SUMMARY
Oncogenic transformation by adenovirus small e1a depends on simultaneous interactions with the host lysine acetylases p300/CBP and the tumor suppressor RB. How these interactions influence cellular gene expression remains unclear. We find that e1a displaces RBs from E2F transcription factors and promotes p300 acetylation of RB1 K873/K874 to lock it into a repressing conformation that interacts with repressive chromatin-modifying enzymes. These repressing p300-e1a-RB1 complexes specifically interact with host genes that have unusually high p300 association within the gene body. The TGFβ-, TNF-, and interleukin-signaling pathway components are enriched among such p300-targeted genes. The p300-e1a-RB1 complex condenses chromatin in a manner dependent on HDAC activity, p300 lysine acetylase activity, the p300 bromodomain, and RB K873/K874 and e1a K239 acetylation to repress host genes that would otherwise inhibit productive virus infection. Thus, adenovirus employs e1a to repress host genes that interfere with viral replication.
INTRODUCTION
Adenovirus (Ad) E1A is a classic DNA virus oncogene (Weinberg, 2013). When expressed alone, small E1A (hereafter called “e1a”) (Figure 1A) causes G1-arrested cells to enter S phase (Ghosh and Harter, 2003). In cooperation with Ad E1B (Branton et al., 1985) or G12V HRAS (Ruley, 1983), e1a stably transforms rodent cells. Two interactions with host cell proteins are essential for e1a-induced cell transformation in cooperation with G12V HRAS: an interaction with RB family proteins (RB1, RBL1 [p107], and RBL2 [p130], hereafter referred to as “RBs”) and an interaction with the closely related nuclear lysine acetylases (KATs) p300 and CBP (Pelka et al., 2008). (Hereafter we refer to both p300 and CBP simply as “P300.”).
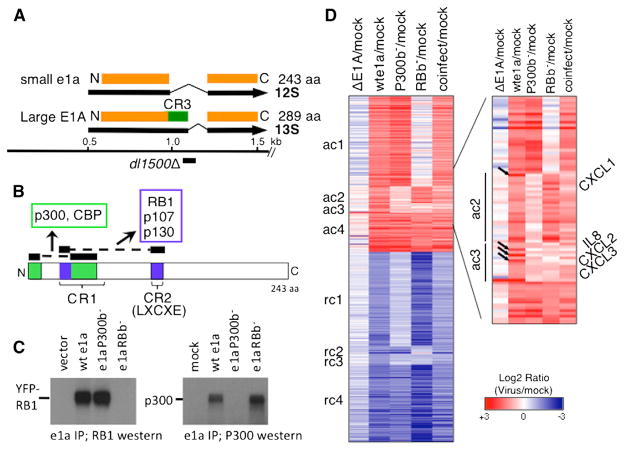
Figure 1. e1a-Regulated Host Cell Gene Expression.
(A) Major E1A early mRNAs and proteins and 9 bp dl1500 deletion.
(B) Regions of e1a that bind to RBs (blue) and p300/CBP (green).
(C) e1a mutant interactions with RB1 and P300. Extracts of HeLa cells transfected with expression vectors for YFP-RB1 (used in Figure 7) and the indicated e1a mutants (left) or infected with Ad vectors for the indicated e1a mutants (right) were immunoprecipited with anti-e1a mAb M73 and immunobloted with anti-RB1 (left) or anti-p300 (right) antibody.
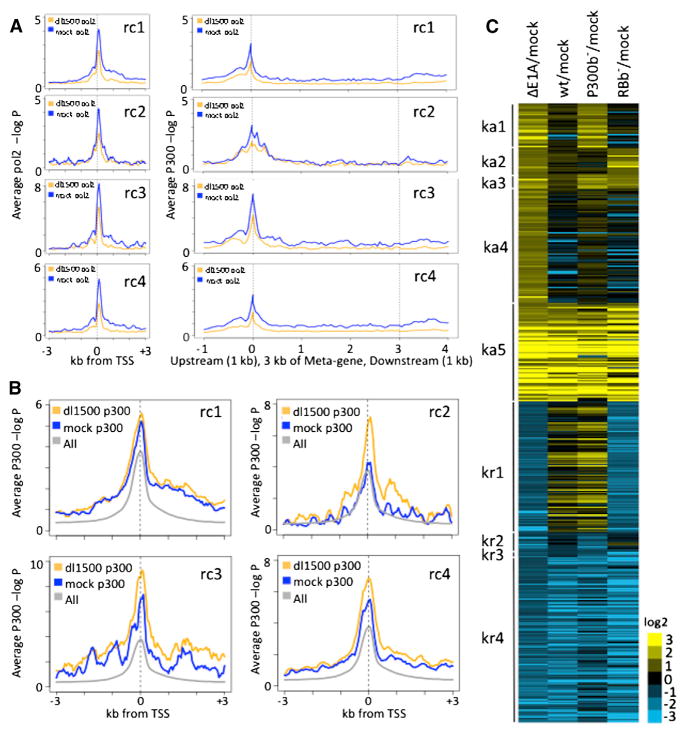
(D) Heat map of RNA increased (red) or decreased (blue) compared to mock-infected cells. See also Figure S1 and Tables S1, S2, S3, and S4.
E2F transcription factors regulate genes required to enter S phase (Dick and Rubin, 2013). In G1, G0 end-differentiated (Chong et al., 2009), and senescent cells (Chicas et al., 2010), RB proteins bind to E2F activation domains (ADs) (Lee et al., 2002; Xiao et al., 2003) repressing E2F-regulated genes by both masking the AD and by inducing repressive chromatin structure through interactions with chromatin-modifying enzymes (Dick and Rubin, 2013). In cycling cells, RBs are phosphorylated by cyclin D-CDK4/6 and cyclin E/A-CDK2, causing them to change conformation and dissociate from E2Fs and repressing chromatin-modifying complexes (Dick and Rubin, 2013), derepressing hundreds of genes required to enter S phase. e1a derepresses these same genes by directly displacing unphosphorylated RBs from E2Fs (Bagchi et al., 1990; Fattaey et al., 1993; Ikeda and Nevins, 1993), which explains why the e1a-RB interaction promotes entry into S phase. In contrast, the explanation for why e1a must bind P300 to transform cells is less clear.
Interestingly, an e1a mutant defective for binding RBs and a second e1a mutant defective for binding P300 do not complement for transformation, suggesting that a single e1a molecule must interact with both P300 and RBs to transform cells (Wang et al., 1995). Although there is negative cooperativity in the binding of both RB1 and a CBP TAZ2 domain to the same e1a molecule (Ferreon et al., 2013), such trimeric complexes form in vivo (Wang et al., 1995) and in vitro (Ferreon et al., 2013). In such complexes, e1a promotes acetylation of RB1 K873/K874 by P300, inhibiting binding of cyclin-CDKs to RB1 and hence RB1 phosphorylation during the cell cycle (Chan et al., 2001). P300 also acetylates e1a K239, inhibiting binding of importin-α3 to a NLS at the e1a C terminus (Madison et al., 2002).
How do the e1a-RB, e1a-P300 interactions and RB-e1a-P300 complexes influence cellular gene expression? To address these questions, we constructed Ad vectors for wild-type (WT) e1a and mutants completely defective for interactions with the RBs or P300. We performed RNA sequencing (RNA-seq) when infected cells were entering S phase. Mechanisms underlying the observed changes in expression were explored by chromatin immunoprecipitation sequencing (ChIP-seq) of RNA polymerase II (pol2), p300, RB1, and posttranslationally modified histones. We found that RB1 is enriched at E2F sites of genes activated by the e1a-RB interaction, much more than p130 or p107. This may help to explain why RB1, and not RBL1 or RBL2, is a tumor suppressor. Further, the results suggest why the e1a-P300 interaction is required for transformation. Importantly, e1a did not completely inhibit histone acetylation by P300. Instead, e1a regulated P300 HAT activity differently at different promoters and enhancers. We discovered an unexpected mechanism of e1a repression by targeting hypophosphorylated RB1 and p300 to the gene bodies of repressed genes. Fluorescently tagged proteins allowed direct visualization of e1a-driven chromatin condensation by a p300-e1a-RB1 complex, dependent on p300 KAT activity, the p300 bromodomain, and acetylation of RB1 and e1a. Our data suggest that e1a exploits the RBs displaced from E2Fs, locked into a repressing conformation by P300 acetylation, to repress host cell genes in pathways that would otherwise inhibit viral replication. Further, our data may explain why primate Ads express small e1a as well as large E1A.
RESULTS
Mutants Defective for Binding Either RBs or P300
Small e1a binds RBs through two interactions: one with the N-terminal ~10 aa of conserved region 1 (CR1) and one through the LXCXE region of CR2 (Lee et al., 1998; Liu and Marmorstein, 2007) (Figure 1B, blue). To completely eliminate e1a binding to RBs, we deleted CR2 (e1a aa 112–128) and mutated L43, L46, and Y47 to A, mutations that individually reduce the affinity of e1a CR1 for the RB1-pocket domain by 10-fold or more (Liu and Marmorstein, 2007). To eliminate the e1a-P300 TAZ2 (CH3) domain interaction, we constructed a multisite mutant based on the high-resolution structure of the e1a-CBP TAZ2 complex (Ferreon et al., 2009): R2G, E59A, V62A, F66A, and E68A. These mutations eliminate several e1a-TAZ2 electrostatic and hydrophobic interactions and mutate the N-terminal region that binds to the other side of TAZ2 relative to e1a residues 53–83 in CR1 (Figure 1B). We call the mutants e1aRB-binding minus (e1aRBb−) and e1a P300-binding minus (e1aP300b−) because they fail to coimmunoprecipitate RBs or p300/CBP, respectively, from extracts of transfected HeLa cells or infected IMR90 cells but bind the alternative interacting protein comparably to WT e1a (Figure 1C; Figures 1A–S1G available online). These mutants were incorporated into Ad5-vectors in the dl1500 background with a deletion of the unique E1A 13S mRNA splice site (Montell et al., 1984). Since large E1A is primarily responsible for activating other viral promoters (Montell et al., 1984; Winberg and Shenk, 1984), these vectors express only very low levels of the other viral early regions compared to WT Ad5. The vector expressing WT e1a drove contact-inhibited primary IMR90 fibroblasts into S phase ~20 hr postinfection (p.i.) (Figures S1H and S1I).
e1a-Regulated Cellular mRNA Expression
Contact-inhibited IMR90 cells arrested in G1 were mock infected; infected with dl312, an Ad5 mutant with a deletion of E1A (Jones and Shenk, 1979), with the Ad vectors expressing WT or mutant e1a; or coinfected with the e1aRBb− and e1aP300b− vectors. e1aRBb− accumulated to lower level than e1a WT or e1aP300b− when the vectors were infected at the same multiplicity of infection (MOI). Consequently, the e1aRBb− vector was used at 4-fold higher MOI to achieve nearly equal levels of WT and mutant e1a’s (Figure S1J). RNA-seq was performed with two biological replicates at 24 hr p.i. (Table S1). Genes with a difference in expression between mock- and WT e1a vector-infected cells of 2-fold or more and p < 0.01 for WT e1a between the two experiments are shown in Figure 1D and Table S2. Genes were clustered according to whether the change in expression required the e1a-RB, e1a-P300, both, or neither interaction. Expression of a representative gene and boxplots of expression levels for each cluster are shown in Figures S1K and S1L.
The e1a-RB interaction controlled expression of the largest cluster of activated genes (Figure 1D, ac1). The ac1 gene ontology is overwhelmingly enriched for S phase genes (Figure 2A; Table S3), amply confirming the generalization that most genes required for S phase are regulated by E2F activators repressed in G1 and G0 by RB-proteins. Detailed studies of the time course of changes in host gene expression following infection with WT Ad2 and closely related Ad5 have been performed in G1-arrested IMR90 cells and primary human foreskin fibroblasts using microarrays (Miller et al., 2007; Zhao et al., 2003), as well as RNA-seq at 12 and 24 hr p.i. (Zhao et al., 2012). The most highly induced genes had gene ontologies of DNA replication and cell cycle. These authors suggested this was due to E1A displacement of RBs from E2Fs. Our data show that this is indeed the case. Figure S2A compares genes regulated by WT Ad2 at 24 hr p.i to our data with the WT e1a vector. While there was considerable overlap in genes induced/repressed by the WT e1a vector compared to Ad2, larger numbers of genes were induced/repressed by Ad2. This is probably because of expression of all viral genes in Ad2-infected cells by 24 hr p.i. Also, we used very stringent criteria for classifying genes as induced/repressed (p < 0.01 in duplicate experiments) to maximize the opportunity of detecting similar trends in histone modifications and pol2, p300, and RB1 association among genes in the individual clusters.
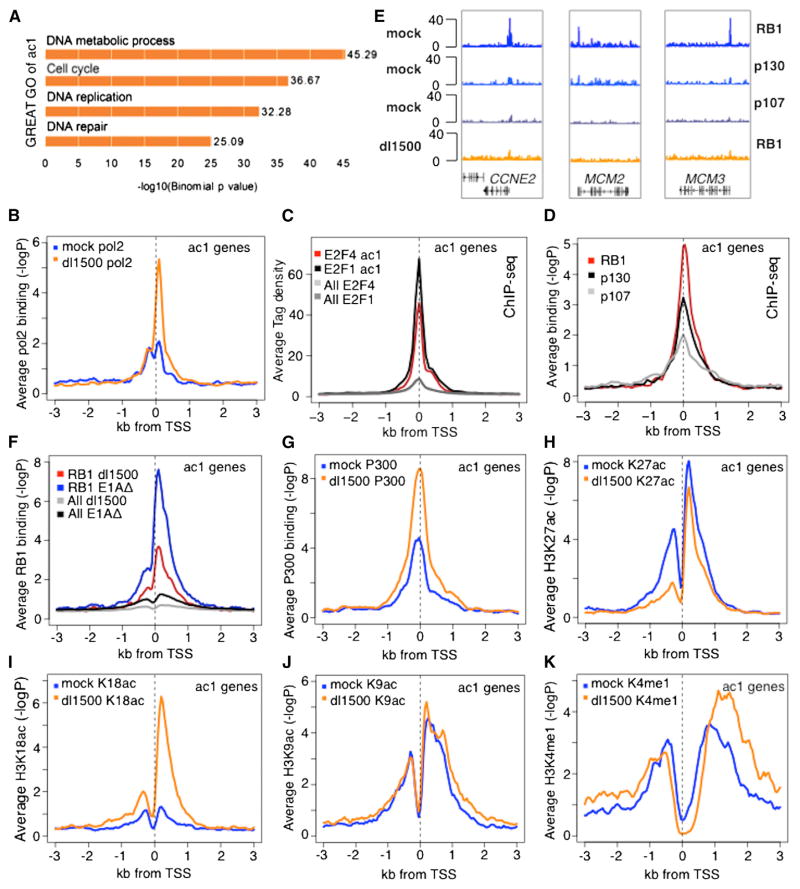
Figure 2. Gene Ontology and ChIP-Seq Data for ac1 Genes.
(A) Gene ontology determined by GREAT.
(B–D and F–K) Plots of average −log10 poissonP or tag density relative to TSS for the indicated proteins for ac1 genes in mock and dl1500-infected cells.
(E) Genome browser maps of ChIP-seq data (seq tags) for RBs for three ac1 genes.
ChIP-seq for pol2 (Figure 2B) showed that at ac1 promoters in mock-infected, G1-arrested cells there were on average small peaks of pol2 at ~+50 to +100 and ~−100 to −200, presumably due to pol2 that initiated transcription in the sense and antisense directions and then paused (Core et al., 2008; Seila et al., 2008). In dl1500-infected cells expressing e1a, there was a large increase in the pol2 peak for sense strand transcription, but not for antisense transcription. This e1a-induced increase in sense-oriented pol2 near the TSS strongly suggests that ac1 mRNAs increase because of increased transcription.
We anticipated that ac1 genes would have a peak of E2Fs near the TSS. Consequently we analyzed available ChIP-seq data for E2Fs 1 and 4 from HeLa cells (Bernstein et al., 2012), because we expected that genes required for S phase would be activated by E2Fs similarly in different human cell types. For example, similar genes are activated in RB1-deficient fibroblasts and pituitary and thyroid tumor cells (Black et al., 2003). Indeed, peaks of E2F1 and E2F4 were observed well above the average for all genes at ac1 promoters (Figure 2C) and not at genes in the other e1a-activated (Figure S2B) or repressed clusters (data not shown). Analysis of our ChIP-seq data for all three RBs in arrested IMR90 cells (Ferrari et al., 2012) showed peaks at ac1 promoters coincident with the E2F peaks (Figures 2C, 2D, and S3). Comparing the significance of peak heights relative to all ChIP data across the genome showed that RB1 association with ac1 promoters was significantly greater than for RBL2 (p130) or RBL1 (p107). Although each of the RB-family members was immunoprecipitated with a different specific antibody, the average ChIP-seq p value across all promoters was similar for each RB (Figure S2C). The much larger signal for RB1 at ac1 genes suggests that RB1 is the predominant RB family member at ac1 promoters. Examples include CCNE2 (cyclin E), the critical regulator of S phase entry, MCM2, and MCM3 (Figure 2E). e1a expression following infection with dl1500 decreased the average signal for RB1 at ac1 promoters more than 2-fold compared to cells infected with the E1A mutant (Figures 2E and 2F), demonstrating that e1a displaces RB1 from E2Fs in vivo, as it does in vitro.
The average peak of p300 at ac1 TSSs doubled in response to e1a (Figures 2G and S3). H3K18 and H3K27 are acetylated primarily by P300 (Horwitz et al., 2008; Jin et al., 2011). Surprisingly, although ac1 genes were repressed by RBs in the G1-arrested cells, H3K27 was acetylated to a significant extent at ac1 promoters (Figure 2H, 3A, and S3). The average H3K27ac downstream peak at ac1 promoters was slightly reduced by e1a, while the upstream peak fell considerably (Figure 2H). This differs from the profile in asynchronous IMR90 cells with ~50% of cells in S phase, where the upstream H3K27ac peak was higher (Hawkins et al., 2010) (Figure S2S). In contrast to ac1 genes, e1a decreased H3K27ac at most other promoters (Figures S2K, S2L, and S4C), including promoters of the other e1a-activated clusters (Figure S4B), intergenic regions, and introns, resulting in extensive global H3K27 deacetylation (Figures 3B–3E), even though there was little change in the sharp peaks of p300 association in intergenic regions (e.g., Figure S5).
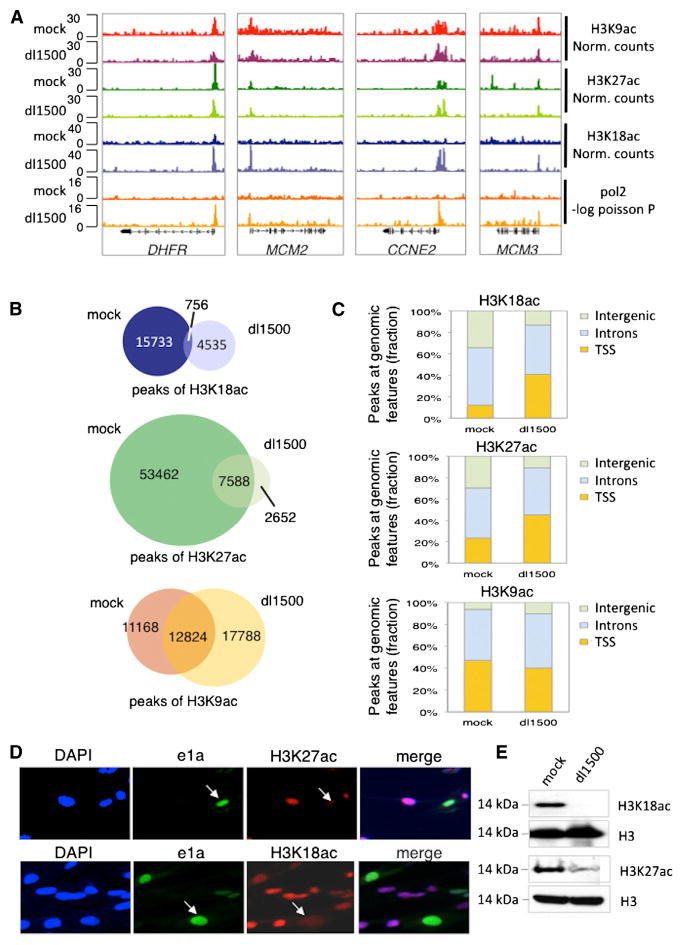
Figure 3. ChIP-Seq for ac1 Genes and Global H3K27 Hypoacetylation in Response to e1a.
(A) Gene browser plots of H3K18ac, H3K27ac, H3K9ac, and pol2.
(B) Venn diagrams of total significant peaks of H3K27ac in mock- and dl1500-infected cells and, for comparison, for H3K18ac and H3K9ac (Ferrari et al., 2012).
(C) Fraction of significant peaks shown in (B)1 at TSSs (3 kb from TSSs), intergenic regions (>3 kb upstream of TSSs and >3 kb downstream of polyA sites), and introns in mock- and dl1500-infected cells.
(D) Immunostaining of H3K27ac and H3K18ac in cells infected at MOI = 0.5.
(E) Western blots of total cell H3K27ac, H3K18ac, and total H3.
H3K18 is the other histone tail lysine acetylated primarily by P300. In contrast to H3K27, H3K18ac was low at ac1 promoters in G1-arrested mock-infected cells and increased greatly in response to e1a, primarily in the downstream direction (Figures 2I, 3A, and S3). Again, this was in contrast to asynchronous IMR90 cells where the upstream H3K18ac peak was higher (Figure S2T). Consequently, acetylation of the two histone tail substrates for P300, H3K18 and H3K27, was regulated differently by e1a at the activated promoters, whereas both H3K18ac and H3K27ac decreased dramatically at repressed promoters, intergenic regions, and introns (Figures 3B–3E, S2M, S2N, S4, and S5).
Like H3K27, H3K9 was acetylated at ac1 promoters in the G1- arrested mock-infected cells (Figure 2J). H3K9ac did not change significantly when the ac1 promoters were derepressed by e1a displacement of RB1. In contrast to H3K27ac and H3K18ac, e1a did not appreciably alter H3K9 at promoters of the other activated gene clusters (Figure S4A) or most intergenic regions (Figures 3B, 3C, and S4D). Similarly, H3K4me1 changes were modest in response to e1a (Figures 2K, S3, and S5).
RNA from ac4 genes increased 2-fold or more in response to WT e1a and both of the e1a mutants (Figures 1D and S1L). These genes may be regulated by e1a interactions with other host proteins besides RBs or P300 (Pelka et al., 2008). In this regard, it is interesting that while E2F binding motifs were highly enriched in ac1 promoters, as expected, other TF binding motifs were enriched in other clusters (Table S4). While E2Fs do not appear to be the major activators for clusters ac2–ac4, they may contribute to activation of some genes in these clusters, accounting for the small reduction in RNA in cells expressing e1aRBb− compared to WT e1a (Figure 1D, ac4), the small peaks of E2F association at the TSS in the average E2F profiles (Figure S2B), and the detection of E2F sites with less significant p values at ac3 and ac4 promoters (Table S4).
e1a Regulation of mRNA Stability
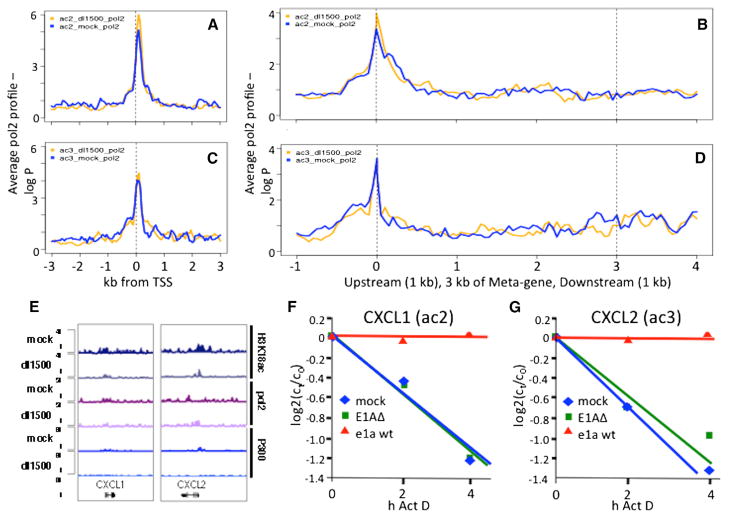
In contrast to ac1 and ac4 genes, e1a did not greatly increase pol2 or modify chromatin at ac2 and ac3 genes (Figures 4A–4E), even though their RNAs increased by >2. This suggests that the increased RNA results from posttranscriptional regulation. Indeed, the stability of mRNA from CXCL1 in ac2 and CXCL2 in ac3 encoding small cytokines increased dramatically in cells expressing e1a, as indicated by Actinomycin D chase experiments (Figures 4F and 4G). CXCL1 was assigned to ac2 because e1aRBb− induced it slightly more than 2-fold, but its activation by WT e1a and the e1a mutants was very similar to that of CXCL2, CXCL3, and IL8, another CXCL cytokine in cluster ac3 (Figure S2E). Remarkably, induction of these cytokines was not observed after infection with the e1aRBb− or e1aP300b− vectors or by coinfection of the two vectors (Figures 1D, right, and S2E). As discussed above, this implies that induction of these cytokines requires RB and P300 binding to the same e1a molecule.
Figure 4. e1a-Induction of CXCL1 and 2 by mRNA Stabilization.
(A–D) Average pol2 ChIP-seq signal from mock- (blue) and dl1500-infected (gold) ac2 ([A] and [B]) and ac3 ([C] and [D]) genes.
(E) Gene browser plots of CXCL1/2.
(F and G) Plots of RNA concentration (log2(ct/c0)) versus time of exposure to actinomycin D in cells infected with the E1AΔ mutant, the WT e1a vector, and mock-infected cells.
e1a-Mediated Repression
e1a significantly repressed slightly more host genes than it activated (Figure 1D). The largest cluster of repressed genes (rc1) required the e1a-P300 interaction for repression. rc1 genes are enriched for secreted glycoproteins comprising the extracellular matrix and were expressed at higher level than any of the other e1a-regulated clusters (Table S3; Figure S1L). Since production of extracellular matrix is a major function of fibroblasts, repression of these genes contributes to the dedifferentiation of cells induced by e1a (Pelka et al., 2008). This repression of abundantly expressed cellular genes may make more of the host cell RNA and protein synthesis capacity available for expression of viral genes. e1a decreased pol2 at the TSS and throughout the transcribed regions of rc1, rc3, and rc4 genes and at the TSS of rc2 genes (Figure 5A). Consequently, e1a repression is largely the result of decreased transcription. p300 association with the repressed promoters was either unchanged or increased (Figure 5B), although H3K18ac and H3K27ac, as well as H3K9ac, were reduced by e1a expression (Figure S4A–S4C).
Figure 5. e1a Repression and Inhibition of Activation and Repression Induced by an E1AΔ Mutant.
(A) Plots of average pol2 ChIP-seq signal relative to TSS and Meta-gene plots from TSS to TTS for e1a-repressed clusters.
(B) Average p300 ChIP-seq −log10 poisson P for e1a-repressed clusters.
(C) Heat map of genes activated (yellow) or repressed (blue) by E1AΔ-infection, clustered by whether the e1a-P300 (ka1), e1a-RB (ka2), both (ka3), or neither (ka4) interactions were required to inhibit induction 2-fold; whether induction was not inhibited by e1a (ka5); whether the e1a interaction with RB (kr1), P300 (kr2), or both (kr3) were required to inhibit repression by E1AΔ; or whether repression was not blocked by e1a (kr4). See also Table S5.
The pol2 ChIP-seq data revealed that the number of genes classified as repressed by WT e1a on the basis of the RNA-seq data is probably an underestimate of the number of transcription-ally repressed genes. For example, for the most abundantly expressed gene in IMR90 cells, COL1A1, pol2 association with the TSS and transcribed region in dl1500-infected cells fell to less than half the level in mock-infected cells, whereas COL1A1 RNA fell less than 2-fold (Figure S6A). This is probably because COL1A1 mRNA is relatively stable, and consequently, the fall in COL1A1 RNA occurs more slowly following inhibition of transcription than for less stable mRNAs. To better estimate the number of repressed and activated genes, we summed the pol2 ChIP-seq signal from the annotated TSS to the TTS from mock- and dl1500-infected cells. Of 24,507 annotated human genes, 3,944 (16%) had a drop in total pol2 ChIP-seq signal across the gene to ≤0.5 the mock-infected level. A total of 1,874 (7.6%) had an increase in total pol2.
e1a Control of Genes Induced in Response to Ad Infection
RNA-seq of cells infected with the E1A deletion mutant dl312 revealed cellular genes induced or repressed by Ad infection in the absence of e1a (Figure 5C; Table S5). e1a prevented induction/repression of the majority, but not all, of these genes. The e1a-regulated genes were clustered according to whether e1aP300b− or e1aRBb− also activated or repressed expression ≥2-fold. Of the genes whose induction was inhibited by e1a dependent on the e1a-P300 interaction (ka1), CDKN1A encoding the p21CIP inhibitor of cyclin-CDKs was the most abundantly expressed. ka1 genes also include SESN2, an mTOR inhibitor (Figures S6B and S6C).
Most of the cellular genes repressed by infection with the E1A mutant are in cluster kr1 (Figure 5C). e1a prevents repression of these genes via the e1a-RB interaction. The gene ontology of kr1 genes is overwhelmingly related to the cell cycle (p = 1.4 × 10−34). These genes probably are repressed by infection with the E1A mutant, because a DNA damage response is activated by the termini of the viral DNA in the absence of E1B and E4 functions (Weitzman and Ornelles, 2005).
High p300 and RB1 throughout the Transcribed Region of Repressed Genes
Repression of the largest cluster of e1a-repressed genes (rc1) required the e1a-P300 interaction (Figure 1D) and was associated with hypoacetylation of H3K18 and H3K27 at their promoters and associated intronic and intergenic regions (Figures 3B, 3C, S4B, S4C, and S5). These results suggest that e1a inhibits P300 HAT activity at repressed promoters and intergenic regions. But since not all genes are repressed, what distinguishes repressed genes?
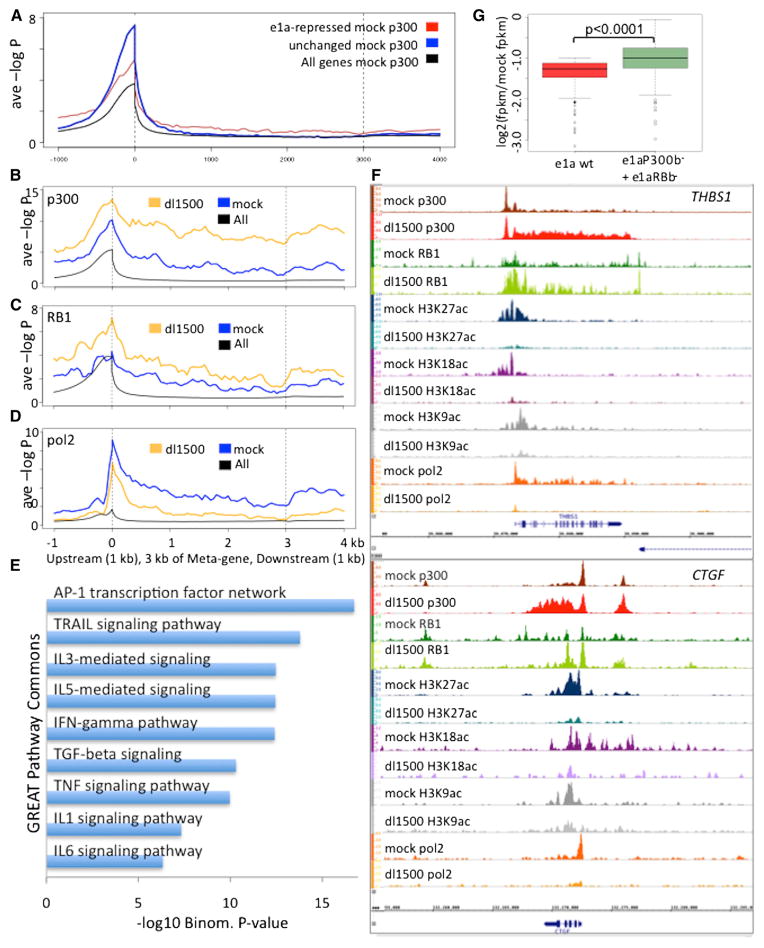
In analyzing p300 association with the e1a-repressed genes, we noted that the average p300-association within their transcribed regions was higher than for a group of genes expressed at similar level but whose expression was not altered by e1a (Figure 6A). This surprising association of p300 throughout the transcribed region of repressed genes, in contrast to the sharp peaks of p300 in most other regions of the genome (e.g., Figure S5), was particularly striking for 76 genes in TRAIL, TNF, TGFβ, and interleukin-signaling pathways that might otherwise inhibit fibroblast cell cycling and viral replication (Figures 6B and 6E; Table S6). e1a induced greater p300 association with these genes at the promoter and a few kb upstream, and even more so throughout the transcribed region (Figure 6B). RB1 also was observed at the promoter and spread throughout the bodies of these genes, and also increased in response to e1a (Figure 6C). At the same time, e1a substantially diminished pol2 association with these genes (Figure 6D). Examples of repressed genes in this group include THBS1 and CTGF, important activators of TGFβ signaling (Figure 6F). The e1aP300b− and e1aRBb− mutants failed to cause the increase in RB1 association (Figure S6B). These results indicate that e1a represses genes that have high P300 association within the gene body before infection and that for genes such as THBS1, CTGF, CYR61, KLF6, and KLF10 (Table S6) in the TGFβ signaling pathway, repression is associated with increased p300 and RB1 throughout the transcribed region.
Figure 6. Repressed Genes with p300 and RB1 throughout the Gene Body.
(A) Average p300 ChIP-seq signal for rc1 genes from mock-infected cells in Meta-gene plots (red), the control group (blue), and the average of all annotated genes (black).
(B–D) Average ChIP-seq data for 76 genes (Table S6) that had high p300 ChIP-seq signal in the gene body in mock-infected cells. Meta-Gene plots of ChIP-seq data for p300 (B), RB1 (C), and pol2 (D).
(E) GREAT gene ontology probabilities categorized as pathways for this cluster of 76 genes.
(F) Examples of Gene Browser plots of ChIP-seq data from mock- and dl1500-infected cells for THBS1 and CTGF.
(G) Boxplots of log2 fold repression of rc1 genes from cells infected with the WT e1a vector (red) or coinfected with the e1aRBb− and e1aP300b− vectors (green). See also Table S6.
In these and subsequent box plots, the horizontal dark line is the median of the distribution; the box includes 50% of the data; the whiskers include 75% of the data. The p value for the significance of the difference between two distributions indicated by brackets was calculated using one-way ANOVA and a Tukey’s HSD post hoc comparison.
This high level of p300 and RB1 association with the transcribed region was most obvious for the 76 genes in Table S6. However, repression of all the genes in the large rc1 cluster was significantly greater in cells infected with the e1a WT vector than in cells coinfected with the e1aP300b− and e1aRBb− vectors (Figure 6G), even though the level of e1a proteins was similar in both groups of cells (Figure S1J). This result suggests that, while repression of rc1 genes required primarily the e1a-P300 TAZ2 domain interaction, the e1a-RB interaction contributed to the full extent of repression by WT e1a. Genes in cluster rc3 required e1a interaction with both P300 and RBs for >2-fold repression (Figures 1D and S1L), including proapoptotic IFIT1 and IFIT2, repressed much less by e1aP300b− plus e1aRBb− than by WT e1a (Figures S6D and S6E).
Chromatin Condensation by a P300-e1a-RB Complex, Dependent on P300 Acetylation of RB1 and e1a
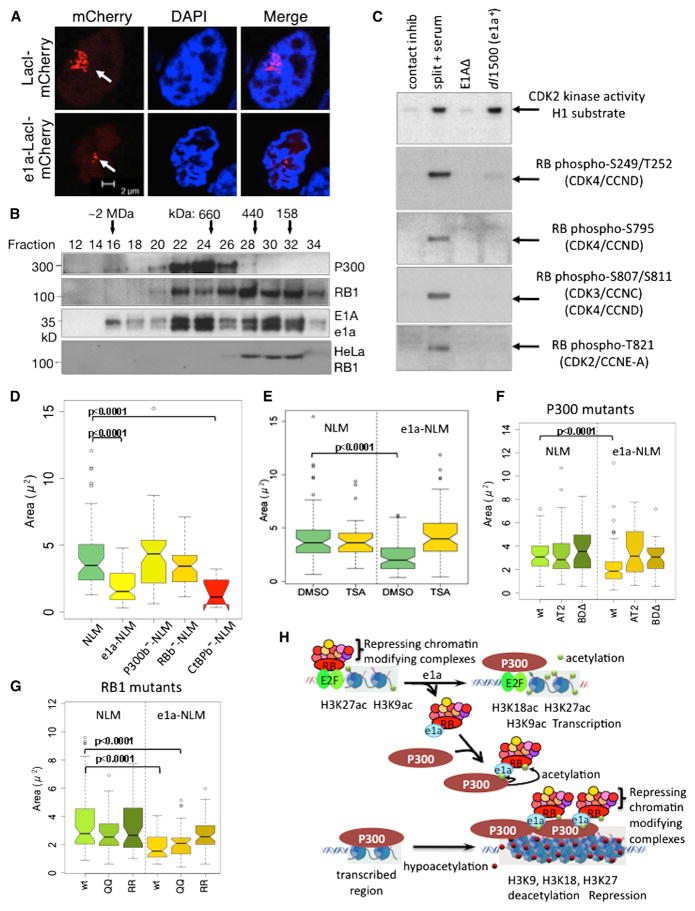
Earlier, using ChIP-chip, we observed that cross-linking of total histone H3 increased in response to e1a at promoter regions of repressed genes with increased p300 and RB1/p130 association (Ferrari et al., 2008). We therefore asked whether e1a causes chromatin condensation, accounting for the increased H3 cross-linking. To assay chromatin condensation, we used a microscopic method that allowed direct observation of chromatin condensation by HP1 (Verschure et al., 2005). The method utilizes CHO cells (RRE.1) engineered to contain ~104 lac operators amplified over a region of ~1 Mb (lacO array) in one chromosome. When LacI-mCherry with an N-terminal SV40 NLS (NLM) was expressed in these cells, the lacO array was visualized by confocal fluorescence microscopy spread through ~5%–10% of the nuclear volume (Figure 7A). When e1a fused to NLS-LacI-mCherry (e1a-NLM) was expressed, the lacO array condensed into a much smaller volume in the confocal slice with the largest area of red fluorescence (Figure 7A). Array areas measured in ~100 cells had a mean value in cells expressing e1a-NLM ~1/2 that of the area in cells expressing NLM, the difference being highly significant (p < 0.0001) (Figure 7D). However, no difference was observed when e1aRBb− or e1aP300b− mutants were fused to NLM, when e1aRBb−-NLM and e1aP300b−-NLM were coexpressed, or when e1a was not fused to LacI (Figures 7D and S7A–S7C). Thus, WT e1a binding to the lacO array caused ~4-fold condensation in volume, dependent on simultaneous e1a interactions with both P300 and RBs. Further, no condensation was observed when e1a with eight ala-nines substituted into the e1a p400 binding region from 25–36 (Fuchs et al., 2001) (e1aP400b−) was fused to NLM (Figure S7D).
Figure 7. e1a-Induced Chromatin Condensation.
(A) Confocal micrographs of mCherry fluorescence in RRE.1 cells transfected with vectors for NLS-LacI-mCherry (NLM) and e1a-NLS-LacI-mCherry (e1a-NLM).
(B) Western blots of Superose 6 column fractions of 293 or HeLa nuclear extract.
(C) Top panel: autoradiogram of gel for CDK2 kinase activity. Lower panels: immunoblots for the indicated phospho-RB1 sites. Lanes from left to right were from mock-infected cells, cells that were split 1 to 3 into fresh media with 20% FBS 24 hr earlier, and cells infected with the E1AΔ mutant or dl1500.
(D–G) Boxplots of mCherry-fluorescent areas in the confocal slice with the largest area.
(H) Model for e1a regulation of host cell gene activation and repression through interactions with RBs and P300. See Discussion and Figure S7.
We found that HDAC activity also was required for chromatin condensation, as treatment of transfected cells with the HDAC inhibitor trichostatin A for 4 hr before fixation reversed e1a-NLM condensation (Figure 7E). Interestingly, a large E1A-NLM fusion failed to condense the lacO array (Figure S7D). This may be because E1A CR3 (Figure 1A) associates with several HATs contributing to CR3-dependent activation (Ablack et al., 2012; Pelka et al., 2009a, 2009b). This may reverse histone hypoacetylation required for e1a-induced condensation.
E1A induces P300 acetylation of RB1 at K873 and K874 near the RB1 C terminus by targeting p300 KAT activity to these lysines in a p300-E1A-RB1 complex (Chan et al., 2001). Acetylation at these sites in RB1 inhibits RB1 phosphorylation by cyclin-CDKs, inhibiting progression into S phase. Consistent with this, we observed coelution of p300, e1a, and RB1 by gel filtration of nuclear extract from e1a-expressing 293 cells, but not from HeLa cells lacking e1a (Figure 7B). Also, in IMR90 cells stimulated to enter S phase by infection with dl1500, RB1 phosphorylation at several cyclin-CDK target sites was inhibited, despite induction of cyclin E (Figure S1K) and high CDK2 kinase activity in extracts of the same cells (Figure 7C). To determine if p300 KAT activity required to acetylate these RB1 sites is required for chromatin condensation, we overexpressed a p300 AT2 mutant having greatly attenuated KAT activity (Kraus et al., 1999). This prevented condensation by e1a-NLM but did not alter the size of the larger array observed with NLM alone (Figure 7F). Interestingly, p300 with a bromodomain deletion also interfered with e1a-induced chromatin condensation (Figure 7F). In addition, expression of RB1 with arginine substitutions at K873/874 also inhibited condensation, but not expression of RB1 K873Q/K874Q, chemical mimics of acetylated lysines (Figure 7G).
P300 acetylates e1a at K239 (Zhang et al., 2000; Madison et al., 2002). e1a K239R also prevented condensation, as well as greatly reducing colocalization of YFP-P300 with e1a-NLM at the array, while mutant K239Q did not (Figures S7E–S7G). e1a K239 acetylation was reported to inhibit binding of corepressors CtBP1/2 to the e1a C terminus (Zhang et al., 2000), although contradictory results have been reported (Madison et al., 2002). In either case, CtBP binding is not required for condensation, since mutation of the e1a PXDLS consensus CtBP binding site (233–237) to ALAAA did not inhibit array condensation (Figure 7D). We conclude that chromatin condensation by a complex of P300/p400-e1a-RB1 requires acetylation of RB1 K873/K874 and e1a K239 by p300 and the p300 bromodomain.
DISCUSSION
This analysis of host cell gene expression in response to Ad small e1a and e1a mutants reveal how the e1a interactions with RBs and the TAZ2 (CH3) domain of p300/CBP (P300) regulate host cell gene expression. For the most part, the two interactions regulate genes independently. Genes in the largest group of activated genes, ac1 (Figure 1D), are activated by the e1a-RB interactions independently of e1a-P300 interactions. The e1a-P300 TAZ2 domain interaction is responsible for most e1a host cell gene repression (Figure 1D, rc1). In addition, e1a activation and repression of a smaller number of host genes requires e1a interactions with both RBs and P300. Many of these are not properly regulated by a combination of e1aRBb− and e1aP300b−, suggesting that e1a regulation of this class of genes requires formation of a trimeric P300-e1a-RB complex (Wang et al., 1995). Also, our mutants may possibly affect one of the many other e1a-host protein interactions reported (Pelka et al., 2008).
e1a interactions with RB1 and CBP are examples of binding-induced folding of an intrinsically unstructured polypeptide (Ferreon et al., 2009, 2013). Nonetheless, there is great complexity in the spatially and chemically complementary interactions at the e1a-RB and e1a-TAZ2 interfaces. Because of the specificity of these interactions, it is likely that genes activated by WT e1a but not by e1aRBb− (ac1 genes, Table S2) comprise the cellular genes regulated principally by E2F-RB complexes in G1-arrested primary IMR90 cells. RB1 is the primary RB-family member binding to E2Fs at promoters of these genes. This helps to explain why RB1 is a tumor suppressor, and not p130 or p107. Further, Chicas et al. (2010) found that induction of senescence is more dependent on RB1 than p130 or p107, likely contributing to its unique function as a tumor suppressor.
e1a Regulates Histone Acetylation by P300 Differently at Different Loci
e1a was reported to inhibit the histone acetylase (HAT) activity of P300 in vitro (Chakravarti et al., 1999). But this is due to competitive inhibition by e1a, which is also a substrate, as opposed to allosteric regulation (Madison et al., 2002). Our results indicate that in the cell, e1a regulation of P300 is subtler than simply inhibiting their HAT activities indiscriminately. H3K18 and H3K27 are acetylated principally by P300 in vivo (Horwitz et al., 2008; Jin et al., 2011). At ac1 promoters, e1a induced hyperacetylation of H3K18, while acetylation of H3K27 was unexpectedly high at ac1 promoters in the G1-arrested cells and remained almost unchanged (Figures 2H and 2J). At ac2, ac3, and ac4 promoters, e1a increased H3K18ac but decreased H3K27ac (Figures S4B and S4C). Finally, e1a caused extensive H3K18 and H3K27 hypoacetylation at most other sites in the genome, including intergenic regions and introns that contain enhancers and at e1a-repressed promoters (Figures 3B, 3D, S5B, and S5C). In e1a-expressing cells, the total number of significant peaks of H3K27ac across the genome fell to only ~17% the level in uninfected cells. H3K18ac also fell in intergenic and intronic regions (Ferrari et al., 2012). How e1a might inhibit P300 HAT activity at some locations, but activate it at others, is presently unclear. We note that although H3K27ac is a mark of active enhancers (Creyghton et al., 2010), RNA- and pol2 ChIP-seq data indicate that for the majority of genes transcription was reduced <2-fold by this extensive reduction in H3K27ac in intergenic and intronic regions where enhancers reside.
Promoter H3K18ac Is Linked to Transcriptional Activation
It is remarkable that transcription of e1a-regulated genes correlated best with H3K18ac at promoters and not with H3K9ac, H3K27ac, or H3K4me1 (Figure S4). Surprisingly, promoter proximal nucleosomes of E2F-RB-regulated genes (cluster ac1) were highly acetylated on H3K9 and H3K27 in the contact-inhibited cells where these promoters are repressed by RB1. These promoters may be in a chromatin structure that is poised for activation in primary fibroblasts, since these genes must be activated rapidly during one of the most important fibroblast functions—wound healing—which requires prompt fibroblast replication. What mechanism would couple final H3K18ac to transcription? TRIM33, a coactivator of SMAD TFs, has a PHD-bromodomain cassette that binds H3K18ac through its bromodomain and a neighboring H3K9me3 through its PHD domain, displacing HP1 to derepress TGFβ target genes (Xi et al., 2011). H3K9 is trimethylated at RB-repressed Cdc6 and CcnA promoters before e1a activation in serum-depleted NIH 3T3 cells (Ghosh and Harter, 2003; Sha et al., 2010), so TRIM33 may bind H3K18ac in these cells. Further studies will be required to identify what interacts with H3K18ac at e1a-activated ac1 promoters in IMR90 cells.
Activation of Small CXCL Cytokine Genes by mRNA Stabilization
e1a increased CXCL1 and 2 mRNAs by stabilizing them (Figure 4). Induction of these genes as well as IL8 and CXCL3 requires the e1a interactions with both RBs and P300 (Figure 1D). All of these homologous cytokines bind and activate the same GPCR, CXCR2 (Rosenkilde and Schwartz, 2004). Stabilization of these mRNAs is a response activated by e1a rather than a cellular response to viral infection because they were not induced by infection with an E1A deletion or the vectors for e1aP300b− or e1aRBb−. It seems surprising that adenovirus would evolve a mechanism to activate expression of these proinflammatory cytokines. However, these cytokines stimulate expression of viral receptors and consequently increase infection of neighboring cells (Lütschg et al., 2011).
e1a probably stabilizes the CXCL cytokine mRNAs indirectly by inducing genes for proteins that stabilize mRNAs with AU-rich 3′ UTRs and repressing genes for proteins that cause degradation of such mRNAs. In this regard, it is interesting that e1a induced an isoform of ELAVL2 (NM_001171197) nearly 10-fold and repressed ZFP36L1 more than 2-fold, since ELAV-family proteins stabilize mRNAs with AU-rich elements and ZFP36-family proteins destabilize them (Mukherjee et al., 2014; Rattenbacher and Bohjanen, 2012). The requirement for e1a binding to both RBs and P300 may be for complete repression of genes that destabilize these mRNAs.
e1a Inhibition of Cellular Responses to Viral Infection may Explain Why the e1a-P300 Interaction and a P300-e1a-RB Complex Are Required for Transformation
Previously, it was not clear why e1a must interact with P300 to transform primary cells. Results from cells infected with the E1AΔ mutant provide potential answers. Genes whose induction by E1AΔ is inhibited by WT e1a, dependent on the e1a-P300 interaction, include CDKN1A and stress-induced SESN2, an mTOR inhibitor (Budanov and Karin, 2008) (Figures S6B and S6C). Genes whose induction by E1AΔ is inhibited dependent on simultaneous interactions between e1a and both P300 and RBs include IFIT1 and IFIT2, interferon-induced proapoptotic proteins (Reich, 2013) (Figures S6D and S6E). e1a’s ability to inhibit induction of these genes following the stress associated with transfection likely contributes to the requirement for these simultaneous interactions for transformation.
A Complex of p300-e1a-RB1 Condenses Chromatin
Perhaps the most unanticipated results in these studies came from searching for a mechanism that accounts for e1a-repression of some, but not all, genes. The largest class of repressed genes requires the e1a-P300 interaction for repression (rc1, Figures 1D and S1L). This might suggest that e1a targets genes for repression that have P300 associated with their control regions. However, this includes virtually all expressed genes (Visel et al., 2009). We noted that e1a-repressed genes have a higher average p300 association within their transcribed regions, the “gene body,” compared to genes expressed at a comparable level that were unchanged in their expression by e1a (Figure 6A). A high level of p300 spread throughout the gene body in uninfected cells was observed in gene browser views of 76 genes (Table S6) enriched for genes of the TRAIL; TGFβ; TNF; and IL1, IL3, and IL5 signaling pathways (Figures 6B and 6E). p300 association with the bodies of these genes increased to high levels in response to e1a. Repression of the TGFβ and TNF pathways by Ad2 and Ad5 infection was noted earlier (Zhao et al., 2012 and references therein). Remarkably, many of the same genes showed similar association of p300 with the gene body when they were induced in serum-starved T98G glioblastoma cells (Ramos et al., 2010) (Figure S6G). However, this was associated with strong activation in T98G cells, whereas these same genes were repressed by e1a in IMR90 cells. This is probably because e1a also induced RB1 association, H3 hypoacetylation, and reduced pol2 within these gene bodies (Figures 6B, 6D, 6F, and S4A–S4C). Based on these data, we suggest that one mechanism to target e1a-repression is for e1a to associate with P300 in genes that have a high level of P300 throughout the gene body, coupled with e1a cobinding to hypophosphorylated, repressing RB proteins associated with repressing chromatin modifying enzymes. The association of P300 and RB1 with these gene bodies in uninfected IMR90 and T98G cells, and the increase in p300 and RB1 in response to serum in uninfected T98G cells (Figures 6B, 6C, 6F, and S6G), suggest that similar mechanisms regulate these genes in uninfected cells. Ad may exploit this cellular mechanism to target hypophosphorylated RBs to these genes, which inhibit cell cycling and promote apoptosis and cytokine secretion, repressing them.
While this high level of p300 and RB1 in gene bodies was observed clearly at only 76 genes, including three lncRNAs of unknown function (Table S6), we note that repression of the large rc1 cluster was significantly greater for WT e1a than for e1aP300b− plus e1aRBb− expressed at similar level (Figure 6G). This suggests that, although the e1a-P300 interaction is primarily responsible for e1a repression of rc1 genes, assembly of P300-e1a-RB complexes contributes to their maximal repression by WT e1a. The stronger repression of rc1 and rc4 genes by e1aRBb− compared to WT e1a (Figure 1D) may have resulted from the higher level of this mutant in these experiments (Figure S1J) or because the absence of negative cooperativity in the cobinding of RB and P300 (Ferreon et al., 2013) results in binding of the e1aRBb− mutant to a larger fraction of total cellular P300.
To explore the mechanism of e1a repression further, we employed a microscopic, cell biological method for directly visualizing chromatin condensation (Figure 7A). When WT e1a was bound to a large, extended array of lacO sites in RRE.1 CHO cells (Verschure et al., 2005) by expression of an e1a-NLS-LacI-mCherry fusion (e1a-NLM), the volume occupied by the lacO array condensed to ~1/4 the volume visualized with NLM alone (Figures 7A and 7D). Moreover, this ability to condense chromatin required interaction of both P300 and RBs with the same e1a molecule, as well as the e1a interaction with p400, HDAC activity, the KAT activity of P300, the P300 bromo domain, and P300 acetylated lysines in RB1 and e1a. RB1 remained hypophoshorylated even when CDK2-cyclin E activity was induced, presumably because e1a-induced P300 acetylation of RB1 K873/K874 inhibits RB1 phosphorylation by cyclin-CDKs (Chan et al., 2001). This is expected to maintain RB1 (and probably the other RBs) in a repressing conformation that interacts with repressing chromatin-modifying enzymes (Dick and Rubin, 2013). We propose that this causes chromatin associated with P300/p400-e1a-RB to become hypoacetylated and condensed, repressing transcription. In this way Ad appears to exploit RB repression complexes released from E2Fs by e1a by targeting them to genes that would otherwise inhibit viral replication (Figure 7H). P400 is a SWI/SNF chromatin remodeler. SWI/SNF complexes also function in other examples of repression (Martens and Winston, 2003). The requirement of the e1a p400 binding region (aa 25–36) (Fuchs et al., 2001) for direct visualization of e1a-mediated chromatin condensation (Figure S7D) is consistent with the model that remodelers can “close” chromatin as well as “open” it.
Importantly, large E1A did not induce chromatin condensation (Figure S7D), revealing an activity of small e1a that is not shared with large E1A, despite the presence of all of the small e1a sequence in the larger protein. This may provide an explanation for why all primate Ads express both proteins via alternative RNA splicing and why a mutant virus that cannot express small e1a because of a point mutation in the 12S mRNA 5′ splice site replicates as well as wild-type virus in cycling primary cells, but not in G1-arrested cells (Spindler et al., 1985).
The finding that the p300 bromo domain is required for e1a-mediated chromatin condensation is interesting in light of the required p300 acetylation of e1a and RB1. It is unlikely that the p300 bromo domain is required for binding to acetylated histones, since the repressed genes become extensively hypoacetylated (Figure 6F). P300 bromo domains might bind e1a K239ac and/or RB1 K873ac/K874ac to form of a lattice of multiple P300-e1a-RB complexes through such interactions in addition to those diagrammed in Figure 1B (Figure 7H). Such a network of interactions might help to explain how p300 and RB1 virtually “coat” the genes in Table S6. It might also explain why YFP-P300 showed reduced association with e1a-K239R at the lacO array in RRE cells (Figures S7F and S7G). Further studies will be required to test these ideas.
EXPERIMENTAL PROCEDURES
Cell Culture
IMR-90 primary human fetal lung fibroblasts (ATCC Number: CCL-186) were obtained from the ATTC and Sigma-Aldrich. They were grown at 37°C in Dulbecco’s modified Eagle’s medium plus 10% FBS, penicillin, and streptomycin in a 5% CO2 incubator until they reached confluence. Cells were then incubated 2 days more and were either mock infected or infected with the indicated Ad5-based vectors.
RNA-Seq
Low-passage IMR-90 cells were mock-infected or infected with Ad5 E1A-E1B-substituted, E3-deleted vectors expressing WT Ad2 small E1A proteins from the dl1520 deletion removing the 13S E1A mRNA 5′ splice site (Montell et al., 1984) as described in the text, 2 days after reaching confluence. RNA was isolated 24 hr p.i. using QIAGEN RNeasy Plus Mini Kit. Eluted RNA was treated with Ambion DNase Treatment and Removal reagent and then Ambion TRIzol reagent, precipitated with isopropanol, and dissolved in sterile water. RNA concentration was measured with a Qubit fluorometer. One microgram of RNA was copied into DNA and PCR amplified with bar-coded primers for separate samples to prepare sequencing libraries using the Illumina TruSeq RNA Sample Preparation procedure. Libraries were sequenced using the Illumina HIseq-2000 to obtain 50-base-long reads. Sequences were aligned to the hg19 human genome sequence using TopHat v2. Fpkm (fragments per kb per million mapped reads) for each annotated hg19 RefSeq transcript was determined using Cuffdiff v2 from Cufflinks RNA-Seq analysis tools at http://cufflinks.cbcb.umd.edu.
ChIP-Seq
Preparation of cross-linked chromatin free of RNA, sonication, and immunoprecipitation was as described in (Ferrari et al., 2012). ChIP of RB1 was done using formaldehyde and DSG cross-linking as described (Chicas et al., 2010). Sequencing libraries were constructed from 1 ng of immunoprecipitated and input DNA using the NuGen Ovation Ultralow DR Multiplex System 1–8 kit. Analysis of sequence data was as described in Ferrari et al. (2012), except that the genome was tiled into 50 bp windows.
Confocal Microscopy of Transfected RRE.1 Cells
RRE.1.1 cells (Verschure et al., 2005) were plated on fibronectin-coated glass bottom 35 mm dishes (MatTek Corporation) and transfected with 2 μg expression vector for e1a or e1a mutant-NLS-LacI-mCherry, or cotransfected with 2 μg WT e1a-NLS-LacI-mCherry and 2 μg YFP-p300 or YFP-P300 mutant, or YFP-RB1, or YFP mutant RB1 in the pCDNA expression vector using the CMV IE enhancer/promoter with Lipofectamine 2000 (Invitrogen). After 24 hr cells were fixed in 1.6% formaldehyde, washed in 1× PBS, mounted onto slides, and imaged for colocalization of mCherry and YFP using a Leica TCS SP2 AOBS single-photon confocal microscope using a 63×1.4-numerical-aperture oil immersion objective. Micrographs were analyzed with ImageJ software to subtract all background that was not at least 25% of maximum fluorescence and subjected to particle analysis in ImageJ to identify foci of fluorescence and measure their areas in μm2. The bromodomain deletion in YFP-P300 BDΔ included p300 amino acids 1,071–1,241. p values for differences between the distributions of data shown in boxplots was calculated using Kaleidograph to perform one way ANOVA with a Tukey’s HSD post hoc comparison. The “p400b−” mutant fused to SV40 NLS-lac I-mCherry (NLM) used in Figure S7D has e1a mutations E25A, E26A, V27A, L28A, D30A, L32A, P35A, and S36A. The “CtBPb−” mutant fused to NLM used in Figure 7D has e1a mutations P233A, D235A, L236A, and S237A.
Supplementary Material
Acknowledgments
Supported by CA25235 to A.J.B., NIH Director’s Innovator Award (1-DP2-OD006515) to S.K.K., GM095656 to M.P., NRSAs T32HG02536 to M.N., T32GM007185 to S.J., and T32AI060567 to N.Z. We thank Professor Ulf Pettersson (Uppsala University) for providing bam files from Zhao et al. 2012.
Footnotes
ACCESSION NUMBERS
All RNA- and ChIP-seq data has been uploaded to NCBI GEO accession number GSE59693.
Supplemental Information includes seven figures, six tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2014.10.004.
AUTHOR CONTRIBUTIONS
All authors contributed to experimental design and data interpretation. G.J. constructed the Ψ5* expression vectors. D.G. performed RNA-seq and ChIP-seq for RB1, R.F. for p300, T.S. for H3K27ac and H3K4me1, and M.N. for pol2. R.F., M.P., and N.Z. performed bioinformatic analysis; A.Y. and M.N. performed gel filtration westerns of Figure 7C; and S.J. performed the microscopy of Figures 7 and S9.
References
- Ablack JN, Cohen M, Thillainadesan G, Fonseca GJ, Pelka P, Torchia J, Mymryk JS. Cellular GCN5 is a novel regulator of human adenovirus E1A-conserved region 3 transactivation. J Virol. 2012;86:8198–8209. doi: 10.1128/JVI.00289-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi S, Raychaudhuri P, Nevins JR. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A transactivation. Cell. 1990;62:659–669. doi: 10.1016/0092-8674(90)90112-r. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black EP, Huang E, Dressman H, Rempel R, Laakso N, Asa SL, Ishida S, West M, Nevins JR. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res. 2003;63:3716–3723. [PubMed] [Google Scholar]
- Branton PE, Bayley ST, Graham FL. Transformation by human adenoviruses. Biochim Biophys Acta. 1985;780:67–94. doi: 10.1016/0304-419x(84)90007-6. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JL, Wenzel PL, Sáenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattaey AR, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Su T, Li B, Bonora G, Oberai A, Chan Y, Sasidharan R, Berk AJ, Pellegrini M, Kurdistani SK. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc Natl Acad Sci USA. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Ghosh MK, Harter ML. A viral mechanism for remodeling chromatin structure in G0 cells. Mol Cell. 2003;12:255–260. doi: 10.1016/s1097-2765(03)00225-9. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda MA, Nevins JR. Identification of distinct roles for separate E1A domains in disruption of E2F complexes. Mol Cell Biol. 1993;13:7029–7035. doi: 10.1128/mcb.13.11.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N, Shenk T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc Natl Acad Sci USA. 1979;76:3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Lee C, Chang JH, Lee HS, Cho Y. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 2002;16:3199–3212. doi: 10.1101/gad.1046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007;21:2711–2716. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütschg V, Boucke K, Hemmi S, Greber UF. Chemotactic antiviral cytokines promote infectious apical entry of human adenovirus into polarized epithelial cells. Nat Commun. 2011;2:391. doi: 10.1038/ncomms1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison DL, Yaciuk P, Kwok RP, Lundblad JR. Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J Biol Chem. 2002;277:38755–38763. doi: 10.1074/jbc.M207512200. [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Miller DL, Myers CL, Rickards B, Coller HA, Flint SJ. Adenovirus type 5 exerts genome-wide control over cellular programs governing proliferation, quiescence, and survival. Genome Biol. 2007;8:R58. doi: 10.1186/gb-2007-8-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Courtois G, Eng C, Berk A. Complete transformation by adenovirus 2 requires both E1A proteins. Cell. 1984;36:951–961. doi: 10.1016/0092-8674(84)90045-x. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Jacobs NC, Hafner M, Kennington EA, Nusbaum JD, Tuschl T, Blackshear PJ, Ohler U. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014;15:R12. doi: 10.1186/gb-2014-15-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka P, Ablack JN, Fonseca GJ, Yousef AF, Mymryk JS. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J Virol. 2008;82:7252–7263. doi: 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka P, Ablack JN, Shuen M, Yousef AF, Rasti M, Grand RJ, Turnell AS, Mymryk JS. Identification of a second independent binding site for the pCAF acetyltransferase in adenovirus E1A. Virology. 2009a;391:90–98. doi: 10.1016/j.virol.2009.05.024. [DOI] [PubMed] [Google Scholar]
- Pelka P, Ablack JN, Torchia J, Turnell AS, Grand RJ, Mymryk JS. Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP. Nucleic Acids Res. 2009b;37:1095–1106. doi: 10.1093/nar/gkn1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos YF, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van Galen M, van Dam H, van Ommen GJ, den Dunnen JT, Zantema A, ‘t Hoen PA. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010;38:5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenbacher B, Bohjanen PR. Evaluating posttranscriptional regulation of cytokine genes. Methods Mol Biol. 2012;820:71–89. doi: 10.1007/978-1-61779-439-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich NC. A death-promoting role for ISG54/IFIT2. J Interferon Cytokine Res. 2013;33:199–205. doi: 10.1089/jir.2012.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkilde MM, Schwartz TW. The chemokine system — a major regulator of angiogenesis in health and disease. APMIS. 2004;112:481–495. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Ghosh MK, Zhang K, Harter ML. E1A interacts with two opposing transcriptional pathways to induce quiescent cells into S phase. J Virol. 2010;84:4050–4059. doi: 10.1128/JVI.02131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler KR, Eng CY, Berk AJ. An adenovirus early region 1A protein is required for maximal viral DNA replication in growth-arrested human cells. J Virol. 1985;53:742–750. doi: 10.1128/jvi.53.3.742-750.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HG, Moran E, Yaciuk P. E1A promotes association between p300 and pRB in multimeric complexes required for normal biological activity. J Virol. 1995;69:7917–7924. doi: 10.1128/jvi.69.12.7917-7924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. The Biology of Cancer. New York, NY: Garland Science; 2013. [Google Scholar]
- Weitzman MD, Ornelles DA. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene. 2005;24:7686–7696. doi: 10.1038/sj.onc.1209063. [DOI] [PubMed] [Google Scholar]
- Winberg G, Shenk T. Dissection of overlapping functions within the adenovirus type 5 E1A gene. EMBO J. 1984;3:1907–1912. doi: 10.1002/j.1460-2075.1984.tb02066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Spencer J, Clements A, Ali-Khan N, Mittnacht S, Broceño C, Burghammer M, Perrakis A, Marmorstein R, Gamblin SJ. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc Natl Acad Sci USA. 2003;100:2363–2368. doi: 10.1073/pnas.0436813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Granberg F, Elfineh L, Pettersson U, Svensson C. Strategic attack on host cell gene expression during adenovirus infection. J Virol. 2003;77:11006–11015. doi: 10.1128/JVI.77.20.11006-11015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Dahlö M, Isaksson A, Syvänen AC, Pettersson U. The transcriptome of the adenovirus infected cell. Virology. 2012;424:115–128. doi: 10.1016/j.virol.2011.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.