Abstract
Objective
Our aim is to map chromosomal regions that harbor loci that increase susceptibility to bipolar disorder.
Methods
We analyzed 644 bipolar families ascertained by the National Institute of Mental Health Human Genetics Initiative for bipolar disorder. The families have been genotyped with microsatellite loci spaced every approximately 10 cM or less across the genome. Earlier analyses of these pedigrees have been limited to nonparametric (model-free) methods and thus, information from unaffected subjects with genotypes was not considered. In this study, we used parametric analyses assuming dominant and recessive transmission and specifying a maximum penetrance of 70%, so that information from unaffecteds could be weighed in the linkage analyses. As in previous linkage analyses of these pedigrees, we analyzed three diagnostic categories: model 1 included only bipolar I and schizoaffective, bipolar cases (1565 patients of whom approximately 4% were schizoaffective, bipolar); model 2 included all individuals in model 1 plus bipolar II patients (1764 total individuals); and model 3 included all individuals in model 2 with the addition of patients with recurrent major depressive disorder (2046 total persons).
Results
Assuming dominant inheritance the highest genome-wide pair-wise logarithm of the odds (LOD) score was 3.2 with D16S749 using model 2 patients. Multipoint analyses of this region yielded a maximum LOD score of 4.91. Under recessive transmission a number of chromosome 20 markers were positive and multipoint analyses of the area gave a maximum LOD of 3.0 with model 2 cases.
Conclusion
The chromosome 16p and 20 regions have been implicated by some studies and the data reported herein provide additional suggestive evidence of bipolar susceptibility genes in these regions.
Keywords: bipolar disorder, chromosome 16, chromosome 20, complex disorder, genetics, gene mapping, linkage analysis, parametric analysis
Introduction
Bipolar I disorder (BPI), also termed manic-depression, is a severe and often disabling neuropsychiatric disorder that afflicts approximately 1% of most populations (Belmaker, 2004). The direct and indirect costs of this disorder to the American society are tens of billions of dollars per year (Kleinman et al., 2003). Approximately 10% of the individuals with BPI disorder will die by suicide. Although pivotal neurobiological underpinnings of BPI disorder have yet to be unraveled, genetics is of major etiological importance, with an estimated heritability of 80% (Merikangas and Low, 2004). Results of some, but not all, segregation analyses implicate major loci underlying the disease (Curtis et al., 1993; Pauls et al., 1995). Family, twin, adoption and segregation studies, however, indicate that bipolar disorder is a complex genetic disease characterized by incomplete penetrance, unknown mode of transmission, phenocopies (etiological heterogeneity), and likely genetic heterogeneity.
Over 20 genome-wide scans have been reported for bipolar disorder, and the results have been recently reviewed (DePaulo, 2004; McQueen et al., 2005) as well as used for two meta-analyses (Badner and Gershon, 2002; Segurado et al., 2003). Analyses have implicated chromosomes 6q, 8q, 13q, 22q, 18p, 18q, and 22q. One meta-analysis of linkage data from 1067 families (n = 5179 individuals) yielded evidence for bipolar loci on 6q [logarithm of the odds (LOD) = 4.15] and 8q (LOD = 3.32)(McQueen et al., 2005). Genome-wide linkage analyses of uniquely large pedigrees have also implicated a number of chromosomal regions including 4p, 4q, 9q, 12q, 21q, and Xq (Blackwood et al., 1996; Adams et al., 1998; Ekholm et al., 2003; Green et al., 2005; Shink et al., 2005; Venken et al., 2005). Most of the linkage findings in the field, however, have not been thoroughly replicated, most likely because of the necessity of large sample sizes for replication (Suarez, 1994). It is, however, clear that possible loci for bipolar disorder identified to date account for only a small proportion of variance in risk. Although genome-wide association will soon come into play, linkage analyses of large bipolar family data sets or uniquely large bipolar pedigrees may still be fruitful in mapping additional bipolar loci. Linkage, unlike association, is robust to allelic heterogeneity and is not dependent on linkage disequilibrium. Given that both rare and common variants likely contribute many if not most complex disorders, both linkage and association will be needed to map the full range of bipolar susceptibility loci (Zwick et al., 2000; Reich and Lander, 2001; Pritchard and Cox, 2002).
In this study we analyzed 644 families ascertained by the National Institute of Mental Health Human (NIMH) genetics initiative. These families, currently the largest pedigree series, which are freely available to qualified researchers worldwide, are also known in the literature as ‘waves 1, 2, 3, and 4 pedigrees’ (Detera-Wadleigh et al., 1997; Edenberg et al., 1997; Dick et al., 2003; Kassem et al., 2006). The 644 pedigrees contain 1565 individuals with BPI disorder or schizoaffective (SA) disorder, bipolar type, 199 persons with bipolar II (BPII) disorder, 282 patients with recurrent major depressive disorder (MDD), and 876 unaffected persons with genotypes. Wave 1, 2, 3, and 4 families have been genotyped, at various intervals, with microsatellite loci spaced every approximately 10 cM or less across the genome. Linkage studies of these families have been performed in four ‘waves’ (McInnes et al., 1996; Detera-Wadleigh et al., 1997; Edenberg et al., 1997; Rice et al., 1997; Stine et al., 1997; Dick et al., 2003; McInnis et al., 2003; Kassem et al., 2006). Implicated regions include 1q, 5q, 6q, 7p, 10p, 16p, 16q, 17q, and 22q. Earlier analyses of these pedigrees have been primarily limited to nonparametric or model-free methods and thus, unaffected subjects – many of whom presumably carry disease alleles without expression of disease – were not considered. In this study, we used parametric analysis, a method that allows for the specification of phenocopies, reduced penetrance, and transmission models. We chose a model-based approach – dominant and recessive models – because simulation studies show that, formulating a genetic model that approximates the true inheritance, may have more power than nonparametric analyses (Abreu et al., 1999; Durner et al., 1999). As in earlier linkage studies of these pedigrees, three diagnostic models were used: model 1 included only BPI or SA, bipolar type cases (1565 cases); model 2 included model 1 and BPII patients (1764 patients); and model 3 included model 2 and recurrent MDD individuals (2046 cases). Of the model 1 cases fewer than 4% were SA, bipolar type. Our strategy was to first carry out genome-wide two-point analyses and then multipoint analyses in the regions of interest, defined as LODs of 1.5 or greater. Pairwise analysis, which considers evidence of linkage at various recombination fractions, can be more robust when there are misspecifications in the model, marker location anomalies, and/or (undetected) genotyping errors (Risch and Giuffra, 1992; Pal et al., 2001).
Methods
Family ascertainment
All families were ascertained as part of the NIMH Bipolar Genetics Initiative, which has been continuously active for the past 15 years. As part of the four-site collaborative effort (Indiana University, Washington University, Johns Hopkins and the NIMH intramural program) that existed between 1989 and 1997, 153 multiplex families were ascertained. The original four-site consortia families are known as waves 1 and 2. Wave 1 families (n = 97) were first ascertained, genotyped, and analyzed (Edenberg et al., 1997). Subsequently, the four-site consortia ascertained an additional 56 kindreds that were genotyped and analyzed (Dick et al., 2003; Willour et al., 2003; Zandi et al., 2003). In 1998 the project was expanded as a 10-site collaborative team (comprising four initial sites plus the University of Pennsylvania, University of Iowa, University of Chicago, University of California–San Diego, University of California–San Francisco, University of Michigan) to ascertain at least 500 families containing at least two siblings with either BPI or SA-bipolar type (Dick et al., 2003; Kassem et al., 2006) during a 4-year period from 1998 to 2002; it exceeded the original aims of the grant and collected 546 pedigrees. The 10-site collaborative families are denoted as waves 3 and 4. Wave 3 families (n = 250) were ascertained, genotyped, and analyzed (Dick et al., 2003) before wave 4 families (n = 309) (Kassem et al., 2006). In all, 699 families were ascertained, and none of the waves contained overlapping pedigrees. Some of the ascertained families were eliminated from genotyping or final linkage analyses because of lack of informativeness for linkage and mispaternity. Over 95% of the pedigrees are European Caucasian.
Detailed procedures for ascertainment and diagnoses have been reported earlier (Edenberg et al., 1997; Dick et al., 2002). In brief, the Diagnostic Interview for Genetic Studies (DIGS) was used to interview patients (Nurnberger et al., 1994). Additional diagnostic information was collected from multiple family informants using the Family Interview for Genetic Studies and from medical records when available. Diagnoses was assigned by two non-interviewing clinicians, who reviewed the DIGS, Family Interview for Genetic Studies, and medical records in a best estimate procedure (Leckman et al., 1982). Diagnostic disagreements were resolved by a third, independent reviewer if needed (less than 2% of cases). Diagnostic and Statistical Manual of Mental Disorders-III-R criteria were used for BPI and SA, bipolar. Research diagnostic criteria were used for BPII with the additional requirement that patients also had to have recurrent episodes of major depression. Research diagnostic criteria were used for Unipolar Depression, Recurrent. Although DSM-IV criteria were published in 1990, the initial diagnostic criteria were used during all four waves of the study to maximize diagnostic consistency.
All of this material, including DNA and cell lines, best estimate final diagnoses, detailed clinical data from the DIGS, pedigree relationship information, and genotypic data from a complete genomic survey, is available to qualified investigators through application to the Center for Genetic Studies – www.nimhgenetics.org.
Genotyping
Waves 1, 2, 3, and 4 families were genotyped separately over several years. Wave 1 and 2 families (23% of the pedigrees) were genotyped by the University sites and NIMH intramural program using microsatellite loci derived from early marker maps. Wave 3 and 4 families (77% of the kindreds) were genotyped at the Center for Inherited Disease Research in two separate samples. The genotype data waves 1, 2, 3, and 4 were cleaned and combined in a project coordinated by Melvin McInnis and Peter Zandi at Johns Hopkins University (Goes et al., 2007). A number of methods that have been described in detail earlier were used to check the genotype data for quality control. The programs UNKNOWN and PEDCHECK were used to check for inheritance errors (Lathrop et al., 1984, O’Connell and Weeks, 1998). The PREST software (http://linkage.rockefeller.edu/soft/) was used to check for unlikely relationships (Sun et al., 2002). Finally, the output from CRIMAP (http://linkage.rockefeller.edu/soft/) was used to check for unlikely double recombinants (Lander and Green, 1987). Identified errors that could not be resolved were removed from the data set. A common genetic map was constructed for the markers using the deCode map as a framework. For microsatellite loci not on the deCode map their relative physical distances were used to interpolate them into the genetic map. Altogether 685 microsatellite loci were genotyped, 366 of which were genotyped in more than one wave (Goes et al., 2007).
Diagnostic models for linkage
Three diagnostic models were used: model 1 included only BPI and SA, bipolar type cases, model 2 included model 1 and BPII patients, and model 3 included model 2 individuals and recurrent MDD patients.
Linkage analyses
Parametric analyses were carried using the FASTLINK 4.1 modifications to the LINKAGE (http://linkage.rockefeller.edu/soft/) software program. We chose a model-based approach (dominant and recessive models) because simulation studies show formulating a genetic model that approximates the true inheritance may have more power than nonparametric analyses (Abreu et al., 1999; Durner et al., 1999). With parametric analyses one can weigh evidence of linkage that some unaffected individuals harbor the disease allele, but did not express illness by using penetrance functions. Family and twin studies indicate that many, if not most, susceptibility alleles for bipolar disorder have reduced penetrance. For the dominant model, we assumed a disease allele frequency of 0.01, a penetrance of 0.7 for the heterozygote and homozygote and a penetrance of 0.001 for the normal allele. For the recessive model we assumed a disease allele frequency of 0.10, a penetrance of 0.7 for the homozygote and a penetrance of 0.001 for the normal allele. These parameters are conservative (e.g. penetrance should not be overestimated) and approximate those in the literature for parametric analyses. Genome-wide two-point calculations were first carried out using the MLINK subroutine. Multipoint analyses were then performed in regions of interest, defined as an area yielding a two-point LOD score of 1.5 or higher.
Linkage results
Pairwise
A summary of the genome-wide pairwise analyses is presented in Table 1. Only scores above 1.5 are presented. D16S748, located 22.65 cM from pter, yielded the highest genome-wide LOD score, 3.29 (θ = 0.3) assuming dominant transmission and using model 2 cases. Flanking markers also produced positive scores using model 2 cases: D16S749, LOD 1.57 at θ = 0.2 (Tables 1 and 2). Evidence of linkage decreased under model 3, which included 282 cases of recurrent unipolar depression: (D16S748, max LOD of 1.97 at 0.3 θ). One explanation for this is that recurrent unipolar disorder is more etiologically and genetically heterogeneous and thus, having these cases in the linkage analysis would include more phenocopies. The second highest genome-wide pairwise LOD score was obtained with D4S2390 using model 1 cases only and assuming dominant transmission (LOD 2.01 at 0.1 θ). D4S2390 is located 208.07cM from the pter. Under recessive transmission and using model 1 cases D4S2390 yielded a LOD score of 1.67 (0.2 θ).
Table 1.
Two-point LOD scores on all chromosomes using all models of transmission that are greater than 1.5
| Chromosome | Marker | Location of marker (cM)a | Modelc | θ | LOD score |
|---|---|---|---|---|---|
| 1 | D1s462 | 97.72 | UP-Dom | 0.2 | 1.87 |
| 4 | D4s2390 | 208.07 | BPI-Dom | 0.1 | 2.01 |
| 4 | D4s2390 | 208.07 | BPI-Rec | 0.2 | 1.67 |
| 6 | D6s495 | 147–153b | BPII-Dom | 0.2 | 1.65 |
| 8 | D8s1771 | 50.05 | BPI-Rec | 0.3 | 1.57 |
| 8 | D8s1771 | 50.05 | UP-Dom | 0.3 | 1.77 |
| 11 | D11s3163 | 0.00 | UP-Dom | 0.2 | 1.89 |
| 12 | D12s395 | 136.82 | UP-Dom | 0.4 | 1.59 |
| 16 | D16s748 | 22.65 | BPII-Dom | 0.3 | 3.29 |
| 16 | D16s749 | 39.04 | BPII-Dom | 0.2 | 1.57 |
| 16 | D16s748 | 22.65 | UP-Dom | 0.3 | 1.97 |
| 20 | D20s162 | 24.70 | BPII-Rec | 0.2 | 1.72 |
| 20 | D20s473 | 9.53 | BPII-Rec | 0.3 | 1.60 |
| 20 | D20s601 | 50.81 | UP-Rec | 0.2 | 1.90 |
LOD, logarithm of the odds.
Taken from deCode map.
Location of this marker has not been identified on either the deCode or Marshfield maps, so the range given is between the locations for the two flanking markers on the deCode map.
BPI includes bipolar disorder type I and schizoaffective disorder, bipolar type. BPII includes both bipolar disorder type I and bipolar disorder type II.
UP includes bipolar disorder type I, bipolar disorder type II, and unipolar depressive disorder.
Table 2.
Chromosome 16 two-point LOD scores at all values of theta using a bipolar II – dominant transmission model
| Marker | Mba | θ = 0 | θ = 0.01 | θ = 0.05 | θ = 0.1 | θ = 0.2 | θ = 0.3 | θ = 0.4 |
|---|---|---|---|---|---|---|---|---|
| D16s3401 | N/A | − 114.10 | − 85.60 | − 46.40 | − 26.40 | − 8.90 | − 2.20 | − 0.10 |
| ATA67b07 | 1.94 | − 45.59 | − 33.15 | − 17.25 | − 9.20 | − 2.40 | − 0.23 | 0.15 |
| D16s2618 | 3.19 | − 67.70 | − 51.40 | − 28.90 | − 16.60 | − 5.50 | − 1.30 | − 0.10 |
| D16s2622 | 3.65 | − 56.01 | − 41.09 | − 20.80 | − 10.53 | − 2.12 | 0.39 | 0.60 |
| D16s2616 | 5.68 | − 142.58 | − 103.87 | − 53.22 | − 27.98 | −7.08 | − 0.49 | 0.60 |
| D16s687 | 9.25 | − 84.30 | − 62.70 | − 33.40 | − 18.70 | − 5.80 | − 1.10 | 0.20 |
| D16s677 | 10.21 | − 34.24 | − 26.12 | − 14.08 | −7.66 | − 2.05 | − 0.17 | 0.14 |
| D16s748 | 12.05 | − 166.97 | − 118.38 | − 54.85 | − 24.24 | − 1.37 | 3.29 | 1.90 |
| D16s2619 | 13.65 | − 32.40 | − 24.50 | − 12.80 | − 6.50 | − 1.30 | 0.10 | 0.20 |
| D16s764 | 16.55 | − 48.20 | − 35.30 | − 17.50 | − 8.40 | − 1.20 | 0.70 | 0.60 |
| D16s3103 | 17.38 | − 131.51 | − 93.39 | − 45.04 | − 21.83 | − 3.81 | 0.71 | 0.62 |
| D16s749 | 19.75 | − 18.27 | − 12.94 | − 4.94 | − 0.97 | 1.57 | 1.47 | 0.65 |
| D16s403 | 22.95 | − 83.00 | − 58.80 | − 29.50 | − 15.20 | − 3.70 | − 0.20 | 0.20 |
| D16s769 | 26.07 | − 105.68 | −78.14 | − 40.46 | − 21.05 | − 4.95 | − 0.15 | 0.47 |
| D16s540 | 47.64 | − 90.60 | − 66.17 | − 32.97 | − 16.44 | − 3.23 | − 0.25 | 0.37 |
| D16s3396 | 49.75 | − 136.38 | − 100.11 | − 51.71 | − 27.12 | − 6.67 | − 0.47 | 0.42 |
| D16s757 | 50.11 | − 58.35 | − 44.54 | − 25.26 | − 14.63 | − 4.96 | − 1.40 | − 0.30 |
| D16s3253 | 56.11 | − 197.47 | − 148.70 | − 80.02 | − 44.17 | − 13.15 | − 2.48 | 0.12 |
| D16s752 | 69.89 | − 67.59 | − 48.59 | − 24.39 | − 12.59 | − 3.09 | − 0.18 | 0.23 |
| D16s2624 | 70.29 | − 170.50 | − 126.49 | − 66.06 | − 35.35 | − 9.73 | − 1.62 | 0.00 |
| D16s3096 | 77.60 | − 104.66 | −77.37 | − 40.00 | − 21.02 | − 5.25 | − 0.51 | 0.13 |
| D16s750 | 78.22 | − 27.08 | − 21.06 | − 11.79 | − 6.51 | − 1.66 | − 0.06 | 0.13 |
| D16s3091 | 81.54 | − 141.02 | − 107.06 | − 59.34 | − 33.96 | − 11.53 | − 3.35 | − 0.76 |
| D16s539 | 84.94 | − 179.83 | − 131.89 | − 66.75 | − 34.08 | −7.90 | − 0.55 | 0.26 |
LOD, logarithm of the odds.
All marker locations were taken from National Center for Biotechnology Information-STS when available. Marker locations for D16s2616 and D16s3253 were extrapolated from flanking markers. Marker D16s3401 is located at the pter end of the chromosome, so extrapolation was not possible.
Three markers on chromosome 20 produced positive scores assuming recessive transmission (Tables 1 and 3). D20S601 (50.81 cM from the pter) generated a LOD score of 1.90 using model 3 cases. Using model 2 cases D20S473 (9.53 from the pter) yielded a LOD score of 1.71 at D20S162 (24.7 cM from the pter) produced a LOD score of 1.60.
Table 3.
Chromosome 20 two-point LOD scores at all values of theta using a bipolar II – recessive transmission model
| Marker | Mba | θ = 0 | θ = 0.01 | θ = 0.05 | θ = 0.1 | θ = 0.2 | θ = 0.3 | θ = 0.4 |
|---|---|---|---|---|---|---|---|---|
| D20s103 | 0.51 | − 248.7 | − 200.11 | − 107.61 | − 56.65 | − 15.51 | − 3.01 | − 0.27 |
| D20s473 | 3.41 | − 56.00 | − 43.20 | − 20.50 | − 8.40 | 0.30 | 1.60 | 0.70 |
| D20s482 | 4.45 | − 233.80 | − 187.70 | − 98.72 | − 49.80 | − 11.13 | − 0.58 | 0.54 |
| D20s95 | 5.66 | − 45.29 | − 39.98 | − 19.65 | − 9.75 | − 1.82 | 0.18 | 0.20 |
| D20s603 | 6.65 | − 26.00 | − 20.40 | − 10.00 | − 4.30 | 0.00 | 0.80 | 0.40 |
| D20s851 | 8.81 | − 285.30 | − 232.30 | − 127.50 | − 68.25 | − 19.33 | − 3.93 | − 0.37 |
| D20s162 | 9.98 | − 17.54 | − 12.64 | − 4.58 | − 0.58 | 1.72 | 1.37 | 0.45 |
| D20s604 | 12.53 | − 306.60 | − 246.60 | − 131.30 | − 66.81 | − 15.16 | − 1.02 | 0.51 |
| D20s470 | 17.32 | − 355.80 | − 288.30 | − 156.80 | − 82.81 | − 22.07 | − 3.59 | − 0.01 |
| D20s477 | 22.36 | − 309.40 | − 248.50 | − 133.50 | − 69.60 | − 17.48 | − 2.02 | 0.44 |
| D20s601 | 34.07 | − 22.28 | − 17.64 | − 9.06 | − 4.24 | − 0.42 | 0.42 | 0.22 |
| D20s478 | 36.67 | − 351.20 | − 287.20 | − 160.50 | − 87.81 | − 26.33 | − 5.93 | − 0.62 |
| D20s481 | 43.20 | − 367.20 | − 299.80 | − 167.10 | − 90.73 | − 26.22 | − 5.34 | − 0.47 |
| D20s480 | 50.03 | − 263.20 | − 212.90 | − 115.60 | − 59.53 | − 14.96 | − 1.88 | 0.27 |
| D20s1085 | 52.12 | − 42.78 | − 36.35 | − 22.48 | − 13.69 | − 5.24 | − 1.69 | − 0.32 |
| D20s1082 | 53.29 | − 29.75 | − 25.15 | − 14.59 | − 8.12 | − 2.45 | − 0.50 | − 0.03 |
| D20s451 | 56.09 | − 258.90 | − 209.10 | − 112.00 | − 57.81 | − 14.12 | − 1.54 | 0.34 |
| D20s171 | 57.24 | − 30.90 | − 26.10 | − 15.80 | − 9.50 | − 3.40 | − 1.00 | − 0.10 |
LOD, logarithm of the odds.
All marker locations were taken from National Center for Biotechnology Information-STS.
Multipoint
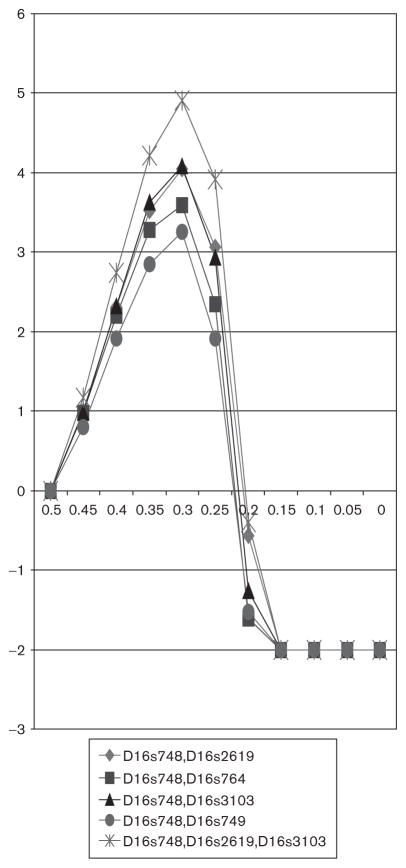
Multipoint linkage analyses were carried in areas that generated two-point scores above 1.5. The most significant multipoint linkage signals occurred on chromosome 16 (dominant transmission) and 20 (recessive inheritance) with the model 2 phenotype (Figs 1 and 2). For chromosome 16 the highest multipoint score was 4.91 derived using three contiguous chromosome 16 loci: D16S748, D16S2619, and D16S3103 (Fig. 1). Three-point analyses yielded maximum LODs of four or greater (Fig. 1). Assuming recessive inheritance and using model 2 cases chromosome 20 multipoint with D20S473 and D20S604 produced a maximum LOD of just over 3.0. Although chromosome 4 had a suggestive two-point LOD score of 2.01 at loci D4s2390, multipoint linkage analysis on chromosome 4 did not generate an increase in the maximum LOD score at this loci, and the maximum multipoint LOD generated in these analyses was 1.56 using D4s3335 and D4s2390.
Fig 1.
Chromosome 16 maximum multipoint logarithm of the odds scores using a bipolar II – dominant transmission model.
Fig 2.
Chromosome 20 maximum multipoint logarithm of the odds scores using a bipolar II – recessive transmission model.
Discussion
Genome-wide parametric analyses were carried out between bipolar disorder phenotypes using 644 families and 685 microsatellite loci. Earlier studies of the 644 pedigrees have been primarily limited to nonparametric analyses and thus, information from unaffecteds was not considered for most analyses. Although Avramopoulos et al. (2004) used parametric methods, only 56 pedigrees were analyzed. The strategy for this study was to first carry out genome-wide parametric two-point analyses and then multipoint analyses in regions yielding LODs of 1.5 or greater. Two-point analyses, which consider evidence for linkage at various recombination fractions, can be more robust when there are misspecifications in the model, marker location anomalies, and/or (undetected) genotyping errors. Three interdependent definitions of illness were used: model 1 included BPI cases and SA disorder (bipolar type) cases; model 2 consisted of individuals with model 1 phenotypes and BPII disorder; and model 3 comprised model 2 cases and recurrent major depression patients. In this study, both dominant and recessive transmission models were implemented using a maximum penetrance of 70%. These parameters are conservative (e.g., penetrance should not be overestimated) and approximate those in the literature for parametric analyses. The LOD scores were not corrected for multiple testing given that the genetic models are interdependent and thus, not totally independent tests. Thus, the linkage results should be interpreted with caution.
Genome-wide, the most positive results were on chromosomes 16 and 20 using model 2 cases (Tables 1–3; Figs 1 and 2). D16S748 yielded the highest two-point LOD score (3.2) assuming dominant inheritance (Table 1). D16S748 is located approximately 23cM from 16 pter. Flanking markers also gave positive pairwise LOD scores (Table 2). A series of three-point and four-point analyses were then carried out. A multipoint analysis of three fixed chromosome 16 markers (D16S748, D16S2619 at 28.30 cM from the pter, and D16s3103 at 37.97 cM from the pter) yielded the highest maximum LOD score 4.91 (Fig. 1). Three-point analyses with various chromosome 16 markers yielded LODs of approximately 3.2–4 (Fig. 1). These findings reinforce the results of analyses carried out before on the initial waves of the NIMH genetics initiative. Edenberg et al. (1997) analyzed 97 wave 1 NIMH bipolar kindred sib pairs that yielded positive scores (P = 0.006) between the model 2 phenotype and D16s2610 (28cM from 16 pter). Dick et al. (2002) carried out nonparametric linkage analyses of 97 wave 1 and 56 wave 2 pedigrees and reported a LOD score of 2.8 with D16s2619 (28cM from the 16 pter) using model 3 cases. Chromosome 16p has also been implicated in studies of other samples. Ewald et al. (1995) studied two Danish bipolar pedigrees and reported a LOD score of 2.5 with D16S510 under the assumption of recessive transmission and using all phenotypic definitions of bipolar illness (BPI, BPII, and recurrent major depression). D16S510 maps approximately 10cM from 16 pter. In one large Costa Rica family segregating bipolar disorder McInnes et al. (1996) found a LOD of 1.46 with D16S521 assuming dominant inheritance and using BPI cases only. D16S521 localizes to the telomeric end of 16p (about 1 cM from 16 pter). In the Finnish population Ekholm et al. (2003) ascertained 40 pedigrees with bipolar disorder and reported a three-point LOD of 2.7 between D16S769 (41.96cM from the 16 pter), and D16S3093 (37.97cM from 16 pter) using cases equivalent to model 1. Finally, Kassem et al. (2006) recently examined ‘polarity at onset’ using 507 waves 3 and 4 NIMH Genetics Initiative pedigrees. Linkage analyses restricted to those patients displaying mania at onset yielded a maximum multipoint LOD of 4.5 with D16S748, the marker that also yielded the highest LOD score in this study. It is likely that both studies have detected the same signal as waves 3 and 4 pedigrees comprise 77% of the kindreds studied herein. In a subsequent analysis, McMahon and colleagues carried out linkage analyses of all four waves using LODPAL, a multipoint affected relative pair program implemented in the SAGE (http://linkage.rockefeller.edu/soft/) package (version 4.4) that is capable of incorporating clinical covariates to adjust the relative risks associated with sharing identity by descent alleles, and yielded a Lodpal score of 5.1 with mania at onset and as a covariate (Francis McMahon personal communication to William Byerley).
The next most positive region was chromosome 20. Using model 2 cases D20S473 (9.53 cM from the pter) yielded a LOD score of 1.71 and D20S162 (30.34cM from the pter) produced a LOD score of 1.60. D20S601 (50.81 cM from the pter) produce a LOD score of 1.90 using model 3 cases. Using model 2 cases multipoint analyses with the D20S473, D20s482 and D20s162 marker loci produced a maximum LOD of just over 3.0. Chromosome 20 has been implicated by two published studies. Radhakrishna et al. (2001) found a multipoint LOD score of 4.35 between bipolar disorder and chromosome 20 markers assuming dominant transmission. The chromosome 20 markers D20s482 and D20s162 map 13.21 and 30.34 cM from the pter. Using wave 1 and 2 families Willour et al. (2003) reported a LOD of 1.82 using model 3 cases and assuming recessive transmission.
Other pairwise scores greater than 1.5 included D1S462 (about 97 cM from the pter), D4S2390 (about 208.07 from the pter), D6S495 (about 150cM from the pter), D8S1771 (about 50 cm from the pter), D11S3163 (about 0cM from the pter), and D12S395 (about 137 cm from the pter). Many of these regions have been implicated by other studies (Avramopoulos et al., 2004; Green et al., 2005; McQueen et al., 2005; Shink et al., 2005; Le Hellard et al., 2007). Multipoint analyses, however, did not increase evidence of linkage in these regions (data not shown).
Although genome wide linkage analysis has been the major method for mapping disease loci for over two decades, genome wide association (GWA) is now being used for gene mapping. Interestingly, two published analyses of these studies have implicated regions on chromosomal regions 16p and 20p in bipolar disorder. The first GWA study of bipolar disorder used 461 unrelated probands drawn from the NIMH genetics initiative, and 563 matched controls, and identified 88 SNPs with association signals and small effect sizes, that were detected in both the initial sample as well as the replicate sample of 772 bipolar I patients recruited from consecutive hospital admissions in Germany. Three SNPs were identified on chromosome 16, rs1818290 and rs7204975 at 16p13.2, and rs10500336 at 16p13.3. The SNP rs4813030 on chromosomal region 20p13 was also identified in both the initial and replication samples in this study (Baum et al., 2008).
A recently completed GWA funded by the Wellcome Trust (http://www.perlegen.com/index.htm?newsroom/pr/2005/2005_10_05_Wellcome_Trust_Affy_Press_Release.htm) examined seven major diseases including bipolar disorder, with 2000 affected subjects for each disorder and 3000 controls. Results from this study identified the chromosomal region 16p12 at 23.3–23.62Mb as a region of strong association for bipolar disorder with a P value of 6.29×10−8. The region on chromosme 20p13 from 3.70 to 3.73Mb was also identified as showing moderate evidence for association in bipolar disorder, with a genotypic P value of 6.71×10−6 (The Wellcome Trust Case Control Consortium, 2007).
At least one other large GWA funded by the Pritzker Foundation (http://www.pritzkerneuropsych.org/news/news.aspx) is underway. Although GWA may be more powerful for mapping common variants, underlying disease linkage analysis does not depend on the presence of linkage disequilibrium and is more robust for detecting rare variants, especially loci having a wide range of allelic heterogeneity. Of note, many Mendelian disorders average ~10 alleles at one locus. As a result, both linkage and association will likely be needed to map the full range of susceptibility loci for complex traits as both common and rare variants contribute to disease liability.
Acknowledgments
This study was supported by Dr Ross’ Training Next Generation Mental Health Researchers Grant MH 060482 from the National Institute of Mental Health.
References
- Abreu PC, Greenberg DA, Hodge SE. Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet. 1999;65:847–857. doi: 10.1086/302536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams LJ, Mitchell PB, Fielder SL, Rosso A, Donald JA, Schofield PR. A susceptibility locus for bipolar affective disorder on chromosome 4q35. Am J Hum Genet. 1998;62:1084–1091. doi: 10.1086/301826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramopoulos D, Willour VL, Zandi PP, Huo Y, Mackinnon DF, Potash JB, et al. Linkage of bipolar affective disorder on chromosome 8q24: follow-up and parametric analysis. Mol Psychiatry. 2004;9:191–196. doi: 10.1038/sj.mp.4001388. [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmaker RH. Bipolar disorder. N Engl J Med. 2004;351:476–486. doi: 10.1056/NEJMra035354. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, He L, Morris SW, Mclean A, Whitton C, Thomson M, et al. A locus for bipolar affective disorder on chromosome 4p. Nat Genet. 1996;12:427–430. doi: 10.1038/ng0496-427. [DOI] [PubMed] [Google Scholar]
- Curtis D, Brynjolfsson J, Petursson H, Holmes S, Sherrington R, Brett P, et al. Segregation and linkage analysis in five manic depression pedigrees excludes the 5HT1a receptor gene (HTR1A) Ann Hum Genet. 1993;57:27–39. doi: 10.1111/j.1469-1809.1993.tb00884.x. [DOI] [PubMed] [Google Scholar]
- DePaulo JR., Jr Genetics of bipolar disorder: where do we stand? Am J Psychiatry. 2004;161:595–597. doi: 10.1176/appi.ajp.161.4.595. [DOI] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Yoshikawa T, Sanders AR, Goldin LR, Turner G, et al. Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 4, 7, 9, 18, 19, 20, and 21q. Am J Med Genet. 1997;74:254–262. doi: 10.1002/(sici)1096-8628(19970531)74:3<254::aid-ajmg4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Edenberg HJ, Miller M, Bowman E, Rau NL, et al. Apparent replication of suggestive linkage on chromosome 16 in the NIMH genetics initiative bipolar pedigrees. Am J Med Genet. 2002;114:407–412. doi: 10.1002/ajmg.10380. [DOI] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, et al. Genome-wide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA. Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet. 1999;64:281–289. doi: 10.1086/302181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, Conneally PM, Sorbel JJ, Carr K, Crose C, et al. Initial genomic scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 3, 5, 15, 16, 17, and 22. Am J Med Genet. 1997;74:238–246. [PubMed] [Google Scholar]
- Ekholm JM, Kieseppa T, Hiekkalinna T, Partonen T, Paunio T, Perola M, et al. Evidence of susceptibility loci on 4q32 and 16p12 for bipolar disorder. Hum Mol Genet. 2003;12:1907–1915. doi: 10.1093/hmg/ddg199. [DOI] [PubMed] [Google Scholar]
- Ewald H, Mors O, Flint T, Koed K, Eiberg H, Kruse TA. A possible locus for manic depressive illness on chromosome 16p13. Psychiatr Genet. 1995;5:71–81. doi: 10.1097/00041444-199522000-00005. [DOI] [PubMed] [Google Scholar]
- Goes FS, Zandi PP, Miao K, Mcmahon FJ, Steele J, Willour VL, et al. Mood-incongruent psychotic features in bipolar disorder: familial aggregation and suggestive linkage to 2p11-q14 and 13q21-33. Am J Psychiatry. 2007;164:236–247. doi: 10.1176/ajp.2007.164.2.236. [DOI] [PubMed] [Google Scholar]
- Green E, Elvidge G, Jacobsen N, Glaser B, Jones I, O’Donovan MC, et al. Localization of bipolar susceptibility locus by molecular genetic analysis of the chromosome 12q23-q24 region in two pedigrees with bipolar disorder and Darier’s disease. Am J Psychiatry. 2005;162:35–42. doi: 10.1176/appi.ajp.162.1.35. [DOI] [PubMed] [Google Scholar]
- Kassem L, Lopez V, Hedeker D, Steele J, Zandi P, Mcmahon FJ. Familiality of polarity at illness onset in bipolar affective disorder. Am J Psychiatry. 2006;163:1754–1759. doi: 10.1176/ajp.2006.163.10.1754. [DOI] [PubMed] [Google Scholar]
- Kleinman L, Lowin A, Flood E, Gandhi G, Edgell E, Revicki D. Costs of bipolar disorder. Pharmacoeconomics. 2003;21:601–622. doi: 10.2165/00019053-200321090-00001. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hellard S, Lee AJ, Underwood S, Thomson PA, Morris SW, Torrance HS, et al. Haplotype analysis and a novel allele-sharing method refines a chromosome 4p locus linked to bipolar affective disorder. Biol Psychiatry. 2007;61:797–805. doi: 10.1016/j.biopsych.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM. Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch Gen Psychiatry. 1982;39:879–883. doi: 10.1001/archpsyc.1982.04290080001001. [DOI] [PubMed] [Google Scholar]
- McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S, et al. A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci U S A. 1996;93:13060–13065. doi: 10.1073/pnas.93.23.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis MG, Dick DM, Willour VL, Avramopoulos D, Mackinnon DF, Simpson SG, et al. Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health genetics initiative bipolar pedigrees. Biol Psychiatry. 2003;54:1265–1273. doi: 10.1016/j.biopsych.2003.08.001. [DOI] [PubMed] [Google Scholar]
- McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Low NC. The epidemiology of mood disorders. Curr Psychiatry Rep. 2004;6:411–421. doi: 10.1007/s11920-004-0004-1. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863–864. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal DK, Durner M, Greenberg DA. Effect of misspecification of gene frequency on the two-point LOD score. Eur J Hum Genet. 2001;9:855–859. doi: 10.1038/sj.ejhg.5200724. [DOI] [PubMed] [Google Scholar]
- Pauls DL, Bailey JN, Carter AS, Allen CR, Egeland JA. Complex segregation analyses of old order Amish families ascertained through bipolar I individuals. Am J Med Genet. 1995;60:290–297. doi: 10.1002/ajmg.1320600406. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant … or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Senol S, Herken H, Gucuyener K, Gehrig C, Blouin JL, et al. An apparently dominant bipolar affective disorder (BPAD) locus on chromosome 20p11.2-q11.2 in a large Turkish pedigree. Eur J Hum Genet. 2001;9:39–44. doi: 10.1038/sj.ejhg.5200584. [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Rice JP, Goate A, Williams JT, Bierut L, Dorr D, Wu W, et al. Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 1, 6, 8, 10, and 12. Am J Med Genet. 1997;74:247–253. doi: 10.1002/(sici)1096-8628(19970531)74:3<247::aid-ajmg3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Risch N, Giuffra L. Model misspecification and multipoint linkage analysis. Hum Hered. 1992;42:77–92. doi: 10.1159/000154047. [DOI] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI, Jr, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet. 2003;73:49–62. doi: 10.1086/376547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shink E, Morissette J, Sherrington R, Barden N. A genome-wide scan points to a susceptibility locus for bipolar disorder on chromosome 12. Mol Psychiatry. 2005;10:545–552. doi: 10.1038/sj.mp.4001601. [DOI] [PubMed] [Google Scholar]
- Stine OC, Mcmahon FJ, Chen L, Xu J, Meyers DA, Mackinnon DF, et al. Initial genome screen for bipolar disorder in the NIMH genetics initiative pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet. 1997;74:263–269. [PubMed] [Google Scholar]
- Suarez BK. Problems of replicating linkage claims in psychiatry. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Sun L, Wilder K, Mcpeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- The Welcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common disease and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken T, Claes S, Sluijs S, Paterson AD, Van Duijn C, Adolfsson R, et al. Genome-wide scan for affective disorder susceptibility Loci in families of a northern Swedish isolated population. Am J Hum Genet. 2005;76:237–248. doi: 10.1086/427836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Zandi PP, Huo Y, Diggs TL, Chellis JL, Mackinnon DF, et al. Genome scan of the fifty-six bipolar pedigrees from the NIMH genetics initiative replication sample: chromosomes 4, 7, 9, 18, 19, 20, and 21. Am J Med Genet B Neuropsychiatr Genet. 2003;121:21–27. doi: 10.1002/ajmg.b.20051. [DOI] [PubMed] [Google Scholar]
- Zandi PP, Willour VL, Huo Y, Chellis J, Potash JB, Mackinnon DF, et al. Genome scan of a second wave of NIMH genetics initiative bipolar pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet B Neuropsychiatr Genet. 2003;119:69–76. doi: 10.1002/ajmg.b.10063. [DOI] [PubMed] [Google Scholar]
- Zwick ME, Cutler DJ, Chakravarti A. Patterns of genetic variation in Mendelian and complex traits. Annu Rev Genomics Hum Genet. 2000;1:387–407. doi: 10.1146/annurev.genom.1.1.387. [DOI] [PubMed] [Google Scholar]