Summary
The basic determinant of chromosome inheritance, the centromere, is specified in many eukaryotes by an epigenetic mark. Using gene targeting in human cells and fission yeast, chromatin containing the centromere-specific histone H3 variant CENP-A is demonstrated to be the epigenetic mark that acts through a two-step mechanism to identify, maintain and propagate centromere function indefinitely. Initially, centromere position is replicated and maintained by chromatin assembled with the centromere-targeting domain (CATD) of CENP-A substituted into H3. Subsequently, nucleation of kinetochore assembly onto CATD-containing chromatin is shown to require either CENP-A’s amino- or carboxy-terminal tails for recruitment of inner kinetochore proteins, including stabilizing CENP-B binding to human centromeres or direct recruitment of CENP-C, respectively.
Introduction
The centromere is the fundamental unit for ensuring chromosome inheritance. Centromeres have a distinct type of chromatin in which histone H3 is replaced by a conserved homologue initially identified in humans and named CENP-A1,2. While specific chromosomal regions containing α-satellite repeats3 are the sites of centromere formation and CENP-A association in humans, α-satellite DNA sequences are neither sufficient nor essential for centromere identity4. Rather, except for budding yeast, it is well accepted that in almost all other organisms, including fission yeast, centromeres depend on one or more as yet unidentified epigenetic marks5. Among the strongest evidence for an epigenetically defined centromere was the discovery in humans of migration of a functional centromere from an initial location to a new site on the same chromosome6-9. These loci, referred to as neocentromeres, are stably loaded with CENP-A8 and form at previous euchromatic DNA sites without α-satellite repeats6,10,11.
Epigenetic inheritance implies that the mark must be self-templating and be stably maintained across the cell cycle. Centromere-bound CENP-A molecules are indeed quantitatively re-distributed to both sister centromeres during centromeric DNA replication, yielding each daughter centromere with half the level of the initially loaded CENP-A12. Furthermore, in human12 and Drosophila13,14 cells loading of new CENP-A into centromeric chromatin occurs once per cell cycle only during or upon exit from mitosis.
Although the precise nature of centromeric chromatin is highly controversial15-24, CENP-A is a central component in all models. Reduction or mutation in CENP-A25-28 has proven it to be essential for continuing centromere function27. Indeed, forced loading of CENP-A by tethering it or components that bind it to specific DNA loci has produced partial ectopic centromere formation29-32 and in one case with full kinetochore function after deletion of the authentic centromere33. These reports reinforce the sufficiency of CENP-A for recruitment of some centromere and kinetochore components, but offer no insight into how centromere identity and function is epigenetically specified and maintained. Approaches25-28 to relying on RNAi-mediated mRNA degradation to identify the epigenetic mark have suffered from incomplete silencing, especially for proteins such as CENP-A with long half lives. To overcome these limitations, we now use gene targeting in human diploid cells and in fission yeast to demonstrate that CENP-A-containing chromatin is the epigenetic mark that can identify, maintain and propagate human or yeast centromere function indefinitely.
Results
Conditional CENP-A gene deletion in diploid human cells
Using a single stranded Adeno Associated Virus 2 (AAV2) vector to enhance homologous recombination34,35, both human CENP-A alleles were targeted in non-transformed diploid human retinal epithelial (RPE1) cells immortalized with human telomerase. One allele was replaced with a CENP-A null allele in which a neomycin resistance gene (flanked by FRT sites to facilitate its later removal) replaced exons 3 and 4 of the CENP-A gene [encoding the essential CENP-A centromere targeting domain (CATD) that is comprised of loop 1 and the α2 helix of CENP-A and which when substituted into histone H3 is sufficient to restrict H3 targeting to centromeres36]. The other allele was specifically targeted with an AAV2 “floxed” construct containing 34 base pair loxP recombination sites flanking exons 3 and 4 (Fig. 1a). This yielded a final cell line with one null allele and one “floxed” CENP-A allele, the latter of which can be converted into a null allele by action of Cre recombinase (Fig. 1a and Supplementary Fig. S1a). A CENP-AF/+ cell line was also produced as a control. CENP-A−/F cells grew at a rate indistinguishable from the control CENP-A+/+ or CENP-AF/+ with only a slight reduction of total pool of CENP-A protein and no reduction seen at centromeres (Supplementary Fig. S1b,c,d).
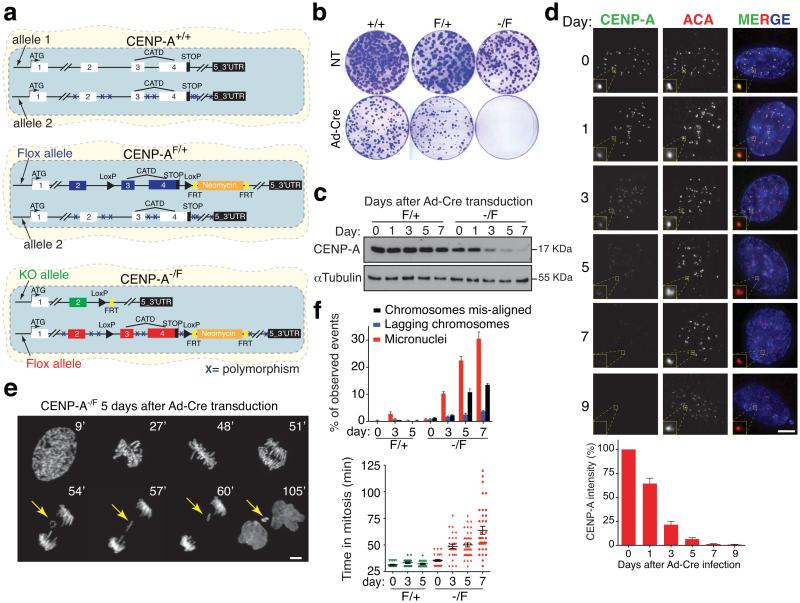
Figure 1.
CENP-A conditional disruption in human cells. (a) Schematic illustration of the final CENP-A cell lines after gene targeting. Positions of the exons, the CATD domain, LoxP sites, FRT sites, the Neomycin cassette and the start-stop codons are indicated. Xs indicate polymorphic mutations between the initial two CENP-A alleles. (b) Clonogenic cell survival assays (visualized with crystal violet) for the indicated cell lines with (Ad-Cre) or without (NT, not treated) treatment. (c) CENP-A protein level is determined by immuno-blotting extracts from CENP-AF/+ and CENP-A−/F cells at the indicated days after Ad-Cre addition. (d) Representative fluorescence images of CENP-A localization and intensity at the indicated days after Ad-Cre addition. (Bottom) Quantification of CENP-A protein levels from fluorescence images. Bars represent the mean of six independent experiments (> 30 cells per experiment). Error bars represent the SEM (standard error of the mean). ACA (anti-centromere antibody) staining was used to identify centromeres. (e) Time-lapse images of CENP-A−/F RPE1 cells stably expressing histone H2B-mRFP 5 days after Ad-Cre addition. Time (in minutes) after nuclear envelope breakdown (NEBD) is shown at the top. Yellow arrows point to a lagging chromosome that ultimately forms a micronucleus. (f) Bars represent the mean of > 50 cells per condition. Each individual point represents a single cell. Error bars represent the SEM of three independent experiments. Time in mitosis was defined as the period from NEBD to chromosome decondensation. Uncropped image of blots are shown in Supplementary fig. S7. Scale bar = 5 μm.
Transient expression of Cre recombinase [using a replication defective adenoviral vector, Ad-Cre to produce excision of exons 3 and 4 of the floxed allele (Supplementary Fig. S1e)] suppressed cell growth in the CENP-A−/F cells (Fig. 1b). PCR analysis of the few surviving clones revealed that they had escaped inactivation of the second CENP-A allele (Supplementary Fig. S1f). To eliminate confounding effects from these “escapers”, a LoxPRFP-STOPLoxP-GFP cassette was inserted (by lentiviral integration) into the CENP-AF/+ and CENP-A−/F cells (Supplementary Fig. S1g). Addition of Cre recombinase converts cells from red to green fluorescence by simultaneous excision of the RFP gene and activation of the GFP gene (Supplementary Movie S1). CENP-A depleted cells were then collected by fluorescent cell sorting (FACS) to recover green fluorescent cells. After depletion of both alleles of CENP-A, cells duplicated at a similar rate as control cells for the first 5 days (~6 divisions), then exhibited slowed cycling from extended mitosis and stopped dividing only 9-11 days (~8-9 divisions) after initial CENP-A gene inactivation (Supplementary Fig. S1h).
CENP-A excision led to progressive loss of accumulated CENP-A and its levels decreased by approximately half each cell cycle (Fig. 1c,d), consistent with the expected redistribution of centromere-bound CENP-A to sister centromeres during DNA replication but without addition of new CENP-A12. Only 1% of the initial CENP-A level was detectable within 7 days (Fig. 1d) (as expected for the 1/27 dilution = 0.8%). No centromere-bound CENP-A could be detected 9 days following excision (Fig. 1d). Lowered levels of CENP-A corresponded with increased rates of chromosome segregation defects and increased mitotic duration, accompanied by a significant increase in micronuclei formation from failure in initial kinetochore attachment to spindle microtubules or initially aligned chromosomes lagging in anaphase (Fig. 1e,f and Supplementary Movie S2).
Disrupted kinetochore nucleation requires almost complete loss of CENP-A
CENP-A loading has been proposed to be at the foundation of recruitment (direct and indirect) of all of the components of the core centromere26,37-39. Three patterns of centromere/kinetochore protein loss after inactivation of the conditional CENP-A allele in the CENP-A−/F RPE1 cells were identified in asynchronous cycling cells.
The first group included: CENP-C and CENP-N, two primary components of the constitutive centromere-associated network (CCAN) that has been shown to directly interact with CENP-A chromatin37,38 and CENP-P, a more distal kinetochore protein39. Loss of centromere-bound CENP-C, CENP-N and CENP-P was initially in proportion to loss of CENP-A (Fig. 2a,b,d and Supplementary Fig. S2a), consistent with direct binding to CENP-A chromatin or, in the case of CENP-P, through a CENP-N/CENP-C-dependent complex. Surprisingly, a substantial proportion (~30%) of CENP-C remained even after CENP-A depletion to <1% of its initial level (Fig. 2a,d).
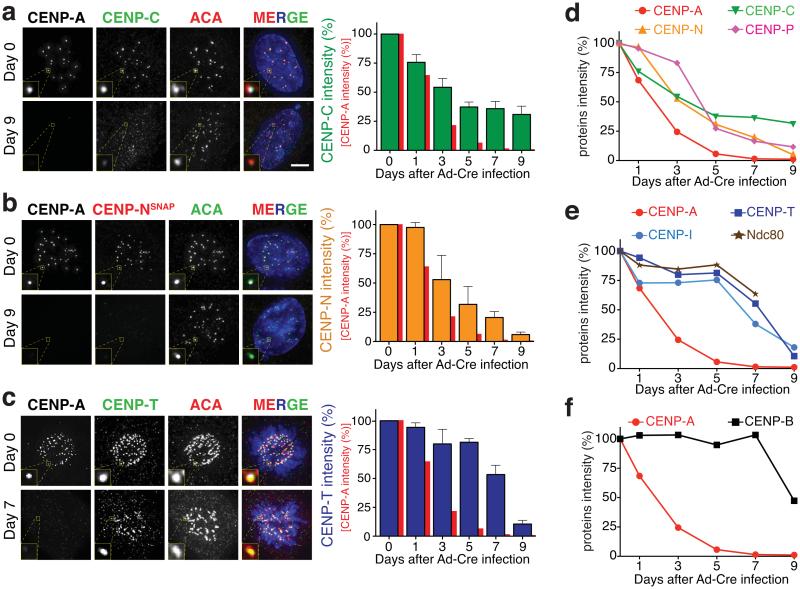
Figure 2.
Disrupted centromere positioning and kinetochore nucleation requires almost complete loss of CENP-A. (a-c) Representative fluorescence images show the localization and intensity of CENP-A and (a) CENP-C, (b) CENP-N, and (c) CENP-T at day 0 and 7 or 9 after Ad-Cre addition to inactivate the remaining CENP-A allele in CENP-A−/F cells. Bar graphs (the mean of four independent experiments; > 30 cells per experiment) show (red) CENP-A, (green) CENP-C, (orange) CENP-N, and (blue) CENP-T levels with days after Ad-Cre addition. Error bars represent the SEM. ACA staining was used to identify centromeres. CENP-N was tracked by covalent labeling with rhodamine-benzyl guanine after stable expression of a gene encoding SNAP-tagged CENP-N. (d-f) Kinetics of loss from centromeres as CENP-A levels diminish defines three groups with similar kinetics of loss: (d) proteins lost in proportion to CENP-A, (e) those lost more slowly and whose depletion requires almost complete loss of CENP-A, and (f) CENP-B, whose binding is reduced by half by complete loss of CENP-A. Statistics source data are in supplementary table S2. Scale bar = 5 μm.
A second pattern included: CENP-T, a component of the CENP-T/W/S/X complex and whose histone fold domains bind to chromatin adjacent to centromeric CENP-A nucleosomes40,41; CENP-I, which has been proposed to act in complex with CENP-H as an additional mediator of CENP-A deposition42; and Ndc80, a direct microtubule-binding component of the outer kinetochore43. Surprisingly, all three of these centromere/kinetochore proteins were almost fully maintained at centromeres (Fig. 2c,e and Supplementary Fig. S2b,c) until CENP-A levels dropped below ~1% of the initial level. Only after additional loss of CENP-A were these kinetochore components then rapidly lost (Fig. 2e).
A final group was represented by the direct DNA binding protein CENP-B that recognizes a 17 bp sequence (CENP-B box) within α-satellite DNA44. As expected, CENP-B binding at centromeres was not initially affected by reduction in CENP-A levels. Quite unexpectedly, however, CENP-B binding dropped by half as centromeric CENP-A was completely depleted (Fig. 2f), thereby identifying a previously unrecognized partial dependency on CENP-A for CENP-B binding at centromeres.
Overall, assessment of loss of kinetochore proteins as centromere-bound CENP-A was depleted identified that kinetochore assembly is not an all or nothing event and that a remarkably small amount of CENP-A is sufficient to nucleate assembly of a kinetochore with partial centromere function.
CENP-A’s CATD maintains centromere position and templates its replication
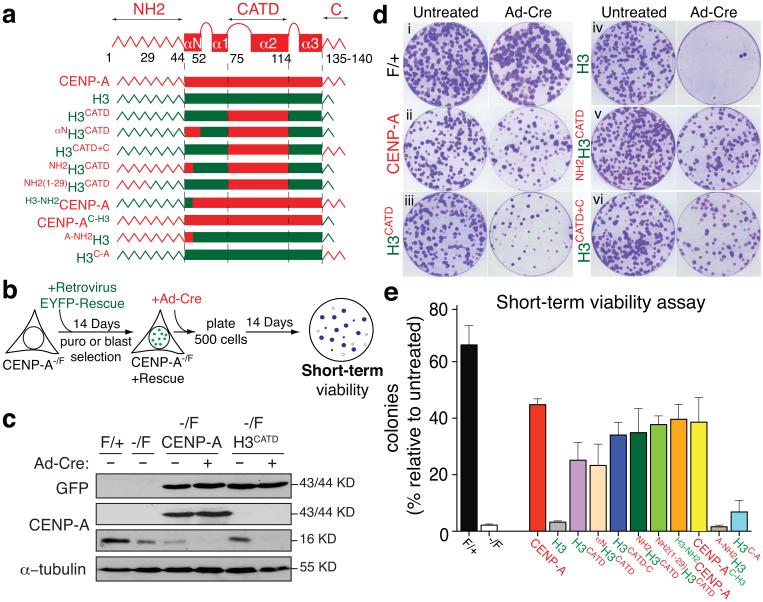
Next we tested whether CENP-A in centromeric chromatin can indefinitely maintain centromere identity by templating loading of new CENP-A onto replicated DNA of natural human centromeres and if so, the mechanism underlying its ability to do this. A series of histone H3 or CENP-A rescue variants were created with EYFP (enhanced yellow fluorescent protein) tags and stably expressed (by retroviral integration) in CENP-A−/F cells (Fig. 3a,b and Supplementary Fig. S3a,b). The remaining CENP-A allele was then inactivated by addition of Ad-Cre. As expected, full-length CENP-A accumulated to the endogenous CENP-A level (Fig. 3c) and was able to support cell viability [Fig. 3d(i-ii),e]. H3CATD rescued complete deletion of CENP-A with an efficiency 2/3 of CENP-A itself, while H3 alone conferred no survival [Fig. 3d(iii-iv),e]. H3CATD continued to be loaded at centromeres at a constant level even 5-6 generations after reduction of CENP-A to below 1% of its normal level (see Discussion) (Fig. 4a-c).
Figure 3.
Short-term rescues of centromere maintenance and function following CENP-A gene depletion. (a) Schematic representation of the rescue constructs. Each CENP-A domain is in red; H3 in green. CENP-A domains and amino acid positions are also indicated. Each construct is tagged with an amino-terminal EYFP (enhanced yellow fluorescent protein) (b) Schematic outlining the construction of cell lines expressing each rescues construct and final clonogenic assay. (c) Immuno-blots of cell extracts with antibodies to GFP, CENP-A and α-tubulin to determine the level of expression of the indicated rescue constructs. (d) Clonogenic survival experiment for the indicated cell lines with or without addition of Ad-Cre. (e) Quantitation of clonogenic assays for each rescue construct shown as the mean of the percentage of Ad-Cre-surviving colonies relative to the untreated condition. Each column represents the average of five independent experiments and error bars represent the SEM. Uncropped image of blots are shown in Supplementary fig. S7.
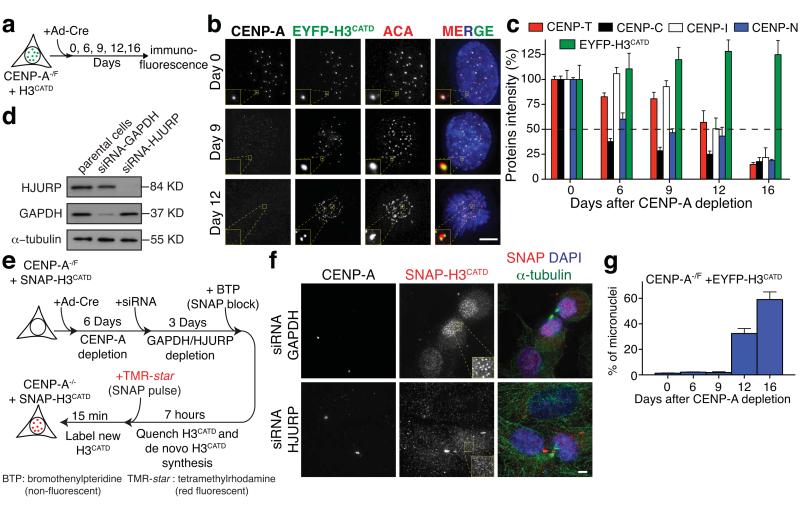
Figure 4.
The H3CATD is sufficient for centromere identity and to template its chromatin replication. (a) Representing schematic of experimental design in (b). (b) Representative fluorescence images of the localization and intensity of endogenous CENP-A and EYFP-H3CATD at day 0, 9 and 12 after Ad-Cre addition. ACA staining was used to identify centromeres. (c) Bar graphs indicate proteins intensity at centromeres of CENP-C, CENP-T, CENP-I, CENP-N and EYFP-H3CATD (in b) at the indicated days after Ad-Cre treatment. Columns represent the mean of five independent experiments (> 30 cells per experiment) and error bars represent the SEM. (d) HJURP and GAPDH levels in cells with or without siRNA treatment for GAPDH or HJURP, determined by immuno-blotting. α-tubulin was used as a loading control. (e) Representing schematic of experimental design in (f). (f) Representative fluorescence images from the experiment in (e) to determine the localization and intensity of CENP-A and SNAP-H3CATD after siRNA of GAPDH or HJURP. α-tubulin staining was used to identify telophase-early G1 cells. (g) Columns are the mean of the percentage of micronuclei formation in CENP-A−/F cells rescued with EYFP-H3CATD at the indicated days after Ad-Cre addition. Error bars represent the SEM of three independent experiments (> 100 cells per experiment). Statistics source data are in supplementary table S2. Uncropped image of blots are shown in Supplementary fig. S7. Scale bar = 5 μm.
To test if CATD-dependent centromere identity was maintained through replication of centromeric chromatin at mitotic exit12 and in an HJURP-dependent manner45,46 in the absence of endogenous CENP-A, a SNAP-tagged H3CATD construct was stably expressed in CENP-A−/F cells. In cells treated with a control siRNA-GAPDH, assembly of new SNAP-H3CATD molecules at centromeres was observed only at mitotic exit. This loading was completely dependent on HJURP, as its reduction abolished new SNAP-H3CATD assembly (Fig. 4d-f). Furthermore, in agreement with recent findings47, the continued HJURP-dependent loading of H3CATD missing Ser68 demonstrated that it is not necessary for CENP-A recognition and loading by HJURP, in contrast with a previous hypothesis48.
While cell cycle-dependent loading of new H3CATD at centromeres continued after CENP-A depletion, other centromeric proteins including CENP-I, CENP-N, CENP-C and CENP-T were inefficiently assembled at the CENP-A depleted H3CATD-containing centromeres, although all but CENP-C were maintained longer compared to cells with no rescue construct (Fig. 4c and Supplementary Fig. S3d). Failure to maintain or recruit kinetochore proteins caused a drastic increase in chromosome segregation defects (Fig. 4g) that prevented long-term survival (>100 generations) (Fig. 5a,b). Thus, substituting the CATD into histone H3 enables it to mediate epigenetic inheritance of centromere position through a mechanism requiring its cell cycle-dependent recruitment by the chaperone/loader HJURP, but H3CATD-containing centromeric chromatin is not sufficient to nucleate assembly of a functional kinetochore.
Figure 5.
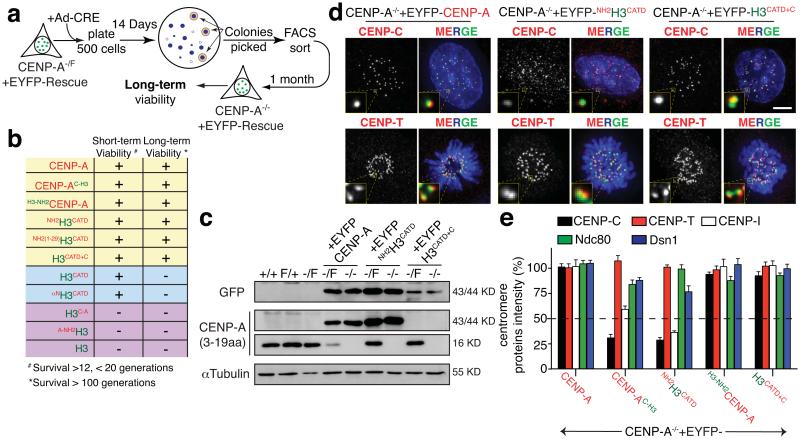
The carboxyl tail of CENP-A nucleates kinetochore assembly. (a) Schematic of the long-term viability assay used to isolate survival clones after endogenous CENP-A depletion. (b) Table of viability of various CENP-A-histone H3 chimera for conferring short (12 to 19 generations) and long term (>100 generations) survival. (c) Immuno-blot to quantify levels of each EYFP-tagged rescue variant or endogenous CENP-A, visualized with GFP or CENP-A antibodies. α-tubulin was used as a loading control. (d) Representative fluorescence images show the localization and intensity of CENP-C or CENP-T in the indicated cell lines. ACA was used to mark centromeres. (e) Column graphs quantifying centromere intensities from experiments like those in (d) to measure CENP-C, CENP-T, CENP-I, Ndc80 and Dsn1 protein intensity in the indicated cell lines. Columns represent the mean of three independent experiments (> 30 cells per experiment). Error bars represent the SEM. Uncropped image of blots are shown in Supplementary fig. S7. Scale bar = 5 μm.
Amino or carboxy tails of centromere-bound CENP-A enable long-term centromere rescue
We then investigated how CENP-A confers long-term centromere function. The αN helix of CENP-A has been shown to be important for contacting DNA as it exits a CENP-A-containing nucleosome19,49 and the six amino acid C-terminal tail of CENP-A can directly recruit CENP-C and drive aspects of kinetochore assembly in in vitro extracts29. We therefore created stable cell lines expressing the H3CATD with the addition of CENP-A’s αN helix (aNH3CATD), carboxy tail (H3CATD+C) or all or part of its amino tail (NH2H3CATD or NH2(1-29)H3CATD) (Fig. 3a and Supplementary Fig. S3a). Addition to H3CATD of either CENP-A tail rescued not only growth in the short-term assay - similar to the full-length CENP-A [Fig. 3d(v-vi),e and Supplementary Fig. S3c] – but also supported growth indefinitely (Fig. 5b), with only a mild slow growth phenotype relative to cells still containing the endogenous CENP-A (Supplementary Fig. S4a). Centromere function continued in the complete absence of CENP-A, as deletion of both endogenous CENP-A genes was confirmed by immuno-blotting, PCR and immunofluorescence (Fig. 5c and Supplementary Fig. S4b,c). As expected, loading of the rescue constructs occurred only after mitotic exit and was dependent on HJURP (Supplementary Fig. S4d-f). Redundancy of the amino- and carboxy-terminal tails of CENP-A in supporting long-term viability was further confirmed by full rescue with CENP-A constructs deleted in either domain (H3-NH2CENP-A and CENP-AC-H3) (Fig. 5b).
The CENP-A carboxy-terminal tail directs CENP-C binding, but is not essential for kinetochore assembly
Our findings suggested that full kinetochore assembly was nucleated by recruitment by either tail domain of one or more components. Indeed, consistent with direct CENP-C recruitment by the CENP-A carboxy-terminal tail29, centromere-associated CENP-C was reduced in cells lacking the CENP-A C-terminus (CENP-AC-H3 and NH2H3CATD) (Fig. 5d,e). Nevertheless, a fraction of CENP-C remained centromere-bound in cells rescued long-term in the absence of the carboxy tail, demonstrating the existence of CENP-A carboxy-terminal-independent loading of CENP-C, possibly through an existing CENP-C that templates new CENP-C loading. This remaining CENP-C was essential for centromere function, as depletion of it with siRNA produced catastrophic chromosome mis-segregation (Supplementary Fig. S4g). Similar reduction was also observed for CENP-I, suggesting its stabilization by CENP-C (Fig. 5e and Supplementary Fig. S5a). In any event, centromeres rescued with CENP-A variants missing its carboxy-terminal tail (CENP-AH3-C and NH2H3CATD) were sufficient for long-term kinetochore function, with normal assembly of associated centromere proteins thereby maintaining centromere function (Fig. 5d,e and Supplementary Fig. S5a).
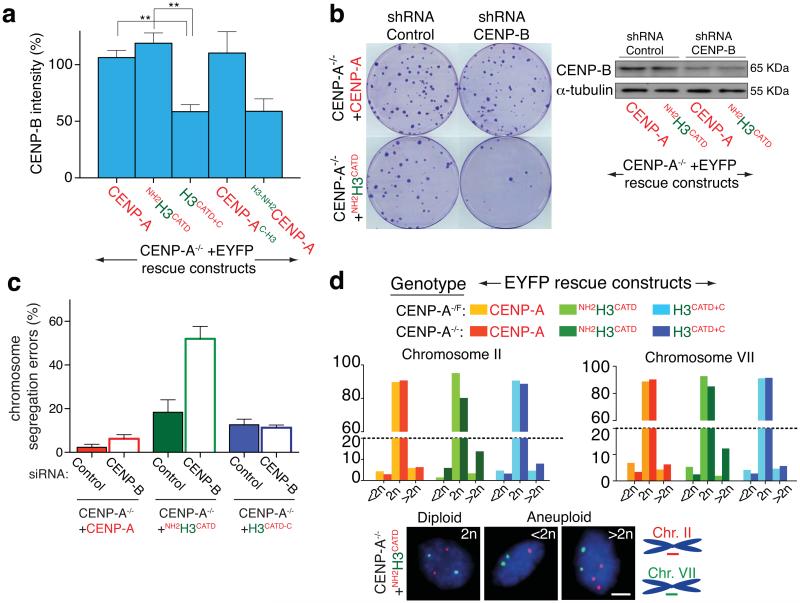
The CENP-A amino-terminal tail controls CENP-B levels at centromeres
CENP-C, CENP-T, CENP-N, Ndc80 and Dsn1 were fully maintained at centromeres following long-term rescue with each variant missing the amino-terminal CENP-A tail (H3-NH2CENP-A and H3CATD+C) (Fig. 5d,e). Surprisingly, however, the CENP-A amino-terminal tail was required for proper loading of the direct DNA binding protein CENP-B. Without that amino-terminal tail, only 50% of the normal centromeric amount of CENP-B was loaded at centromeres (Fig. 6a and Supplementary Fig. S5b). This CENP-A-dependent CENP-B binding was essential for centromere function. Indeed, stable reduction of total CENP-B by half in CENP-A deleted cells rescued by NH2H3CATD drastically increased chromosome mis-segregation and nearly eliminated colony survival (Fig. 6b,c and Supplementary Fig. S5c). Similar reduction in CENP-B did not affect rescue with full-length CENP-A, presumably because kinetochore functionality was still maintained through carboxy tail-mediated recruitment of CENP-C (Fig. 6b,c). Indeed, further reduction of CENP-B did not affect kinetochore function and survival of the H3CATD-C cells (Fig. 6c). This finding implicates CENP-B stabilization as one important component of kinetochore assembly nucleated in a CENP-A amino-terminal tail-dependent manner.
Figure 6.
The CENP-A amino terminal tail controls CENP-B levels at centromeres. (a) Column graphs represent the means of the percentage of CENP-B intensity in the indicated cell lines. Error bars represent the SEM of three independent experiments (> 30 cells per experiment. ** p < 0.006). (b) (left) Clonogenic survival assays after stable insertion to express a control shRNA (scramble) or against CENP-B. (Right) Immuno-blot of cell extracts to measure total CENP-B levels. α-tubulin was used as a loading control. (c) Percentage of cells that undergo chromosome mis-segregation after GAPDH or CENP-B depletion by siRNA in the indicated cell line. Column graphs represent the means of three independent experiments and error bars represent the SEM (d) Faithfulness of chromosome segregation after growth for 30 generations measured with Fluorescence In Situ Hybridization (FISH) for centromere regions of chromosomes 2 and 7 (CEN2 and CEN7, respectively) in the presence (CENP-A−/F) or absence (CENP-A−/−) of endogenous CENP-A. Bars represent the mean of duplicate experiments in the indicated cell line (> 100 cells per experiment). (Bottom) Diploid or aneuploid cells identified by localization of (red) CEN2 and (green) CEN7 after growth of the CENP-A−/−+NH2H3CATD cells. Statistics source data are in supplementary table S2. Uncropped image of blots are shown in Supplementary fig. S7. Scale bar = 5 μm.
Accurate chromosome segregation with centromere-targeted histone H3 carrying either CENP-A tail domain
The degree of complementation of normal chromosome segregation in cells deleted in CENP-A and rescued with CENP-A or H3CATD including either CENP-A tail domain was measured using fluorescence in situ hybridization (FISH) to visualize chromosomes 2 and 7. No increase in aneuploidy was observed in CENP-A or H3CATD-C rescued cells and only a minor increase was found in NH2H3CATD rescued cells (Fig. 6d). Direct visualization of chromosome segregation using time-lapse microscopy confirmed almost normal segregation in all rescued lines (Supplementary Fig. S5d,e). Overall, long-term, faithful centromere replication and kinetochore function was mediated by CATD-containing centromeric chromatin with either tails of CENP-A.
The principles of epigenetic centromere inheritance and function are conserved in fission yeast
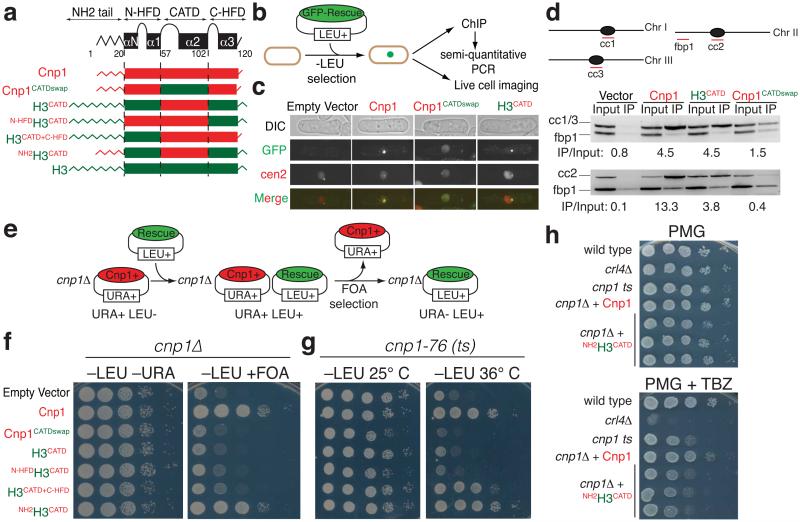
We next tested the generality of our findings by examining if the CATD and tail domains of CENP-A similarly specify centromere identity and kinetochore assembly in the fission yeast S. pombe, an organism whose epigenetically defined centromeres contain repeat motifs that are reminiscent of the repetitive arrays found at most higher eukaryotic centromeres50. Various histone H3-CENP-Acnp1 rescue genes were constructed in order to test the contribution of the fission yeast CATD [that lies within the CENP-ACnp1 histone fold domain (HFD)], the short CENP-ACnp1 N-terminal tail (19 aa) and the remaining regions of the CENP-ACnp1 HFD that were not included in the CATD [for simplicity we refer to these as N-HFD and C-HFD] (Fig. 7a). GFP-tagged chimeric CENP-ACnp1 variants were expressed in a yeast strain bearing a tetO array inserted at centromere 2, as well as expressing a tetR-Tomato fusion protein that allows visualization of centromere 2 (Fig. 7c, supplementary Table 1 and fig. S6). H3CATD co-localized with centromere, whereas a Cnp1 whose CATD was exchanged with the corresponding domain of histone H3 (Cnp1CATDswap) was found scattered throughout the nucleus (Fig. 7b,c). Chromatin immuno-precipitation (ChIP) confirmed enrichment of H3CATD, but not Cnp1CATDswap, specifically at centromeres relative to non-centromeric regions (Fig. 7b,d).
Figure 7.
The fission yeast CATD is necessary and sufficient for centromere identity, but requires addition of the CENP-ACnp1 amino-terminus to provide long-term centromere function. (a,b) Schematic of (a) the CENP-Acnp1-histone H3 gene constructs and (b) experimental approach used to test maintenance of centromere position and function in the fission yeast S. pombe. CENP-ACnp1 domains are red; H3 in green. (c) Representative live cell imaging of GFP-tagged constructs as outlined in (b). Positions of centromere of chromosome 2 was marked by binding of a tetR-tomato fusion protein to a tetO array inserted at the central core of cen263. (d) Chromatin immuno-precipitation using GFP antibody of cells from (b). Wild type cells were transformed with GFP-tagged constructs as indicated. Enrichment at centromeric central cores (cc1-3 and cc2 products) was compared with input DNA relative to the control non-centromeric locus fbp1. The depiction of fission yeast chromosomes is not drawn to scale. (e) Schematic of plasmid shuffling assay for testing CENP-ACnp1 rescue constructs in cnp1Δ cells. (f,g) Rescue experiment of cnp1Δ cells using the plasmid-shuffling assay outlined in (e) or of cnp1-76 cells. (h) Serial dilution of cnp1Δ cells containing NH2H3CATD or CENP-ACnp1 as the unique source of CENP-ACnp1. Growth was assayed in pombe minimal medium with glutamate (PMG) with or without the addition of spindle poison drugs thiabendazole (TBZ) (15µg/ml). Wild type, clr4Δ (heterochromatin defective mutant) and cnp1-ts controls are also shown. In (f, g, h), a ten-fold dilution series is shown for each strain.
The ability of the various Cnp1 rescue constructs to complement the complete absence of CENP-A was tested by employing a plasmid-shuffling assay (Fig. 7e). H3CATD was unable to confer cell viability, but addition of the Cnp1 amino-terminal tail to H3CATD did sustain long-term viability (Fig. 7f). Similar results were observed when the rescue constructs were then introduced in cells harboring a CENP-ACnp1 temperature-sensitive mutant allele (cnp1-76)51 as the unique source of CENP-A (Fig. 7g). There was no noticeable difference in growth rates among control cells expressing full-length Cnp1 or H3CATD with the Cnp1 amino tail. Only a slight sensitivity was seen for cells rescued with the latter construct and exposed to drugs that drive spindle disassembly, indicating that centromere function is largely restored (Fig. 7h).
In contrast with human, CENP-ACnp1 has only one amino acid carboxy-terminal extension beyond the end of the HFD52. The corresponding rescue construct (H3CATD+C-HFD) did not maintain growth in the complete absence of CENP-ACnp1, although it did restore growth of the cnp1-76 mutant at the restrictive temperature (Fig. 7f,g), probably facilitating the recruitment of thermo-sensitive cnp1-76 mutant to centromere that in turn may recruit kinetochore components through its amino-terminal tail. Thus, in fission yeast the CATD requires the addition of the Cnp1 amino tail to provide long-term viability, as in human cells.
Discussion
A long-standing question in chromosome inheritance is the epigenetic mark of centromere identity. By artificially targeting components to chromatin29-31,33, several groups have demonstrated that large artificial arrays of CENP-A-containing chromatin are sufficient to generate partially functional centromeres, but only after completely abrogating any epigenetic component. One earlier effort had used reduction in CENP-A level with siRNA27. Such approaches fail to test the epigenetic question as they are plagued by partial suppression that precludes testing epigenetic sufficiency. Indeed, we now demonstrate that as little as 1% of the original CENP-A level is sufficient for retention of at least partial centromere function and assembly of kinetochore proteins.
Our use of conditional gene inactivation in human cells and in fission yeast has overcome the prior technical limitations and has now established that the CATD of CENP-A when substituted into histone H3 can template its cell cycle-dependent, HJURP/Scm3-dependent centromeric loading at a constant level in the complete absence of CENP-A. Consideration of the number of molecules of CENP-A at the normal centromere offers strong support for this conclusion. The number of CENP-A molecules at the 40-500 kb chicken centromeres has been reported to be between 25-4053. Increasing that by 10 fold to account for the increased centromere size in humans yields a maximal estimate of ~250-400 CENP-A molecules per centromere, with a corresponding prediction of <1 molecule remaining per centromere within 9 divisions after CENP-A gene inactivation (400 molecules/centromere × 1/29 = 0.8 molecules/centromere). Therefore, our evidence demonstrates that H3CATD continues to be loaded at its initial level at each centromere for 4-5 generations after CENP-A level has fallen below ~1 molecule/centromere. It is important to note that initial H3CATD assembly occurs in the presence of endogenous CENP-A, therefore our evidence offers no insight into de novo centromere formation.
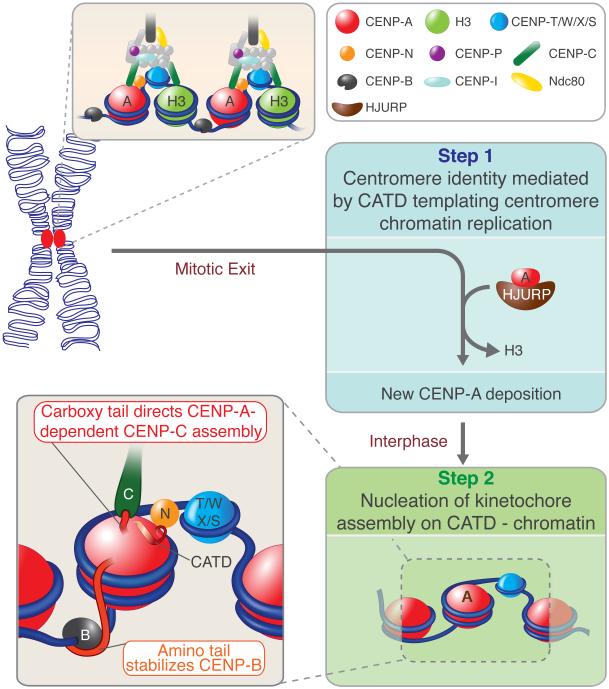
Despite its necessary and sufficiency for maintaining centromere identity in the absence of CENP-A, we have shown that H3CATD is not sufficient for long-term centromere function and cell viability because it does not nucleate kinetochore assembly. Rather, we have identified two redundant pathways that function to initiate kinetochore assembly onto an epigenetically defined chromatin core containing the CATD. Our evidence establishes that CENP-A-containing chromatin is the epigenetic mark that can identify, maintain and propagate human centromere function indefinitely through a conserved two-step mechanism in which it templates its own CATD-dependent replication and nucleates subsequent kinetochore assembly (Fig. 8). Furthermore, we demonstrate that the principles of epigenetic centromere inheritance and function are conserved from human to fission yeast.
Figure 8.
Model for centromeric identity, maintenance and function through a two-step mechanism. In the first step centromere identity is achieved by the CATD-containing chromatin through its loading mediated by HJURP at the mitotic exit. Subsequently, new and old CENP-A can nucleate kinetochore assembly on the CATD chromatin through the action of the CENP-A’s amino tail that stabilize CENP-B and by the direct recruitment of CENP-C by the carboxy tail of CENP-A.
The most plausible model is that the CATD establishes the epigenetic mark by physically modifying centromeric chromatin54 mediated by its interaction with histone H421 and by the exposure of the positively charged Loop1 of the CATD21,22. Conformationally constrained CENP-A-containing chromatin and the exposed Loop1 can then attract centromeric components like CENP-N or the nucleosome–like CENP-T/W/S/X complex. Indeed, recognition of these chromatin features offers an explanation for the slow loss of CENP-N or CENP-T from H3CATD–defined centromeres.
Further, earlier evidence had shown that the carboxy-terminal CENP-A tail can recruit CENP-C in vitro and that this is sufficient to initiate assembly of kinetochore components that are capable of microtubule capture29. To that earlier work, our evidence has established that the human CENP-A carboxy-tail not only recruits CENP-C, but also is sufficient to nucleate functional kinetochore assembly required for high fidelity chromosome segregation indefinitely when linked to a histone H3 variant with the CATD. Nevertheless, our evidence has also demonstrated that CENP-A rescue construct lacking the CENP-A carboxy tail still supports kinetochore assembly. Indeed, a fraction of CENP-C remains bound to centromeres even in absence of CENP-A’s carboxy-terminus, possibly through its proposed DNA binding domain or interaction with histone H3 or CENP-B55-57. This underscores that the C-terminus of CENP-A is not essential for centromere function as previously suggested29, although in its absence kinetochore function is slightly impaired, as observed by an increase in chromosome segregation errors and modestly increased aneuploidy.
Regarding a previously unrealized influence of CENP-B on kinetochore assembly, binding of CENP-B had previously been reported to affect the formation of heterochromatin58. Added to this, use of chromatin fibers and super-resolution microscopy had revealed CENP-A nucleosomes to be interspersed with chromatin containing H3K4me259 or H3K9me353 histone H3 modifications. Additionally, evidence for CENP-B binding to the CENP-B box was demonstrated to include an effect on positioning of CENP-A nucleosomes on alphoid DNA60,61. Our evidence has uncovered that at least a fraction of the centromeric CENP-B that is dependent on the first 29 amino acids of CENP-A amino-terminus is required for accurate chromosome segregation (in the absence of the parallel kinetochore formation pathway mediated by the CENP-A carboxy-terminal tail). This provides direct evidence in support of a role for the amino tail of CENP-A in centromere function and for CENP-B in kinetochore assembly.
Finally, we note that an essential impact of CENP-B on reinforcing CENP-A chromatin offers an explanation for the previous finding that the density of CENP-B boxes influences the assembly and maintenance of CENP-A chromatin in human cells62. Our findings, coupled with evidence that direct targeting of CENP-C to a specific DNA site is sufficient to recruit CENP-A to that site in chicken cells33 (but not in human cells32, at least when the endogenous centromere is still present), are consistent with epigenetic centromere identity which is stabilized by CENP-C and CENP-B.
Online methods
Cell culture
hTERT RPE-1 (ATCC) cells were maintained at 37°C in a 5% CO2 atmosphere with 21% oxygen. hTERT RPE-1 cells were maintained in DMEM:F12 medium containing 10% fetal bovine serum (Clontech), 0.348% sodium bicarbonate, 100 U/ml penicillin, 100 U/ml streptomycin and 2 mM L-glutamine. Monastarol was used at 100 μM for 10 hours.
CENP-A gene targeting
To generate the CENP-A−/F cell line, two rounds of gene targeting were performed. To generate the first conditional allele the left homology arm (encompassing a total of 1373 bp of the genomic CENP-A locus from 43 bp upstream of CENP-A exon 2 through 134 bp downstream of exon 4) was PCR amplified from BAC clone RP11-106G13, carrying the entire genomic CENP-A locus (Invitrogen) and cloned into the BamHI / SpeI sites of the Bluscript derivative pNY35. A loxP site was inserted 102bp upstream of exon 3 such that exon 3 and 4 were flanked between this loxP and a loxP present in pNY. The 1042bp right homology arm was amplified from the same BAC clone and runs from 351 bp upstream of exon 5 through 691 bp downstream of the beginning of exon 5 and was subcloned as an XhoI/KpnI fragment into the corresponding sites of pNY. The whole cassette encompassing the left arm, loxP, FRT, Neo cassette, FRT, right arm was cloned into the NotI sites of pAAV-LacZ, replacing the lacZ gene. The CENP-A knockout (KO) targeting construct was derived from this “Floxed” construct by transformation into Cre expressing E. coli, thereby resulting in site-specific removal of exons 3 and 4. A second “Floxed” allele was created by gene synthesis (GenScript) to create 9 polymorphic variants and inserted into the pBluescript derivative pNY using the same approaches as described above. The entire insert was then excised via NotI digestion and ligated to a pAAV vector backbone. Procedures for preparation of infectious AAV particles, transduction of hTERT-RPE1 cells and isolation of properly targeted clones was performed as described previously35.
Generation of stable cell lines, siRNA and SNAP-tag
The different trans-genes used in this study were introduced by retroviral delivery as described previously64. Stable integrates were selected in 5 μg/ml puromycin or 10 μg/ml blasticidin S and single clones isolated using fluorescence activated cell sorting (FACS Vantage; Becton Dickinson, Franklin Lakes, NJ). An RFP-STOP-GFP was integrated by lentiviral infection. siRNAs were introduced using Lipofectamine RNAiMax (Invitrogen). A pool of four siRNAs directed against CENP-C (GCGAAUAGAUUAUCAAGGA,GAACAGAAUCCAUCACAAA,CGAAGUUGAUAGAGGAUGA,UCAGGAGGAUUCGTGAUUA), CENP-B (CCAACAAGCUGUCUCCCUA,GGACAUCAAAG CUGAGUCA,GGAGGGUGAUGUUGAUAGU,GGCGGGAGUUCGAGGUCUU) or HJURP (targeting nucleotides 1135-1153, 1225-1243, 1815-1833 and 2017-2033 of the HJURP open reading frame) and a single siRNA directed against GAPDH (UGGUUUACAUGAUCCAAUA) were purchased from Dharmacon. shRNA against CENP-B and 2 negative controls (an empty and a non-effective 29-mer scramble) were obtained from OriGene. SNAP labeling was conducted as described previously12.
Clonogenic colony assay and Adeno-Cre treatment
Cells were plated in a 12-well plate at 4 × 104. The next day, cells were washed 3 times in DMEM:F12 medium containing 2% fetal bovine serum. Ad-Cre virus was added at MOI 250 in 400μl of DMEM:F12 medium containing 2% fetal bovine serum. After 3.5 hours, cells were washed 3 times with DMEM:F12 medium containing 10% fetal bovine serum. After 2 days, 500 cells were plated in triplicate on a 10cm2 dish. After 14 additional days, colonies were fixed 10 min in methanol and stained for 10 min using a crystal violet staining solution (1% Crystal violet, 20% EtOH). The percentage of clonogenic survival was determined by dividing the number of colonies formed in the Ad-Cre treated condition versus the untreated cells. For live cell microscopy, immuno-blot analysis or immuno-fluorescence staining, cells were plated at 4 × 105 on a 10cm2 dish.
Immuno-blotting
For immuno-blot analysis protein samples were separated by SDS-PAGE, transferred onto nitrocellulose membranes (BioRad) and then probed with DM1A (α-tubulin, 1:5000), CENP-A (Cell Signaling, 1:1000), GFP (Roche, 1:500), HJURP (Covance, 1:100046), CENP-B (Abcam and Upstate, 1:1000) or GAPDH (Abcam, 1:10000). Proteins for fission yeast were extracted with 0.7N NaOH solution and probed with GFP (Roche, 1:500) and β-actin (ACTB, proteintech, 1:4000).
Immuno-fluorescence and live-cell microscopy
Cells were fixed in 4% formaldehyde at room temperature or in methanol at 20°C for 10 min. Incubations with primary antibodies were conducted in blocking buffer for 1 hr at room temperature using the following antibodies: CENP-A (Abcam, 1:1500), CENP-T (1:5000), CENP-C (Covance, 1:1000), CENP-B (Abcam, 1:1000), ACA (Antibodies Inc, 1:500), Ndc80 (Abcam, 1:1000), CENP-I (a gift from Song-Tao Liu, University of Toledo, OH), Dsn1 (1:1000) and α-tubulin (1:2000). Immuno-fluorescence images were collected using a Deltavision Core system (Applied Precision). For quantification of centromere signal intensity, un-deconvolved 2D maximum intensity projections were saved as un-scaled 16-bit TIFF images and signal intensities determined using MetaMorph (Molecular Devices). A 15 × 15 pixel circle was drawn around a centromere (marked by ACA staining) and an identical circle drawn adjacent to the structure (background). The integrated signal intensity of each individual centromere was calculated by subtracting the fluorescence intensity of the background from the intensity of the adjacent centromere. ~20 centromeres were averaged to provide the average fluorescence intensity for each individual cell. Centromere signal intensity was also quantified using an automated system65. Aliquots treated with Ad-Cre were compared to untreated conditions (Day 0). For CENP-N and CENP-P –SNAP, untreated controls were processed at the same time of the Ad-Cre treatment (day 1 to day 9). For high-resolution spinning-disk fluorescence microscopy, cells were imaged using a 60× 1.4 NA PlanApochromat oil lens on a spinning disk confocal mounted on a Nikon TE2000-E inverted microscope equipped with a solid-state laser combiner (ALC) – 491 nm and 561 nm lines – a Yokogawa CSU10 head and a CCD Clara camera (Andor Technology). Acquisition parameters, shutters, and focus were controlled by iQ 1.10.0 software (Andor Technology). 5 × 2 mM z sections were acquired at 5 min time intervals for RFP and/or EYFP and maximum intensity projection created using MetaMorph. Movies were assembled and analyzed using QuickTimeTM (Apple).
FISH
FISH was performed as previously described28 with the following alterations: centromere specific probes for chromosome 2 and 7 were labeled with Tetramethyl-Rhodamine-5-dUTP and Fluorescein-12-dUTP (Roche, Indianapolis, IN), respectively and coverslips were washed in 2X SSC containing 60% formamide. More than 100 cells were analyzed for each data point.
Yeast strain and plasmid construction
Standard procedures were used for bacterial and fission yeast growth, genetics and manipulations66. CENP-Acnp1/H3 chimeric constructs were obtained by a combination of DNA synthesis (GenScript), PCR and subsequent subcloning into standard LEU2-marked pREP81 plasmids. The codon usage of all constructs was optimized to the initial CENP-Acnp1 gene codon usage (Leto software). S. pombe strains used in this study are described in Supplementary Table S1. Cells were grown at 30°C in all the experiments unless indicated otherwise. To create a cnp1? strain, a standard auxotrophic strain P013 was transformed with an ura4+-marked plasmid pREP82-cnp1+ thereby producing P162. P162 was subsequently transformed with a NAT cassette replacing the entire cnp1 ORF and thus obtaining a ura+ NATr strain, P164, which was checked by DNA blot and transformed with the aforementioned pREP81-based plasmids harboring chimeric constructs. Finally, transformants were employed in plasmid shuffling assays by plating serial dilution in PMG –LEU + FOA (containing 50 µg/ml uracil) and PMG –LEU –URA as control plates. For the cnp1-76 (ts) complementation assay, P482 strain was transformed with the mentioned pREP81-based plasmids. leu+ transformants were isolated and analyzed by serial dilution assays on PMG –LEU at 25°C and 36°C. In order to isolate cnp1? harboring pREP81-based plasmids as unique source providing CENP-ACnp1, FOA resistant colonies were picked directly from the plasmid shuffling assay plate and subjected to two rounds of streaking onto PMG –LEU + FOA. Loss of the ura+ marker (i.e., cnp1+ plasmid) was confirmed. Subsequently, novel isolated strains were assessed by dilution serial assays on PMG containing thiabendazole (TBZ) at 15 µg/ml 4 days at 30°C.
S. pombe live-imaging
Cells were grown overnight until logarithmic phase in minimal medium PMG –LEU at 30°C and then mounted in PMG 2% agarose as described. Cells were imaged on a spinning disk microscope with a 100X 1.4 NA PlanApochromat oil lens on a spinning disk confocal mounted on a Nikon TE2000-E inverted microscope equipped with a solid-state laser combiner (ALC) – 491 nm and 561 nm lines – a Yokogawa CSU10 head and a CCD Clara camera (Andor Technology). Fourteen 0.5-μm z sections were acquired and further image processing, including maximum intensity projections was performed using ImageJ (National Institutes of Health).
Chromatin immunoprecipitation (ChIP)
Cells were grown at 30°C in PMG –LEU. 1µl of anti-GFP (A11122, Invitrogen) was used for each IP. ChIP was performed as described67 with the following modifications. After incubating the samples (i.e., crude extracts and beads) with 250 µl of TES for 6h at 65°C, 220 µl of TE and 30µl of proteinase K (10 mg/ml) were added for subsequent overnight incubation at 37C. At the following day, 3 volumes of PB buffer (Qiagen) were added and samples were purified with Qiagen PCR purification kit as indicated by the manufacturer.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank P. Jallepalli (Sloan-Kettering, New York, NY) for helpful suggestions, J.F. Mata and M.C.C. Silva (Gulbenkian Institute, Oeiras, Portugal), T. Panchenko (University of Pennsylvania, Philadelphia) for technical help, D. Foltz (University of Virginia, Charlottesville, VA), A. Straight (Stamford university, CA), P. Maddox (Universite’ de Montreal, Canada), Song-Tao Liu (University of Toledo, OH), A. Miyawaki (RIKEN, Japan), Robin Allshire (WTCCB, Edinburgh), Yoshi Watanabe (University of Tokyo, Japan) and Oliver Limbo and Paul Russell (TSRI, La Jolla) for providing reagents. We also thank B.E. Black and C. Bartocci (TSRI, La Jolla) for helpful comments on the manuscript, Elena Khaliullina (elenprints@gmail.com) for drawing the model in Figure 8, the Neuroscience Microscopy Shared Facility (P30 NS047101, University of California, San Diego) and the FACS facility in the HESCCF (Sanford Consortium for Regenerative Medicine, La Jolla, CA). This work was supported by a grant (GM 074150) from the National Institutes of Health to D.W.C., who receives salary support from the Ludwig Institute for Cancer Research. D.F. was supported by a European Molecular Biology Organization (EMBO) long-term fellowship.
Literature Cited
- 1.Palmer, DK, O'Day, K, Wener, MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 3.Cleveland, DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 4.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 5.Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 6.du Sart D, et al. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 7.Ventura M, et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amor DJ, et al. Human centromere repositioning "in progress". Proc Natl Acad Sci U S A. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 10.Depinet TW, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 11.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 12.Jansen, LE, Black, BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuh, M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 14.Mellone BG, et al. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimitriadis, EK, Weber, C, Gill, RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci U S A. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krassovsky, K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci U S A. 2012;109:243–248. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuguchi, G, Xiao, H, Wisniewski, J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Conde e Silva N, et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 20.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekulic, N, Bassett, EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regnier V, et al. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, ST, Rattner, JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 28.Fujita Y, et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Guse, A, Carroll, CW, Moree, B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnhart MC, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendiburo, MJ, Padeken, J, Fulop, S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 32.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hori, T, Shang, WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berdougo, E, Terret ME, Jallepalli PV. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol Biol. 2009;545:21–37. doi: 10.1007/978-1-60327-993-2_2. [DOI] [PubMed] [Google Scholar]
- 36.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 37.Carroll, CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll, CW, Silva, MC, Godek, KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 40.Nishino T, et al. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 43.Cheeseman, IM, Chappie, JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Masumoto, H, Masukata, H, Muro, Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 46.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassett EA, et al. HJURP Uses Distinct CENP-A Surfaces to Recognize and to Stabilize CENP-A/Histone H4 for Centromere Assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panchenko T, et al. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci U S A. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castillo AG, et al. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007;3:e121. doi: 10.1371/journal.pgen.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torras-Llort, M, Moreno-Moreno O, Azorin F. Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J. 2009;28:2337–2348. doi: 10.1038/emboj.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ribeiro SA, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Black, BE, Brock, MA, Bedard, S, Woods, VL, Jr., Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trazzi S, et al. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 2009;4:e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugimoto, K, Yata, H, Muro Y, Himeno M. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J Biochem. 1994;116:877–881. doi: 10.1093/oxfordjournals.jbchem.a124610. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki N, et al. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J Biol Chem. 2004;279:5934–5946. doi: 10.1074/jbc.M306477200. [DOI] [PubMed] [Google Scholar]
- 58.Okada T, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hasson D, et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka Y, et al. Human centromere protein B induces translational positioning of nucleosomes on alpha-satellite sequences. J Biol Chem. 2005;280:41609–41618. doi: 10.1074/jbc.M509666200. [DOI] [PubMed] [Google Scholar]
- 62.Okamoto, Y, Nakano, M, Ohzeki, J, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakuno, T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- 64.Shah JV, et al. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 65.Bodor, DL, Rodriguez, MG, Moreno N, Jansen LE. Analysis of Protein Turnover by Quantitative SNAP-Based Pulse-Chase Imaging. Curr Protoc Cell Biol. 2012;8 doi: 10.1002/0471143030.cb0808s55. Chapter 8, Unit8. [DOI] [PubMed] [Google Scholar]
- 66.Moreno, S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 67.Folco, HD, Pidoux, AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.