Abstract
Vinculin binding to actin filaments is thought to be critical for force transduction within a cell, but direct experimental evidence to support this conclusion has been limited . In this study, we found mutation (R1049E) of the vinculin tail impairs its ability to bind F-actin, stimulate actin polymerization, and bundle F-actin in vitro. Further , mutant (R1049E) vinculin expressing cells are altered in cell migration, which is accompanied by changes in cell adhesion, cell spreading, and cell generation of traction forces, providing direct evidence for the critical role of vinculin in mechanotransduction at adhesion sites. Lastly, we herein discuss the viability of models detailing the F-actin-binding surface on vinculin in context of our mutational analysis.
Keywords: vinculin, actin, focal adhesion, force, adhesion
Introduction
Cell migration is a complex process that is critical for embryonic development, wound healing, and maintenance of tissue integrity [1]. The current model of cell migration is a multi-step process involving polarization in the direction of a stimulus, extension or protrusion of a membrane, contraction of the cell body and retraction of the cell rear. In recent years, our understanding of cell migration has been significantly advanced by the identification of proteins that govern lamellipodial protrusion [2-8]. This information combined with studies of adhesion plaques has revealed that, once formed, lamellipodial protrusions attach to the substratum by transient adhesion complexes. Some of these transient structures anchor the protrusion and subsequently mature into adhesive structures known as focal adhesions, which support the generation of traction forces necessary to pull the cell body forward and break older adhesions at the cell rear.
The adhesive complexes of migrating cells are rich in the highly conserved focal adhesion protein vinculin. Vinculin consists of a head domain, a short proline rich linker and a tail domain [9, 10]. In its “closed” conformation, the vinculin head interacts with its tail. Binding of proteins to the head domain releases this intramolecular interaction allowing vinculin to adopt an “open” conformation. In this open conformation, the vinculin tail binds actin [11]. Vinculin can bind to and modify existing actin bundles and stimulate the formation of actin bundles and networks [12], making it an ideal candidate for establishing new actin assemblies and linking them to the existing cytoskeleton.
Multiple models detailing where actin binds the vinculin tail have emerged [13-15]. Work by Janssen et al. [1 3 ] suggests that vinculin binds to F - actin via two distinct binding surfaces within vinculin tail – an upper and a lower actin monomer binding site. In this model, the R1049 residue is a contact point in the lower site, located just beyond the 5th alpha-helix, within vinculin tail. Janssen et al. also implicate the region near the C-terminal loop as important for vinculin dimerization, a finding supported by the recent work of Shen et al. Clustered charge-to-alanine experiments [16] are fully consistent with the binding sites in Janssen et al., and support the structural conclusions that the top site is mostly hydrophobic in nature, whereas the one at the bottom contains a significant amount of electrostatic interactions.
Recent work by others has challenged the vinculin tail:actin interface proposed by Janssen. These studies reveal regions important for actin binding and bundling located outside of the proposed Janssen sites (I997, V1001; [15, 17]). In support, full-length vinculin harboring either I997A or V1001A mutations compromise focal adhesion size, number, and cell spreading [15].
With these studies in mind, we tested the Janssen hypothesis that R1049 directly participates in actin binding. We report here that mutating vinculin R1049 directly impacts actin binding, which in turn compromises cell migration, adhesion, spreading, and traction force generation.
Experimental Procedures
Purification of VT or RE
pET21a (Novagen) was engineered with a 6His tag and TEV protease cleavage site fused to the N-terminal region of chicken vinculin amino acids 881-1066 to generate pET21a-VT (vinculin tail). pET21a-VT was mutagenized using the Stratagene Quickchange protocol to generate pET21a-RE (R1049E). Both VT and RE proteins were prepared as follows: for expression of the vinculin tail (VT), E. coli containing this plasmid were grown to early log phase, and induced with 1mM IPTG for 3 hours at 37 °C. Bacteria pellets were resuspended in lysis buffer (50mM phosphate, 300mM NaCl, 20mM imidazole, 5% glycerol, aprotinin (10µg/ml), pepstatin A (2.5µg/ml), lysozyme (0.2µg/ml) and DNAseI (0.02µg/ml). The bacterial pellet was sonicated twice for 20 seconds, then insoluble debris was removed by centrifugation at 12,000 × g for 15 minutes. The supernatant was filtered through 0.45µm syringe filters and incubated for 1 hour at 4 °C with Ni-NTA-agarose pre-washed in lysis buffer. The beads were washed twice in lysis buffer, then twice with 50mM Tris pH 7.6, 150mM NaCl. Agarose was resuspended in a suitable volume of 250mM imidazole in Tris-Buffered Saline (TBS), pH 7.6 The agarose was allowed to settle and the supernatant was passed over a PD-10 column equilibrated with TEV buffer (50mM Tris, pH7.5, 150mM NaCl, 0.5mM EDTA, 1mM DTT). The 6His tags were cleaved from the VT proteins by the addition of 6His-TEV protease (0.16mg/ml). The 6His-VT proteins were incubated with 6His-TEV overnight at 4 °C. Following the incubation, 6His-TEV and remaining 6His-VT were bound to Ni-NTA-agarose beads, and the supernatant was passed over a PD-10 column to exchange the buffer to 50mM Tris, pH 7.6, 50mM NaCl. This buffer was diluted with a final concentration of 20% glycerol and the proteins were stored at −80 °C until use. Protein concentration was determined by A280, using the molar extinction coefficient 18,470 M−1cm−1.
Circular Dichroism and Dynamic Light Scattering
For Circular Dichroism (CD) analyses, spectra of VT or RE in 50mM Tris, pH7.6, 50mM NaCl, 20% glycerol were recorded from 190 to 260nm at 0.1nm increments on a JASCO J-815 machine. For Dynamic Light Scattering (DLS) analyses, VT or RE were purified by FPLC. The purified VT or RE samples were then subjected to DLS using a DynaPro NanoStar instrument (Wyatt Technology). Ten measurements were taken and averaged to produce an “average measurement”; five average measurements per experiments were then averaged to yield the “overall average” hydrodynamic radius. Three overall averages were then averaged to yield the mean values reported here.
Chemical Crosslinking
Chemical crosslinking experiments were carried out as in [18]. Briefly, solutions of VT or RE in Buffer CL were crosslinked with 100µM disuccinimidyl suberate (DSS) for 20 minutes at room temperature. The reactions were quenched by the addition of 50mM Tris, pH 7.6.
Purification of yeast actin, pyrene fluorescence and actin-VT cosedimentation assays
Actin was purified from the lysates of frozen yeast cells by a combination of DNaseI affinity chromatography, DEAE anion exchange chromatography, and polymerization/ depolymerization cycles as described in [12]. Pyrene fluorescence and co-sedimentation assays were performed as described in [12]. Cosedimentation assays were spun at either 20,000 or 80,000 rpm in a Beckman TL-100 centrifuge using a TLA 100 rotor.
Electron microscopy
Reactions for visualization using TEM were generally prepared as in [12]. Briefly, for low ionic strength conditions: Reactions containing 1µM G-actin were combined with varying concentrations of VT or RE, allowed to reach steady state, then 3µl of the reaction was pipetted onto a 400 mesh Formvar TEM grid. The grids were stained with 1% uranyl acetate, and visualized using a JEOL TEM 1230. For physiological salt conditions: The reactions were prepared in the same manner as for low salt conditions, except 2 mM MgCl2 and 50 mM KCl was added following the addition of VT or RE.
Generation of stable cell lines
Vinculin-null mouse embryonic fibroblasts (MEFs) were obtained from vinculin-null mice and were the generous gift of Eileen Adamson (Burnham Institute) and maintained in DMEM plus 10% FBS, 1% penicillin/streptomycin, and 400 µg/ml G418. The vector pLPCX-GFP (Clontech) was engineered to include a GFP tag with a restriction site at its 3’ end, and full-length WT vinculin was cloned into this construct to generate pLPCX-GFP-WTvin. Site-directed mutagenesis was carried out on pLPCX-GFP-WTvin using the Stratagene QuickChange protocol to generate pLPCX-GFP-REvin. The entire coding sequence of vinculin was sequenced to confirm the R1049E mutation. The plasmids were transfected into 293-GPG cells using Lipofectamine reagent (Invitrogen) as previously described [19, 20]. Virus was collected and added to vinculin-null MEFs. Infected MEFs were selected in 2µg/ml puromycin, and mass populations of cells were sorted by FACS to achieve the level of expression desired. Expression levels were assayed by immunoblotting using antibodies against vinculin (hVIN1, Sigma) at a concentration of 1:1000 or against the p34-Arc subunit of the Arp2/3 complex has been previously described [21].
Cell spreading assays
Glass coverslips were coated with 10µg/ml fibronectin in PBS overnight at room temperature. The coverslips were rinsed twice with PBS, and blocked with 2% BSA in PBS for 2 hours at room temperature, then rinsed twice with PBS. Cells were seeded on the coverslips and allowed to adhere for 25 minutes or 4 hours, then fixed in 3.7% formaldehyde. For 25 minute assays, Nomarski images were taken. The outlines of at least thirty cells from three independent experiments were traced in Adobe Photoshop CS4 and converted to cell area. For 4 hour assays, coverslips were permeabilized for 20 minutes in 0.5% TritonX-100 in Universal Buffer (UB: 150 mM NaCl, 50 mM Tris pH 7.6, 0.01% NaN3), blocked for 45 minutes in 1% BSA in UB, then stained with DAPI (500ng/ml) and Texas-Red phalloidin (1:750; Invitrogen) for 45 minutes. Coverslips were rinsed twice in PBS, twice with water, then mounted onto slides using Mowiol (Fisher Scientific).
Cell adhesion assays
Glass coverslips were coated with 10µg/ml fibronectin as before. Cells were seeded on the coverslips at 50,000 cells per coverslip and allowed to adhere for 25 minutes. Wells of the 24-well plate containing coverslips were vigorously washed three times in PBS, then fixed in the same manner as described. Nine non-overlapping 10x fields were taken of each coverslip at 10x magnification. Each experiment was performed on duplicate coverslips per condition, and the number of cells relative to the GFP (vinculin-null) control for each experiment was recorded.
Cell migration assays
Glass coverslips were coated with 10µg/ml fibronectin as before. Cells were seeded on the coverslips and allowed to adhere overnight. Cells were imaged on a Zeiss Axiovert 200M in an enclosed stage heater and a humidified, 5% CO2 environment. Images were taken every 5 minutes for a total of 10 hours. Only cells visible for a minimum of 4 hours were included in the analysis. Tracks were analyzed using NIH ImageJ Manual Cell Tracker plugin.
Traction force assays
Microfabricated post array deflection device (mPAD) silicon masters were prepared as described previously [22]. In brief, elastomeric micropost arrays were fabricated using PDMS replica molding. To make microfabricated post array templates, 1:10 PDMS prepolymer was cast on top of silanized mPAD silicon masters, cured at 110 °C for 30 min, peeled off gently, oxidized with oxygen plasma (Plasma-Preen; Terra Universal), and silanized overnight with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane (Sigma-Aldrich) vapor under vacuum. To make the final PDMS mPAD device, 1:10 PDMS prepolymer was cast on the template, degassed under vacuum for 20 min, and cured at 110 °C for 20 h and gently peeled off the template on a #1 25 mm diameter circular coverslip (Electron Microscopy Services). Peeling-induced collapse of the mPADs was rectified by sonication in 100% ethanol, followed by supercritical drying in liquid CO2 using a critical point dryer (Samdri-PVT-3D; Tousimis), as described previously [22]. Flat PDMS stamps were generated by casting 1:30 PDMS prepolymer on flat silanized silicon wafers. Stamps were coated in saturating concentration of fibronectin (Invitrogen) (50 µg/mL in PBS) for 1 h. These stamps were washed in distilled water and dried under a stream of nitrogen gas. FN-coated stamps were placed in contact with surface-oxidized mPAD substrates (UVO-Model 342; Jelight). mPAD substrates were labeled with 5 µg/mL of Δ9-DiI (Invitrogen) in distilled water for 30 min. mPAD substrates were subsequently transferred to a solution of 0.2% Pluronics F127 (Sigma-Aldrich) for 30 min, to prevent nonspecific protein absorption. WT, RE, and null eGFP-vinculin MEF cells were seeded in growth medium and then allowed to spread overnight. mPAD substrates were transferred to an aluminum coverslip holder (Attoflour Cell Chamber; Invitrogen) for live cell microscopy and placed in a stage top incubator that regulated temperature, humidity, and CO2 (Live Cell; Pathology Devices). Confocal images were taken with a Nikon C2-Confocal Module connected to a Nikon Eclipse Ti inverted microscope using a high magnification objective (CFI Plan Apochromat total internal reflection fluorescence (TIRF) 60× oil, N.A. 1.45; Nikon). Post images were captured using a 561-nm laser with a 595/50 filter, and vinculin images were captured using a 488 nm laser and 525/50 filter. For force measurements, the top and bottom of the posts were sequentially imaged and the deflection measured.
The resulting force, F, was calculated using Euler-Bernoulli beam theory, in which E, D, L, and δ are the Young’s modulus, post diameter, post height, and post deflection:
Results
In this study, we were motivated to test the consequence of mutating R1049, a vinculin residue implicated in actin binding, on cell migration, adhesion, spreading, and traction force generation.
The R1049E mutant vinculin tail is structurally similar to wild type vinculin tail
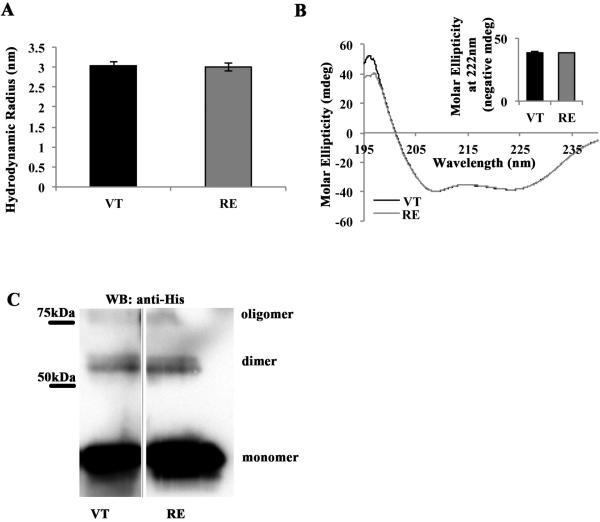
For our in vitro analyses, we used site-specific mutagenesis to generate an R to E substitution at position 1049 in a His-tagged fusion protein containing the wild type vinculin tail (VT) residues 881-1066. Except where noted, the His tag was cleaved from the protein. We first examined if mutation of R1049E, herein referred to as RE vinculin, had any effect on vinculin tail structure. For this, both RE and VT purified tail domains were examined by dynamic light scattering, which estimates the hydrodynamic radius of a protein. The average hydrodynamic radii for VT and RE were 3.0±0.1 nm and 3.0±0.1 nm, which do not differ significantly (Figure 1A). The alpha-helical content of both VT and RE were also examined by circular dichroism (CD). In this method, alpha-helical content is characterized by valleys in the spectra around 208 and 222 nm. We used a correction factor to set the spectra equal at 208 nm, then corrected every other value by this number and examined the difference in spectra at 222 nm. The spectra track nearly identically (Figure 1B), and the difference in ellipticity was virtually zero (Figure 1B inset). Taken together, these data indicate there are no major structural changes of alpha-helices in RE compared to VT.
Figure 1. R1049E is structurally similar to wild type VT.
(A) VT and RE have similar size and shape in solution. Solutions of either VT or RE were subjected to Dynamic Light Scattering (DLS) analysis, which yields estimates of hydrodynamic radii. Shown is the average hydrodynamic radius predicted from three independent trials of VT and RE, with error bars representing SEM. There is no statistical difference between VT and RE hydrodynamic radii. (B) VT and RE have similar alpha-helical composition. Solutions of VT or RE were subjected to analysis by Circular Dichroism (CD), at wavelengths from 195-240nm. The alpha-helical content of VT and RE is characterized by valleys at 208 and 222nm. Spectra from three independent trials of VT and RE were averaged then set equal at 208nm and graphed. They track almost identically along the entire spectrum of wavelengths examined. The inset panel shows the negative molar ellipticity of VT and RE at 222nm (negative values used for ease of data presentation), which do not differ statistically from one another. (C) VT and RE are crosslinked to a similar degree in solution. Preparations of 15mM VT-His or RE-His in Buffer CL were crosslinked with 100mM disuccinimidyl suberate (DSS) for 30 minutes. Half of the reactions were run on 15% acrylamide gels and blotted with anti-His antibody. Shown are samples from the same western blot. Crosslinked monomer bands are visible below 50kDa, dimer bands are visible just above 50kDa, and the presence of a higher molecular weight oligomer is observed near 75kDa.
There is speculation that R1049 may contribute to vinculin dimerization. Hence we sought to determine if RE was a dimerization mutant. Work from the Craig laboratory demonstrated that vinculin tail can dimerize and is cross-linked into higher order oligomers [18]. Hence, we next asked whether RE could be cross-linked to the same extent as VT. Using the same cross-linking approach as Johnson and Craig [18], we found that the relative amounts of dimer formed when VT or RE are cross-linked was virtually the same (Figure 1C). Thus, RE does not affect dimerization of the vinculin tail. Taken together, these data demonstrate that there are no major structural changes in RE compared to VT.
R1049E is an actin-binding and bundling mutant in physiological ionic strength conditions
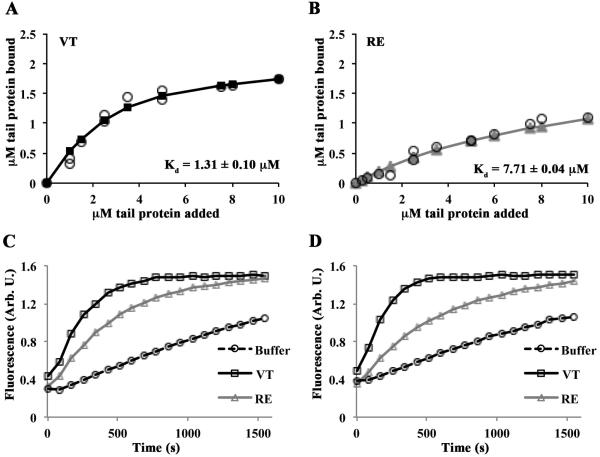
We next tested if RE was defective in actin binding under physiological salt concentrations. To assay vinculin tail binding to actin filaments, VT or RE was pre-incubated with 1 µM G-actin, and then F-salt added to initiate actin polymerization. Completed reactions were centrifuged at a speed sufficient to pellet all polymerized actin as well as actin-bound vinculin tail peptide (co-sedimentation). Quantified analysis of pelleted vinculin tail peptide was fit to the quadratic binding equation as previously described [12] and estimates of dissociation constants (Kd) were generated. Binding data from 80K co-sedimentation assays demonstrated a six-fold difference in actin binding between VT (Kd, = 1.31±0.10 µM) and RE (Kd, = 7.71±0.04 µM) (Figure 2A and 2B). As RE exhibited weaker actin binding, we assessed the ability of VT or RE to stimulate actin filament formation. F-salt was added to reactions containing 1 µM pyrene-labeled G-actin mixed with varying concentrations of VT or RE and then actin polymerization measured by the increase in fluorescence over time. At concentrations of either 0.25 µM or 0.5 µM vinculin tail, VT induces actin nucleation more rapidly than RE (Figures 2C and 2D). Together, these data indicate that RE vinculin is compromised in its ability to bind and stimulate actin polymerization under physiological salt concentrations.
Figure 2. RE is an actin binding and polymerization mutant in physiological salt conditions.
(A, B) VT and RE bind actin with different affinities. Reactions containing VT or RE and G-actin were polymerized by the addition of F-salts, then centrifuged at 80,000 rpm to pellet actin filaments. Twenty-five percent of the supernatant and the entire pellets were run on separate gels. The amount of tail protein in the pellet of cosedimentation reactions was plotted as a function of the tail concentration added to the sample. Estimates of Kd were generated by fitting the data to the quadratic binding equation and are noted in each panel. The error on the Kd is the error of the fit. Open circles represent individual observations; solid lines with squares (VT; A) or triangles (RE; B) represent the fitted data. (C,D) Varying concentrations of VT, RE, or an equivalent volume of buffer was added to 1µM pyrene labeled G-actin in G-buffer, then polymerized with F-salts, and the change in fluorescence was monitored over time. Representative graphs are shown. RE fails to stimulate actin polymerization to the same extent as VT at 0.25µM (C) or 0.5µM (D).
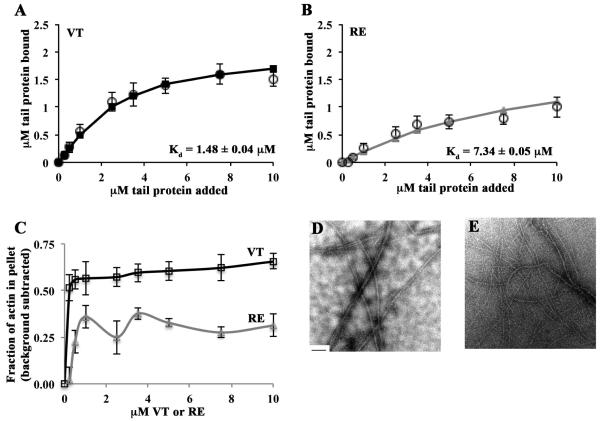
Next, we examined the extent of vinculin-driven actin bundling. VT or RE was pre-incubated with 1 µM G-actin, and then F-salt concentrations added to initiate actin polymerization. Here, completed reactions were centrifuged at a speed sufficient to pellet bundled actin and actin-bound vinculin peptide. Small amounts of thin, two filament thick actin bundles also co-sediment in this approach [23]. Binding data from 20K co-sedimentation assays yielded vinculin tail:actin dissociation constants that were nearly identical to those generated from 80K centrifugations (Kd, = 1.48±0.04 µM for VT and 7.34±0.05 µM for RE) (Figure 3A and 3B). Furthermore, VT exhibited ~35% greater bundling activity than RE (Figure 3C). Since RE was not completely deficient in actin bundling, we examined the samples by transmission electron microscopy (TEM). Reactions containing VT had thick, tightly packed bundles, while those containing RE had fewer, thinner, more loosely packed bundles (Figures 3D and 3E). This data supports the conclusion that RE is defective in both quantity and quality of actin bundling activity under physiological salt conditions.
Figure 3. RE is an actin bundling mutant in physiological salt conditions.
(A, B) RE does not bind or bundle actin as well as VT. Reactions containing VT or RE and G-actin were polymerized by the addition of F-salts, then centrifuged at 20,000 rpm to pellet actin bundles. Twenty-five percent of the supernatant and the entire pellets were run on separate gels. The amount of tail protein recovered in the pellet was plotted as a function of the tail concentration added to the sample. Estimates of Kd were generated by fitting the data to the quadratic binding equation and are noted in each panel. Open circles represent data from at least 3 independent experiments. Error bars are SEM. Solid lines with squares (VT; A) or triangles (RE; B) represent the fitted data. The error on the Kd is the error of the fit. (C) RE does not bundle actin as well as VT. The amount of actin recovered in the pellet was plotted as a function of the total tail concentration added to the sample. The percent of actin that was pelleted in the control experiment containing only actin was subtracted from each data point. Shown is the mean of at least 3 independent experiments ± SEM. (D) VT bundles actin in physiological salt conditions. VT (2.5µM) and actin (1µM) were combined then polymerized by the addition of F-salts. A fraction of the reactions was spotted on Formvar-coated TEM grids and imaged at 12,000x magnification. Bundles several filaments thick are visible. (E) RE does not bundle actin as well as VT. When RE (2.5µM) and actin (1µM) were combined, thinner and more loosely packed bundles than those in (D) result. The scale bar in (D) is 200nm.
R1049E is an actin-binding and bundling mutant in low ionic strength conditions
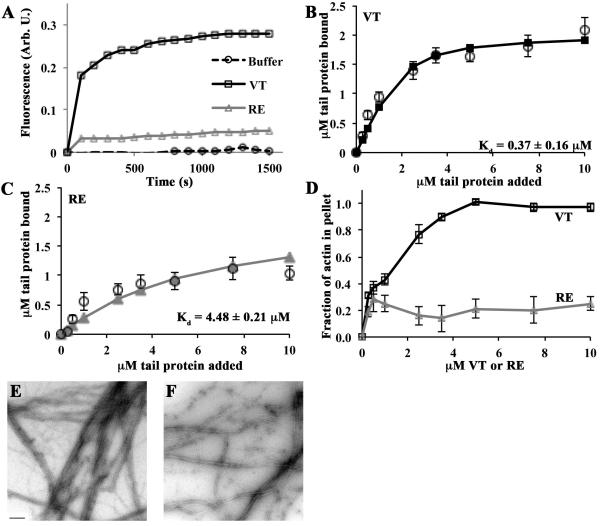
We have previously shown that VT is also capable of stimulating actin polymerization under low ionic strength conditions [12]. To assess the ability of RE to trigger filament formation in low salt, we combined varying concentrations of VT or RE with 1 µM pyrene-labeled G-actin in the absence of F-salt and then measured polymerization through the increase in fluorescence over time. VT stimulated more actin polymerization than RE at all vinculin tail concentrations examined (i.e. 0.25 µM to 10 µM; Figure 4A).
Figure 4. RE is an actin binding, polymerization, and bundling mutant in low ionic strength conditions.
(A) RE is defective in stimulating actin polymerization in low ionic strength conditions. Varying concentrations of VT, RE, or an equivalent volume of buffer was added to 1mM pyrene labeled G-actin in G-buffer, and the change in fluorescence was monitored over time. A representative graph at 2.5µM is shown. (B, C) RE does not bind or bundle actin as well as VT. Reactions containing VT or RE and G-actin were allowed to go to completion, then centrifuged at 20,000 rpm to pellet actin bundles. Twenty-five percent of the supernatant and the entire pellets were run on separate gels. The amount of tail protein recovered in the pellet was plotted as a function of the tail concentration added to the sample. Estimates of Kd were generated by fitting the data to the quadratic binding equation and are noted in each panel. Open circles represent data from at least 3 independent experiments. Error bars are SEM. Solid lines with squares (VT; B) or triangles (RE; C) represent the fitted data. The error on the Kd is the error of the fit. (D) RE does not bundle actin as well as VT. The amount of actin recovered in the pellet was plotted as a function of the total tail concentration added to the sample. Shown is the mean of at least 3 independent experiments ± SEM. (E) VT bundles actin in low salt conditions. VT (2.5µM) and G-actin (1µM) were combined in a low salt buffer. A fraction of the reactions was spotted on Formvar-coated TEM grids and imaged at 12,000x magnification. Bundles several filaments thick are visible. (F) RE does not bundle actin as well as VT. When RE (2.5µM) and actin (1µM) were combined, thinner and more loosely packed bundles than those in (E) result. The scale bar in (E) is 200nm.
Next, VT or RE was incubated with 1 µM G-actin in G-buffer, and the resulting product centrifuged at a speed sufficient to pellet actin bundles. In accordance to actin polymerization activity produced in Figure 4A, under the low ionic strength condition the Kd for VT:actin was calculated to be 0.37±0.16 µM and for RE:actin to be 4.48±0.21 µM (Figure 4B and 4C). Furthermore, incubation of actin with VT resulted in 25-40% bundling at concentrations of 0.25 µM, 0.5 µM and 1 µM, which increased to 80% and higher above 2.5 µM VT (Figure 4D). In contrast, the bundling by RE was 20-30% for all concentrations examined. To confirm and visualize this difference in actin bundling, samples were analyzed by TEM. The bundles formed when VT is present are several filaments thick and the filaments are tightly packed (Figure 4E). In contrast, those bundles formed when RE is present are thinner, and the filaments are loosely packed (Figure 4F). Along with the studies in Figures 2 and 3, these data demonstrate that RE vinculin tail is compromised in its ability to bind and stimulate the polymerization and bundling of actin filaments in multiple ionic strength conditions.
Vinculin R1049E mutant cells display decreased spreading and adhesion but enhanced migration
We sought to determine if the interaction of vinculin R1049 with actin was a physiologically relevant interaction. Hence, we next sought to examine the consequence of re-expressing GFP-tagged full-length R1049E vinculin (vinRE) in mouse embryonic fibroblasts (MEFs) harvested from the vinculin null mouse [24]. As controls, we also generated lines expressing GFP-tagged full-length WT vinculin (vinWT) or cytoplasmic GFP (GFP).
After generating and validating the three cell lines by immunoblotting (Figure 5A), we first asked how defective actin binding and bundling affects cell spreading. We performed spreading assays on fibronectin-coated glass at two different time points. Cells were seeded on surfaces coated with fibronectin and allowed to adhere for 25 minutes or 4 hours before fixation and imaging. Consistent with previous studies, we found that cells expressing vinWT spread more robustly and had more prominent actin stress fibers than cells expressing GFP alone at both time points (Figure 5B) [24]. We measured the area of the spread cells and found that at 25 minutes, vinRE cells exhibit an intermediate ability to spread between that of vinWT and GFP cells (Figure 5C). After 4 hours, vinRE cells are still spread out to an intermediate degree, ~20% less than vinWT cells, but are more than double the size of the GFP cells (Figure 5B and 5D). The shape of the vinRE cells is comparable to vinWT in that the cells are approximately as long as they are wide, but appeared to have less prominent stress fibers (Figure 5B). This contrasts sharply with the phenotype of the GFP cells, which are typically more than three-fold longer in one axis than the other (Figure 5B).
Figure 5. Cells expressing R1049E are deficient in spreading, adhesion, and migration.
Vinculin null mouse embryo fibroblasts expressing GFP, or a fusion of GFP and wild type vinculin (vinWT) or GFP and R1049E vinculin (vinRE) were examined. (A) Expression levels of the various vinculins and GFP. Total cell lysates were harvested from the indicated cells and blotted with antibodies against vinculin to show the level of re-expressed protein or a loading control (p34-Arc). (B-D) RE expressing cells do not spread as well as WT expressing cells. The indicated cell lines were allowed to spread on fibronectin coated surfaces for 25 minutes (C) or 4 hours (B and D). Representative images of cells spreading at 4 hours and stained with DAPI and phalloidin are shown in B, scale bar =25µm. (C and D) At least 30 cells from each of three independent were analyzed to determine cell area – the means ± SEM are shown. (E) RE cells do not adhere as well as WT cells to fibronectin. Cells were seeded on glass coverslips coated with 10µg/ml fibronectin and allowed to adhere for 25 minutes. The coverslips were washed and adherent cells were counted in nine non-overlapping fields. Fold-adhesion is reported relative to the total number of GFP cells adhered, which is set equal to 1.0. Shown is the mean of at least 3 independent experiments ± SEM. (F) RE cells display a migration speed that is intermediate between WT and GFP cells. Cells were seeded on fibronectin and allowed to adhere overnight. Cells were imaged for a total of 10 hours, and only those cells visible for a minimum of 4 hours were included in the analysis. Cells from at least three independent experiments were tracked using NIH ImageJ Manual Cell Tracker plugin; a minimum of 16 cells total were analyzed. The asterisks represent the following: * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001; One-way ANOVA with Tukey correction for multiple comparisons.
Actin stress fibers anchor adhesions to the cytoskeleton, providing stability and support. We hypothesized that the defect in spreading observed in vinRE cells might coincide with a defect in the ability of the cells to adhere. Therefore, we asked what effect the R1049E mutation had on the ability of vinculin re-expressing cells to adhere to a fibronectin-coated substrate. Cells were seeded on fibronectin coated surfaces, allowed to adhere, and washed. The number of adherent cells was counted and reported relative to the number of adherent GFP cells, which was set equal to one. In this assay, twice as many vinWT cells adhered as vinRE or GFP cells (Figure 5E). This assay did not find a statistical difference between adhesion of vinRE and GFP cells, though vinRE cells tended to adhere better than GFP cells. Thus, the adhesiveness of the vinRE cells is markedly reduced compared to vinWT cells.
As cell spreading and adhesion are both crucial for proper cell migration, we analyzed the migration of the three cell lines on fibronectin-coated surfaces. Cells were seeded and allowed to adhere overnight then imaged every 5 minutes for a total of 10 hours. The movies were analyzed using NIH ImageJ Manual Cell Tracker plugin. It is well known that vinculin-null cells display enhanced migration compared to those expressing vinculin, and our results here agree with the published data [21, 24]. As expected, the GFP cells displayed enhanced migration on fibronectin compared to vinWT cells, with an average velocity of 1.3±0.09 µm/min. The vinWT cells migrated significantly slower with an average velocity of 0.81±0.05 µm/min (Figure 5F). The vinRE cells migrated at an intermediate velocity, 1.0±0.07 µm/min. We also noted that many of the vinRE cells appeared to have “trailing tails” and some were “snapped back” by these tails after attempts to crawl forward.
Vinculin R1049E mutant cells exert less traction force than WT cells
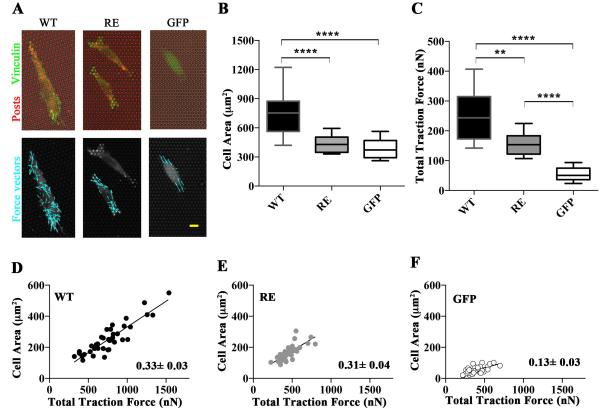
Cell migration depends upon a balance of forces at the leading and trailing edges, requiring that old adhesions must be disassembled in order for cells to move forward. The intermediate phenotype observed in vinRE cells led us to next examine if the traction force exerted by vinRE cells differed from vinWT cells. We used microfabricated post array deflectors (mPADs) to measure the traction forces generated by the three cell lines [25]. Cells were seeded overnight onto fibronectin-coated mPADs and developed focal adhesions (Figure 6A). The vinWT cells spread more than both vinRE and GFP cells (Figure 6B). Knowing the micropost stiffnesses, we measured post deflections for vinWT, vinRE, and GFP cell lines to quantify cell traction forces. Figure 6A shows images of the fibronectin-coated posts (red), with the cell outline visible, and the recruitment of GFP-vinculin (green) to focal adhesions. Force vectors (cyan) were calculated from the deflection of the microposts. As observed previously [26], the magnitude of the forces varied across the cell, with the highest forces located at the cell periphery. Figure 6C presents box-whisker plots for the total traction force per cell, which represents the sum of the magnitudes of the force vectors for each cell. As observed previously the vinculin-null (GFP) cells do generate traction force (~50 nN), confirming that vinculin is not required for force transmission at focal adhesions. Expression of WT vinculin increased the traction forces five-fold compared with the GFP controls, demonstrating that vinculin enhances transmission of traction forces. The expression of the R1049E mutant (vinRE cells) increased the traction force three-fold compared to the GFP cells, indicating that the R1049E mutant vinculin is capable of enhancing traction forces, though not to the degree of WT vinculin. Taken together, these data demonstrate that actin-binding deficient vinculin R1049E protein inadequately restores cellular traction forces.
Figure 6. Cells expressing R1049E are defective in generating traction forces.
(A) Vinculin regulates traction forces. The three cell lines (vinWT, vinRE, or GFP) were seeded overnight on fibronectin-coated MPADs (posts labeled red; upper panel). GFP is visible in green (upper panel), and the cell outlines are visible (upper and lower panels). Force vectors are indicated with cyan arrows (lower panel). (B) Vinculin regulates traction forces and area. Box-whisker plots for area of cells seeded on MPAD devices (mean, 10th, 25th, 75th, and 90th percentile for cell area; >62 cells per condition). (C) Box-whisker plots for traction force generated by cells seeded on mPADs (mean, 10th, 25th, 75th, and 90th percentile for cell area; minimum 31 cells per condition). (D) Linear regression between traction force and area of WT cells with slope of the fit line shown. (E) Linear regression between traction force and area of RE cells with slope of the fit line shown. (F) Linear regression between traction force and area of GFP cells with slope of the fit line shown. In Figure 6, asterisks represent the following: ** p<0.01, **** p<0.0001; Kruskal-Wallis test.
We also generated traction force-cell area plots (Figure 6 D, E, and F), as Dumbauld et al. [26] had previously demonstrated cell area and cytoskeletal tension coupling. We evaluated traction force-cell area coupling as another metric for assessing how vinculin expression modulates traction force generation. There is strong correlation between cell area and traction force generation for null, vinRE, and vinWT cells. As observed previously, vinculin null cells exhibit a linear relationship between cell area and traction force, indicating that vinculin is not required for cell area-traction force coupling. This result also suggests other focal adhesion components can play a role in traction force generation, such as talin-actin force transfer [27]. Both vinRE and vinWT expression, however, significantly enhance coupling between cell area and traction force generation, as demonstrated by the nearly three-fold increase in the regression slope for both vinRE and vinWT compared to the null cells. Based on the similar traction force-cell area slopes between vinRE and vinWT, this also suggests that the vinRE spreading defect contributes to the weaker total traction force generation compared to the vinWT cells.
Discussion
In 2006 Janssen et al. proposed a model for how vinculin binds actin [13]. A more recent model for actin binding to vinculin [15] called into question the validity of the Janssen model. In the current study, we determined the consequence of mutating a vinculin residue identified by Jansen et al. as critical for actin binding. Our in vitro data shows that this mutant form of vinculin disrupts actin binding and bundling. Furthermore, expression of our mutant vinculin in cells lacking vinculin leads to a reduced ability of cells to spread, adhere, and generate traction forces; these cells migrate at enhanced rates. Based on this new information, we prioritize which aspects of the current vinculin tail:actin models are most likely.
This study is the first to show that the actin binding and bundling by vinculin is required for the generation of traction forces. A previous study showed that a mutant form of vinculin deficient in actin binding was compromised in its ability to stiffen when forces were applied to integrins [15]. Collectively, these data suggest that the actin binding and bundling properties of vinculin are required for force transmission. In our study (Figures 5 and 6), cells with defects in force transmission migrated faster than the vinculin-null MEFs re-expressing wild type vinculin. We found that the vinRE cells migrated faster than wild type re-expressers, a phenotype reminiscent of parental vinculin null MEFs. Previously it has been shown that migration speed exhibits a biphasic dependency on adhesive force [31]. Hence it is possible that the increased migration rate in the RE cells results from changes in their adhesivity.
In the traction force studies, we found some striking differences between the spreading of cells on fibronectin-coated mPAD devices that measure traction forces when compared to the same cells on fibronectin-coated glass surfaces. On the stiff substrate glass, we consistently observed that vinRE cells show a spreading phenotype that was intermediate between vinWT and GFP cells (Figure 5C and 5D). In contrast, on the mPADs the difference between vinWT and vinRE are more pronounced with the vinRE more closely mirroring the GFP cells (Figure 6). This observation is consistent with previous work showing that fibroblasts spread more on stiffer substrates whereas cells on softer substrates tend to adopt a more spherical phenotype [32]. Consequently, the differences we observe in cell spreading on the two substrates can likely be attributed to their pliability.
It is striking that in all of the biological events we measured, the RE mutant form of vinculin is not completely void of activity. Rather, it produces phenotypes that are intermediate between vinWT and vinculin-null (GFP) cells. These intermediate phenotypes are likely due to two possibilities. First, we speculate that the intermediate phenotype could be due to retention of some actin binding and bundling capability in R1049E. The examination of other mutations in vinculin, I997A and V1001A, [15], supports the idea that greater the actin binding capacity, the better cell spreading. A second possible explanation for the intermediate phenotypes lies in the ability of vinculin to bind proteins that modulate actin dynamics. Vinculin recruits the Arp2/3 complex an actin nucleating protein) and VASP (an anti-capper of actin filaments) to focal adhesions. Cells re-expressing P878A vinculin that fails to bind Arp2/3 displayed a number of defects, including decreased cell spreading and lamellipodial extension, an increased length to width ratio that is more similar to vinculin-null cells, and enhanced migration [21]. As is the case with our R1049E mutant, the phenotypes of the P878A mutant are intermediate between WT and vinculin-null cells, indicating that it, too, preserves some of the other functions of vinculin.
We considered the possibility that our R1049E mutation might affect vinculin binding to other tail ligands. However, the binding sites on VT for PIP2, Raver1, and paxillin are all distant from R1049. In the case of the focal adhesion protein paxillin, the binding site is within VT residues 881-1000 [28]. The VT-binding portion of Raver1, a heterogenous nuclear ribonucleoprotein, has been co-crystallized with VT, and the four residues that form the interface are within helices two and three of VT – distant from R1049 [29]. Finally, VT associates with PIP2 [30], but it is the N-terminal strap and nearby region of VT that is thought to be important for this interaction. As before, R1049E is on the back side of the VT protein, distant from the N-terminal strap. Based on this information, in conjunction with our studies demonstrating that the secondary structure and dimerization capability of R1049E are not perturbed, it is unlikely that our mutation disrupts binding to these VT-associated proteins.
The surface on vinculin involved in actin binding is the subject of much discussion in the field, and several models have appeared in the literature over the past ten years. The first, the Janssen Model [13], describes the interaction between chicken vinculin and rabbit skeletal muscle actin, and is based on the reconstruction of negative stain electron microscopy and diffraction data. The Janssen Model proposes two distinct surfaces on vinculin tail as contact points for an upper and lower actin monomer of a filament. In this model, R1049 lies within the lower actin monomer binding site.
Second and third models were proposed by Thompson et al. [15]. To generate these models, the crystal structure of vinculin tail (PDB code 1QKR) and the atomic model of F-actin (PDB code 3MFP) were manually docked into negatively stained 3D EM reconstruction of the rabbit skeletal muscle actin filaments decorated with chicken vinculin tail (Thompson Manual Fit Model) or manually docked and refined using discrete molecular dynamics (DMD; Thompson DMD Model). These newer models propose an alternative interface for vinculin binding to actin. Mutational studies of this alternative vinculin interface (i.e. I997 or V1001) identified actin binding defects [17]. Thompson et al. also mutated a vinculin residue (I948) and found no actin binding deficiency in this mutant. These observations led them to challenge the Janssen model despite the fact, that although I948 is adjacent to the Janssen predicted interface, this residue has less than 15% probability of interacting with F-actin in the Janssen model [13] and was not considered to be part of the interface. Charge-to-alanine mutation clusters scanning studies [16] do not show any significant effects on actin binding in the regions indicated by Thompson et al. as an alternative interface, but clusters that do show significant effects on actin binding match well with regions of high interaction probability predicted by Janssen et al. [13].
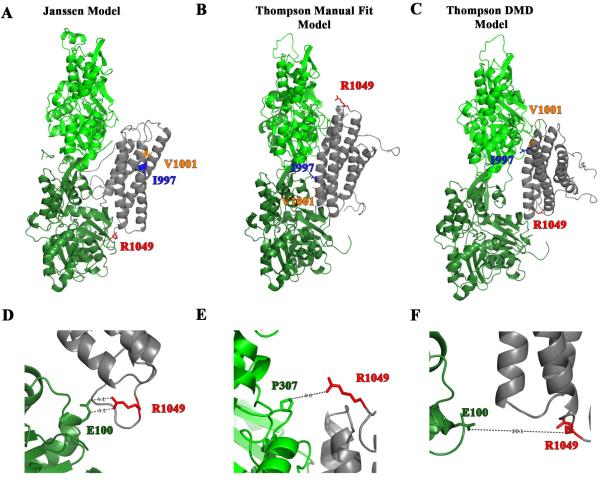
In this study, we present evidence that mutation of a residue predicted to be in the interface with over 50% probability by the Janssen Model (i.e. R1049) diminishes actin binding and bundling and has profound effects on the events required for cell migration. Based on this new information, we have re-evaluated which of the existing models most likely reflect how vinculin binds F-actin. In Figure 7A-C, we show the three models with the actin filaments aligned in the same orientation and with the I997, V1001, and R1049 residues highlighted. In all three models one molecule of the vinculin tail (gray) is interacting with two actin monomers (light green and dark green). In the Janssen Model, the side chain of R1049 is 5.1 Å away from E100 on the actin monomer suggesting that it is poised to form an electrostatic interaction (Figure 7D). In further support of the existence of such an interaction vinculin binds actin approximately 4 times tighter under low ionic strength conditions (Figure 4B, Kd=0.37±0.16µM) than physiological salt conditions (Figure 2A and 3A Kd=1.31±0.10µM, Kd=1.48±0.04µM). Finally, in support of the Janssen Model, R1049 vinculin is proposed to be in direct contact with actin monomer in a Molecular Dynamics simulation of vinculin binding to actin [14]. The Thompson Manual Fit and Thompson DMD Model both have R1049 more than 9.0 or 20.1 Å away from the nearest residue side chain on the actin monomer (Figure 7E and 7F). Moreover, the nearest residue in the Manual Fit Model is a proline residue, which is not expected to form an electrostatic interaction. Hence, our data agree with the Janssen Model more than either of the models proposed by Thompson et al. [15].
Figure 7. R1049E is a residue predicted to contact actin by the Janssen Model.
(A-C) The three different proposed models for vinculin tail (gray) binding to actin monomers (green). The I997 and V1001 residues are shown as ribbon-and-stick in blue and orange, respectively. The residue R1049 is red and represented as a ribbon-and-stick residue. The ribbon diagram in A shows the Janssen Model and was derived from PDB coordinates that were supplied by Dr. Niels Volkmann of the Burnham Institute [13]. The ribbon diagrams in B and C depict a more recent model put forth by Thompson et al. [15] and were downloaded from the Supplementary data. The models were created by Thompson et al. by manually docking the crystal structures of VT and actin filaments onto a 20 Å EM structure of vinculin bound to F-actin. The Manual Fit Model in B was manual docked using Chimera and the DMD Model in C was manual docked and then computation refinement approaches using discrete molecular dynamics (DMD) were applied. (D-E) An enlargement in the vicinity of R1049 of the above models. In the Janssen Model in Figure D, R1049 is poised 5.1 Å from E100 in one of the actin monomers suggesting the possibility that an electrostatic interaction forms. The enlargements of the Thompson Manual Fit Model in E and the Thompson DMD Model in F show that R1049 is not in close enough proximity to form electrostatic interactions. Moreover in the Manual Fit Model the nearest residue is a proline, which is not expected to form an electrostatic interaction with R1049.
Collectively, these data [13, 15-17, 33] raise the question as to which model more accurately represents vinculin binding to F-actin. Data from four groups, including ours [13, 16, 33] support residues or regions identified as important for the vinculin:actin interaction by Janssen [13]. Two other groups have published data that implicates residues outside of the Janssen Model as important for the vinculin:actin interaction [15, 17]. Clearly, the picture is more complex than originally thought, and further mutational studies are needed to resolve how vinculin binds actin with an emphasis on determining if there are multiple binding conformations.
Summary Statement.
Vinculin transduces force and orchestrates mechanical signaling at cell-cell and cell-matrix adhesions. Cells expressing a mutant vinculin deficient in actin binding and bundling display migration and traction force defects. Vinculin binding to actin is critical for cell migration and force generation.
Acknowledgements
We would like to thank Eileen Adamson for the generous gift of reagents. We are especially grateful to Niels Volkmann, Dorit Hanein, and Peter Rubenstein for their thoughtful discussions and critical comments on the work. We thank Kuo Kuang Wen for experimental advice during the initial phases of this project. We thank Lokesh Gakar for assistance with DLS analyses. We thank Xu Liu for assistance with CD analyses and Ernie Fuentes for help with Pymol and discussion of structural models.
Funding
This work is supported by National Science Foundation grant no.1120478 (to K.A. DeMali) and NIH R01 GM065918 (to A. J. García), and NSF Graduate Research Fellowship award (to D. W. Zhou). Research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA086862 to the Holden Comprehensive Cancer Center at the University of Iowa. The data presented herein were obtained in the Central Microscopy and Flow Cytometry Facilities, which are Carver College of Medicine / Holden Comprehensive Cancer Center core research facilities at the University of Iowa. These Facilities are funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran's Administration Medical Center.
Footnotes
Author Contribution
K.M.J. designed and performed experiments, analyzed data, and drafted the manuscript. S.M.E. and D.W.Z. designed and performed experiments, analyzed data, and edited the manuscript. S.C. and D.J.C. performed and analyzed experiments and edited the manuscript. A.J.G. and K.A.D. conceived experiments and edited the manuscript. All authors approved the manuscript in its final form.
References
- 1.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Current biology : CB. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 4.Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Current biology : CB. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 5.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 6.Schuldt A. Spire: a new nucleator for actin. Nature cell biology. 2005;7:107. doi: 10.1038/ncb0205-107. [DOI] [PubMed] [Google Scholar]
- 7.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nature cell biology. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bristow JM, Sellers MH, Majumdar D, Anderson B, Hu L, Webb DJ. The Rho-family GEF Asef2 activates Rac to modulate adhesion and actin dynamics and thereby regulate cell migration. J Cell Sci. 2009;122:4535–4546. doi: 10.1242/jcs.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakolitsa C, de Pereda JM, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- 10.Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 12.Wen KK, Rubenstein PA, DeMali KA. Vinculin nucleates actin polymerization and modifies actin filament structure. The Journal of biological chemistry. 2009;284:30463–30473. doi: 10.1074/jbc.M109.021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen ME, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Molecular cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Golji J, Mofrad MR. The interaction of vinculin with actin. PLoS computational biology. 2013;9:e1002995. doi: 10.1371/journal.pcbi.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson PM, Tolbert CE, Shen K, Kota P, Palmer SM, Plevock KM, Orlova A, Galkin VE, Burridge K, Egelman EH, Dokholyan NV, Superfine R, Campbell SL. Identification of an Actin Binding Surface on Vinculin that Mediates Mechanical Cell and Focal Adhesion Properties. Structure. 2014;22:697–706. doi: 10.1016/j.str.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. The Journal of biological chemistry. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 17.Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemljic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, Waterman CM. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. The Journal of cell biology. 2013;202:163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson RP, Craig SW. Actin activates a cryptic dimerization potential of the vinculin tail domain. The Journal of biological chemistry. 2000;275:95–105. doi: 10.1074/jbc.275.1.95. [DOI] [PubMed] [Google Scholar]
- 19.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. International review of cell and molecular biology. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. The Journal of cell biology. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang MT, Fu J, Wang YK, Desai RA, Chen CS. Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nature protocols. 2011;6:187–213. doi: 10.1038/nprot.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook R, Blake W, Rubenstein PA. Removal of the amino-terminal acidic residues of yeast actin. The Journal of biological chemistry. 1992 [PubMed] [Google Scholar]
- 24.Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 25.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nature methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumbauld DW, Lee TT, Singh A, Scrimgeour J, Gersbach CA, Zamir EA, Fu J, Chen CS, Curtis JE, Craig SW, Garcia AJ. How vinculin regulates force transmission. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9788–9793. doi: 10.1073/pnas.1216209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424:334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 28.Wood CK, Turner CE, Jackson P, Critchley DR. Characterisation of the paxillin-binding site and the C-terminal targeting sequence in vinculin. Journal of cell science. 1994;107:709–717. [PubMed] [Google Scholar]
- 29.Lee JH, Rangarajan ES, Yogesha SD, Izard T. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure. 2009;17:833–842. doi: 10.1016/j.str.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer SM, Playford MP, Craig SW, Schaller MD, Campbell SL. Lipid binding to the tail domain of vinculin: specificity and the role of the N and C termini. The Journal of biological chemistry. 2009;284:7223–7231. doi: 10.1074/jbc.M807842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palecek SP, Loftus JC, Ginsberg MH, Lauffenberger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 32.Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. Journal of applied physiology. 2005;98:1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 33.Shen K, Tolbert CE, Guilluy C, Swaminathan VS, Berginski ME, Burridge K, Superfine R, Campbell SL. The vinculin C-terminal hairpin mediates F-actin bundle formation, focal adhesion, and cell mechanical properties. The Journal of biological chemistry. 2011;286:45103–45115. doi: 10.1074/jbc.M111.244293. [DOI] [PMC free article] [PubMed] [Google Scholar]