Abstract
Tumor drug delivery is a complex phenomenon affected by several elements in addition to drug or delivery vehicle’s physico-chemical properties. A key factor is tumor microvasculature with complex effects including convective transport, high interstitial pressure and enhanced vascular permeability due to the presence of “leaky vessels”. Current in vitro models of the tumor microenvironment for evaluating drug delivery are oversimplified and, as a result, show poor correlation with in vivo performance. In this study, we report on the development of a novel microfluidic platform that models the tumor microenvironment more accurately, with physiologically and morphologically realistic microvasculature including endothelial cell lined leaky capillary vessels along with 3D solid tumors. Endothelial cells and 3D spheroids of cervical tumor cells were co-cultured in the networks. Drug vehicle screening was demonstrated using GFP gene delivery by different formulations of nanopolymers. The synthetic tumor network was successful in predicting in vivo delivery efficiencies of the drug vehicles. The developed assay will have critical applications both in basic research, where it can be used to develop next generation delivery vehicles, and in drug discovery where it can be used to study drug transport and delivery efficacy in realistic tumor microenvironment, thereby enabling drug compound and/or delivery vehicle screening.
INTRODUCTION
In recent years, myriad delivery technologies have been employed to deliver novel cancer therapeutics ranging from antibodies, cytokines, gene therapy and traditional chemical drugs to tumors. Furthermore, drug delivery vehicles ranging from viral (e.g., adenovirus, lentivirus) and non-viral vectors (e.g., polymers, liposomes, nanoparticles) have been developed [1–3] to enhance the delivery performance. The efficacy of any new therapeutic in eradicating tumors depends critically on uniform and effective delivery of the drugs [4–6] to all the tumor cells. The possibility of even a single cell to not come in contact with the drug can lead to regeneration of tumors and even worse, one that is drug-resistant [6–9].
High-efficiency drug delivery to tumors is a daunting challenge and rendered difficult primarily due to the complexity of the tumor microenvironment. The tumor microenvironment [9,10] is highly heterogeneous comprising of tumor and stromal cells (e.g., fibroblasts, inflammatory cells) embedded in an extracellular matrix connected to a vascular supply for nutrients. It also has gradients of cell proliferation and differential regions of hypoxia and acidity. In addition, solid tumors which account for more than 85% of the cancers have less than 10% of blood vessels. One of the unique features of the tumor vasculature is their leakiness as a result of the discontinuity of the endothelium [11, 12]. Studies using in vivo data have shown that the pore size of the leaky vessels ranges from 100s of nanometer to a few microns in a mouse mammary carcinoma [13]. In comparison, the vascular permeability in normal tissues is typically less than 6 nm [14] with the largest size of 150 nm in spleen endothelium [15].
Several in vivo techniques have been developed to study tumor drug delivery. A commonly used model employs windowed chambers in dorsal skin [16–18] or brain models [19–20] to study drug distribution. A relatively new method is the use of systems like the IVIS® optical imaging system (PerkinElmer, Waltham, MA) that can detect non-invasively fluorescent tags in live animals. However, such in vivo studies are expensive and require skilled personnel due to the use of live animals.
In contrast, in vitro models are a cost-effective means to study and screen drug delivery vehicles. In classical studies, the delivery vehicle containing the therapeutic of interest (drug/fluorescent tags) is incubated with the tumor cells in culture. At regular time points, the cells are analyzed either for uptake of the fluorescent tags or reduction in cell proliferation as a measure of delivery efficacy. Improvements to monolayer experiments in tissue culture have led to the development of in vitro methods which use multicellular tumor spheroids [21–23]. However, these static methods [24] do not account for transport across the vascular endothelium and the complex microvascular network structure observed in vivo. Furthermore, depending of the model, they rely exclusively on diffusion for the drugs to permeate the tumors and do not allow real-time visualization to study the diffusion of the delivery vehicle and/or drugs due to the use of semipermeable membrane. Recent research has focused on the development of microfluidic devices to study cellular behavior under fluidic conditions [25–28]. Studies incorporating angiogenesis, tumor growth, invasion and tumor-endothelial cell interactions have also been reported [29–35]. However, all of these devices are not well-suited for the study of tumor drug delivery vehicles in conditions representing in vivo scenarios.
In this study, we report on the development of a microfluidics based synthetic vasculature assay that models the tumor microenvironment observed in vivo. This synthetic tumor network builds upon our previous work where we developed a novel methodology for reproducing microvascular networks digitized from in vivo images of rodent vasculature onto a microfluidic device [36–38]. The microfluidic device recreates the in vivo tumor microenvironment encompassing (a) circulatory flow in the vessels derived from in vivo morphology, (b) transport across the leaky vessel walls based on engineered barriers between the vascular and the tumor cells, and (c) delivery to 3D culture of tumor cells across the interstitial space. The combination of these features distinguishes the present synthetic tumor network model from other in vitro models discussed above. Two nanopolymeric based gene delivery systems were tested and the results were compared with in vivo rodent data highlighting the predictive ability of the microfluidic device and assay.
MATERIALS AND METHODS
Design of Synthetic Tumor Network
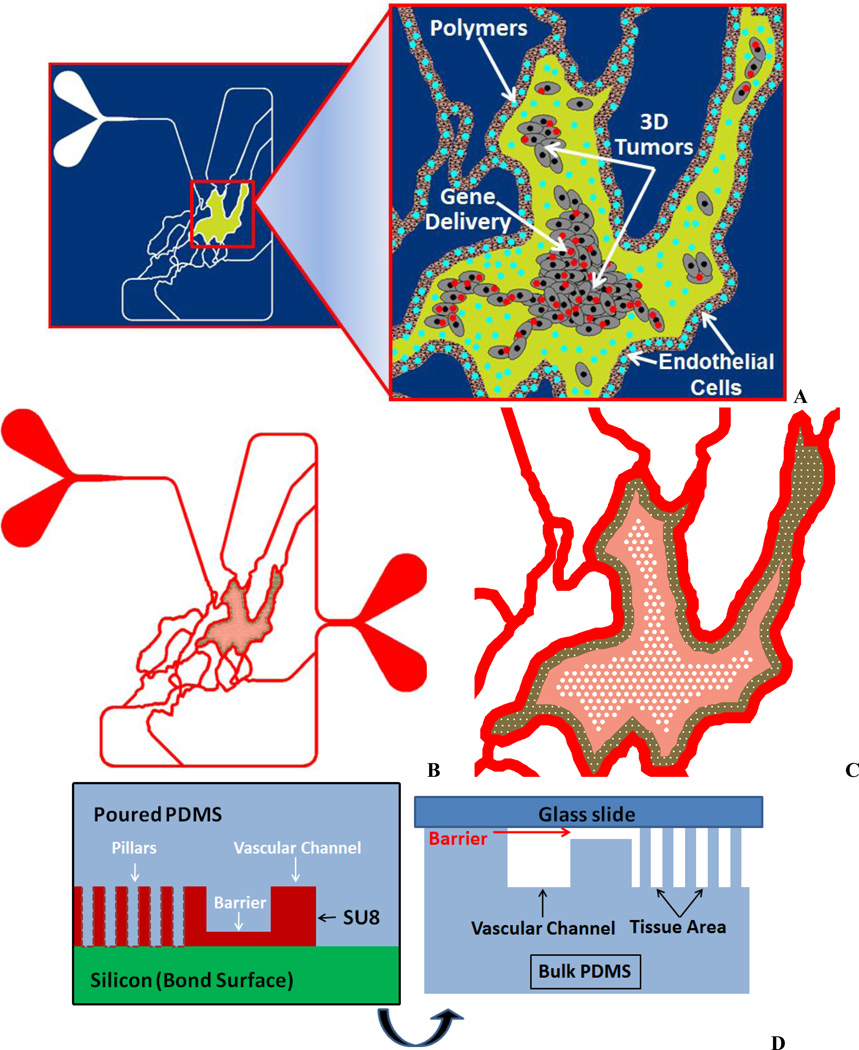
The microvascular network digitized previously [37] was modified to include regions for growth of tumors and the leaky gaps between the vessel lumen and the tumor growth region. The largest tissue area from the network was selected and the vessel wall adjacent to the tumor growth region was modified in AutoCAD to include 2 µm size leaky gaps, typical pore size found in MCa-IV mouse mammary carcinomas vessel walls [13]. A cylindrical micro pillar array with prescribed dimensions of 50 µm diameter, 100 µm height and 50 µm spacing was designed to create a scaffold for 3D tumor growth in the tissue area. Figure 1A show a schematic of the Synthetic Tumor Network and Figure 1B–C shows the image highlighting the microfabricated pillars for 3D culture. Figure 1D shows the side view schematic of the 2 µm leaky gaps and the microfabricated scaffolds.
Figure 1.
Design of Synthetic Tumor Network. A. Concept. B. Schematic showing the vascular channels for culturing endothelial cells and the tissue compartment for culturing tumor cells.
C. Magnified view of the tissue chamber showing the scaffolds for the 3D tumor.
D. Concept with side view showing the 2 µm leaky gaps
Microfabrication of Synthetic Tumor Network
The designed devices were fabricated using PDMS based soft-lithography. The tumor area was separated from the vascular channels using the barrier method shown in Figure 1D. The barrier is structured on SU-8 by patterning an extra layer in addition to the fluidic layer, which contained the pillars, channels and access port holes, to form a thin slab between the tissue area and the vascular channels. The two step fabrication process for the soft lithography masters was as follows: (a) 500 µm thick, 4″ diameter p-type Si wafers were organically cleaned and dehydrated @ 200 °C for 5 min, (b) SU8 spin deposition to obtain 2 µm film, (c) Hot plate @ 65 °C for 1 min → 95 °C for 2 min, (d) Exposure at ~250 mJ/cm2, (e) Hot plate @ 65 °C for 1 min → 95 °C for 1 min (allow to cool for 10 min), (f) Develop in PGMEA (SU8 developer) until field clears (<1 min), (g) Spin coat fluidic SU8 layer @ 100 µm over existing features, (h) Hot plate @ 95 °C for 30 min, (i) Exposure @ ~250 mJ/cm2, (j) Hot plate @ 65 °C for 1 min → 95 °C for 5 min (allow to cool for 10 min), (k) Develop in PGMEA (SU8 developer) until field clears (5 min) and finally rinse with IPA. SEM images of the SU-8 masters were acquired using a Hitachi S-2600N (Hitachi High Technologies America, Inc., Pleasanton, CA) scanning electron microscope. Samples were coated with 50 nm of Au using a Hummer 6.2 sputtering system (Anatech Ltd., Union City, CA). An acceleration voltage of 15 kV was used for subsequent imaging.
Sylgard 184 PDMS (Dow Corning) was poured over the developed master to generate devices in PDMS and cured at 60 °C overnight in an oven, following which the PDMS was peeled off from the master. Through holes, defining the inlets and outlets, were punched using a 1.5 mm biopsy punch. For injection of tumor cells, a 30 gauge blunt and sharpened needle was used to punch holes in the tumor area using a stereo microscope for proper alignment of the access port. The surfaces of the PDMS and a pre-cleaned glass slide were cleaned using oxygen plasma treatment prior to bonding. Tygon Microbore tubing with an outside diameter of 0.06 inch and inner diameter of 0.02 inch served as the connecting ports for fluidic interface.
Fluidic Testing
A fluorescent marker (FITC) was used to visualize the leakiness of the fabricated synthetic tumor network. FITC at a concentration of 10 µg/ml was injected into the network using a syringe pump (PHD 2000, Harvard Apparatus, MA) at a flow rate of 1 µl/min. An image of the entire device was acquired using an automated stage (LEP Ltd) mounted on an inverted fluorescence microscopy system (NIKON, Melville, NY). Images were visualized using NIKON Elements software. In order to test the leakiness of the 2 µm barriers, 1 µm and 5 µm fluorescent particles (Fisher Scientific, Pittsburgh, PA) were injected into the vascular chamber and their penetration into the tumor chamber was visualized.
Co-Culture of Endothelial and Tumor Cells in Synthetic Tumor Network
We utilized an immortalized endothelial cell line, RBE4 (courtesy of Dr. Michael Aschner, Vanderbilt University Medical Center, Nashville, TN) to represent the vascular cells while the commonly used tumor cell line (HeLa-cervical cancer) was chosen to represent the tumor cells in the synthetic tumor network. RBE4 cells were cultured in Eagle’s Minimum Essential Medium and Ham’s F-10 media (1:1) supplemented with 10% FBS, 1% Pen/Strep, 2 mM L-glutamine and G418 (300 µg/mL). Cells were incubated at 37 °C, 95% humidity and 5% CO2 until confluent. HeLa cells were obtained from ATCC (#CCL-2™) and maintained in DMEM media with 10% serum supplemented with, 4 mM L-glutamine and 100 U/ml penicillin/streptomycin on T25 tissue culture flask at 37 °C in 5% CO2. Confluent cells for both types were trypsinized and sub-cultured at a ratio of 1:3 until ready for experiments.
HeLa cells (~107/ml) were harvested and mixed in a ratio of 1:3 with cold Matrigel™ for a total volume of 50 µl. The solution was mixed uniformly and 10 µl of the solution injected slowly into the tumor area access port of the device. The device was kept on ice bath until this process was completed. Sterile cell culture media without serum was continuously perfused at a flow rate of 10 µl/min in the vascular channel side to flush out any HeLa cells entering the vessel lumen. The device was then incubated overnight at 37 °C, 95% humidity and 5% CO2 until confluent. The next morning, fibronectin at a concentration of 50 µg/ml and flow rate of 1 µl/min was injected into the vascular channels for 30 min followed by incubation for another 30 min. Endothelial cells were trypsinized and injected into the vascular channels at concentration of 5×106 cells/ml. Flow was stopped for 30 min by clamping the inlet and outlet for 2 hour. At the end of two hours, fresh media was injected into the channels overnight (RBE4 media mixed with HeLa media at 1:1 and allowed to perfuse overnight at flow rate of 0.1 µl/min. RBE4 cells and HeLa cells were allowed to grow together for additional 24 hour prior to initiation of the delivery system screening experiments. Co-cultured RBE4 cells and HeLa cells were assayed using Calcein AM (Life Technologies, Carlsbad, CA), a cell-permeant dye used to determine cell viability.
Delivery System Screening in Synthetic Tumor Network
We compared two nanopolymer based gene delivery systems: (1) PPC, and (2) Express-In in the Synthetic Tumor Network. Express-In is a commercially available polymer based transfection reagent that has been shown to produce very high transfection activity in a variety of cell types in vitro [39]. PPC is a polymeric delivery system that has been shown to efficiently deliver plasmid in vivo [40] and has been tested in clinically for the delivery of IL-12 plasmid in ovarian cancer patients with recurrent platinum resistant ovarian cancer [41–42]. When tested in vivo (intraperitoneal delivery), Express-In is associated with relatively high levels of toxicity in contrast to PPC which has shown to be well tolerated in both pre-clinical and clinical studies [40–42]. The two polymers labeled with Rhodamine (fluorescent tag) at a ratio of 3.6:1 wt/wt were complexed with GFP encoding DNA for a total concentration of 10 µg/ml. This concentration is the optimal concentration utilized for the transfection studies.
Assays were conducted in two ways. In the first assay, HeLa cell transfection was monitored following polymer injection from the vascular channel. In the second assay, HeLa cell transfection was monitored by injecting the complexed polymers directly at the tumor site. To ensure that at least 3× volume of the polymer/GFP complex was circulated in the networks, the complexes were injected into the network for 30 min at a flow rate of 0.5 µl/min. At the end of 30 min, flow was immediately switched to cell culture medium comprising of 1:1 of RBE4 and HeLa culture mediums. A circulating flow was maintained for 24 hours before GFP expression was measured in the tumor area.
RESULTS
Fabrication and Testing of Synthetic Tumor Network
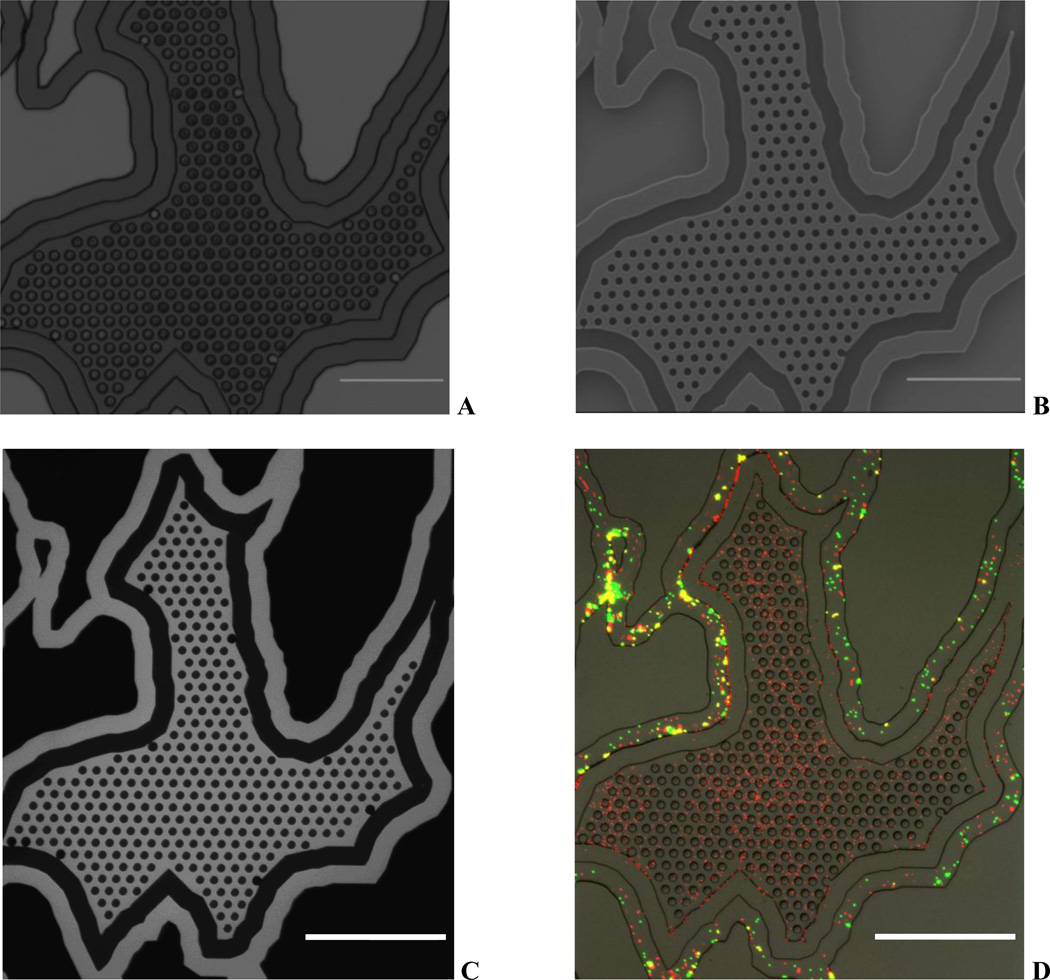
Figure 2A shows the image of the microfabricated synthetic tumor network highlighting the vascular channel, walled barrier and the tumor chamber. Figure 2B shows the SEM image with detailed pattern of the microfabricated pillars, which are used as scaffolds for 3D culture of tumor cells. Figure 2C shows network perfused with the fluorescent dye and Figure 2D shows image perfused with the particles highlighting the intact barrier between the tumor and vascular channels. In addition, the images demonstrate that the tumor area with microfabricated scaffolds for the 3D culture of tumor cells is fully functional with the 2µm leaky vasculature.
Figure 2.
Microfabricated Synthetic Tumor Network. A. Optical image of SU-8 master with wall barrier. B. SEM Image showing the tissue area in the network with microfabricated pillars for tumor growth. C. FITC perfused device indicating fluidically connected vascular and tissue chambers. D. Particles (1 µm – red; 5 µm – green) perfused device indicating a leaky barrier. 1 µm particle freely perfuse to the tumor area whereas 5 µm are restricted to vascular channel.
Scale bars: 500 µm.
Co-Culture of Endothelial and Tumor Cells in Synthetic Tumor Network
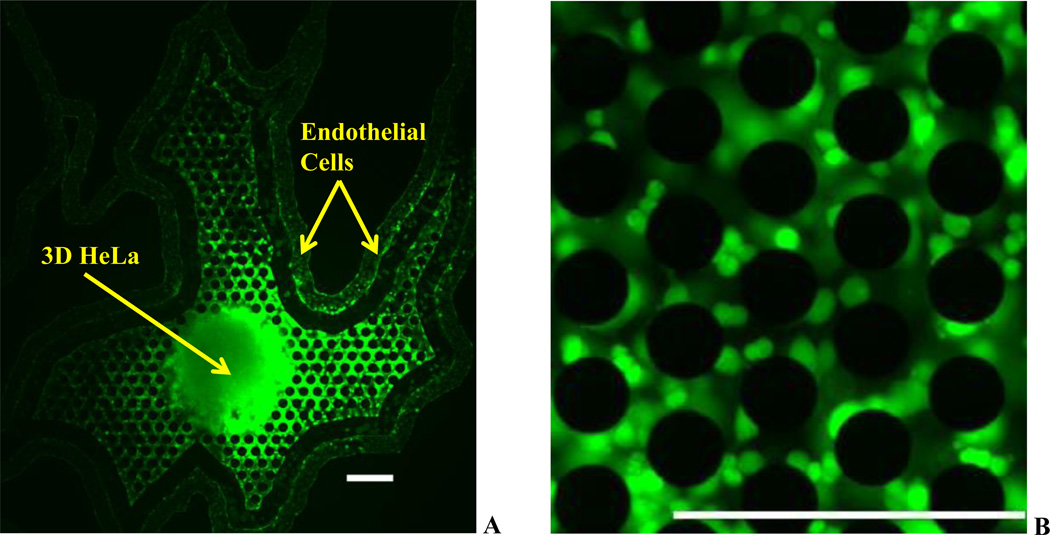
Figure 3 shows the network with HeLa cells cultured in 3D in the tumor region and endothelial cells in the vascular region stained with calcein AM. As can be seen from the images, the cells (endothelial and tumor cells) were in healthy condition. In addition, several smaller 3D spheroids were observed growing around the micropillars with varying number of cellular colonies at each of the location. Uniform calcein AM labeled cells indicate a fully active co-culture system of endothelial cells and tumor cells in the network.
Figure 3.
Co-culture of Tumor (HeLa) and Vascular (Endothelial) Cells in Synthetic Tumor Networks. A. Co-culture of 3D HeLa Cells cultured on microfabricated scaffolds and endothelial cells cultured in the vascular lumen seperated by the walled barrier. B. Magnified view of the microfabricated scaffolds showing daughter HeLa cell colonies.
Scale bars: 250 µm
Delivery System Screening in Synthetic Tumor Network
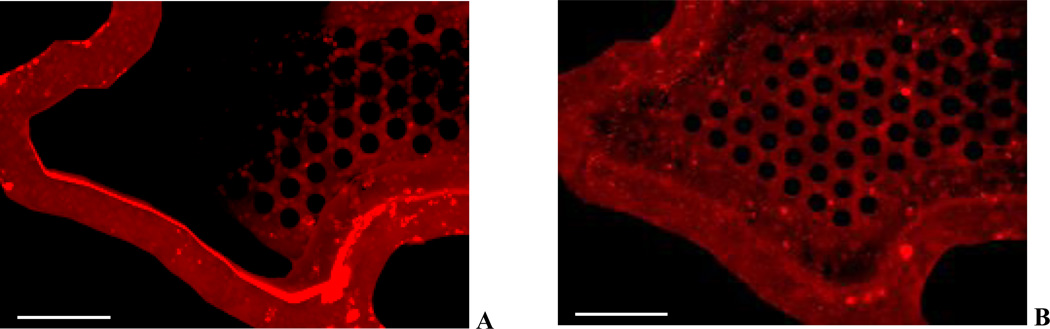
Figure 4 show images of Rhodamine labeled Express-In (Figure 4A) and Rhodamine labeled PPC polymers (Figure 4B) in the synthetic tumor network. As can be observed, Rhodamine signal for Express-In is more intense in the vessel lumen compared to PPC which is more uniform. The increased fluorescence intensity is indicative of particle aggregation, which is not seen in the case of PPC, where minimal aggregation of particles is observed. In addition, Express-In is found to have more aggregation near the tortuous bends and turns of the network compared to linear sections of the network. Finally, PPC is found to be more uniformly dispersed in the tissue chamber compared to Express-In.
Figure 4.
Polymers in Synthetic Tumor Network. A. Express-In shows non-uniform distribution and significant aggregation. B. PPC shows uniform distribution and minimal aggregation.
Scale bars: 250 µm
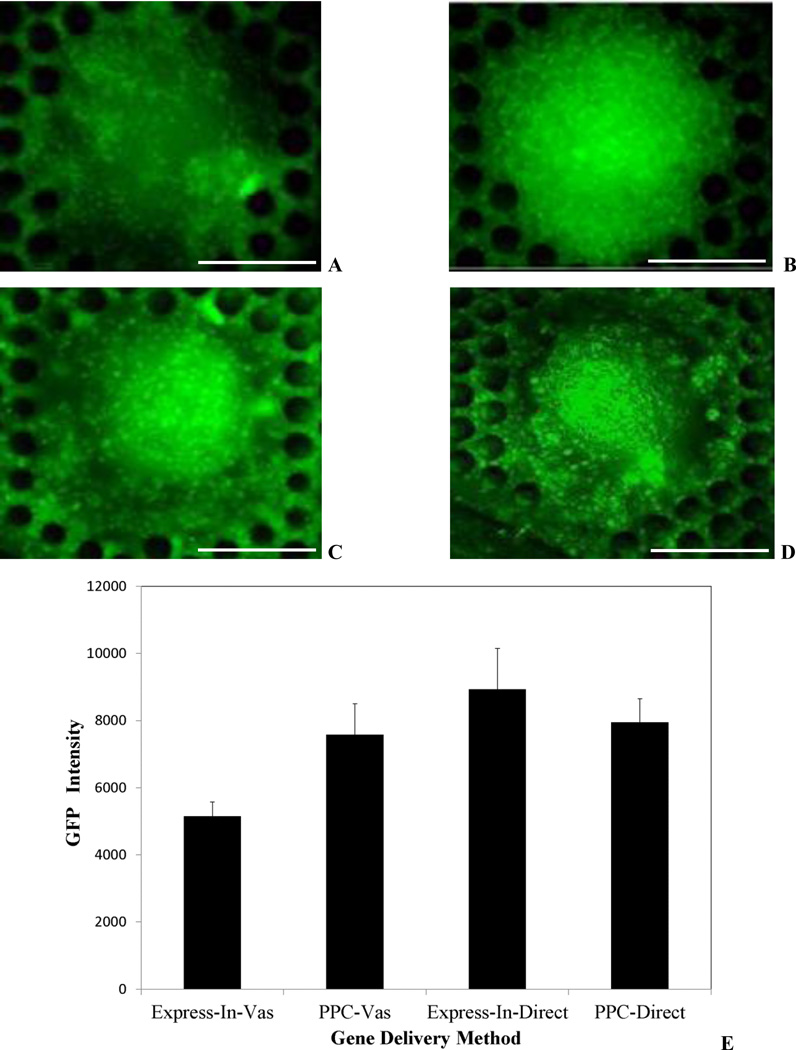
Figure 5A shows GFP expression of the 3D tumor mass using Express-In while Figure 5B shows GFP expression of the 3D tumor mass using PPC polymers following vascular injection. PPC polymer based GFP transfection is more uniform showing a relatively constant amount of expression across the entire tumor. However, Express-In shows non-uniform GFP expression and the core of the tumor is poorly transfected.
Figure 5.
Delivery System Screening. A. Express-In based GFP transfection following vascular injection. Non-uniform and minimal GFP expression only on the periphery of the tumor with untransfected core is observed. B. PPC based GFP transfection following vascular injection. Uniform and intense GFP expression and transfected core is observed. C. Express-In based GFP transfection following direct injection. D. PPC based GFP transfection following direct injection. Both polymers demonstrate uniform expression. E. GFP intensity comparison for PPC and Express-In following vascular and direct injection. PPC and Express-In perform similarly following direct injection. PPC performs significantly better than Express-In following vascular injection. Data is shown as mean ± S.D with experiments performed in triplicates. Scale Bars: 250 µm
Surprisingly, when injected directly to the tumor site, Express-In based GFP expression was found to be more intense, although both of the polymers exhibited GFP expression (Figure 5C–D). Figure 5E shows the quantitative intensity values of the GFP expression using PPC and Express-In polymers for both the direct and vascular injection test conditions. Furthermore, these findings match the in vivo delivery performance of several plasmid based approaches including PPC and Express-In where intra-tumoral injection have shown uniform transfection while intraperitoneal injection have shown poor transfections [43]. These findings also serve as positive controls indicating that the cells, the polymer and the GFP DNA complex are functional. These results clearly establish the fact that our device is able to predict the drug vehicle characteristics in vivo based on the drug injection route [40–42].
DISCUSSION
The efficacy of a drug reaching its desired location is dependent upon the attributes of the delivery system. Hence, it’s imperative that the delivery system is able to maintain its functional properties in the context of the in vivo environment. Highly complex physical and biological conditions exist in this environment including flow, cell-cell and cell-particle interactions. Unfortunately, standard in vitro tests comprising of static well plate incubation severely misrepresent the in vivo scenario and thus cannot adequately predict or provide a realistic understanding of the properties and behavior of a molecule or particle in vivo.
In this study, a clinical grade polymer PPC and an in vitro grade polymer Express-In were used for transfection of 3D tumors by complexing with GFP expressing plasmid DNA. In vivo preclinical studies following intra-tumoral injection have shown that these systems behave similarly [43] while intra-peritoneal injection and subsequent clinical studies showed that only PPC is fully effective [40–42]. Well plate studies, in contrast, while allowing for direct injection (data not shown) cannot reproduce intraperitoneal or vascular injection scenarios for comparison with in vivo data. Assays in the Synthetic Tumor Network reproduced the exact scenario observed in vivo. Vascular injection of polymers demonstrated higher efficiency for PPC while direct tumor injection showed similar results for both the polymers, although Express-In based GFP expression signal was brighter.
The poor efficacy of Express-In can be attributed due to the fact that serum proteins under flow interact with Express-In, causing it to aggregate to an extent that presents a steric obstacle to uniform transfection. On the other hand, PPC, which remains relatively aggregation- free, is able to flow freely and transfect cells uniformly. The translational diffusion coefficient values can be calculated using the Stokes-Einstein equation and viscosity of cell medium at 0.78cP [45] and are between 11.7 and 3.69 µm2/s for Express-In (diameter range of 50–150 nm) and 11.7 and 3.16 µm2 for PPC (diameter range of 50–175 nm). The diffusivity of these polymers are comparable with those reported in the literature [44–45]. Both of these delivery systems are highly cationic due to the polyethyleneimine (PEI) core structure which allows for the condensation of plasmid DNA into nanoparticles. However, PPC is further modified by the addition of a polyethylene glycol (PEG) which improves serum stability through molecular shielding of the cationic charge [40–42]. The high toxicity of Express-In in vivo (data not shown) may be presumably due to interaction with blood proteins, opsinization and aggregation of the nanoparticle complexes which is significantly attenuated with PPC. In addition, flow and polymer interaction with the cells may express receptors on the cell surface for uptake of GFP which is again not possible to test in static well plate conditions. Detailed studies need to be conducted to understand these cell-receptor and delivery system ligand interactions. Although, in the current study, none of the polymers were targeted specifically to the tumor, directed approaches will allow more focused delivery of the drugs or genes to the desired location. In addition, optimization of delivery system receptor type and density to maximize binding strength can be readily tested in the developed assay.
Drug delivery systems come in all shapes and sizes. Recent studies have shown that rod shaped particles have greater binding affinity than spheres for both micro and nano sized delivery systems [47–49]. In addition, even simple flow based systems have shown differences in binding affinities compared to static well plate assays [50–51]. The synthetic tumor networks developed in this study can be used to optimize the size and shape of delivery systems in conjugation with targeted receptors.
Drug toxicity is of critical importance in evaluating drugs for efficacy. In this study, we did not focus on the toxicity of the delivery systems to the tumors or the normal cells (endothelial). Studies incorporating toxicity analysis for delivery systems, drugs, etc. will be pursued in the future. A key interesting study will be to investigate the difference between a bolus injection of drug vs. a constant infuse and the tradeoff between efficacy and toxicity. Conditions of gradients of nutrients and oxygen diffusion can also be tested in these systems which will again allow more realistic test conditions similar to in vivo. A significant advantage of using microfluidic based systems is the savings in reagents and time compared to standard well plate assays.
The developed synthetic tumor network device and assay provides an ideal in vitro platform to test the efficiency of delivery systems under conditions mimicking physiological situations. Different from other microfluidic in vitro tumor models reported in the literature [25–35], the developed Synthetic Tumor Network model replicates the morphology, fluidics and leaky vasculature observed in vivo, specifically (a) in vivo based vascular morphology, (b) engineered leaky gaps between the vessels and the tumor, and (c) 3D culture of tumor cells. The leakiness of the vasculature used in this study was 2µm. However, this can be readily modified from a few nanometers to several tens of micrometers to account for heavily leaky vessels or non-leaky portions of the vasculature. The developed synthetic tumor network model can be used to study the mechanisms of drug delivery vehicle transport, drug-cell interactions, tumor transfection, and tumor-endothelium interactions.
CONCLUSION
Well plate assays routinely used to assess performance of drug delivery systems do not predict in vivo responses. In this study, Synthetic Tumor Networks clearly demonstrated its utility in accurately predicting in vivo behavior. Both the GFP gene delivery nanopolymers studied here – PPC and Express-In – showed similar high efficiency transfection results using intra-tumoral injection. In contrast, intra-peritoneal administration in vivo showed uniform transfection for PPC and poor transfection for Express-In similar to the results obtained from the Synthetic Tumor Network assays.
Synthetic Tumor Network assay allows replication of in vivo conditions comprising of morphology from in vivo vascular networks, co-culture of endothelial cells under physiological fluid flow and 3D culture of tumor cells, as well as the leakiness of the tumor vasculature in an in vitro model. The developed system and assay can be used to study cell-cell and cell-particle interactions and will have significant applications in basic and applied research, where it can be used to characterize and develop next generation delivery vehicles, and in drug discovery where it can be used to study the efficacy of the drug in realistic tumor microvascular networks.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support from National Institutes of Health under grant number 1R43CA139841-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zhang G, Zeng X, Li P. Nanomaterials in cancer-therapy drug delivery system. J Biomed Nanotechnol. 2013;9:741–750. doi: 10.1166/jbn.2013.1583. [DOI] [PubMed] [Google Scholar]
- 2.Vilar G, Tulla-Puche J, Albericio F. Polymers and drug delivery systems. Curr Drug Deliv. 2012;9:367–394. doi: 10.2174/156720112801323053. [DOI] [PubMed] [Google Scholar]
- 3.Khare R, Chen CY, Weaver EA, Barry MA. Advances and future challenges in adenoviral vector pharmacology and targeting. Curr Gene Ther. 2011;11:241–258. doi: 10.2174/156652311796150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langer R. Drug delivery and targeting. Nature. 1998;392:5–10. [PubMed] [Google Scholar]
- 5.Jain RK. Barriers to drug delivery in solid tumors. Sci Am. 1994;271:58–65. doi: 10.1038/scientificamerican0794-58. [DOI] [PubMed] [Google Scholar]
- 6.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 7.Grantab R, Sivananthan S, Tannock IF. The penetration of anticancer drugs through tumor tissue as a function of cellular adhesion and packing density of tumor cells. Cancer Res. 2006;66:1033–1039. doi: 10.1158/0008-5472.CAN-05-3077. [DOI] [PubMed] [Google Scholar]
- 8.Trédan O, Galmarini CM, Patel K, Tannoc IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 9.Jang SH, Wientjes MG, Lu D, Au JL. Drug delivery and transport to solid tumors. Pharm Res. 2003;20:1337–1350. doi: 10.1023/a:1025785505977. [DOI] [PubMed] [Google Scholar]
- 10.Kuszyk BS, Corl FM, Franano FN, Bluemke DA, Hofmann LV, Fortman BJ, Fishman EK. Tumor transport physiology: implications for imaging and imaging-guided therapy. AJR Am J Roentgenol. 2001;177:747–753. doi: 10.2214/ajr.177.4.1770747. [DOI] [PubMed] [Google Scholar]
- 11.Fukumura D, Jain RK. Tumor microenvironment abnormalities: causes, consequences, and strategies to normalize. J Cell Biochem. 2007;101:937–949. doi: 10.1002/jcb.21187. [DOI] [PubMed] [Google Scholar]
- 12.Ribatti D, Nico B, Crivellato E, Vacca A. The structure of the vascular network of tumors. Cancer Lett. 2007;248:18–23. doi: 10.1016/j.canlet.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Hashizume H, Baluk P, Morikawa S, McLean JW, Thurston G, Roberge S, Jain RK, McDonald DM. Openings between defective endothelial cells explain tumor vessel leakiness. Am J Pathol. 2000;156:1363–1380. doi: 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firrell JC, Lewis GP, Youlten LJ. Vascular permeability to macromolecules in rabbit paw and skeletal muscle: a lymphatic study with a mathematical interpretation of transport processes. Microvasc Res. 1982;23:294–310. doi: 10.1016/s0026-2862(82)80002-2. [DOI] [PubMed] [Google Scholar]
- 15.Lum H, Malik AB. Regulation of vascular endothelial barrier function. Am J Physiol. 1994;267:L223–L241. doi: 10.1152/ajplung.1994.267.3.L223. [DOI] [PubMed] [Google Scholar]
- 16.Fukumura D, Duda DG, Munn LL, Jain RK. Tumor microvasculature and microenvironment: novel insights through intravital imaging in pre-clinical models. Microcirculation. 2010;17:206–225. doi: 10.1111/j.1549-8719.2010.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 18.Dreher MR, Liu W, Michelich CR, Dewhirst MW, Yuan F, Chilkoti A. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J Natl Cancer Inst. 2006;98:335–344. doi: 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- 19.Yuan H, Goetz DJ, Gaber MW, Issekutz AC, Merchant TE, Kiani MF. Radiation-induced up-regulation of adhesion molecules in brain microvasculature and their modulation by dexamethasone. Radiat Res. 2005;163:544–551. doi: 10.1667/rr3361. [DOI] [PubMed] [Google Scholar]
- 20.Gaber MW, Sabek OM, Fukatsu K, Wilcox HG, Kiani MF, Merchant TE. Differences in ICAM-1 and TNF-alpha expression between large single fraction and fractionated irradiation in mouse brain. Int J Radiat Biol. 2003;79:359–366. doi: 10.1080/0955300031000114738. [DOI] [PubMed] [Google Scholar]
- 21.LaBarbera DV, Reid BG, Yoo BH. The multicellular tumor spheroid model for high-throughput cancer drug discovery. Expert Opin Drug Discov. 2012;7:819–830. doi: 10.1517/17460441.2012.708334. [DOI] [PubMed] [Google Scholar]
- 22.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J Biomol Screen. 2004;9:273–285. doi: 10.1177/1087057104265040. [DOI] [PubMed] [Google Scholar]
- 23.Kyle AH, Huxham LA, Chiam AS, Sim DH, Minchinton AI. Direct assessment of drug penetration into tissue using a novel application of three-dimensional cell culture. Cancer Res. 2004;64:6304–6309. doi: 10.1158/0008-5472.CAN-04-1099. [DOI] [PubMed] [Google Scholar]
- 24.Breslin S, O'Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2013;18:240–249. doi: 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Kim L L, Toh YC, Voldman J, Yu H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 26.Farokhzad OC, Khademhosseini A, Jon S, Hermmann A, Cheng J, Chin C, Kiselyuk A, Teply B, Eng G, Langer R. Microfluidic system for studying the interaction of nanoparticles and microparticles with cells. Anal Chem. 2005;77:5453–5459. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 27.Kusunose J, Zhang H, Gagnon MK, Pan T, Simon SI, Ferrara KW. Microfluidic system for facilitated quantification of nanoparticle accumulation to cells under laminar flow. Ann Biomed Eng. 2013;41:89–99. doi: 10.1007/s10439-012-0634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alves CS, Burdick MM, Thomas SN, Pawar P, Konstantopoulos K. The dual role of CD44 as a functional P-selectin ligand and fibrin receptor in colon carcinoma cell adhesion. Am. J. Physiol. Cell Physiol. 2008;294:C907–C916. doi: 10.1152/ajpcell.00463.2007. [DOI] [PubMed] [Google Scholar]
- 29.Liu T, Li C, Li H, Zeng S, Qin J, Lin B. A microfluidic device for characterizing the invasion of cancer cells in 3-D matrix. Electrophoresis. 2009;30:4285–4291. doi: 10.1002/elps.200900289. [DOI] [PubMed] [Google Scholar]
- 30.Hsiao AY, Torisawa YS, Tung YC, Sud S, Taichman RS, Pienta KJ, Takayama S. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials. 2009;30:3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidi PA, Maleki T, Ochoa M, Wang L, Clark SM, Leary JF, Lelièvre SA. Disease-on-a-chip: mimicry of tumor growth in mammary ducts. Lab Chip. 2014;14:172–177. doi: 10.1039/c3lc50819f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verbridge SS, Chakrabarti A, DelNero P, Kwee B, Varner JD, Stroock AD, Fischbach C. Physicochemical regulation of endothelial sprouting in a 3D microfluidic angiogenesis model. J Biomed Mater Res A. 2013;101:2948–2956. doi: 10.1002/jbm.a.34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zervantonakis IK, Kothapalli CR, Chung S, Sudo R, Kamm RD. Microfluidic devices for studying heterotypic cell-cell interactions and tissue specimen cultures under controlled microenvironments. Biomicrofluidics. 2011;5:13406. doi: 10.1063/1.3553237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moya M, Tran D, George SC. An integrated in vitro model of perfused tumor and cardiac tissue. Stem Cell Res Ther. 2013;4(Suppl 1):S15. doi: 10.1186/scrt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc Natl Acad Sci U S A. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prabhakarpandian B, Shen MC, Pant K, Kiani MF. Microfluidic devices for modeling cell-cell and particle-cell interactions in the microvasculature. Microvasc Res. 2011;82:210–220. doi: 10.1016/j.mvr.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prabhakarpandian B, Pant K, Scott RC RC, Patillo CB, Irimia D, Kiani MF, Sundaram S. Synthetic microvascular networks for quantitative analysis of particle adhesion. Biomed Microdevices. 2008;10:585–595. doi: 10.1007/s10544-008-9170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosano JM, Tousi N, Scott RC, Krynska B, Rizzo V, Prabhakarpandian B, Pant K, Sundaram S, Kiani MF. A physiologically realistic in vitro model of microvascular networks. Biomed Microdevices. 2009;11:1051–1057. doi: 10.1007/s10544-009-9322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matar MM, Slobodkin G, Ramsey A, Brunhoeber E, Fewell JG, Anwer K. Synthesis and Characterization of Low Molecular Weight Linear Polyethylenimines for Gene Delivery. Journal of Biomed Nanotech. 2006;2:53–61. [Google Scholar]
- 40.Fewell JG, Matar MM, Rice JS, Brunhoeber E, Slobodkin G, Pence C, Worker M, Lewis DH, Anwer K. Treatment of disseminated ovarian cancer using nonviral interleukin-12 gene therapy delivered intraperitoneally. J Gene Med. 2009;11:718–728. doi: 10.1002/jgm.1356. [DOI] [PubMed] [Google Scholar]
- 41.Anwer K, Barnes MN, Fewell J, Lewis DH, Alvarez RD. Phase-I clinical trial of IL-12 plasmid/lipopolymer complexes for the treatment of recurrent ovarian cancer. Gene Ther. 2010;17:360–369. doi: 10.1038/gt.2009.159. [DOI] [PubMed] [Google Scholar]
- 42.Anwer K, Kelly FJ FJ, Chu C C, Fewell JG, Lewis D, Alvarez RD. Phase I trial of a formulated IL-12 plasmid in combination with carboplatin and docetaxel chemotherapy in the treatment of platinum-sensitive recurrent ovarian cancer. Gynecol Oncol. 2013;131:169–173. doi: 10.1016/j.ygyno.2013.07.081. [DOI] [PubMed] [Google Scholar]
- 43.Lasek W, Zagożdżon R, Jakobisiak M. Interleukin 12: still a promising candidate for tumor immunotherapy? Cancer Immunol Immunother. 2014;63:419–435. doi: 10.1007/s00262-014-1523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sparks J, Slobodkin G, Matar MM, Congo R, Ulkoski D, Rea-Ramsey A, Pence C, Rice J, McClure D, Polach KJ, Brunhoeber E, Wilkinson L, Wallace K, Anwer K, Fewell JG. Versatile cationic lipids for siRNA delivery. J Control. Release. 2012;158:269–276. doi: 10.1016/j.jconrel.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Bacabac RG, Smit TH, Cowin SC, Van Loon JJ, Nieuwstadt FT, Heethaar R, Klein-Nulend J. Dynamic shear stress in parallel-plate flow chambers. J Biomech. 2005;38:159–167. doi: 10.1016/j.jbiomech.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Vijayanathan V, Thomas T, Antony T, Shirahata A, Thomas TJ. Formation of DNA nanoparticles in the presence of novel polyamine analogues: a laser light scattering and atomic force microscopic study. Nucleic Acids Res. 2004;32:127–134. doi: 10.1093/nar/gkg936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barua S, Yoo JW, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc Natl Acad Sci U S A. 2013;110:3270–3275. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Champion JA, Katare YK, Mitragotri S. Particle shape: a new design parameter for micro- and nanoscale drug delivery carriers. J Control Release. 2007;121:3–9. doi: 10.1016/j.jconrel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doshi N, Prabhakarpandian B, Rea-Ramsey A, Pant K, Sundaram S, Mitragotri S. Flow and adhesion of drug carriers in blood vessels depend on their shape: a study using model synthetic microvascular networks. J Control Release. 2010;146:196–200. doi: 10.1016/j.jconrel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decuzzi P, Gentile F, Granaldi A, Curcio A, Causa F, Indolfi C, Netti P, Ferrari M. Flow chamber analysis of size effects in the adhesion of spherical particles. Int J Nanomedicine. 2007;2:689–696. [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng C, Zhao L, Chen G, Zhou Y, Pang Y, Huang Y. Quantitative study of the dynamic tumor-endothelial cell interactions through an integrated microfluidic coculture system. Anal Chem. 2012;84:2088–2093. doi: 10.1021/ac2032029. [DOI] [PubMed] [Google Scholar]