Abstract
Background
Interferon gamma (IFN-γ) is a key regulatory cytokine, which plays an important role in antiviral defense of an infected host. However, the association between the IFN-γ +874T/A gene polymorphism and hepatitis virus-related diseases is heterogeneous.
Methods
Based on the Preferred Reporting Items for Systematic Reviews and Meta-analyses statement, a comprehensive literature search of eligible studies in Embase, Pubmed, and the Cochrane Library was undertaken through November 2014. Odds ratios (ORs) and the corresponding 95% confidence intervals (CIs) were used to measure the strength of the models.
Results
Seventeen case-control articles, including 24 studies with 5503 individuals, met the inclusion criteria. The results indicated a statistically significant association between the IFN-γ +874T/A polymorphism and hepatitis virus—related diseases in a recessive gene model (AA vs. TT+TA: OR=1.350, 95% CI=1.101-1.657, P=0.004, I2%=54.3, and PQ=0.001 for heterogeneity), especially in Asians (OR=1.407, 95% CI=1.035-1.911, P=0.029, I2%=61.9, and PQ=0.005 for heterogeneity) and hepatitis B virus (HBV)–related disease (OR=1.486, 95% CI=1.195–1.849, P=0.000, I2%=40.4, and PQ=0.053 for heterogeneity).
Conclusions
The evidence suggests that the IFN-γ +874T/A polymorphism increases the risk of hepatitis virus—related diseases, especially in Asians and HBV—related diseases. Further studies on this topic in different ethnicities, especially genome-wide association studies, should be conducted to strengthen our results.
Introduction
Liver disease can have various causes, including infection with viruses, such as the hepatitis B virus (HBV) and the hepatitis C virus (HCV), especially in developing countries [1]. Interferon gamma (IFN-γ) is a key regulatory cytokine, which plays an important role in antiviral defense by an infected host. IFN-γ can exert antiproliferative and antitumor activity by binding with a specific cell-surface receptor (IFN-γR) [2]. IFN-γ is thought to play a major role in combating chronic hepatitis (CH), liver cirrhosis (LC), and hepatocellular carcinoma (HCC)[3].
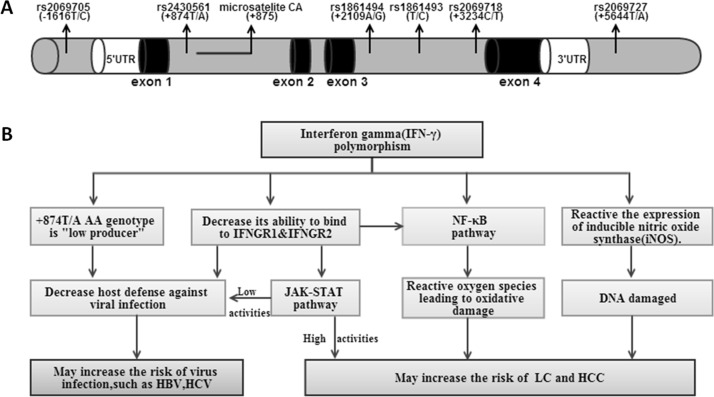
Genetic susceptibility is an important factor in the development of diseases. The IFN-γ gene on chromosome 12q24 spans approximately 5.4 kb and is composed of four exons, with three intervening regions; many single nucleotide polymorphisms (SNPs) are located in this region [4] (Fig 1). However, only the SNP (+874T/A, rs2430561) of the gene is widely studied. This polymorphism results in the replacement of the T allele with an A allele, which is located at the translation start site in the first intron of the IFN-γ gene. The TT genotype is associated with increased IFN production when the immune system responds to stimuli [5]. In contrast, the AA and TA genotypes are linked to low IFN production [5]. The release of IFN-γ by CD4+ Th1 cells, natural killer cells, and other lymphocytes increases T-cell cytotoxicity and natural killer cell activity and enhances cellular immune responses. It appears that the +874T/A polymorphism can influence IFN-γ expression, and in this way, affects immune response.
Fig 1. The position of +847A/T (rs2430561) locus and other SNPs in the neighborhood of IFN-γ gene on chromosome 12q24(A) and the hypothesis of IFN-γ gene polymorphism pathway increase the risk of hepatitis virus-related diseases(B).
The figure includes the 5’flanking region (white column), exons (dark column) interrupted by introns (gray column) and 3’flanking region. IFNGR, interferon gamma receptor; JAK, Janus kinase; STAT, Signal Transducer and Activator of Transcription; NF, nuclear factor.
Many studies have mainly focused on the IFN-γ +874T/A polymorphism and virus-related disease risk, especially liver diseases, such as CH, LC, and HCC, which are caused by persistent hepatitis virus infection. However, the literature on the impact of the IFN-γ gene polymorphism on the risk of hepatitis virus—related diseases has provided contradictory results [6–16]. Since the IFN-γ +874T/A polymorphism is strongly associated with circulating IFN-γ concentrations, it is assumed that this polymorphism might be closely related to the risk of the hepatitis virus—related diseases. Our aim was to examine the association between the IFN-γ +874T/A polymorphism and the risk of hepatitis virus-related diseases in published studies using a meta-analysis.
Materials and Methods
This meta-analysis was conducted according to Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement, including the search strategy, selection criteria,data extraction and data analysis [17].
Identification of eligible studies
We used the following terms to search for articles published in Embase, PubMed, and the Cochrane Library up to November 30, 2014: “interferon,” “IFN” “polymorphism,” “polymorphisms,” “mutation,” “variant,” “hepatitis,” “cirrhosis,” and “hepatocellular carcinoma.” Two investigators (Yifan Sun and Yu Lu) conducted an extensive independent literature search. Articles in reference lists were hand-searched. Only English-language articles and human studies were searched.
Inclusion and exclusion criteria
The following criteria were used to select suitable studies: (1) case-control or cohort design studies; (2) contained data that could be extracted to calculate the odds ratio (OR), 95% confidence intervals (CIs), and Hardy—Weinberg equilibrium (HWE); (3) stated the DNA genotyping method and source of cases and controls. The following were excluded: review articles, letters, case reports, editorials, conference abstracts, and family-based studies.
Data extraction
Two investigators (Yifan Sun and Yu Lu) independently extracted data from the included studies. The data included the first author’s name, publication date, country, ethnicity, total sample size, virus type and liver disease type, genotyping method, genotype frequencies of cases and controls, source of the case group and control group, age and sex ratio, source of specimens used to determine the genotypes, and the estimated HWE. If the literature did not provide sufficient data, the investigators tried to contact the author to get the original data by email.
In the subgroup analysis, ethnicities from Europe, West Asia, North Africa, and the Americas were categorized as Caucasian and those from East or Southeast Asia as Asian. The hepatitis virus type and liver disease type were also categorized. If the data in the study were related to various liver diseases, the study was treated as a separate study in the meta-analysis. To determine the accuracy of the extracted information, the data extracted by the two investigators were expected to be the same. In cases of a dispute, they checked their data again. If they could not reach an agreement, the dispute was resolved by a third reviewer (Xue Qin).
Quality score assessment
Two investigators (Yifan Sun and Shan Li) assessed the quality of the selected studies independently, following the criteria predefined by Thakkinstian et al. [18] and used by Ye et al. [19](S1 Table). The criteria were the sources of the cases and controls, the total sample size, the source of the specimens, and the HWE of the controls. According to the quality score assessment, a study scoring <10 was considered a “low-quality” study, whereas one with a score ≥10 was classified as a “high-quality” study. The lowest score was 0, and the highest score was 15 [19].
Statistical analysis
The association between the IFN-γ +874T/A polymorphism and hepatitis virus-related diseases was assessed with various comparison models, including an allelic model (A vs. T), a co-dominant model (TA vs. TT and AA vs. TT), a dominant model (TA+AA vs. TT), and a recessive model (AA vs. TA+TT). Because the studies lacked details of the environmental factors, unadjusted ORs and the corresponding 95% CIs were used to measure the strength of the models.
In common with previous studies [20, 21], heterogeneity was assessed with a chi-squared Q test and I-squared statistics. If P Q<0.1 or I 2≥50%, the heterogeneity was considered significant. The random-effects model used the DerSimonian and Laird method. Otherwise, the summary OR and the corresponding 95% CI were calculated with a fixed-effects model (the Mantel—Haenszel method). We also conducted a subgroup analysis by ethnicity, genotyping method, source of controls, hepatitis virus type, liver disease type, and quality assessment score. A meta-regression that included the following covariates was performed: ethnicity, genotyping method, source of controls, and quality scores. Covariates with values of P<0.05 were considered the main sources of heterogeneity. Galbraith plot analysis was performed for further exploration of the heterogeneity. Sensitivity analysis was conducted to examine such influence by removing studies one by one and recalculating the pooled OR and 95% CI.
A Begg’s funnel plot and an Egger’s test were used to investigate publication bias in the meta-analysis. P<0.05 indicated that the result was statistically significant.
All tests in this meta-analysis were conducted with STATA software (version 12.0; Stata Corporation, College Station, Texas, USA).
Results
Literature selection and study characteristics
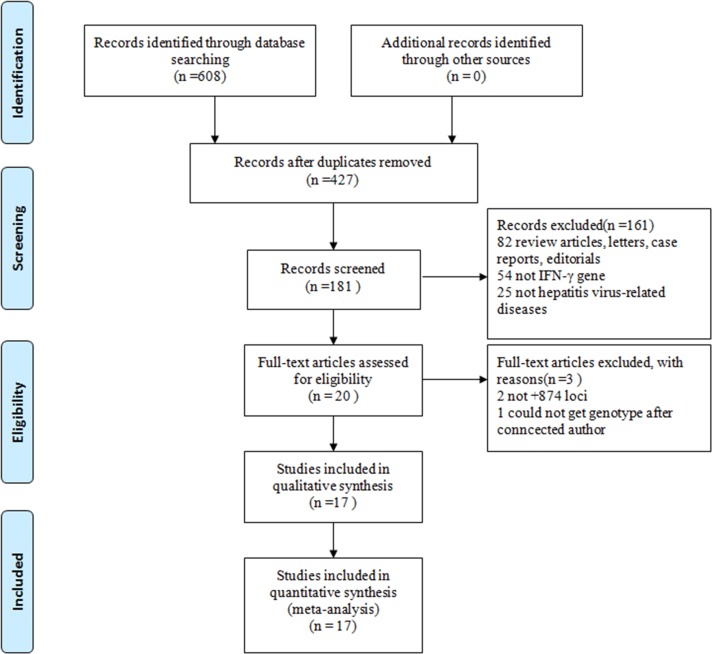
Based on the search terms, 17 articles, which included 24 case-control studies containing 2,607 cases and 2,896 controls, were identified as suitable for a meta-analysis [3, 6, 7, 9, 12–14, 16, 22–30] (Fig 2). The primary characteristics of the 24 studies are shown in Table 1. Nine of the articles focused on Caucasians and eight on Asians. Sixteen studies focused on HBV-related diseases, five on HCV-related diseases, one on HDV-related disease, one on HEV-related disease, and one on mixed virus (HBV and HCV)-related HCC. When the articles were categorized by liver disease, two focused on HBV infection (only HBV carrier), nine focused on chronic hepatitis B (CHB), and seven focused on LC/HCC. The genotyping methods included the polymerase chain reaction-amplification refractory mutation system (ARMS-PCR), polymerase chain reaction-sequence specific primers (PCR-SSP), competitively differentiated-polymerase chain reaction (CD-PCR), and DNA sequencing. The HWE of the control population was calculated according to the genotypes. The control population did not fall into HWE in three articles [13, 25, 29]. According to the quality scores, 13 articles were “high-quality” studies. The source of the control population was divided into hospital based and population based.
Fig 2. Flow diagram of included studies for this meta-analysis.
Table 1. Characteristics of Studies Included in This Review.
| Study | Year | Country | Enthicity | Liver virus | Liver diseases | Genotyping method | Source of control | Case sequence | Control sequence | P * (HWE) | Quality scores | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AT | TT | AA | AT | TT | ||||||||||
| Teixeira | 2013 | Brazil | Caucasian | HBV | HCC | SSP-PCR | PB | 40 | 50 | 21 | 79 | 82 | 41 | 0.024 | 11 |
| Arababadi | 2011 | Iran | Caucasian | HBV | HBV infection | AMRS-PCR | PB | 18 | 25 | 14 | 28 | 47 | 25 | 0.554 | 9 |
| Basturk | 2008 | Turkey | Caucasian | HBV | CHB | SSP-PCR | PB | 19 | 23 | 8 | 26 | 24 | 10 | 0.282 | 10 |
| Ben-Ari | 2003 | Israel | Caucasian | HBV | CHB | SSP-PCR | HB | 33 | 13 | 10 | 18 | 24 | 6 | 0.644 | 10 |
| Ben-Ari | 2003 | Israel | Caucasian | HBV | LC/HCC | SSP-PCR | HB | 17 | 10 | 4 | 18 | 24 | 6 | 0.644 | 10 |
| Bouzgarrou | 2009 | Tunisia | Caucasian | HCV | HCV infection | SSP-PCR | PB | 22 | 10 | 10 | 33 | 47 | 33 | 0.074 | 11 |
| Bouzgarrou | 2010 | Tunisia | Caucasian | HCV | LC/HCC | SSP-PCR | PB | 17 | 21 | 20 | 33 | 47 | 33 | 0.074 | 11 |
| Cheong | 2006 | South Korea | Asian | HBV | CHB | Sequencing | HB | 314 | 94 | 5 | 151 | 47 | 3 | 0.760 | 11 |
| Conde | 2013 | Brazil | Caucasian | HBV | CHB | SSP-PCR | PB | 27 | 19 | 7 | 51 | 37 | 9 | 0.547 | 11 |
| Falleti | 2007 | Italy | Caucasian | HCV | LC/HCC | AMRS-PCR | PB | 12 | 28 | 10 | 30 | 51 | 15 | 0.382 | 11 |
| Gao | 2009 | China | Asian | HBV | HBV infection | SSP-PCR | PB | 25 | 35 | 9 | 14 | 53 | 7 | <0.000 | 8 |
| Gao | 2009 | China | Asian | HCV | HCV infection | SSP-PCR | PB | 23 | 27 | 5 | 14 | 53 | 7 | <0.000 | 8 |
| Karatayli | 2014 | Turkey | Caucasian | HDV | CHD | SSP-PCR | HB | 9 | 33 | 22 | 11 | 28 | 15 | 0.753 | 9 |
| Karatayli | 2014 | Turkey | Caucasian | HBV | CHB | SSP-PCR | HB | 37 | 47 | 34 | 11 | 28 | 15 | 0.753 | 9 |
| Korachi | 2013 | Turkey | Caucasian | HBV | CHB | Sequencing | PB | 38 | 57 | 5 | 16 | 56 | 27 | 0.147 | 12 |
| Korachi | 2013 | Turkey | Caucasian | HCV | CHC | Sequencing | PB | 20 | 50 | 30 | 16 | 56 | 27 | 0.147 | 12 |
| Mishra | 2011 | India | Asian | HEV | HEV infection | Sequencing | PB | 24 | 84 | 28 | 90 | 178 | 106 | 0.370 | 12 |
| Nieters | 2004 | China | Asian | Mixed | HCC | SSP-PCR | HB | 86 | 155 | 94 | 164 | >0.200 | 10 | ||
| Saxena | 2014 | India | Asian | HBV | HCC/LC | SSP-PCR | PB | 36 | 56 | 27 | 52 | 77 | 17 | 0.180 | 12 |
| Saxena | 2014 | India | Asian | HBV | CHB | SSP-PCR | PB | 31 | 20 | 13 | 52 | 77 | 17 | 0.180 | 12 |
| Srivastava | 2014 | India | Asian | HBV | LC/HCC | SSP-PCR | PB | 44 | 53 | 29 | 14 | 55 | 7 | <0.000 | 9 |
| Srivastava | 2014 | India | Asian | HBV | CHB | SSP-PCR | PB | 32 | 61 | 13 | 14 | 55 | 7 | <0.000 | 9 |
| Peng | 2007 | China | Asian | HBV | CHB | CD-PCR | HB | 247 | 89 | 4 | 65 | 33 | 2 | 0.545 | 10 |
| Migita | 2005 | Japan | Asian | HBV | HCC | SSP-PCR | HB | 41 | 7 | 0 | 157 | 31 | 0 | 0.218 | 11 |
Abbreviation: CHB, chronic hepatitis B;CHD, chronic hepatitis D; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HBV /HCV/HEV infection, only hepatitis virus carriers; SSP-PCR, polymerase chain reaction-sequence specific primers; AMRS-PCR, polymerase chain reaction-amplification refractory mutation system; CD-PCR, competitively differentiated-polymerase chain reaction; HB, Hospital-based; PB, Population-based; Mixed, HBV and HCV infection; HWE, Hardy—Weinberg equilibrium.
*P value of HWE of control
Allele frequencies in different ethnicities
Allele frequencies in different ethnicities were calculated according to the original data of the studies. The IFN-γ +874T/A loci A allele had a higher representation in the controls of the Asian group than in the Caucasian group (65.5% vs. 55.6%, χ 2 = 47.07,P = 0.000). On average, the frequency of AA, TA, and TT as a proportion of 1 was 0.38, 0.41, and 0.21 respectively in the Asian population controls and 0.32, 0.48, and 0.20 in the Caucasian population; a significant difference was observed between the two ethnicities (χ 2 = 14.467, P = 0.001), as well as in the cases (χ 2 = 56.53, P = 0.000). The allele frequencies in the African group were not analyzed because of the sample size was too small.
Meta-analysis results
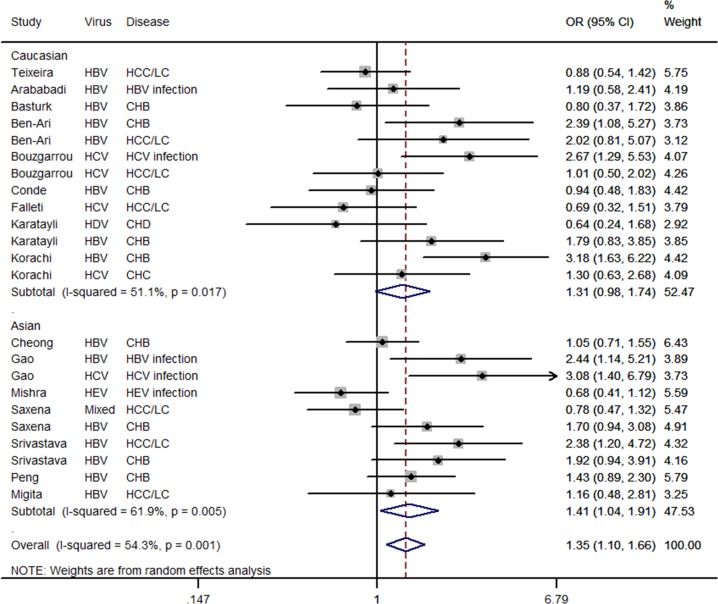
The results of the meta-analysis of the IFN-γ +874T/A polymorphism and hepatitis virus—related disease risk are listed in Table 2. In the pooled analysis using the random-effects model, there was a significant increased risk of hepatitis virus-related disease in the recessive model (AA vs. TT+TA: OR = 1.350, 95% CI = 1.101–1.657, P = 0.004, I 2% = 54.3, and P Q = 0.001 for heterogeneity; see Fig 3).
Table 2. Meta-analysis of interferon gamma +874T/A polymorphisms and hepatitis virus-related diseases.
| Group | N | A vs. T | AA vs. TT | TA vs. TT | AA vs. TT+TA | N* | AA+TA vs. TT | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P Q ▲ | OR(95%CI) | P Q ▲ | OR(95%CI) | P Q ▲ | OR(95%CI) | P Q ▲ | OR(95%CI) | P Q ▲ | |||
| Overall | 23 | 1.121(0.993–1.265) | 0.022 | 1.106(0.845–1.446) | 0.032 | 0.789(0.597–1.042) | 0.003 | 1.284(1.126–1.464) | 0.001 | 23 | 0.914(0.738–1.133) | 0.031 |
| Ethnicity | ||||||||||||
| Asian | 10 | 1.082(0.953–1.228) | 0.246 | 0.906(0.659–1.246) | 0.464 | 0.634(0.363–1.105) | 0.002 | 1.407(1.035–1.911) | 0.005 | 10 | 0.790(0.559–1.117) | 0.051 |
| Caucasian | 13 | 1.134(0.931–1.380) | 0.008 | 1.210(0.820–1.787) | 0.013 | 0.944(0.747–1.194) | 0.137 | 1.306(0.979–1.743) | 0.017 | 13 | 1.052(0.846–1.308) | 0.107 |
| Virus-related | ||||||||||||
| HBV | 15 | 1.196(1.065–1.343) | 0.169 | 1.262(0.878–1.815) | 0.067 | 0.763(0.503–1.158) | 0.004 | 1.486(1.195–1.849) | 0.053 | 14 | 0.931(0.651–1.333) | 0.020 |
| HCV | 5 | 1.144(0.845–1.550) | 0.078 | 1.182(0.783–1.785) | 0.275 | 0.766(0.525–1.117) | 0.999 | 1.488(0.863–2.565) | 0.030 | 5 | 0.909(0.643–1.285) | 0.869 |
| Liver disease | ||||||||||||
| CHB | 9 | 1.245(1.009–1.538) | 0.045 | 1.424(0.791–2.564) | 0.012 | 0.861(0.476–1.557) | 0.006 | 1.498(1.133–1.980) | 0.070 | 9 | 1.063(0.620–1.821) | 0.011 |
| LC/HCC | 7 | 0.914(0.774–1.079) | 0.547 | 0.748(0.528–1.059) | 0.581 | 0.623(0.386–1.007) | 0.081 | 1.094(0.782–1.530) | 0.068 | 7 | 0.751(0.550–1.024) | 0.186 |
| Source of control | ||||||||||||
| HB | 15 | 1.221(1.036–1.438) | 0.652 | 1.259(0.821–1.933) | 0.851 | 0.692(0.469–1.020) | 0.880 | 1.475(1.190–1.828) | 0.106 | 15 | 0.919(0.709–1.191) | 0.988 |
| PB | 8 | 1.070(0.906–1.265) | 0.006 | 1.058(0.738–1.517) | 0.004 | 0.827(0.567–1.205) | 0.000 | 1.235(0.940–1.622) | 0.002 | 8 | 0.915(0.658–1.273) | 0.001 |
| Score≥10 | 16 | 1.110(0.944–1.304) | 0.006 | 1.111(0.771–1.601) | 0.006 | 0.889(0.621–1.274) | 0.003 | 1.213(0.964–1.526) | 0.006 | 16 | 0.990(0.753–1.303) | 0.014 |
Abbreviation: N, number of studies; OR, odds ratio; CI, confidence interval; P Q, chi-squared Q test value, CHB, chronic hepatitis B;LC, liver cirrhosis;
HCC, hepatocellular carcinoma; HB, Hospital-based; PB, Population-based; Score, quality scores
▲If P Q<0.1,using a random-effects model; If P Q≥0.1,using a fixed- effects model.
*number of studies: AA+TA vs. TT mode
Fig 3. Forest plot for the association between IFN-γ +874T/A polymorphism and hepatitis virus-related diseases risk stratified by ethnicity in a recessive model (AA vs. TT+TA) using a random-effects mode.
The squares and horizontal lines correspond to the study-specific OR and 95%CI. The diamond represents the summary OR and 95%CI.
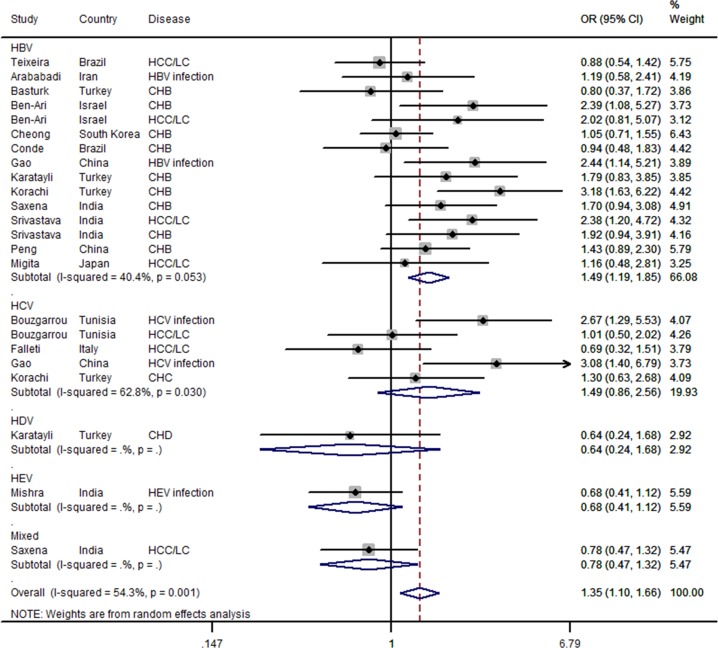
For the recessive model, the results of the subgroup analysis showed that the IFN-γ +874T/A polymorphism significantly increased the risk of hepatitis virus-related disease in Asians (OR = 1.407, 95% CI = 1.035–1.911, P = 0.029, I 2% = 61.9 and P Q = 0.005 for heterogeneity; see Fig 3). Similar results were found for the HBV—related diseases group (OR = 1.486, 95% CI = 1.195–1.849, P = 0.000, I 2% = 40.4, and P Q = 0.053 for heterogeneity; see Fig 4), CHB patient group (OR = 1.498, 95%CI = 1.133–1.980, P = 0.005, I 2% = 44.8, and P Q = 0.070 for heterogeneity), SSP-PCR method group (OR = 1.449,95%CI = 1.124–1.867, P = 0.004, I 2% = 53.8, and P Q = 0.007 for heterogeneity), and hospital-based population (OR = 1.475, 95% CI = 1.126–1.464, P = 0.000, I 2% = 39.3, and P Q = 0.106 for heterogeneity).
Fig 4. Forest plot for the association between IFN-γ +874T/A polymorphism and hepatitis virus-related diseases risk stratified by hepatitis virus type in a recessive model (AA vs. TT+TA) using a random-effects mode.
The squares and horizontal lines correspond to the study-specific OR and 95%CI. The diamond represents the summary OR and 95%CI.
For the allelic model (allele A vs. allele T), the IFN-γ +874T/A polymorphism significantly increased the risk of hepatitis virus—related disease in the HBV-related diseases group (OR = 1.196,95%CI = 1.065–1.343, P = 0.003, I 2% = 25.9,and P Q = 0.169 for heterogeneity). Similar results were observed for the CHB patient group (OR = 1.245, 95% CI = 1.009–1.538, P = 0.004, I 2% = 49.0, and P Q = 0.045 for heterogeneity) and the hospital-based population (OR = 1.221, 95%CI = 1.036–1.438, P = 0.017, I 2% = 0.0, and P Q = 0.652 for heterogeneity).
Other results indicated a lack of statistical significance between the IFN-γ +874T/A polymorphism and hepatitis virus—related disease risk (Table 2)
Heterogeneity analysis
All the models revealed statistical heterogeneity for the IFN-γ +874T/A polymorphism in the overall population, with I 2 values of heterogeneity greater than 50% and P Q values lower than 0.100. Heterogeneity still existed in some studies following the subgroup analysis according to ethnicity, virus genotyping method, sources of control, quality score assessment, hepatitis virus type, and liver disease type. A meta-regression of the sources of heterogeneity revealed that the genotype methods were the main sources of heterogeneity (P = 0.005, 95% CI = 0.299–1.470). A Galbraith plot analysis confirmed that the studies by Korachi et al. (HBV), Gao et al. (HCV), Bouzgarrou et al. (HCV), and Mishra et al. (HEV) were responsible for the heterogeneity in the recessive model. After these four studies were excluded, the summary OR value did not change significantly (OR = 1.251, 95% CI = 1.034–1.513, P = 0.021, I 2% = 25.8, P Q = 0.078 for heterogeneity). In the allelic model, the studies by Korachi et al. (HBV) [27] and Saxena et al. (HCC/LC) [3] were the outliers. In the co-dominant model and the dominant model, the summary OR value did not change significantly after these two studies were excluded. However, following their exclusion, the I 2 values were lower than 50%, and the P Q value was larger than 0.10 (data not shown).
Sensitivity analysis
The control groups in the studies by Teixeira et al. [29], Gao et al. [25], and Srivastava et al. [14] were out of HWE (Table 1), and these three studies were excluded to perform a sensitivity analysis of the pooled ORs for the IFN-γ (+874T/A) polymorphism. Further sensitivity analysis was performed by excluding the studies by Karatayli et al. [26] and Mishra et al. [12], in which the study virus types were HDV and HEV, respectively. Three articles that used the DNA sequencing method to obtain the genotype were also excluded one by one [7, 12, 27]. Finally, the corresponding pooled ORs were not qualitatively altered with or without including these studies (data not shown).
Publication bias
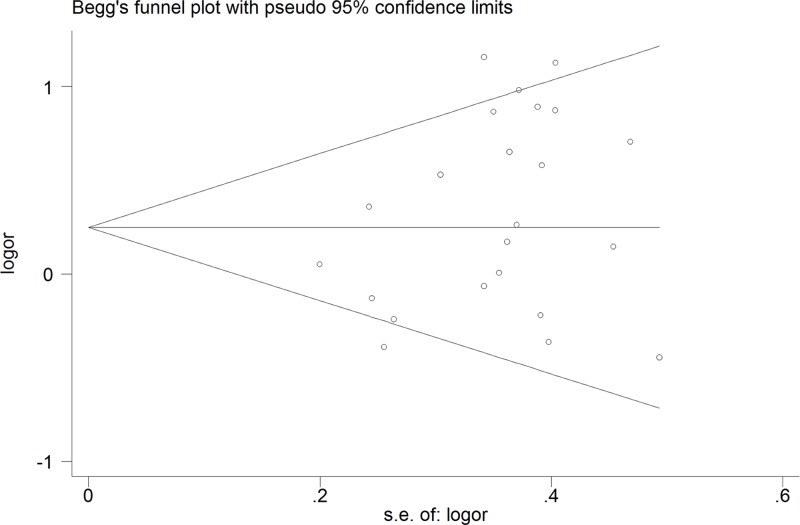
A Begg’s funnel plot and an Egger’s test were used to investigate the publication bias in the meta-analysis (Fig 5). No significant publication bias was detected with the funnel plot in the overall population in the recessive model. The statistical results of the Egger’s test also provided evidence of funnel plot symmetry (P Egger’s test = 1.840; P = 0.08).
Fig 5. Begg’s funnel plot for contrast in a recessive model (AA vs. TT+TA).
Each point represents a separate study for the indicated association. Size graph symbol by weights. LogOR natural logarithm of OR. Horizontal line mean effect size.
Discussion
Various environmental and dietary factors are responsible for liver diseases, but hepatitis infection is the main cause of CH, LC, and HCC [31]. The association between the IFN-γ +874T/A polymorphism and the risk of hepatitis virus—related diseases has been studied extensively because of the importance of IFN-γ in immune responses [3, 9, 13, 16, 25, 29]. However, previous studies provided inconsistent results. To clarify this issue, we conducted a meta-analysis, and the results showed that there was a significant association between the IFN-γ +874T/A polymorphism and risk of hepatitis virus—related disease in the pooled analysis in the recessive model, with the +874 AA genotype associated with a 1.350-fold increased risk of hepatitis virus related—disease, especially in Asian and HBV—related diseases
The results indicate that the IFN-γ +874T/A polymorphism may play an important role in hepatitis virus—related disease. The following pathways are considered the main way in which the IFN-γ +874T/A polymorphism affects the development of hepatitis virus infection. First, the IFN-γ +874T/A genotype TT, which produces a high level of IFN-γ, aids the host’s antiviral defense system. In contrast, the AA and TA genotypes result in low IFN-γ production, potentially increasing the risk of hepatitis virus infection [32]. Second, as noted earlier, IFN-γ binds to a specific cell-surface receptor (IFN-γR), which plays a significant role in multiple types of cancers and stimulates cell signaling pathways (JAK-STAT). The IFN-γ +874T/A polymorphism leads to dysfunction of IFN-γR, potentially increasing the risk of liver diseases [2]. In addition, the DNA sequence containing the +874 T allele is the specific binding site for the nuclear factor Kappa B(NF-κB) transcription factor. If the NF-κB pathway is affected, it may lead to oxidative damage, which can also increase the risk of LC and HCC [32]. The possible pathway of the IFN-γ polymorphism increase in hepatitis virus—related diseases risk is shown in Fig 1.
The stratified analysis by ethnicity in the present study suggested that the IFN-γ +874T/A polymorphism influenced the risk of hepatitis virus-related diseases in Asians but not in Caucasians. Many factors may contribute to this finding. First, hepatitis virus related—disease is a complex entity, with multiple determinants other than virus infection, such as diet and the environment. Some studies noted significant gene—gene and gene—environmental interactions in patients with hepatitis virus related—diseases [33–35]. Second, as shown in previous studies, the gene frequency of the IFN-γ +874T/A polymorphism varies in ethnicities [36, 37], and the Asian population had a higher rate of the A allele than that in the Caucasian population in our study. Third, the inclusion of study subjects from different populations, together with the various genotyping methods and the different sample sizes used in these studies, means it is not possible to reach a definitive conclusion, and the results remain conflicting.
The subgroup analysis showed that the significant results are mainly reflected in the HBV—related group. Evidence suggests that IFN-γ can suppress viral replication/clear HBV without causing liver injury, the non-cytolytic HBV suppression effect of IFN-γ is believed to have an important role in the control of HBV viral activity in patients with CHB [38]. Regarding CHB, IFN-γ +874 was seen to play a functional role in relation to viral load, consistent with its known role inhibiting and replicating of HBV-infected cells [16]. IFN-γ can also induce expression of HLA class II [39], and a genome-wide association study (GWAS) showed that HLA contributes to the risk of persistent HBV infection [40].
As we now known, the IFN-γ +874T/A TT genotype produces a high level of IFN-γ, and the AA and TA genotypes result in low IFN-γ production. Thus we can suppose that the AA+TA vs. TT model may have significant association in the analysis. However, the ORs of AA vs. TT+TA and AA+TA vs. TT are opposite in our meta-analysis. These confusing results suggested that the T allele is a protective gene in liver disease; moreover, this allele has more power than the A allele, therefore, only the population with the AA genotype has higher risk of hepatitis virus-related liver disease. Indeed, similar results happened in the association between the IFN-γ +874T/A polymorphism and cervical cancer with human papillomavirus (HPV) infection [41] and leprosy caused by Mycobacterium leprae[42]. A meta-analysis also indicated that the +874 T allele of IFN-γ showed a protective significant association with tuberculosis susceptibility [43].
Studies have also reported the role of the IFN-γ gene in the response to drug treatment in cancer [44]. Sarvari et al. [13] indicated that the +2109 locus of the IFN-γ gene, which is also the specific binding site for NF-κB similar to the +874 locus, influenced HCV therapy. Moreover, Oxenkrug et al. [45] found that T alleles at the +874 locus of the IFN-γ gene were a risk factor in depressed patients with HCV who were treated with IFN-alpha. The IFN-γ gene polymorphism may further affect the development of HCC and the response to cancer drugs by increasing the risk of hepatitis virus-related diseases, although we did not observe a significant association between the (+874T/A) polymorphism and HCC risk in this study, which is worthy of further investigation in the future.
We noticed that the meta-analysis by Ge et al.[46] also suggested that the +874T/A polymorphism may not contribute to HCC susceptibility with three studies, which was verified by our meta-analysis with six studies. In addition, Sun et al. [47] also performed a meta-analysis of the +874T/A polymorphism and CHB risk with five studies, and their results indicated the TT genotype and the T allele reduce the risk of chronic HBV infection in Asian individuals, which was similar to our meta-analysis of nine studies. However, we did not observed a significant association in co-dominant models. These differences may be because more studies were involved in our study, therefore; therefore, we believe our results are more robust with more statistical power.
Heterogeneity is the most common problem when explaining the results of a meta-analysis. In the current meta-analysis, we assessed heterogeneity using various statistical methods, including subgroup analysis, meta-regression, and Galbraith plot analysis. Finally, we concluded that the heterogeneity in the meta-analysis was due to the genotyping method, mixed infection, and various virus types. Sensitivity analysis was performed by excluding the studies not in HWE. The studies of HDV infection and HEV infection and those that used different genotyping methods, especially the DNA sequencing method, were also excluded one by one. The corresponding pooled OR value did not differ significantly from that of the overall meta-analysis. However, a Begg’s funnel plot and an Egger’s test (P>0.05) showed no publication bias. Therefore, we consider that the results are credible, although there are some confounding factors in the articles.
GWASs have evolved over the last 10 years into a powerful tool for investigating the genetic architecture of human disease. Recently, many GWASs determined that SNPs at HLA-DPA1, HLA-DPB1, HLA-DQB1–HLA-DQA2 and HLA-DQB2 are associated with chronic HBV infection in Japanese and Korean populations [40, 48, 49], and Hu et al. identified two new loci associated with chronic HBV infection: rs3130542 at 6p21.33 and rs4821116 at 22q11.21 in Han Chinese [50]. In addition, rs17401966 in KIF1B on chromosome 1p36.22 locus confers susceptibility to HBV-related HCC [51]. Our results suggested that rs2430561 in IFN-γ on chromosome 12q24 contributes to susceptibility to HBV-related diseases based on traditional genotyping methods, and these results have motivated us to carry out a GWAS focus on the SNPs on chromosome 12 to discover novel susceptibility loci for HBV-related diseases in the future.
Several limitations of the present meta-analysis should be considered when interpreting the results. First, most of the included studies had small sample sizes, which led to insufficient statistical power to explore the interaction between the IFN-γ +874T/A polymorphism and hepatitis virus-related diseases. The lack of association in the other ethnicity or genetic model may be most likely because of insufficient studies. Further studies on this topic in different ethnicities are expected to be conducted to strengthen our results. Second, because a lack in the original data of the reviewed studies, a more precise analysis could not conducted of individual information including other covariates such as age and sex. In spite of these, our meta-analysis had some advantages. First, a substantial number of cases and controls were pooled from different studies, which greatly increased the statistical power compared with individual studies. Second, the results of the sensitivity analyses were not materially altered and did not draw different conclusions, indicating that our results were moderately robust.
Conclusion
In summary, this meta-analysis indicated that the IFN-γ +874T/A polymorphism increases the risk of hepatitis virus—related diseases, especially in Asians and HBV-related disease. We expect more relevant studies to be published on this topic in the future to strengthen our conclusion, as it remains a topic of concern and interest.
Supporting Information
(DOC)
(DOC)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nature reviews Gastroenterology & hepatology 2010, 7(8):448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Z-E, Reiner SL, Zheng S, Dalton DK, Locksley RM. CD4+ effector cells default to the Th2 pathway in interferon gamma-deficient mice infected with Leishmania major. The Journal of experimental medicine 1994, 179(4):1367–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saxena R, Chawla YK, Verma I, Kaur J. IFN-gamma (+874) and not TNF-alpha (-308) is associated with HBV-HCC risk in India. Molecular and cellular biochemistry 2014, 385(1–2):297–307. 10.1007/s11010-013-1827-z [DOI] [PubMed] [Google Scholar]
- 4. Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annual review of biochemistry 1998, 67(1):227–264. [DOI] [PubMed] [Google Scholar]
- 5. Schena FP, Cerullo G, Torres DD, Scolari F, Foramitti M, Amoroso A, et al. Role of interferon-γ gene polymorphisms in susceptibility to IgA nephropathy: a family-based association study. European journal of human genetics 2006, 14(4):488–496. [DOI] [PubMed] [Google Scholar]
- 6. Bouzgarrou N, Hassen E, Farhat K, Bahri O, Gabbouj S, Maamouri N, et al. Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis C severity. Hum Immunol 2009, 70(4):230–236. 10.1016/j.humimm.2009.01.019 [DOI] [PubMed] [Google Scholar]
- 7. Cheong JY, Cho SW, Chung SG, Lee JA, Yeo M, Wang HJ, et al. Genetic polymorphism of interferon-gamma, interferon-gamma receptor, and interferon regulatory factor-1 genes in patients with hepatitis B virus infection. Biochemical genetics 2006, 44(5–6):246–255. [DOI] [PubMed] [Google Scholar]
- 8. Dai CY, Chuang WL, Chang WY, Chen SC, Lee LP, Hsieh MY, et al. Polymorphisms in the interferon-gamma gene at position +874 in patients with chronic hepatitis C treated with high-dose interferon-alpha and ribavirin. Antiviral research 2005, 67(2):93–97. [DOI] [PubMed] [Google Scholar]
- 9. Falleti E, Fabris C, Toniutto P, Fontanini E, Cussigh A, Caldato M, et al. Genetic polymorphisms of inflammatory cytokines and liver fibrosis progression due to recurrent hepatitis C. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research 2007, 27(3):239–246. [DOI] [PubMed] [Google Scholar]
- 10. Kim HJ, Chung JH, Shin HP, Jeon JW, Park JJ, Cha JM, et al. Polymorphisms of interferon gamma gene and risk of hepatocellular carcinoma in korean patients with chronic hepatitis B viral infection. Hepato-gastroenterology 2013, 60(125):1117–1120. 10.5754/hge11333 [DOI] [PubMed] [Google Scholar]
- 11. Liu M, Cao B, Zhang H, Dai Y, Liu X, Xu C. Association of interferon-gamma gene haplotype in the Chinese population with hepatitis B virus infection. Immunogenetics 2006, 58(11):859–864. [DOI] [PubMed] [Google Scholar]
- 12. Mishra N, Arankalle VA. Association of polymorphisms in the promoter regions of TNF-alpha (-308) with susceptibility to hepatitis E virus and TNF-alpha (-1031) and IFN-gamma (+874) genes with clinical outcome of hepatitis E infection in India. Journal of hepatology 2011, 55(6):1227–1234. 10.1016/j.jhep.2011.03.023 [DOI] [PubMed] [Google Scholar]
- 13. Sarvari J, Norozian H, Fattahi MR, Pirbonyeh N, Moattari A. The Role of Interferon Gamma Gene Polymorphism (+874A/T, +2109A/G, and -183G/T) in Response to Treatment Among Hepatitis C Infected Patients in Fars Province, Southern Iran. Hepatitis monthly 2014, 14(1):e14476 10.5812/hepatmon.14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Srivastava M, Ranjan A, Choudhary JK, Tripathi MK, Verma S, Dixit VK, et al. Role of Proinflammatory Cytokines (Interferon Gamma) and Anti-Inflammatory Cytokine (Interleukin-10) Gene Polymorphisms in Chronic Hepatitis B Infection: An Indian Scenario. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu QR, Ge YL, Gu SQ, Yu H, Wang JS, Gu XH, et al. Relationship between cytokines gene polymorphism and susceptibility to hepatitis B virus intrauterine infection. Chinese medical journal 2005, 118(19):1604–1609. [PubMed] [Google Scholar]
- 16. Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. The American journal of gastroenterology 2003, 98(1):144–150. [DOI] [PubMed] [Google Scholar]
- 17. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Annals of internal medicine 2009, 151(4):264–269. [DOI] [PubMed] [Google Scholar]
- 18. Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, et al. Systematic review and meta-analysis of the association between β2-adrenoceptor polymorphisms and asthma: a HuGE review. American journal of epidemiology 2005, 162(3):201–211. [DOI] [PubMed] [Google Scholar]
- 19. Ye X-H, Bu Z-B, Feng J, Peng L, Liao X-B, Zhu X-L, et al. Association between the TP53 polymorphisms and lung cancer risk: a meta-analysis. Molecular biology reports 2013:1–13. 10.1007/s11033-012-1930-3 [DOI] [PubMed] [Google Scholar]
- 20. Higgins J, Thompson SG. Quantifying heterogeneity in a meta‐analysis. Statistics in medicine 2002, 21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 21. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ: British Medical Journal 2003, 327(7414):557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arababadi MK, Pourfathollah AA, Jafarzadeh A, Hassanshahi G, Daneshmandi S, Shamsizadeh A, et al. Non-association of IL-12 +1188 and IFN-gamma +874 polymorphisms with cytokines serum level in occult HBV infected patients. Saudi journal of gastroenterology: official journal of the Saudi Gastroenterology Association 2011, 17(1):30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Basturk B, Karasu Z, Kilic M, Ulukaya S, Boyacioglu S, Oral B. Association of TNF-alpha -308 polymorphism with the outcome of hepatitis B virus infection in Turkey. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 2008, 8(1):20–25. [DOI] [PubMed] [Google Scholar]
- 24. Conde SR, Feitosa RN, Freitas FB, Hermes RB, Demachki S, Araujo MT, et al. Association of cytokine gene polymorphisms and serum concentrations with the outcome of chronic hepatitis B. Cytokine 2013, 61(3):940–944. 10.1016/j.cyto.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 25. Gao QJ, Liu DW, Zhang SY, Jia M, Wang LM, Wu LH, et al. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World journal of gastroenterology: WJG 2009, 15(44):5610–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karatayli SC, Ulger ZE, Ergul AA, Keskin O, Karatayli E, Albayrak R, et al. Tumour necrosis factor-alpha, interleukin-10, interferon-gamma and vitamin D receptor gene polymorphisms in patients with chronic hepatitis delta. Journal of viral hepatitis 2014, 21(4):297–304. 10.1111/jvh.12139 [DOI] [PubMed] [Google Scholar]
- 27. Korachi M, Ceran N, Adaleti R, Nigdelioglu A, Sokmen M. An association study of functional polymorphic genes IRF-1, IFNGR-1, and IFN-gamma with disease progression, aspartate aminotransferase, alanine aminotransferase, and viral load in chronic hepatitis B and C. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases 2013, 17(1):e44–49. 10.1016/j.ijid.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 28. Peng XM, Lei RX, Gu L, Ma HH, Xie QF, Gao ZL. Influences of MxA gene -88 G/T and IFN-gamma +874 A/T on the natural history of hepatitis B virus infection in an endemic area. International journal of immunogenetics 2007, 34(5):341–346. [DOI] [PubMed] [Google Scholar]
- 29. Teixeira AC, Mendes CT Jr, Marano LA, Deghaide NH, Secaf M, Elias J Jr, et al. Alleles and genotypes of polymorphisms of IL-18, TNF-alpha and IFN-gamma are associated with a higher risk and severity of hepatocellular carcinoma (HCC) in Brazil. Hum Immunol 2013, 74(8):1024–1029. 10.1016/j.humimm.2013.04.029 [DOI] [PubMed] [Google Scholar]
- 30. Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection—association between TGF-beta1 polymorphisms and hepatocellular carcinoma. Journal of hepatology 2005, 42(4):505–510. [DOI] [PubMed] [Google Scholar]
- 31. Chuang SC, La Vecchia C, Boffetta P. Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer letters 2009, 286(1):9–14. 10.1016/j.canlet.2008.10.040 [DOI] [PubMed] [Google Scholar]
- 32. Pravica V, Perrey C, Stevens A, Lee J-H, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-γ gene:: Absolute correlation with a polymorphic CA microsatellite marker of high IFN-γ production. Human immunology 2000, 61(9):863–866. [DOI] [PubMed] [Google Scholar]
- 33. Kordi Tamandani MK, Sobti RC, Shekari M, Mukesh M, Suri V. Expression and polimorphism of IFN-gamma gene in patients with cervical cancer. Experimental oncology 2008, 30(3):224–229. [PubMed] [Google Scholar]
- 34. Gangwar R, Pandey S, Mittal RD. Association of interferon-gamma +874A polymorphism with the risk of developing cervical cancer in north-Indian population. BJOG: an international journal of obstetrics and gynaecology 2009, 116(12):1671–1677. 10.1111/j.1471-0528.2009.02307.x [DOI] [PubMed] [Google Scholar]
- 35. Lee JE, Lee SJ, Namkoong SE, Um SJ, Sull JW, Jee SH, et al. Gene-gene and gene-environmental interactions of p53, p21, and IRF-1 polymorphisms in Korean women with cervix cancer. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society 2004, 14(1):118–125. [DOI] [PubMed] [Google Scholar]
- 36. Delaney NL, Esquenazi V, Lucas DP, Zachary AA, Leffell MS. TNF-alpha, TGF-beta, IL-10, IL-6, and INF-gamma alleles among African Americans and Cuban Americans. Report of the ASHI Minority Workshops: Part IV. Hum Immunol 2004, 65(12):1413–1419. [DOI] [PubMed] [Google Scholar]
- 37. Poli F, Nocco A, Berra S, Scalamogna M, Taioli E, Longhi E, et al. Allele frequencies of polymorphisms of TNFA, IL‐6, IL‐10 and IFNG in an Italian Caucasian population. European journal of Immunogenetics 2002, 29(3):237–240. [DOI] [PubMed] [Google Scholar]
- 38. Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 1996, 4(1):25–36. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez T, Mendez R, Del Campo A, Aptsiauri N, Martin J, Orozco G, et al. Patterns of constitutive and IFN-γ inducible expression of HLA class II molecules in human melanoma cell lines. Immunogenetics 2007, 59(2):123–133. [DOI] [PubMed] [Google Scholar]
- 40. Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Human molecular genetics 2011, 20(19):3884–3892. 10.1093/hmg/ddr301 [DOI] [PubMed] [Google Scholar]
- 41. von Linsingen R, Bompeixe EP, Maestri CA, Carvalho NS, da Graca Bicalho M. IFNG (+874 T/A) polymorphism and cervical intraepithelial neoplasia in Brazilian women. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research 2009, 29(5):285–288. [DOI] [PubMed] [Google Scholar]
- 42. Cardoso C, Pereira A, Brito-de-Souza V, Dias-Baptista I, Maniero V, Venturini J, et al. IFNG+ 874 T> A single nucleotide polymorphism is associated with leprosy among Brazilians. Human genetics 2010, 128(5):481–490. 10.1007/s00439-010-0872-x [DOI] [PubMed] [Google Scholar]
- 43. de Albuquerque AC, Rocha LQ, de Morais Batista AH, Teixeira AB, Dos Santos DB, Nogueira NA. Association of polymorphism +874 A/T of interferon-gamma and susceptibility to the development of tuberculosis: meta-analysis. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 2012, 31(11):2887–2895. [DOI] [PubMed] [Google Scholar]
- 44. Wiechec E, Hansen LL. The effect of genetic variability on drug response in conventional breast cancer treatment. European journal of pharmacology 2009, 625(1–3):122–130. 10.1016/j.ejphar.2009.05.033 [DOI] [PubMed] [Google Scholar]
- 45. Oxenkrug G, Perianayagam M, Mikolich D, Requintina P, Shick L, Ruthazer R, et al. Interferon-gamma (+874) T/A genotypes and risk of IFN-alpha-induced depression. Journal of neural transmission (Vienna, Austria: 1996) 2011, 118(2):271–274. 10.1007/s00702-010-0525-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ge YZ, Wang YD, Xu Z, Xu LW, Wang YP, Gu MH, et al. Lack of association between interferon gamma +874 T/A polymorphism and cancer risk: an updated meta-analysis. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 2014, 35(7):6405–6414. 10.1007/s13277-014-1861-9 [DOI] [PubMed] [Google Scholar]
- 47. Sun XR, Wu J, Tang KF. The interferon-gamma (IFN-gamma) +874T allele reduces the risk of hepatitis B infection in an Asian population. Journal of viral hepatitis 2014, 21(4):281–287. 10.1111/jvh.12140 [DOI] [PubMed] [Google Scholar]
- 48. Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009, 41(5):591–595. 10.1038/ng.348 [DOI] [PubMed] [Google Scholar]
- 49. Nishida N, Sawai H, Matsuura K, Sugiyama M, Ahn SH, Park JY, et al. Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PloS one 2012, 7(6):e39175 10.1371/journal.pone.0039175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nature genetics 2013. [DOI] [PubMed] [Google Scholar]
- 51. Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet 2010, 42(9):755–758. 10.1038/ng.638 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.