Abstract
Nuclear factor erythroid-2 related factor 2 (Nrf2) is a master transcription factor that controls the basal and inducible expression of a battery of antioxidant genes and other cytoprotective phase II detoxifying enzymes. While knockout of Nrf2 exaggerates cardiac pathological remodeling and dysfunction in diverse pathological settings, pharmacological activation of Nrf2 protects against cardiomyocyte injury and cardiac dysfunction. In contrast, there is also a concern that the chronic activation of Nrf2 secondary to oxidative stress is a contributing mechanism for the reductive stress-mediated heart failure. However, a direct link between cardiac specific activation of Nrf2 and cardiac protection or dysfunction in vivo remains to be established. Therefore, we investigated the effect of cardiomyocyte-specific transgenic activation of Nrf2 (Nrf2ctg) on cardiac pathological remodeling and dysfunction. We found that the cardiomyocyte-specific activation of Nrf2 suppressed myocardial oxidative stress as well as cardiac apoptosis, fibrosis, hypertrophy, and dysfunction in a setting of sustained pressure overload induced by transverse aortic arch constriction (TAC) in mice. Notably, the constitutive activation of Nrf2 increased the steady level of autophagosomes while decreasing the ubiquitinated protein aggregates in the heart after TAC. Nrf2 gene gain- and loss-of-function approaches revealed that Nrf2 enhances autophagosome formation and autophagic flux in cardiomyocytes. Unexpectedly, while Nrf2 minimally regulated apoptosis, it suppressed significantly the proteotoxic necrosis in cardiomyocytes. In addition, Nrf2 attenuated the proteocytotoxicity presumably via enhancing autophagy-mediated clearance of ubiquitinated protein aggregates in cardiomyocytes. Taken together, we demonstrated for the first time that cardiac specific activation of Nrf2 suppresses cardiac maladaptive remodeling and dysfunction most likely by enhancing autophagic clearance of toxic protein aggregates in the heart.
Keywords: Nrf2, Cardiac dysfunction, Autophagy, Proteinopathy, Necrosis, Oxidative stress
1. Introduction
Heart failure is the consequence of sustained, abnormal neurohormonal and mechanical stress and remains a leading cause of death worldwide. Pathological stress, such as hypertension, results in cardiac hypertrophy, myocardial apoptosis and fibrosis, altered microvascular structure, and chamber dilation, culminating in cardiac dysfunction and heart failure [1]. However, the underlying mechanisms are far from clear.
Notably, a causative role of oxidative stress in the pathogenesis of cardiovascular disease has been established [2,3]. However, the antioxidant approaches of non-selective reactive oxygen species (ROS) scavenging for the treatment of cardiovascular disease are ineffective or even harmful [3]. Accordingly, an effective therapy may require more specific targeting of either the source of oxidative stress or the endogenous antioxidant defense system [3]. In this context, we have demonstrated that Nrf2, a master transcription factor in controlling the basal and inducible expression of a battery of antioxidant genes and other cytoprotective phase II detoxifying enzymes, is a negative regulator of cardiac pathological remodeling and dysfunction in diverse pathological settings [4–7]. While it has been documented that Nrf2 plays a mediator role in hydrogen sulfide-mediated cardioprotection [8], we and others have demonstrated that Nrf2 might be a drug target for the treatment of cardiomyocyte injury and cardiac dysfunction [4,7, 9–12]. Despite the prominent contribution of Nrf2 in cardiac protection, a direct link between cardiac specific activation of Nrf2 and cardiac protection in vivo remains to be established.
Recently, emerging evidence has indicated that reductive stress due to an increase in reduced glutathione may causally contribute to cardiomyopathy induced by protein aggregation [13]. In the setting of protein aggregation cardiomyopathy, there is a concern that the chronic activation of Nrf2 secondary to oxidative stress is a contributing mechanism for the reductive stress-mediated heart failure [14]. Although the ‘dark’ side of Nrf2 has not yet been demonstrated, Nrf2 appears to be involved in the regulation of protein aggregation in autophagic substrate selection [15] as well as the macroautophagy (commonly known as autophagy) per se [16–18]. Autophagy is an evolutionarily conserved process that mediates the lysosome-dependent turnover of macromolecules and entire organelles [19]. Importantly, autophagy and the ubiquitin proteasome system (UPS) are the major routes for the complete degradation/clearance of abnormal protein products in cells [20,21]. UPS is usually effective in clearing soluble misfolded or damaged proteins via ubiquitination of the target proteins where as autophagy is generally efficient in clearing less soluble or insoluble ubiquitinated protein aggregates. Upon a functional impairment of UPS, the ubiquitinated soluble abnormal proteins accumulate and aggregate into insoluble and toxic protein aggregates, and then autophagy is activated to be a major clearance route of the ubiquitinated proteins by default. Moreover, myocardial protein aggregation is often associated with cardiac proteasome insufficiency in various cardiomyopathies and heart failure [22,23] while the activation of autophagy may result in either protective or detrimental consequences in the heart [24,25]. These results raise an intriguing question whether Nrf2 plays a role in the control of protein quality via regulating autophagy in the heart thereby contributing to cardiac remodeling and heart failure.
Thus, we sought to investigate the impact of cardiac specific activation of Nrf2 on cardiac maladaptive remodeling and dysfunction and explore the possibility of a novel role for Nrf2 in regulating autophagy-mediated clearance of protein aggregates and preventing cardiac dysfunction. The results herein indicate that constitutive activation of Nrf2 facilitates autophagic clearance of protein aggregates in the heart thereby protecting against cardiac dysfunction. In addition, Nrf2 primarily suppresses necrosis rather than apoptosis in cardiomyocytes in a setting of toxic ubiquitinated protein overload via facilitating autophagic clearance of the toxic protein aggregates.
2. Methods
2.1. Animals
Transgenic mice with cardiomyocyte-specific overexpression of Nrf2 (Nrf2ctg) were generated in a FVB/NJ background using the transgene cassette containing a murine 5.5 kb alpha myosin heavy chain (α-MHC) promoter [26] (a gift from Dr. Jeffrey Robbins at Cincinnati Children’s Hospital Medical Center, Cincinnati, Oho) in the Medical University of South Carolina (MUSC) Transgenic Core. Nrf2 knockout (Nrf2−/−) mice were generated using heterozygote breeding pairs (Nrf2+/− in an ICR/Sv129 background) as previously described [4]. Animals were treated in compliance with the Guide for the Care and Use of Laboratory Animals (National Institution of Health), and all protocols were approved by the University of South Carolina Institutional Animal Care and Use Committee.
2.2. Transverse aortic arch constriction (TAC)
Male mice at 8–9 weeks of age were subjected to sham or TAC operations as described [4]. Mice were humanely euthanized after 4 weeks of TAC. Echocardiography, blood pressure measurement, pathological and biochemical analysis were performed as described [4]. Ubiquitinated protein aggregates were determined by filter trap assay as described [27].
2.3. Cell cultures, virus preparations, infection, autophagy flux, and cell death assay
Rat neonatal cardiomyocytes and mouse adult cardiomyocytes were isolated and cultured as described [4,28]. Rat cardiac myocyte-like H9C2 cells were purchased from ATCC. Virus preparation, stable infectants, autophagic flux assay, and cell death assay were performed as described in the Online Data Supplement.
2.4. Statistics
Data are shown as mean± SEM. Differences between 2 groups were evaluated for statistical significance with the Student t test when the sample size was appropriate and the population was distributed normally. When differences among >3 groups were evaluated, results were compared by ANOVA followed by Bonferroni test for multiple comparisons. Differences were considered significant at p < 0.05.
3. Results
3.1. Generation of Nrf2 cardiomyocyte-specific transgenic (Nrf2ctg) mice
Three founders of Nrf2ctg mice were identified by PCR (Supplementary Fig. 1A – C). Two lines of Nrf2ctg mice with relative high or modest overexpression of inserted human Nrf2 gene have been established by standard breeding (Supplementary Fig. 1D). Nrf2ctg mice with modest overexpression of hNrf2 were expanded. Upregulation of Nrf2 protein expression in the heart of Nrf2ctg mice was confirmed by Western blot (Supplementary Fig. 1E). These mice were viable and fertile, and had no observable health problems when housed in pathogen-free conditions. Adult Nrf2ctg mice did not develop any apparent cardiovascular system functional abnormalities (Supplementary Table 1 and Supplementary Table 2), suggesting that constitutive activation of Nrf2 in cardiomyocytes is not detrimental to the heart.
3.2. Cardiomyocyte-specific overexpression of Nrf2 suppresses cardiac maladaptive remodeling and dysfunction in response to chronic pressure overload
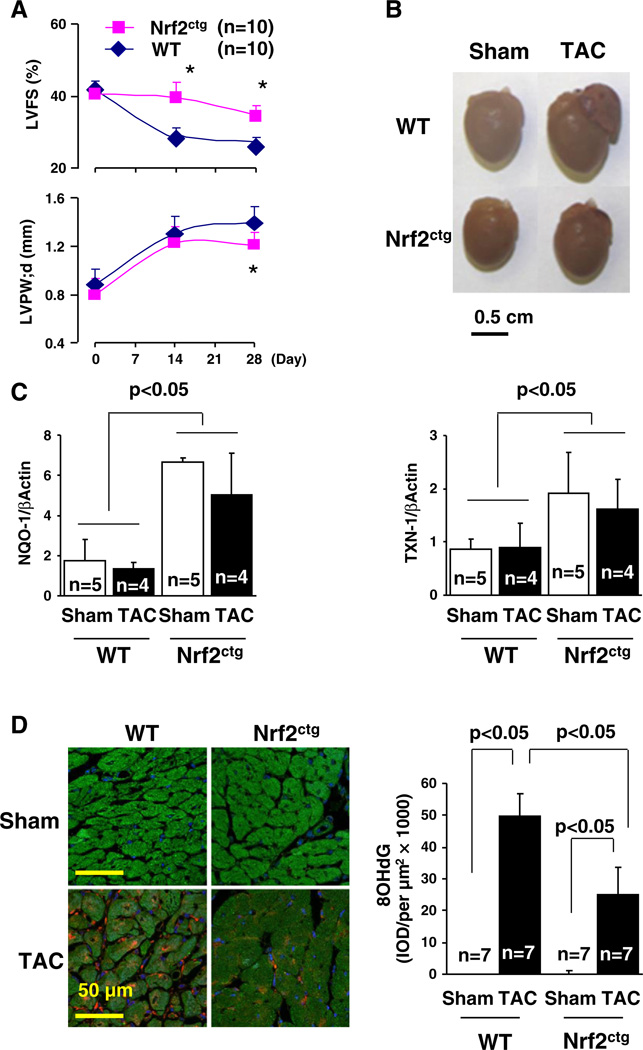
To determine the effect of cardiac specific activation of Nrf2 on cardiac dysfunction, we created TAC in littermates of male wild type (WT) and Nrf2ctg mice. We have previously demonstrated that the TAC-induced pressure overload initially results in an adaptive cardiac remodeling with preserved cardiac function (days 1–14) in mice [3]. However, the ability of the heart to compensate for this sustained elevation in stress is limited and, as a result, the remodeling becomes maladaptive and left ventricular dysfunction ensues (days 14–28) [3]. The decrease in fractional shortening FS (%) appears on day 14, and thereafter is escalated over time [3]. There is a ∼40% decrease in FS (%) in survived 4 week-TAC mice, which develops apparently myocardial oxidative stress, hypertrophy, fibrosis, and cell death [3]. Importantly, compared with TAC WT control, the decreases in FS (%) on day 14 and day 28 were less in TAC Nrf2ctg mice, demonstrating that chronic activation of Nrf2 in cardiomyocytes protects against TAC-induced cardiac dysfunction (Table 1 and Supplementary Table 3). WT mice after TAC developed myocardial oxidative stress, hypertrophy, fibrosis, and apoptosis (Fig. 1, Supplementary Figs. 2–4, and Table 1); however, the increased fibrosis in left ventricles (LVs) (25%) was excessively high compared with the previous findings [7,29]. Of interest, there was a substantial fibrosis (11%) in right ventricles (RAs) (Table 1 and Supplementary Fig. 3), which is usually around 2% in WT C57BL/6J mice 4 weeks after TAC [29,30]. A careful review of previous studies regarding TAC-induced pathological cardiac remodeling and heart failure revealed that the severity of TAC-induced cardiac dysfunction depends on the magnitude of stenosis in aortic arches [31]. Compared with using a 27-gauge needle, TAC-induced heart failure using a 28 or 30-gauge needle is more severe in age-matched C57BL/6J mice [31]. Moreover, TAC-induced severe heart failure with a >1.5-fold increase in lung weight but not moderate heart failure with a <1.5-fold increase in lung weight results in pulmonary hypertension as well as RV hypertrophy and fibrosis (10%) [32]. We noticed that male littermates of WT (FVB/N) and Nrf2ctg (FVB/N) mice are bigger than age-matched male WT (C57BL/6J) mice. Thus, we postulated that the magnitude of TAC-induced aortic stenosis in WT (FVB/N) mice using a 27-gauge needle was equivalent to that in age-match WT (C57BL/6J) mice using a 28 or 30-gauge needle. Indeed, we observed that TAC-induced cardiac dysfunction was associated with a 2.3-fold increase in lung weight in WT controls (Table 1). Accordingly, the excessively increased LV fibrosis and the RV fibrosis in TACWT controls are likely due to the more severe stenosis in aortic arches of those TAC WT controls. Nevertheless, the constitutive expression of Nrf2 specific in cardiomyocytes suppressed these TAC-induced adverse effects, that was associated with an upregulation of Nrf2 downstream antioxidant genes such as NAD(P)H:quinone oxidoreductase (NQO1) and thioredoxin-1 (TXN-1) [4], while it did not cause any functional or structural abnormalities in sham operated mice (Fig. 1, Supplementary Figs. 2 and 4, Table 1, and Supplementary Table 3). These results demonstrate that sustained activation of Nrf2 in cardiomyocytes is not detrimental but cardioprotective, providing direct evidence to support the existence of an axis of Nrf2-antioxidative stress-cardiac protection as described [4–6,8–12].
Table 1.
Echocardiography and pathology of WT and Nrf2ctg after TAC.
| (n) | WT |
Nrf2ctg |
||
|---|---|---|---|---|
| Sham | TAC | Sham | TAC | |
| Echocardiography | (11) | (10) | (11) | (10) |
| LVID;d (mm) | 3.83 ± 0.34 | 3.60 ± 0.32 | 3.57 ± 0.38 | 3.32 ± 0.20 |
| LVID;s (mm) | 2.18 ± 0.48 | 2.63 ± 0.30A | 2.17 ± 0.33 | 2.18 ± 0.20C |
| LVPW;d (mm) | 0.81 ± 0.13 | 1.37 ± 0.15A | 0.93 ± 0.08 | 1.20 ± 0.10BC |
| FS (%) | 42.90 ± 5.20 | 27.26 ± 2.79A | 39.25 ± 2.19 | 33.89 ± 3.02BC |
| EF (%) | 74.43 ±3.97 | 53.82 ± 7.38A | 70.68 ± 1.65 | 63.51 ± 8.79BC |
| Pathology | (11) | (10) | (11) | (10) |
| BW (g) | 29.75 ± 1.49 | 28.55 ± 0.89 | 26.15 ± 1.21 | 27.45 ± 0.75 |
| HW/Tibia (g/cm) | 0.12 ± 0.01 | 0.20 ± 0.03A | 0.10 ± 0.01 | 0.15 ± 0.02BC |
| LW/Tibia (g/cm) | 0.10 ± 0.01 | 0.23 ± 0.06A | 0.09 ± 0.01 | 0.11 ± 0.01C |
| MC CSA (µm2) | 298.4 ± 21.5 (7) | 524.0 ± 65.9A (7) | 264.9 ± 39.8 (7) | 441.7 ± 47.5BC (7) |
| Fibrosis (%area) | ||||
| LV | 0.80 ± 0.23 (7) | 25.43 ± 6.80A (7) | 0.45 ± 0.13 (7) | 10.86 ± 3.54BC (7) |
| IVS | 0.80 ± 0.20 (7) | 12.01 ± 2.77A (7) | 0.64 ± 0.15 (7) | 4.71 ± 1.47BC (7) |
| RV | 1.19 ± 0.42 (7) | 11.31 ± 4.76A (7) | 0.61 ± 0.34 (7) | 4.51 ± 2.41BC (7) |
| TUNEL (% cells) | 0.06 ± 0.01 (7) | 0.49 ± 0.07A (7) | 0.08 ± 0.03 (7) | 0.12 ± 0.03BC (7) |
| mRNAs | (5) | (4) | (5) | (4) |
| ANF | 1.03 ± 0.24 | 9.09 ± 2.90A | 0.97 ± 0.27 | 4.78 ± 0.41BC |
| BNP | 1.13 ± 0.28 | 3.16 ± 0.69A | 0.82 ± 0.23 | 1.78 ± 0.73BC |
| αMHC | 0.95 ±0.13 | 0.61 ± 0.08A | 0.99 ± 0.07 | 0.80 ± 0.08BC |
| βMHC | 1.00 ± 0.27 | 3.96 ± 0.09A | 0.92 ± 0.09 | 2.43 ± 0.33BC |
| SERCA | 1.07 ± 0.15 | 0.61 ± 0.12A | 1.13 ± 0.17 | 0.85 ± 0.09B |
Littermates of male WT and Nrf2ctg mice at ages of 8–10 weeks were subjected to sham and TAC operations. LVID;d, left ventricular internal dimension diastolic; LVID;s, left ventricular internal dimension systolic; LVPW;d, left ventricular posterior wall diastolic; FS, fractional shortening; BW, body weight; HW/BW, heart weight/body weight ratio; MCCSA, left ventricular myocyte cross-sectional area; Lung wt/BW, lung weight/body weight ratio; LV, left ventricle; IVS, interventricular septum; RV, right ventricle; ANF, atrial natriuretic factor; BNP, brain natriuretic factor; βMHC, beta-myosin heavy chain; αMHC, alphamyosin heavy chain; SERCA, sarcoplasmic reticulum calcium ATPase2a. Animal numbers for each group are indicated in the parentheses.
p < 0.05 vs WT sham;
p < 0.05 vs Nrf2ctg sham;
p < 0.05 vs WT TAC.
Fig. 1.
Cardiac hypertrophy and myocardial redox signaling in WT and Nrf2ctg mice after TAC. A, Time-course study of echocardiography in WT and Nrf2ctg mice after TAC. *, p < 0.05 vs. WT, n = 10. B. The representative pictures of hearts of WT and Nrf2ctg mice 4 weeks after TAC. C. qPCR analysis of NQO-1 and TXN-1 expression in the hearts of WT and Nrf2ctg mice 4 weeks after TAC. D. Staining of 8OHdG in the hearts of WT and Nrf2ctg mice 4 weeks after TAC.
3.3. Cardiomyocyte-specific overexpression of Nrf2 increases the steady level of autophagosomes and decreases the ubiquitinated protein aggregates in response to chronic pressure overloaded
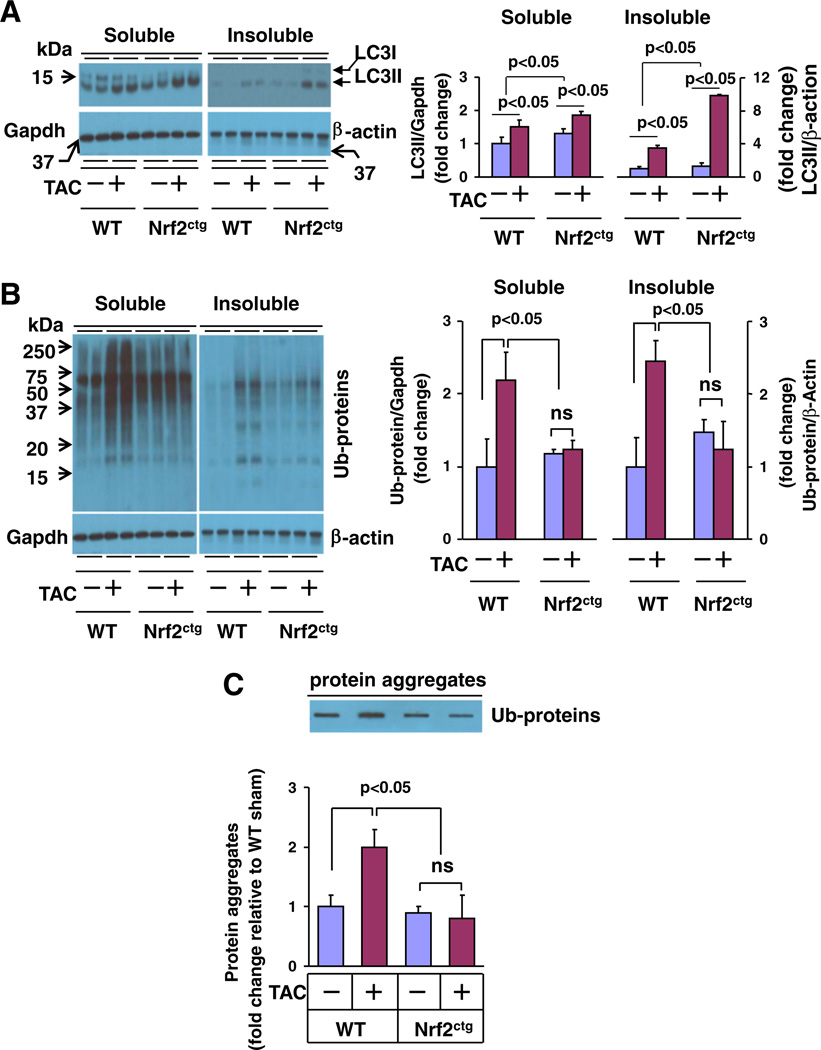
Given the dramatic myocardial protein aggregation in response to hemodynamic stress, pressure overload-induced heart disease has been included in the category of proteinopathies [33]. Previous reports on myocardial proteasomal activity in response to pressure overload are controversial. Either increased or decreased myocardial proteasome activities have been documented in murine TAC model [34,35]. However, given the observed TAC-induced myocardial accumulation of ubiquitinated proteins and protein aggregates [33], it is conceivable that TAC may induce functional insufficiency of myocardial UPS; in which autophagy is activated to be a major clearance route of the ubiquitinated proteins by default aforementioned [20,21]. In addition, since proteasome inhibition results in the activation of autophagy in cardiomyocytes [33], the observed cardioprotection of proteasome inhibitors in murine TAC model [34] may be at least partly achieved by their abilities to activate myocardial autophagy. On the other hand, cardiomyocyte autophagy is activated in response to a variety of pathological settings including chronic pressure overload and plays a critical role in the regulation of maladaptive cardiac remodeling and heart failure [33]. Considering the aforementioned emerging role of Nrf2 in the regulation of autophagy, we questioned whether Nrf2 plays a role in the control of protein quality via regulating autophagy in the heart thereby contributing to cardiac adverse remodeling and heart failure. The autophagy begins with formation of the autophagosome, a double-membrane structure of unknown origin that engulfs cytoplasmic contents, and then fuses it with a lysosome to form autolysosome (known as autophagosome mature), leading to proteolysis of engulfed materials [19]. During the autophagic response, LC3-I, an 16-kDa homologue of Atg8 in yeast, is processed and lipid conjugated, resulting in LC3-II, a 14-kDa active isoform that migrates from the cytoplasm to isolation membranes and autophagosomes [19]. The protein abundance of LC3-II usually reflects the steady level of autophagosomes, which is dependent on a balance between autophagosome synthesis and autophagosome clearance via lysosomes [19]. Thus, we examined the expression of LC3-I and -II, as well as the ubiquitinated proteins in both soluble and insoluble fractions of the hearts from WT and Nrf2ctg mice 4 weeks after TAC. TAC increased the levels of LC3-II and ubiquitinated proteins in both soluble and insoluble fractions, and augmented the amount of protein aggregates in the heart of WT mice (Fig. 2A – C), suggesting that sustained pressure overload results in the accumulation of ubiquitinated toxic protein aggregates as well as the activation of autophagy serving as feedback mechanism to clear the toxic protein aggregates in the heart as previously reported [33]. However, cardiomyocyte specific overexpression of Nrf2 dramatically suppressed the TAC-induced accumulation of ubiquitinated proteins and aggregates while enhancing the amount of LC3-II in both soluble and insoluble fractions in the heart (Figs. 2A – C). Importantly, the Nrf2-induced upregulation of LC3-II was more profound in the insoluble fraction (Fig. 2A). These results suggest that cardiac activation of Nrf2 suppresses the accumulation of toxic ubiquitinated protein aggregates via facilitating the autophagic clearance in the heart in response to hemodynamic stress.
Fig. 2.
The steady level of autophagosomes and protein aggregation in the heart of WT and Nrf2ctg mice 4 weeks after TAC. A, Western blot analysis of myocardial LC3-I & II in soluble and insoluble fractions. B, Western blot analysis of myocardial ubiquitinated proteins in soluble and insoluble fractions. C, Filter trap analysis of myocardial protein aggregates. Left ventricle (LV) tissues from 4 mice for each group were subjected to the analysis as indicated above. ns, non-significant.
3.4. Nrf2 enhances autophagosome formation and autophagic flux
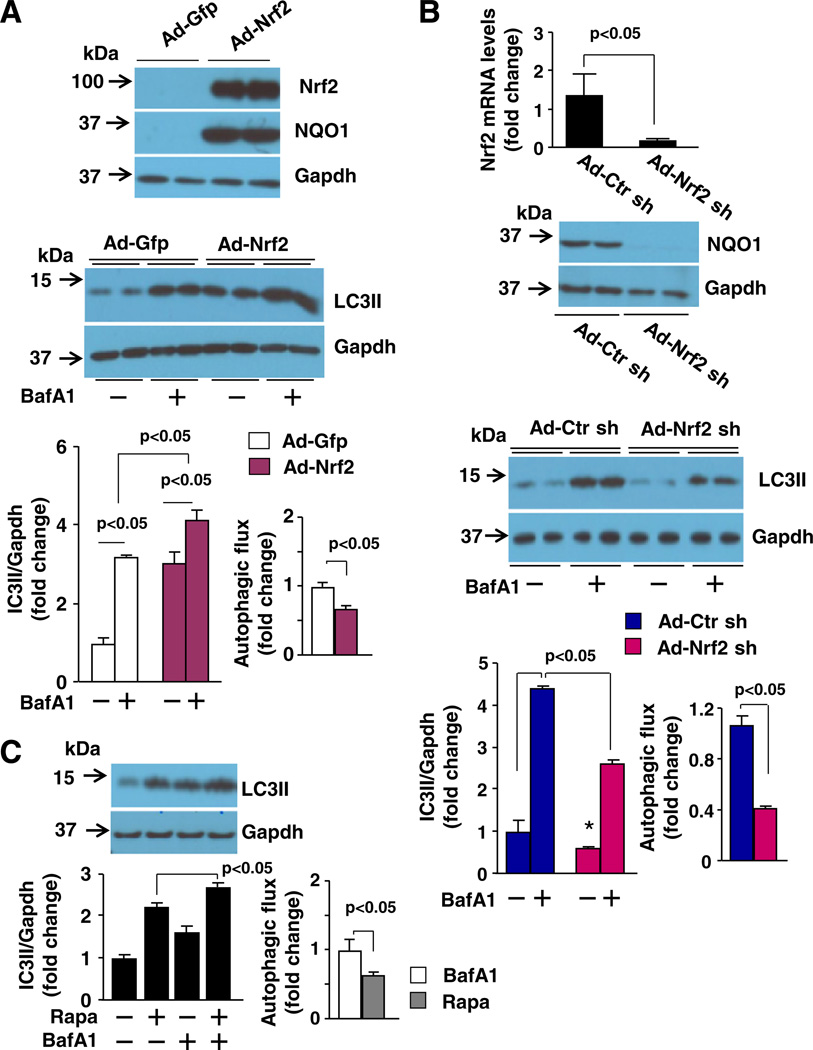
Considering the established role of autophagy in clearing protein aggregates and the increased steady level of autophagosomes that coincides with the enhanced clearance of protein aggregates in the heart with constitutively cardiomyocyte-specific activation of Nrf2, we hypothesized that Nrf2 is a critical mediator of autophagosome formation and autophagosome clearance in cardiomyocytes. Accordingly, we measured autophagic flux, a more accurate assessment of autophagic activity that indicates the capacity of autophagic clearance [19] in cardiomyocytes. In rat neonatal cardiomyocytes, Nrf2 overexpression clearly enhanced LC3-II expression as well as the accumulation of LC3-II by bafilomycin A1 (BafA1), an inhibitor of autophagosome–lysosome infusion (autophagic flux) (Fig. 3A). In contrast, overexpression of Nrf2 shRNA suppressed LC3-II expression and the accumulation of LC3-II by BafA1 (Fig. 3B). The efficacy of adenoviral overexpression of Nrf2 and Nrf2 shRNA was verified by Western blot and/or Q-PCR analysis of Nrf2 and its downstream gene expression in rat neonatal cardiomyocytes (Figs. 3A and B). Therefore, these results indicate that Nrf2 enhances autophagosome formation and autophagosome clearance via lysosomes, i.e., a sufficient autophagy, in cardiomyocytes. Interestingly, we noticed that compared with Ad-Gfp control, despite the increased total amount of LC3-II accumulation, the magnitude of LC3-II accumulation after BafA1 treatment is less in Nrf2 overexpressed cells (Fig. 3A), raising a concern whether the Nrf2-mediated upregulation of LC3-II levels is due to a Nrf2-dependent impairment of autophagic flux, rather than activation of sufficient autophagy. However, it is unlikely true since compared with Ad-scramble control, both the total amount of LC3-II and the magnitude of LC3-II accumulation after BafA1 treatment is less in Nrf2 silenced cells (Fig. 3B), supporting a Nrf2-mediated autophagosome formation and clearance but not an impairment of autophagosome clearance via lysosomes. Given the mechanisms of BafA1-induced impairment of LC3-II degradation differ at different times of its treatment; i.e., BafA1 has its major effects on inhibiting the degradation of LC3-II within existing autolysosomes at early time-point (∼2 h) while impairing the fusion of autophagosomes with lysosomes at later time points (after 6 h) [36], we determined the effects of BafA1 treatment for a time period of 4 h in cultured rat neonatal cardiomyocytes that were treated with or without rapamycin, an autophagy activator [19]. We observed an increase in the steady level of LC3-II by the treatment of rapamycin for 16 h, reflecting an increased autophagosome formation as described elsewhere [19] (Fig. 3C). The rapamycin-induced upregulation of LC3-II was further enhanced by an additional treatment of BafA1 for 4 h while the BafA1 treatment alone resulted in the accumulation of LC3-II (Fig. 3C). Importantly, compared with vehicle-treated cells, the net accumulation of LC3-II after BafA1 treatment is less in rapamycin-treated cells (Fig. 3C). It should be noted that a similar observation has been documented in cultured HL-1 cardiomyocytes [37]. Apparently, it is not true that rapamycin suppressed autophagic flux; instead, it indicates that BafA1 treatment for 4 h predominantly suppresses the degradation of LC3-II in existing autolysosomes in cardiomyocytes although inhibiting a small portion of autophagosome fusion with lysosomes cannot be excluded. Therefore, the total accumulation of LC3-II by BafA1 treatment for 4 h mainly indicates the amount of cellular autolysosomes, reflecting autophagic flux. Under a condition of increased autophagosome formation, such a rapamycin treatment or Nrf2 activation aforementioned, the conventional measurement of a net accumulation of LC3-II by BafA1 as autophagic flux may not precisely reflect the capacity of autophagosome clearance via lysosomes. Collectively, these results suggest that Nrf2 is a critical mediator for activating sufficient autophagy in cardiomyocytes.
Fig. 3.
The effects of Nrf2 and Nrf2 shRNA overexpression on autophagy flux in cardiomyocytes. A, and B, The effect of Nrf2 and Nrf2 shRNA overexpression on the steady levels of LC3-II and bafilomycin A1 (BafA1)-induced accumulation of LC3-II in rat neonatal cardiomyocytes. Upper panel: Efficacy of adenoviral overexpression of Nrf2 (A) and Nrf2 shRNA (B) in rat neonatal cardiomyocytes. The expression of Nrf2 and NQO1, a downstream target gene of Nrf2, inadenovirusofNrf2 (Ad-Nrf2) and Ad-Gfp controlor Ad-scramble control (Ad-Ctr sh) and Ad-Nrf2 sh infected cells was determined by Western blot and/or qPCR analysis. Middle panel: Representatives of Western blot for examining BafA1 (5 nmol/L, 4 h)-induced LC3-II accumulation in Ad-Gfp and Ad-Nrf2 or Ad-Ctr sh and Ad-Nrf2 sh infected cells. Lower panel: Semi-quantified Western blot analysis of BafA1 (5 nmol/L, 4 h)-induced LC3-II accumulation in Ad-Gfp and Ad-Nrf2 or Ad-Ctr sh and Ad-Nrf2 sh infected cells (n = 4). C, BafA1 (5 nmol/L, 4 h)-induced LC3-II accumulation in rat neonatal cardiomyocytes treated with or without rapamycin (Rapa, 0.5 µmol/L, 16 h). Upper panel: Representatives of Western blot analysis of LC3-II expression. Lower panel: Semi-quantified Western blot analysis of the BafA1-induced LC3-II accumulation (n = 4).
Because the LC3-II accumulation can be also caused by a decrease in lysosomes, we investigated the impact of Nrf2 overexpression and deficiency on lysosomal genesis in cardiomyocytes. Overexpression of Nrf2 or Nrf2 shRNA upregulated or downregulated LC3-II expression in H9C2 cardiomyocytes as observed in rat neonatal cardiomyocytes; however, the expression of 2 major lysosomal membrane sialoglycoproteins (LAMP-1 and LAMP-2) was hardly affected by overexpression or knock-down of Nrf2 (Supplementary Fig. 5). These results suggest that Nrf2 minimally regulates lysosomal genesis in cardiomyocytes. Overall, these results demonstrate that Nrf2 plays a critical role in activating sufficient autophagy via enhancing autophagosome synthesis and facilitating autolysosome-mediated autophagosome clearance in cardiomyocytes.
3.5. Nrf2 does not inhibit apoptosis but suppresses the proteotoxic necroptosis in cardiomyocytes via facilitating the clearance of ubiquitinated toxic proteins
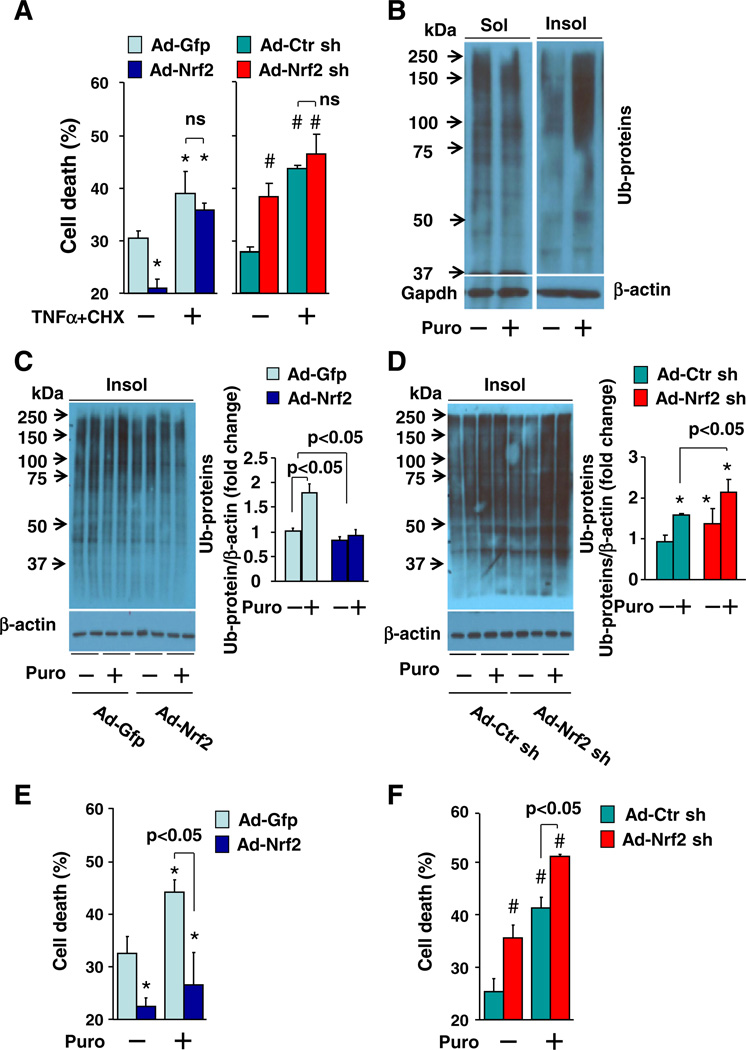
Previously, we affirmed that Nrf2 activation inhibits myocardial apoptosis in vivo [4,7]. To our surprise, however, forced activation of Nrf2 via adenoviral overexpression of Nrf2 in a primary culture of rat neonatal cardiomyocytes did not affect cell death induced by tumor necrosis factor alpha (TNFα) + cycloheximide (CHX), a typical combination to activate pro-apoptotic signaling [38]. However, Nrf2 activation did inhibit the basal level of cell death (Fig. 4A). Knock down of Nrf2 via adenoviral overexpression of Nrf2 shRNA enhanced the basal cell death without affecting TNFα + CHX-induced apoptosis in the cardiomyocytes (Fig. 4A). Similarly, adenoviral overexpression of Nrf2 or Nrf2 shRNA minimally affected cell death in H9C2 cardiomyocytes induced by several established pro-apoptotic stimuli including TNFα + CHX, staurosporine [39], tunicamycin, and thapsigargin [40, 41] (Supplementary Fig. 6). These results suggest that the observed Nrf2-suppressed cell death in cardiomyocytes in vitro is not likely apoptosis but other types of cell death, such as necrosis, which appear to be crucial for cardiac dysfunction secondary to myocardial infarction and pressure overload [42]. Since the terminal deoxynucleotidyl-transferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL) assay could not precisely discriminate apoptosis or necrosis in vivo [43,44], we postulated that the Nrf2 overexpression-inhibited TUNEL positive cells in vivo (Table 1 and Supplementary Fig. 4) are not apoptotic but necrotic cardiac cells.
Fig. 4.
Effect of Nrf2 overexpression and knockdown on apoptosis and proteotoxic necroptosis in cardiomyocytes. A, Effect of TNFα (10 ng/ml,24h) and CHX (10 µg/ml, 24 h)on cell death in rat neonatal cardiomyocytes. n = 4, *p < 0.05 vs. Ad-Gfp (–), #p < 0.05 vs Ad-Ctr sh (–). B, Western blot analysis of puromycin (Puro, 0.1 µmol/L, 6 h)-induced accumulation of ubiquitinated proteins in soluble (Sol) and insoluble (Insol) fractions in rat neonatal cardiomyocytes, (C), in insoluble fractions of rat neonatal cardiomyocytes infected with Ad-Gfp and Ad-Nrf2 or (D) infected with Ad-Ctr sh and Ad-Nrf2 sh. n = 4, *p < 0.05 vs. Ad-Ctr sh (–). E, Effect of puromycin (Puro, 0.1 µmol/L, 24 h) on cell death in rat neonatal cardiomyocytes infected with Ad-Gfp and Ad-Nrf2 or (F), infected with Ad-Ctr sh and Ad-Nrf2 sh. n = 4, *p < 0.05 vs Ad-Gfp (–), #p < 0.05 vs. Ad-Ctr sh (–).
To test this hypothesis, we determined a potential role of Nrf2 in regulating proteotoxic necrosis in cardiomyocytes because of the emerging role of protein aggregation in the pressure overload-induced cardiac dysfunction aforementioned. Proteotoxic necrosis in cardiomyocytes was induced by puromycin, an aminonucleoside antibiotic that causes premature chain termination during translation and promycylated nascent chain release to form insoluble ubiquitinated toxic protein aggregates leading to necrosis [45,46]. A careful dose–response analysis showed that puromycin-induced cell death appears at a dose of 0.2 µg/ml and becomes obvious at doses larger than 0.5 µg/ml in H9C2 cardiomyocytes (Supplementary Fig. 7). Puromycin at amaximumcytotoxic dose of 5 µg/ml induced accumulation of ubiquitinated proteins in insoluble fractions (protein aggregates) in H9C2 cardiomyocytes (Supplementary Fig. 7) as previously reported [46]. Of interest, the puromycin-induced accumulation of insoluble ubiquitinated protein aggregates and cell death were inhibited by the overexpression of Nrf2 whereas they were enhanced by the overexpression of Nrf2 shRNA (Supplementary Fig. 8). Importantly, we observed similar effects of overexpression of Nrf2 or Nrf2 shRNA on puromycin-induce accumulation of protein aggregates as well as cell death in primary cultures of rat neonatal cardiomyocytes (Figs. 4B – F). These results reveal that Nrf2 plays an inhibitor role in the regulation of proteotoxic cell death, most likely as necrosis in cardiomyocytes, via enhancing clearance of toxic ubiquitinated protein aggregates.
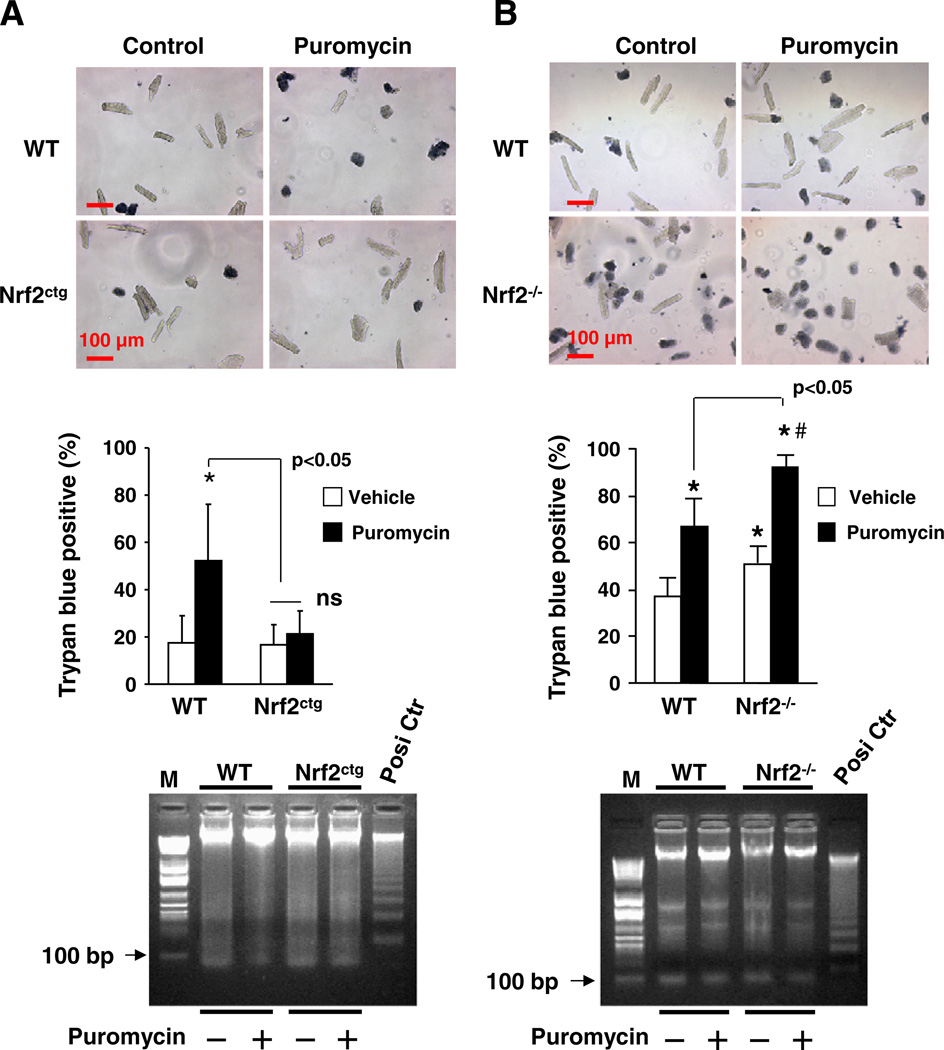
To further establish the inhibitory role of Nrf2 in regulating necrosis in cardiomyocytes, we examined the effect of puromycin on cell death in primary cultures of cardiomyocytes isolated from adult male WT, Nrf2ctg, and Nrf2−/− mice. Since most adult murine ventricular myocytes lost their dysfunction and even died after 24 h in culture, we adapted an improved method for isolation and culture of adult murine ventricular myocytes, which maintains ∼70% of the cultured cells alive for 24 h in culture [28]. Indeed, we observed that 20–40% of the cultured ventricular myocytes isolated from WT mice died within 24 h (Fig. 5). Because these dead cells were positive with trypan blue staining and negative with DNA ladder assessment, it is most likely that they die via necrosis (Fig. 5). Puromycin (5 µg/ml) enhanced the necrosis in adult cardiomyocyte of FVB/NJ and ICR/sv129 WT mice; however, the puromycin-induced cell death was suppressed or enhanced in adult cardiomyocytes of Nrf2ctg or Nrf2−/− mice, respectively (Fig. 5). These results strongly support an anti-necrotic effect of Nrf2 in the heart.
Fig. 5.
Effect of Nrf2 overexpression and knockout on proteotoxic necroptosis in adult cardiomyocytes. Adult cardiomyocytes that were isolated from male WT and Nrf2ctg mice (A) at ages of 12 weeks as well as male WT and Nrf2−/− mice (B) at age of 10 weeks were treated with puromycin (5 µg/ml) for 16 h and the subjected to trypan blue staining (upper panels) and DNA ladder assay (lower panels). n = 3, *, #p < 0.05 vs WT vehicle. M, DNA marker; Posi Ctr, positive control of DNA ladder.
3.6. Nrf2 inhibits the proteotoxic necrosis in cardiomyocytes via facilitating autophagic clearance of toxic ubiquitinated protein aggregates
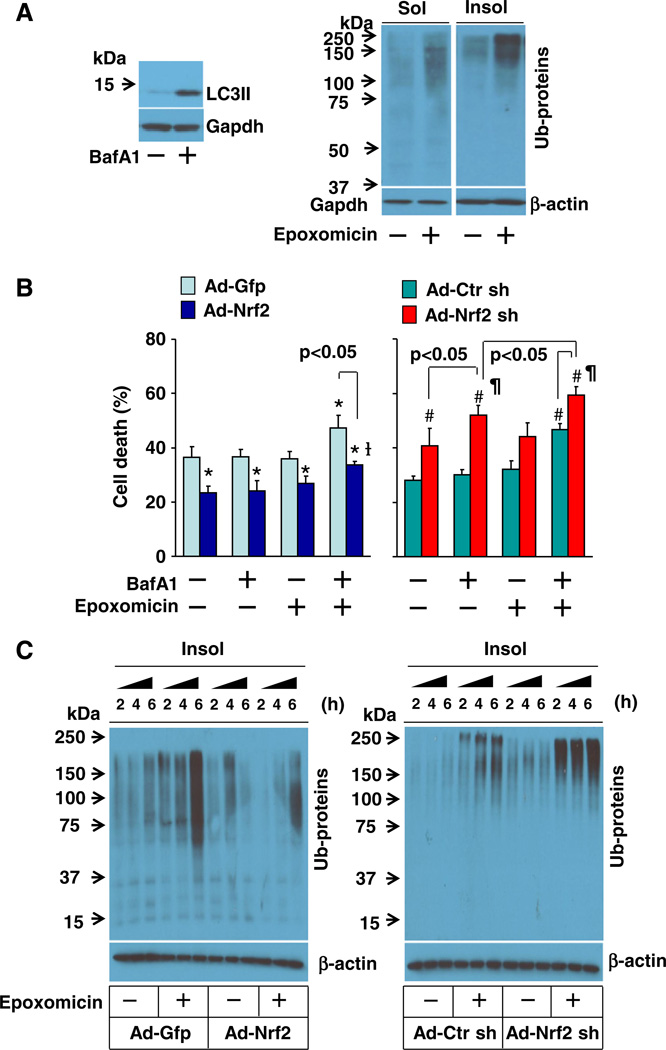
Because cytotoxic protein aggregates are cleared by both UPS and autophagy pathways [20,21], we next determined the contributions of UPS and autophagy to the Nrf2-mediated suppression of proteotoxicity in cardiomyocytes. Epoxomicin, a proteasome inhibitor, was used for suppressing UPS-mediated protein degradation, and the autophagy inhibitor BafA1 was utilized for inhibiting autophagy-mediated protein clearance. BafA1 dose-dependently induced cell death associated with accumulated LC3-II proteins (autophagosomes) in H9C2 cardiomyocytes, which appears at a dose of 2 nmol/L and becomes apparent at doses higher than 5 nmol/L (Supplementary Fig. 9A). Epoxomicin also dose-dependently induced cell death associated with accumulation of ubiquitinated proteins in insoluble fractions (toxic protein aggregates) in H9C2 cardiomyocytes, which appears at a dose of 10 nmol/L and becomes obvious at doses higher than 0.1 µmol/L (Supplementary Fig. 9B). The efficacy of epoxomicin or BafA1 on the accumulation of insoluble ubiquitinated proteins or LC3-II was further confirmed in primary culture of rat neonatal cardiomyocytes (Fig. 6A). Moreover, we carefully titrated sub-toxic doses of BafA1 and epoxomicin and observed a synergistic effect of BafA1 and epoxomicin on cell death in cultured rat neonatal cardiomyocytes (Fig. 6B), indicating a critical interplay of proteasome and autophagy pathways for suppressing proteotoxicity in cardiomyocytes. Overexpression of Nrf2 inhibited whereas overexpression of Nrf2 shRNA exaggerated the synergistic effect of BafA1 and epoxomicin on cardiomyocyte death, revealing an important role of Nrf2 in coordinating proteasome- and/or autophagy-mediated clearance of cytotoxic proteins in cardiomyocytes. Of significance, the sub-toxic dose of BafA1 rather than epoxomicin became toxic in the cardiomyocytes infected with adenovirus of Nrf2 shRNA (Fig. 6B), suggesting that Nrf2 may predominantly regulate autophagy-mediated clearance of cytotoxic proteins in cardiomyocytes. To further address this notion, we examined the effect of Nrf2 overexpression or Nrf2 knockdown on the clearance of ubiquitinated toxic proteins in cardiomyocytes in a setting of UPS impairment induced by epoxomicin at a toxic dose of 0.1 µmol/L, when autophagy is the major clearance route of the ubiquitinated proteins by default [20,21]. The epoxomicin-induced accumulation of ubiquitinated proteins in insoluble fractions was dramatically inhibited by the forced activation of Nrf2 whereas it was enhanced by the loss of Nrf2 function in cardiomyocytes (Fig. 6C), revealing a critical role of Nrf2 in facilitating autophagic clearance of ubiquitinated toxic protein aggregates in cardiomyocytes.
Fig. 6.
Role of autophagy in Nrf2-mediated suppression of cell death and ubiquitinated protein aggregates in cardiomyocytes. A, Left; Western blot analysis of BafA1 (5 nmol/L, 6 h)-induced accumulation of LC3-II in rat neonatal cardiomyocytes. Right; Western blot analysis of epoxomicin (0.1 µmol/L, 6 h)-induced accumulation of ubiquitinated proteins in soluble (Sol) and insoluble (Inso) fractions of rat neonatal cardiomyocytes. B, BafA1 (5 nmol/L, 24h)and epoxomicin (10 nmol/L,24 h)-induced cell death inrat neonatal cardiomyocytes infected with Ad-Gfp, Ad-Nrf2, Ad-Ctr sh, or Ad-Nrf2 sh. n = 4, *p < 0.05 vs Ad-Gfp (–), #p < 0.05 vs Ad-Ctr sh. C, Epoxomicin (0.1 µmol/L)-induced accumulation of ubiquitinated proteins in insoluble fraction in H9C2 cells infected with Ad-Gfp, Ad-Nrf2, Ad-Ctr sh, or Ad-Nrf2 sh.
4. Discussion
Herein, we have demonstrated directly that sustained activation of Nrf2 in cardiomyocytes suppresses cardiac maladaptive remodeling and dysfunction in response to sustained pressure overload. Mechanistically, Nrf2 does not directly inhibit apoptosis but suppresses the proteotoxic necrosis in cardiomyocytes presumably via an enhancement of the autophagic clearance of ubiquitinated toxic proteins. In addition, these novel findings may help explain the controversial observations regarding redox signaling in the regulation of autophagy and cellular defense in the heart.
A careful review of previous studies regarding the Nrf2-regulated cardiomyocyte death [4,12,47–49] revealed that the precise role of Nrf2 in the regulation of apoptotic death in cardiomyocytes is still elusive and a potential role of Nrf2 in regulation of other non-apoptotic death in cardiomyocytes remains to be explored. Previously, it appeared that a mediator role of Nrf2 in the suppression of apoptotic and nonapoptotic cardiomyocyte death had been firmly established. However, Prudom-Dickinson et al. demonstrated that overexpression of Nrf2 did not inhibit doxorubicin-induced increases in caspase 3 activity, Annexin V expression, and DNA degradation in rat neonatal cardiomyocytes ruling out the anti-apoptotic role of Nrf2 [49]. Intrigued by this lack of Nrf2-mediated suppression of apoptosis in cardiomyocytes in vitro, we applied Nrf2 gain- and loss-of-function approach in an H9C2 cardiomyocyte cell line, rat neonatal cardiomyocytes, and adult cardiomyocytes and demonstrated that Nrf2 preferentially regulates non-apoptotic cell death rather than apoptosis in cardiomyocytes. Importantly, we further revealed that the Nrf2-suppressed non-apoptotic cell death is most likely necrosis, especially in a setting of proteotoxic stress. Our results for the first time highlight that the Nrf2-mediated suppression of proteotoxic necrosis in cardiomyocytes may serve as a novel mechanism to protect the heart against cardiac pathological remodeling and dysfunction.
In view of the fact that, following a functional impairment of UPS, autophagy becomes a major clearance route of the ubiquitinated proteins by default [20,21] and that myocardial protein aggregation is often associated with cardiac proteasome insufficiency in various cardiomyopathies and heart failure [22,23], it is highly conceivable that activation of autophagy is cardioprotective. However, paradoxically, the activation of myocardial autophagy could be either detrimental or protective [24, 25]. Although the underlying molecular mechanisms are poorly understood, a plausible explanation is that the biological consequences of autophagy activation are dependent on the functional integrity of autophagic activity. That is, increased autophagosome formation without sufficient fusion with lysosomes and/or the subsequent lysosomemediated clearance of autophagosomes (i.e., an insufficient or frustrated activation of myocardial autophagy) is functionally detrimental to the heart. On the other hand, increased autophagosome formation with sufficient fusion with lysosomes and the subsequent lysosome-mediated clearance of autophagosomes (i.e., a sufficient activation of myocardial autophagy) is functionally integrated and therefore cardiac protective. The critical regulators of sufficient or frustrated activation of autophagy in the heart remain unknown. However, our previous observations that Nrf2 deficiency coincidences with an earlier onset of cardiac maladaptive remodeling and the subsequent development of heart failure [4,6,7], together with the present findings that a sustained activation of Nrf2 in the heart is cardiac protective via its ability to enhance autophagic clearance of toxic protein aggregates in response to pressure overload, identify Nrf2 to be a crucial regulator for the sufficient activation of autophagy in the heart. Since the pressure overload-induced cardiac dysfunction has been situated in the category of proteinopathies [33], our findings have clarified that a sustained activation of Nrf2 in the heart is not likely harmful as previously hypothesized [14], but rather beneficial even in the setting of protein aggregation cardiomyopathy.
There is a sophisticated interaction between oxidative stress and autophagy in the heart [50]. Given a mediator role of ROS in the induction of autophagy, the overproduced ROS could activate autophagy to serve as a predominantly prosurvival mechanism in a setting of oxidative stress; however, the autophagy machinery might also be destructive thereby exaggerating oxidative stress when it is induced rigorously but insufficiently. Indeed, ROS-mediated autophagy has been shown to lead to either survival or death in cardiac cells depending on stimuli and cell types [51–53]. The precise mechanisms for these discrepancies are unknown. Considering the major role of autophagy and Nrf2 in cellular defense as well as the Nrf2-mediated suppression of ROS formation and the Nrf2-mediated autophagic clearance of ubiquitinated toxic protein aggregates, we postulate that Nrf2 enhances autophagy-mediated removal of the damaged proteins or organelles caused by oxidative stress via a mechanism independent of ROS formation thereby preserving ROS-mediated activation of autophagy in the heart. In particular, the observed normal or reserved cardiac function of mice with constitutive activation of Nrf2 specific in cardiomyocytes under a normal physiological state or sustained pressure overload suggests that activation of Nrf2 may preserve the generation of ROS for maintaining normal physiological function while selectively shutting down the source of ROS for oxidative stress in the stressed heart. Accordingly, we interpret the increased steady level of autophagosomes due to Nrf2 deficiency [16,18] to be caused by an impairment of autophagosome clearance rather than an enhancement of ROS-mediated autophagosome formation, thereby leading to an insufficient autophagic activity. In fact, this notion has been partly strengthened by the observed Nrf2-mediated autophagic clearance of ubiquitinated proteins secondary to ROS formation in macrophages [17].
It should note that the ROS independent mechanism by which Nrf2 activates autophagy in the heart remains unknown. We strived to uncover the downstream targets of Nrf2 to activate autophagy by qPCR array analysis of 80 autophagy related genes in the heart utilizing mRNAs isolated from left ventricles of littermates of WT, Nrf2−/− and Nrf2ctg mice. However, we did not observe any dramatically and reciprocally regulated genes in the hearts of Nrf2−/− mice vs. Nrf2ctg mice (data not shown). Subsequent Western blot analysis of autophagy related genes such as Beclin-1 was consistent with the qPCR array results. These results suggest that Nrf2 activates autophagy most likely via a mechanism independent of these autophagy related genes in cardiomyocytes. In addition, Nrf2 has been shown to enhance proteasomal activity via increasing expression of several 20S subunits in response to dietary Nrf2 activators or oxidative stress [54]. However, whether Nrf2 regulates proteasomal activities in the heart remain to be explored. Further investigation of the mechanisms by which Nrf2 regulates autophagy and proteasomal activities in the heart may provide valuable insights for a better understanding of redox signaling in the control of myocardial autophagy as well as suggest potential therapeutic interventions to restore Nrf2-mediated redox homeostasis in cardiac disease.
Supplementary Material
Acknowledgments
We are grateful for the expertise and reagents to generate Nrf2ctg mice provided by Dr. Yibin Wang at David Geffen School of Medicine at UCLA and Dr. Michael J Kern at the Medical University of South Carolina, Charleston.
Source of funding
This work was supported by the National Institute of Health KO1 grant 5KO1DK080884-05, the Shandong University National Qianren Scholar Fund, the Taishan Scholar Fund, and the National Natural Science Foundation of China (No. 81370267).
Abbreviations
- ANF
atrial natriuretic factor
- Ang II
angiotensin II
- αMHC
alpha-myosin heavy chain
- βMHC
beta-myosin heavy chain
- BNP
brain natriuretic factor
- BafA1
bafilomycin A1
- LC3
microtubule-associated protein 1 light chain 3
- NQO1
NAD(P)H:quinone oxidoreductase
- Nrf2
nuclear factor erythroid-2 related factor 2
- ROS
reactive oxygen species
- SERCA
sarcoplasmic reticulum calcium ATPase2a
- TAC
transverse aortic arch constriction
- TXN-1
thioredoxin-1
- UPS
ubiquitin proteasome system
Footnotes
Disclosure statement
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dxx.doi.org/10.1016/j.yjmcc.2014.04.006.
Contributor Information
Dongqi Tang, Email: Dongqi.Tang@uscmed.sc.edu.
Taixing Cui, Email: Taixing.Cui@uscmed.sc.edu.
References
- 1.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 2.Takimoto E, Kass DA. Role of oxidative stress in cardiac hypertrophy and remodeling. Hypertension. 2007;49:241–248. doi: 10.1161/01.HYP.0000254415.31362.a7. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Ichikawa T, Janicki JS, Cui T. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin Ther Targets. 2009;13:785–794. doi: 10.1517/14728220903025762. [DOI] [PubMed] [Google Scholar]
- 4.Ichikawa T, Li J, Meyer CJ, Janicki JS, Hannink M, Cui T. Dihydro-CDDO-trifluoroethyl amide (dh404), a novel Nrf2 activator, suppresses oxidative stress in cardiomyocytes. PLoS One. 2009;4:e8391. doi: 10.1371/journal.pone.0008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Zhang C, Xing Y, Janicki JS, Yamamoto M, Wang XL, et al. Up-regulation of p27(kip1) contributes to Nrf2-mediated protection against angiotensin II-induced cardiac hypertrophy. Cardiovasc Res. 2011;90:315–324. doi: 10.1093/cvr/cvr010. [DOI] [PubMed] [Google Scholar]
- 6.Tan Y, Ichikawa T, Li J, Si Q, Yang H, Chen X, et al. Diabetic downregulation of Nrf2 activity via ERK contributes to oxidative stress-induced insulin resistance in cardiac cells in vitro and in vivo. Diabetes. 2011;60:625–633. doi: 10.2337/db10-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing Y, Niu T, Wang W, Li J, Li S, Janicki JS, et al. Triterpenoid dihydro-CDDO-trifluoroethyl amide protects against maladaptive cardiac remodeling and dysfunction in mice: a critical role of Nrf2. PLoS One. 2012;7:e44899. doi: 10.1371/journal.pone.0044899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvert JW, Elston M, Nicholson CK, Gundewar S, Jha S, Elrod JW, et al. Genetic and pharmacologic hydrogen sulfide therapy attenuates ischemia-induced heart failure in mice. Circulation. 2010;122:11–19. doi: 10.1161/CIRCULATIONAHA.109.920991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Sano M, Shinmura K, Tamaki K, Katsumata Y, Matsuhashi T, et al. 4-Hydroxy-2-nonenal protects against cardiac ischemia-reperfusion injury via the Nrf2-dependent pathway. J Mol Cell Cardiol. 2010;49:576–586. doi: 10.1016/j.yjmcc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, et al. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci U S A. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piao CS, Gao S, Lee GH, Kim do S, Park BH, Chae SW, et al. Sulforaphane protects ischemic injury of hearts through antioxidant pathway and mitochondrial K(ATP) channels. Pharmacol Res. 2010;61:342–348. doi: 10.1016/j.phrs.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Ichikawa T, Jin Y, Hofseth LJ, Nagarkatti P, Nagarkatti M, et al. An essential role of Nrf2 in American ginseng-mediated anti-oxidative actions in cardiomyocytes. J Ethnopharmacol. 2010;130:222–230. doi: 10.1016/j.jep.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajasekaran NS, Varadharaj S, Khanderao GD, Davidson CJ, Kannan S, Firpo MA, et al. Sustained activation of nuclear erythroid 2-related factor 2/antioxidant response element signaling promotes reductive stress in the human mutant protein aggregation cardiomyopathy in mice. Antioxid Redox Signal. 2011;14:957–971. doi: 10.1089/ars.2010.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao VA, Klein SR, Bonar SJ, Zielonka J, Mizuno N, Dickey JS, et al. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem. 2010;285:34447–34459. doi: 10.1074/jbc.M110.133579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011;108:1427–1432. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattart L, Calay D, Simon D, Roebroek L, Caesens-Koenig L, Van Steenbrugge M, et al. The peroxynitrite donor 3-morpholinosydnonimine activates Nrf2 and the UPR leading to a cytoprotective response in endothelial cells. Cell Signal. 2012;24:199–213. doi: 10.1016/j.cellsig.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurode-generation. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 21.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res. 2010;85:253–262. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res. 2010;106:463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottlieb RA, Mentzer RM. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu Rev Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZV, Rothermel BA, Hill JA. Autophagy in hypertensive heart disease. J Biol Chem. 2010;285:8509–8514. doi: 10.1074/jbc.R109.025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 27.Menzies FM, Hourez R, Imarisio S, Raspe M, Sadiq O, Chandraratna D, et al. Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum Mol Genet. 2010;19:4573–4586. doi: 10.1093/hmg/ddq385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou YY, Wang SQ, Zhu WZ, Chruscinski A, Kobilka BK, Ziman B, et al. Culture and adenoviral infection of adult mouse cardiac myocytes: methods for cellular genetic physiology. Am J Physiol Heart Circ Physiol. 2000;279:H429–H436. doi: 10.1152/ajpheart.2000.279.1.H429. [DOI] [PubMed] [Google Scholar]
- 29.O’Connell TD, Ishizaka S, Nakamura A, Swigart PM, Rodrigo MC, Simpson GL, et al. The alpha(1A/C)- and alpha(1B)-adrenergic receptors are required for physiological cardiac hypertrophy in the double-knockout mouse. J Clin Invest. 2003;111:1783–1791. doi: 10.1172/JCI16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Y, Lee K, Li N, Corbett D, Mendoza L, Frangogiannis NG. Characterization of the inflammatory and fibrotic response in a mouse model of cardiac pressure overload. Histochem Cell Biol. 2009;131:471–481. doi: 10.1007/s00418-008-0541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suryakumar G, Kasiganesan H, Balasubramanian S, Kuppuswamy D. Lack of beta3 integrin signaling contributes to calpain-mediated myocardial cell loss in pressure-overloaded myocardium. J Cardiovasc Pharmacol. 2010;55:567–573. doi: 10.1097/FJC.0b013e3181d9f5d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Guo H, Xu D, Xu X, Wang H, Hu X, et al. Left ventricular failure produces profound lung remodeling and pulmonary hypertension in mice: heart failure causes severe lung disease. Hypertension. 2012;59:1170–1178. doi: 10.1161/HYPERTENSIONAHA.111.186072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 35.Tsukamoto O, Minamino T, Okada K, Shintani Y, Takashima S, Kato H, et al. Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Commun. 2006;340:1125–1133. doi: 10.1016/j.bbrc.2005.12.120. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 37.Iwai-Kanai E, Yuan H, Huang C, Sayen MR, Perry-Garza CN, Kim L, et al. A method to measure cardiac autophagic flux in vivo. Autophagy. 2008;4:322–329. doi: 10.4161/auto.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Moissac D, Mustapha S, Greenberg AH, Kirshenbaum LA. Bcl-2 activates the transcription factor NFkappaB through the degradation of the cytoplasmic inhibitor IkappaBalpha. J Biol Chem. 1998;273:23946–23951. doi: 10.1074/jbc.273.37.23946. [DOI] [PubMed] [Google Scholar]
- 39.Ekhterae D, Platoshyn O, Zhang S, Remillard CV, Yuan JX. Apoptosis repressor with caspase domain inhibits cardiomyocyte apoptosis by reducing K+ currents. Am J Physiol Cell Physiol. 2003;284:C1405–C1410. doi: 10.1152/ajpcell.00279.2002. [DOI] [PubMed] [Google Scholar]
- 40.Isodono K, Takahashi T, Imoto H, Nakanishi N, Ogata T, Asada S, et al. PARM-1 is an endoplasmic reticulum molecule involved in endoplasmic reticulum stress-induced apoptosis in rat cardiac myocytes. PLoS One. 2010;5:e9746. doi: 10.1371/journal.pone.0009746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickson P, Toth A, Erhardt P. PUMA is critical for neonatal cardiomyocyte apoptosis induced by endoplasmic reticulum stress. Cardiovasc Res. 2007;73:48–56. doi: 10.1016/j.cardiores.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kung G, Konstantinidis K, Kitsis RN. Programmed necrosis, not apoptosis, in the heart. Circ Res. 2011;108:1017–1036. doi: 10.1161/CIRCRESAHA.110.225730. [DOI] [PubMed] [Google Scholar]
- 43.Gold R, Schmied M, Giegerich G, Breitschopf H, Hartung HP, Toyka KV, et al. Differentiation between cellular apoptosis and necrosis by the combined use of in situ tailing and nick translation techniques. Lab Invest. 1994;71:219–225. [PubMed] [Google Scholar]
- 44.Ohno M, Takemura G, Ohno A, Misao J, Hayakawa Y, Minatoguchi S, et al. “Apoptotic” myocytes in infarct area in rabbit hearts may be oncotic myocytes with DNA fragmentation: analysis by immunogold electron microscopy combined with In situ nick end-labeling. Circulation. 1998;98:1422–1430. doi: 10.1161/01.cir.98.14.1422. [DOI] [PubMed] [Google Scholar]
- 45.Longnecker DS, Shinozuka H, Farber E. Molecular pathology of in-vivo inhibition of protein synthesis. Electron microscopy of rat pancreatic acinar cells in puromycin-induced necrosis. Am J Pathol. 1968;52:891–915. [PMC free article] [PubMed] [Google Scholar]
- 46.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 47.Zhu H, Jia Z, Misra BR, Zhang L, Cao Z, Yamamoto M, et al. Nuclear factor E2-related factor 2-dependent myocardiac cytoprotection against oxidative and electrophilic stress. Cardiovasc Toxicol. 2008;8:71–785. doi: 10.1007/s12012-008-9016-0. [DOI] [PubMed] [Google Scholar]
- 48.He X, Kan H, Cai L, Ma Q. Nrf2 is critical in defense against high glucose-induced oxidative damage in cardiomyocytes. J Mol Cell Cardiol. 2009;46:47–58. doi: 10.1016/j.yjmcc.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Purdom-Dickinson SE, Lin Y, Dedek M, Morrissy S, Johnson J, Chen QM. Induction of antioxidant and detoxification response by oxidants in cardiomyocytes: evidence from gene expression profiling and activation of Nrf2 transcription factor. J Mol Cell Cardiol. 2007;42:159–176. doi: 10.1016/j.yjmcc.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurusamy N, Das DK. Autophagy, redox signaling, and ventricular remodeling. Antioxid Redox Signal. 2009;11:1975–1988. doi: 10.1089/ars.2009.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, et al. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–H479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Younce CW, Kolattukudy PE. MCP-1 causes cardiomyoblast death via autophagy resulting from ER stress caused by oxidative stress generated by inducing a novel zinc-finger protein, MCPIP. Biochem J. 2010;426:43–53. doi: 10.1042/BJ20090976. [DOI] [PubMed] [Google Scholar]
- 53.Marambio P, Toro B, Sanhueza C, Troncoso R, Parra V, Verdejo H, et al. Glucose deprivation causes oxidative stress and stimulates aggresome formation and autophagy in cultured cardiac myocytes. Biochim Biophys Acta. 1802;2010:509–518. doi: 10.1016/j.bbadis.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 54.Chapple SJ, Siow RC, Mann GE. Crosstalk between Nrf2 and the proteasome: therapeutic potential of Nrf2 inducers in vascular disease and aging. IntJ Biochem Cell Biol. 2012;44:1315–1320. doi: 10.1016/j.biocel.2012.04.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.