Oncopeltus fasciatus: an electron microscopic study of the relationship between changes in chromatin structure and transcriptional activity. Cold Spring Harb Symp Quant Biol. 1978;42:723–40. doi: 10.1101/sqb.1978.042.01.074. Pt 2. [DOI] [PubMed] [Google Scholar]
- 25.Hamming J, Arnberg A, Ab G, Gruber M. Electron microscopic analysis of transcription of a ribosomal RNA operon of E. coli. Nucleic Acids Res. 1981;9:1339–50. doi: 10.1093/nar/9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunala S, Brash DE. Excision repair at individual bases of the Escherichia coli lacI gene: relation to mutation hot spots and transcription coupling activity. Proc Natl Acad Sci USA. 1992;89:11031–5. doi: 10.1073/pnas.89.22.11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen SE, Walker GC. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr Biol. 2010;20:80–5. doi: 10.1016/j.cub.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen SE, Lewis CA, Mooney RA, Kohanski MA, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci USA. 2010;107:15517–22. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epshtein V, Kamarthapu V, McGary K, Svetlov V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–7. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem. 1998;273:9197–201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 31.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 32.Veaute X, Delmas S, Selva M, Jeusset J, et al. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–9. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Florés MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol Microbiol. 2005;57:1664–75. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 34.Centore RC, Sandler SJ. UvrD limits the number and intensities of RecA-green fluorescent protein structures in Escherichia coli K-12. J Bacteriol. 2007;189:2915–20. doi: 10.1128/JB.01777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickson ID, Arthur HM, Bramhill D, Emmerson PT. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190:265–70. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- 36.Matson SW. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3' to 5' direction. J Biol Chem. 1986;261:10169–75. [PubMed] [Google Scholar]
- 37.Matson SW, George JW. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987;262:2066–76. [PubMed] [Google Scholar]
- 38.Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–35. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 39.Maluf NK, Ali JA, Lohman TM. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J Biol Chem. 2003;278:31930–40. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 40.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Lee KS, Balci H, Jia H, Lohman TM, et al. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat Commun. 2013;4:1878. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn B. A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol Cells. 2000;10:592–7. doi: 10.1007/s10059-000-0592-5. [DOI] [PubMed] [Google Scholar]
- 43.Manelyte L, Guy CP, Smith RM, Dillingham MS, et al. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair (Amst) 2009;8:1300–10. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korzheva N, Mustaev A, Kozlov M, Malhotra A, et al. A structural model of transcription elongation. Science. 2000;289:619–25. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 45.Arthur HM, Eastlake PB. Transcriptional control of the uvrD gene of Escherichia coli. Gene. 1983;25:309–16. doi: 10.1016/0378-1119(83)90235-4. [DOI] [PubMed] [Google Scholar]
- 46.Dutta D, Shatalin K, Epshtein V, Gottesman ME, et al. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–43. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–66. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 48.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–34. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, et al. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci USA. 1985;82:6774–8. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–8. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burmann BM, Schweimer K, Luo X, Wahl MC, et al. NusE:NusG complex links transcription and translation. Science. 2010;328:501–4. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 52.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–5. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 53.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–9. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumura K, Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984;259:1560–5. [PubMed] [Google Scholar]
- 55.Siegel EC. The Escherichia coli uvrD gene is inducible by DNA damage. Mol Gen Genet. 1983;191:397–400. doi: 10.1007/BF00425753. [DOI] [PubMed] [Google Scholar]
- 56.Easton AM, Kushner SR. Transcription of the uvrD gene of Escherichia coli is controlled by the lexA repressor and by attenuation. Nucleic Acids Res. 1983;11:8625–40. doi: 10.1093/nar/11.24.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schalow BJ, Courcelle CT, Courcelle J. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli. J Bacteriol. 2012;194:2637–45. doi: 10.1128/JB.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, et al. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–93. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 59.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–3. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 60.Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio. 2012;3:e00156–12. doi: 10.1128/mBio.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell. 2013;52:473–84. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 62.Drapkin R, Reardon JT, Ansari A, Huang JC, et al. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–72. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 63.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–54. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 64.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 65.Shepherd NS, Churchward G, Bremer H. Synthesis and activity of ribonucleic acid polymerase in Escherichia coli. J Bacteriol. 1980;141:1098–108. doi: 10.1128/jb.141.3.1098-1108.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crowley DJ, Hanawalt PC. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6-4 photoproducts, in UV-irradiated Escherichia coli. J Bacteriol. 1998;180:3345–52. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]