Summary
Depurinating DNA adducts formed by aromatic hydrocarbons and catechol estrogen quinones play a major role in cancer initiation. Most of these adducts depurinate instantaneously, but some guanine adducts depurinate from DNA with half-lives of hours. We report here, that after 10 h at 37 °C, reaction of estradiol-3,4-quinone (E2-3,4-Q) with ds-DNA to yield N7Gua and N3Ade adducts was complete and more efficient than with ss-DNA. When E2-3,4-Q reacted with t-RNA, no adducts were detected after 10 h, and the level of N3Ade and N7Gua adducts after 10 days was less than half that with ss-DNA after 10 h. Reaction of E2-3,4-Q and dG yielded 4-OHE2-1-N7dG, which spontaneously depurinated to yield 4-OHE2-1-N7Gua. To investigate the mechanism of depurination, E2-3,4-Q was reacted with carbocyclicdeoxyguanosine, in which the ring oxygen of the deoxyribose moiety is substituted with CH2, and depurination was observed. The results from this experiment demonstrate that the oxocarbenium ion mechanism plays the major role in depurination and provides the first experimental evidence for this mechanism. A newly discovered β-elimination mechanism also plays a minor role in depurination. Understanding why the depurinating estrogen-DNA adducts come from DNA, and not from RNA, underscores the critical role that these adducts play in initiating cancer.
Keywords: depurinating adducts; mechanism of depurination; estrogen-DNA adducts; catechol estrogen-3,4-quinones
INTRODUCTION
The first critical step in the cancer initiation process by chemicals is the formation of covalently bonded carcinogen-DNA adducts (1). There are two types of adducts formed: stable adducts that remain in DNA unless removed by repair (e.g., adducts at the exocyclic NH2 group of Ade or Gua) and depurinating adducts that are released from DNA with the formation of apurinic sites (e.g., adducts at the N3 and N7 of Ade, N7 of Gua, and sometimes C8 of Gua) (2–5). The N7 position of Gua and the N3 and N7 positions of Ade are among the most nucleophilic sites in DNA (6). In general, when DNA is exposed to electrophiles, these are the sites where covalent bonds are most abundantly formed.
The loss of Ade or Gua by depurination produces apurinic sites that can generate the mutations leading to cancer initiation (2–5). Evidence that depurinating polycyclic aromatic hydrocarbon (PAH)-DNA adducts and estrogen-DNA adducts play a major role in cancer initiation derives from a correlation between depurinating adducts and oncogenic Harvey (H)-ras mutations in mouse skin papillomas, preneoplastic mouse skin, and preneoplastic rat mammary gland (7–10). That is, if the major depurinating adducts contain Ade, the mutations are at adenines, and if the major depurinating adducts contain Gua, the mutations are at guanines. In general, depurination of PAH and estrogen adducts occurs instantaneously after the adducts are formed, with a few exceptions.
Reaction of benzo[a]pyrene (BP) radical cation (11, 12) or dibenzo[a,l]pyrene (DB[a,l]P) radical cation (13) with dG afforded, among the various adducts, the C8dG adducts, which slowly lost deoxyribose by cleavage of the glycosyl bond to yield the C8Gua adducts. However, when those radical cations reacted with Guo, the C8Guo adducts were formed, but no loss of ribose to yield C8Gua was observed (11–13).
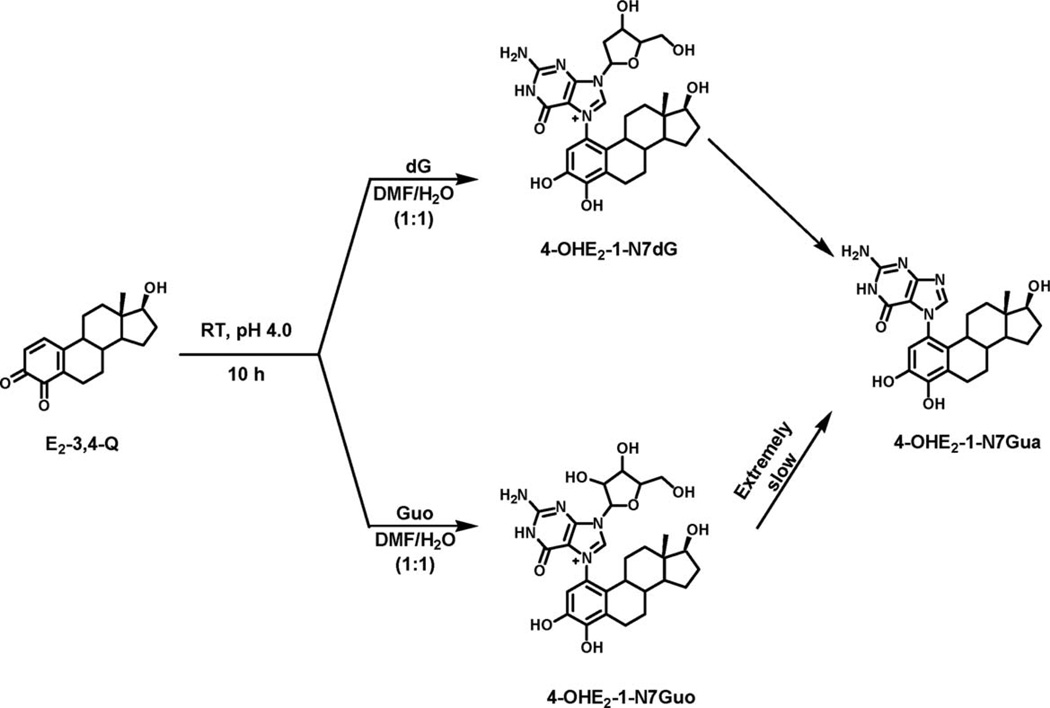
Similarly to the PAH-C8Gua, slow depurination to form the N7Gua adduct was also observed in the reaction of estradiol-3,4-quinone (E2-3,4-Q) with dG (Fig. 1) (14) or DNA (15). As reported here, virtually no depurination occurs after reaction of E2-3,4-Q with Guo (Fig. 1) or RNA. This is consistent with the observation that purine bases in DNA depurinate 500 times more rapidly than in RNA. This effect is a consequence of the 2′-hydroxyl group in RNA (16).
Figure 1.
Reaction of E2-3,4-Q with dG at room temperature and pH 4.0 to form 4-OHE2-1-N7Gua as the major product and 4-OHE2-1-N7dG as the minor product and reaction with Guo form 4-OHE2-1-N7Guo as the major product and 4-OHE2-1-N7Gua as the very minor product.
To investigate the mechanism of depurination, we reacted E2-3,4-Q with dG or carbocyclicdeoxyguanosine (CdG), in which the ring oxygen is replaced with a CH2 moiety, and determined that the depurination product, 4-OHE2-1-N7Gua, is formed in both cases.
EXPERIMENTAL
Chemicals and Reagents
MnO2, dimethylsulfoxide-d6 (DMSO-d6), and dimethylformamide (DMF) were purchased from Aldrich Chemical Co. (Milwaukee, WI). Guo, 2′-dG, and calf thymus DNA were purchased from USB (Cleveland, OH). t-RNA was purchased from Sigma Chemical Co. (St. Louis, MO). A racemic mixture of D-CdG and L-CdG was a kind gift from Dr. J. A. Secrist, Southern Research Institute, Birmingham, AL. 4-Hydroxyestradiol (4-OHE2) was synthesized by a previously reported method (17). High-performance liquid chromatography (HPLC) grade CH3OH was from Merck, and glacial CH3COOH and CH3CN were from Fisher Scientific (Kansas City, MO). All chemicals and reagents were used without further purification.
Instrumentation
Nuclear Magnetic Resonance
Proton and homonuclear two-dimensional chemical shift correlation spectroscopy nuclear magnetic resonance (NMR) spectra were recorded in DMSO-d6 on a Varian Unity-Inova 500 instrument at 500 MHz at 25 °C. Chemical shifts are reported relative to DMSO (2.49 ppm).
Fast Atom Bombardment Mass Spectrometry
Fast atom bombardment tandem mass spectrometry (FAB-MS/MS) was performed at the Nebraska Center for Mass Spectrometry, University of Nebraska-Lincoln, using a MicroMass (Manchester, England) AutoSpec high resolution magnetic sector mass spectrometer. The instrument was equipped with an orthogonal acceleration time-of-flight serving as the second mass spectrometer in the tandem experiment. Xenon was admitted to the collision cell at a level to attenuate the precursor ion signal by 75%. Data acquisition and processing were accomplished using OPUS software that was provided by the manufacturer (Microcasm). Samples were dissolved in 5–10 µL of CH3OH; 1 µL aliquots were placed on the sample probe tip along with 1 µL of a 1:1 mixture of glycerol/thioglycerol.
High-Performance Liquid Chromatography
Preparative HPLC was conducted on a Waters 600E solvent delivery system equipped with a 996 photodiode array detector, using a YMC ODS-AQ column (20 × 250 mm). The column was initially eluted with 10% CH3CN in H2O (0.4% trifluoroacetic acid [TFA]) for 1 min, followed by a linear gradient to 100% CH3CN in 45 min at a flow rate of 6 mL/min. Analyses of the reaction mixtures were conducted on a Waters 2690 Separations Module equipped with a Waters 996 photodiode array detector interfaced to a Digital Venturis Fx 5100 computer, and a reversed phase Phenomenex Luna-2 C-18 column (250 × 4.6 mm, 5 µm, 120 Å, Torrance, CA). The column was first eluted with 10% CH3CN in H2O (0.4% TFA) for 5 min, followed by increasing the proportion of CH3CN linearly up to 80% in the next 30 min at a flow rate of 1 mL/min. Analytical HPLC using electrochemical detection was conducted on a system equipped with dual ESA Model 580 solvent delivery modules, an ESA Model 540 autosampler and a 12-channel CoulArray detector (ESA, Chelmsford, MA). Depurinating adducts from the reaction mixture were separated with two mobile phases, A (CH3CN/CH3OH/H2O/Buffer, 15:5:70:10) and B (CH3CN/CH3OH/H2O/buffer, 50:30:10:10) starting with 90% mobile phase A and 10% phase B and increasing the proportion of mobile phase B linearly up to 90% for more than 50 min with a flow rate of 1 mL/min at 35 °C. The buffer was a mixture of citric acid (0.25 M) and ammonium acetate (0.5 M) in double-distilled water; the pH was adjusted to 3.6 with glacial CH3COOH. The serial array of 12 coulometric electrodes was set at potentials between −35 and 680 mV. The system was controlled and the data were acquired and processed using the CoulArray software package (ESA). Peaks were identified by both retention time and peak height ratios between the dominant peak and the peaks in the two adjacent channels. The depurinating adducts were quantified by comparison of peak response ratios with known amounts of standards. Analyses were conducted by using a reversed phase Luna-2 C-18 column (250 × 4.6 mm, 5 µm; Phenomenex).
LC-MS and LC-MS/MS
Compound confirmation was performed on a Finnigan LCQ ion trap mass spectrometer coupled with a Waters 2695 HPLC pump and autoinjector. A mobile phase gradient of (a) aqueous ammonium formate (NH4OOCH, 0.5 g/L) and (b) methanol (containing 0.5 g/L NH4OOCH) at a flow rate of 0.2 mL/min was used for the separation of the reaction products on a HyPurity C-18 column (250 × 2.1 mm, 5 µm; Thermo Scientific). The initial gradient composition of 10% CH3OH was linearly changed to 30% B at 5 min, then to 80% B at 18 min, and finally to 100% B at 24 min, held for 14 min and then returned to the initial conditions for 7 min to equilibrate the column. The column temperature was 33 °C. Instrument conditions for the liquid chromatography–mass spectrometry (LC-MS) and LC-MS/MS analyses were: electrospray ionization (positive ion), source voltage: 4.5 kV, capillary temp: 200 °C, capillary voltage: 25.0 V, tube lens voltage: 7.0 V, sheath gas flow rate: 75 (arb. unit), auxiliary gas flow rate: 15 (arb. unit), multipole 1 offset voltage: −3.00 V, lens voltage: −9.00 V, multipole 2 offset voltage: −7.00 V, multipole RF amplitude: 500.00 Vp-p. The MS/MS isolation width was maintained at 3.0 m/z with an activation time of 30.00 msec and an activation Q value of 0.250. The normalized collision energy used was 30% for the parent ions of the initial E2-3,4-Q reaction products (m/z = 552 or m/z = 554) and 48% for the parent ion of the depurinated product (m/z = 438). Helium was used as the collision gas.
Reaction of E2-3,4-Q with dG
E2-3,4-Q was reacted with dG as described previously (14, 18). To an efficiently stirred suspension of activated MnO2 (61 mg, 0.70 mmol) in 2 mL of DMF at 0 °C was added a solution of 4-OHE2 (20 mg, 0.07 mmol) in 1 mL of DMF under argon. After stirring for 20 min, the yellowish-green E2-3,4-Q was filtered quickly through a syringe filter into a solution of dG (99 mg, 0.35 mmol) in DMF/H2O (4 mL, 1:3). The final solution contained DMF/H2O (1:1). The pH of the solution was adjusted to 4 with CH3COOH (200 µL). After stirring for 10 h, the solution was filtered and concentrated to 3 mL at reduced pressure and analyzed on HPLC, as described above. Two adducts were eluted at 15.9 and 16.6 min, respectively, and purified by preparative HPLC. The fractions containing adducts were lyophilized and characterized by NMR spectroscopy. The major product was found to be 4-OHE2-1-N7Gua (16.6 min, yield 48%) after comparing spectroscopic data with authentic sample. Spectroscopic data for the minor 4-OHE2-1-N7dG at 15.9 min were not obtained, because the compound lost the deoxyribose moiety and converted to 4-OHE2-1-N7Gua before NMR analysis.
Reaction of E2-3,4-Q with Guo
To a stirred suspension of MnO2 (18.2 mg, 0.21 mmol) in DMF (1 mL) was added 4-OHE2 (10 mg, 0.04 mmol) at 0 °C. Stirring for 20 min yielded a greenish-yellow solution of E2-3,4-Q, which was filtered through a 0.2-µm syringe filter directly into a solution of Guo (49.5 mg, 0.18 mmol) in DMF/H2O (1:2, 3 mL). The final solution contained DMF/H2O (1:1). The pH of the solution was adjusted to 4 with a few drops of CH3COOH. After 10 h, the reaction mixture was filtered and evaporated at low pressure. The residue was dissolved in 1 mL of CH3OH/DMF (1:1) and analyzed on HPLC as described above. Two peaks were observed at 15.6 min (major peak) and 16.6 min (very minor peak). They were purified by preparative HPLC as described above. The major product was collected, lyophilized and found to be 4-OHE2-1-N7Guo (yield 60%); UV: λmax, 254 and 290 nm. 1H NMR, δ (ppm, DMSO-d6): 11.78 and 11.73 (s, 1H, NH-Guo, exchangeable with D2O), 9.75 and 9.71 (s, 1H, Ar-OH, exchangeable with D2O), 9.66 and 9.58 (s, 1H, 8′-H-Guo), 8.86 and 8.84 (s, 1H, Ar-OH, exchangeable with D2O), 7.36 (br s, 2H, NH2-Guo, exchangeable with D2O), 6.78 and 6.74 (s, 1H, 2-H), 5.97 and 5.89 (d, J = 2.9 Hz, 1 H, 1′-H), 5.79 and 5.33 (bs, 1H, OH-ribose, exchangeable with D2O), FAB-MS, [M + H]+, C28H36N5O8 calcd m/z 570.2564; obsd 570.2555. The very minor product (5%) was found to be 4-OHE2-1-N7Gua by comparison with the authentic sample.
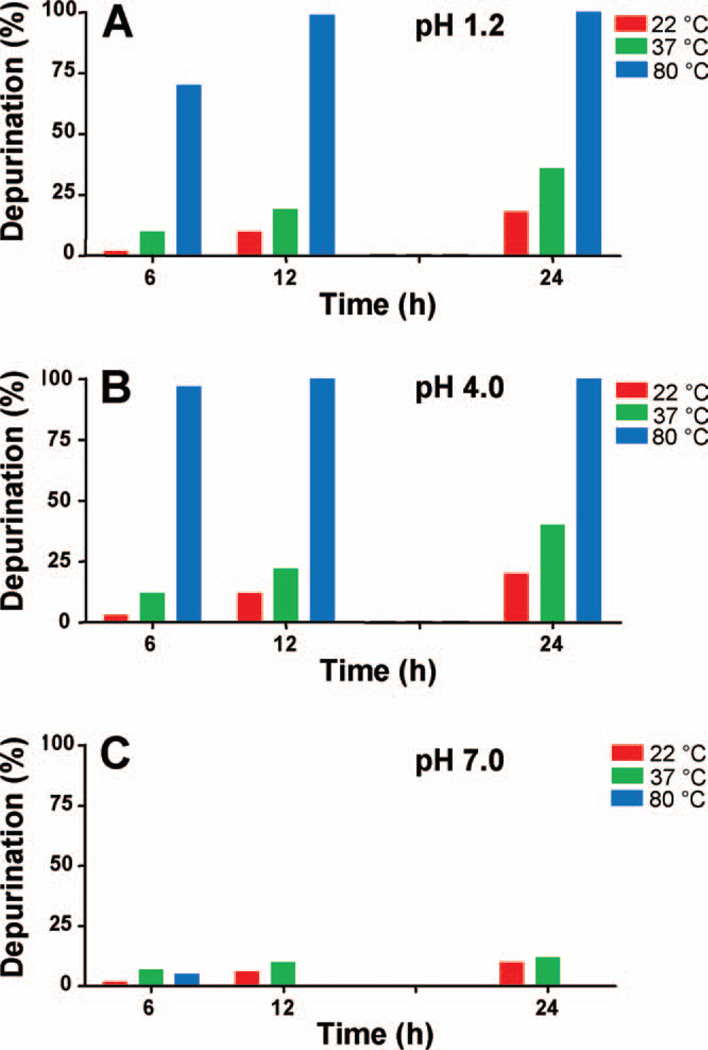
Rate of Loss of the Ribose Moiety from 4-OHE2-1-N7Guo at Different pHs and Temperatures
4-OHE2-1-N7Guo (500 µg) was dissolved in DMF/H2O (1:1, 500 µL) and the pH of the solution was brought quickly to 1.2 or 4.0 with CH3COOH and one or two drops of HCl for pH 1.2. For the reaction at pH 7.0, H2O was replaced with 0.067 M Na–K phosphate at pH 7.0. The solution was stirred at 22, 37, or 80 °C. Aliquots of the solution were taken at different times (6, 12, and 24 h) and analyzed by HPLC as described above (Fig. 2).
Figure 2.
Loss of ribose from 4-OHE2-1-N7Guo at various temperatures and pHs. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Reaction of E2-3,4-Q with CdG
To a stirred solution of 4-OHE2 (4 mg, 0.014 mmol) in 200 µL of CH3CN at 0 °C, activated MnO2 (9 mg, 0.103 mmol) was added under an argon atmosphere. After 10 min, the yellowish green solution was filtered through a Gilman acrodisc directly into a flask containing a solution of CdG (22 mg, 0.08mmol) in 1 mL of DMF:H2O (1:4). To facilitate the Michael addition, the pH of the reaction was adjusted to pH 4 with a few drops of acetic acid. The reaction was stirred at 37 °C and at selected times (1, 10, 24, 168, 336, 504, and 648 h), a small aliquot was taken out and analyzed by analytical HPLC. Analyses of the reaction mixtures were conducted on a Waters 2690 Separations Module equipped with a Waters 996 photodiode array detector interfaced to a Digital Venturis Fx 5100 computer, and a reversed phase Phenomenex Luna-2 C-18 column (250 × 4.6 mm, 5 µm, 120 Å, Torrance, CA). The column was initially eluted with 10% CH3CN in H2O (0.1% TFA) and changed linearly up to 20% CH3CN in 4 min and then from 20 to 28% in 18 min, followed by a linear increase to 100% CH3CN in 30 min at a flow rate of 1 mL/min. Two peaks were observed at 11.6 min (major peak) and 14.2 min (very minor peak). The peaks were collected and analyzed by LC-MS/MS, and their structures were confirmed as the 4-OHE2-1-N7CdG and 4-OHE2-1-N7Gua adducts, respectively.
Depurination of ds-DNA, ss-DNA, and t-RNA after Reaction with E2-3,4-Q
Calf thymus DNA (3 mM in 10 mL of 0.067 M Na–K phosphate, pH 7.0) was treated with freshly prepared E2-3,4-Q (8.7 µmol in 500 µL DMSO, final concentration 0.87 mM). The reaction mixture was incubated at 37 °C for 10 h. DNA was precipitated with two volumes of ethanol and the supernatant was processed for analysis of depurinating adducts after evaporation of solvents and reconstitution of the residue in 1 mL of CH3OH/DMF (1:1). The solution was filtered through a 0.2-µm syringe filter and analyzed on HPLC with an electrochemical detector, as described above. In a time-course study, aliquots of the reaction mixture containing DNA and E2-3,4-Q were analyzed by HPLC at different time points (0.5, 1, 2, 3, 4, 5, 10, 15, and 20 h).
For ss-DNA, 10 mL of 3 mM calf thymus DNA in 0.067 M Na–K phosphate, pH 7.0, was heated at 100 °C for 5 min with occasional shaking. The DNA solution was quick-cooled to 0 °C on ice and treated with freshly prepared E2-3,4-Q (8.7 µmol in 500 µL DMSO, final concentration 0.87 mM). The reaction mixture was incubated at 37 °C for 10 h. DNA was precipitated with two volumes of ethanol, and the supernatant was processed for the analysis of depurinating adducts, as described above.
For RNA, 5 mg of t-RNA was dissolved in 10 mL of 0.067 M Na-K phosphate, pH 7.0. The solution was incubated with freshly prepared E2-3,4-Q (8.7 µmol in 500 µL DMSO, final concentration 0.87 mM) at 37 °C for 10 h. RNA was precipitated with two volumes of chilled ethanol overnight at 4 °C. The supernatant was then processed and analyzed for depurinating adducts, as described above.
RESULTS
Reaction of E2-3,4-Q with dG
HPLC analysis of the reaction mixture containing E2-3,4-Q (11.6 mM) and dG (61.8 mM) at pH 4.0 and 22 °C indicated two adducts at 15.9 min (minor) and 16.6 min (major). An earlier report from our laboratory (14) described the minor adduct at 15.9 min as a labile adduct, 4-OHE2-1-N7dG, which slowly converted into the major depurinating adduct, 4-OHE2-1-N7Gua, eluting at 16.6 min (Fig. 1). Analysis of the reaction mixture by direct infusion on mass spectrometry (data not shown) showed the molecular ions at m/z = 554, after 10 h, corresponding to the labile adduct 4-OHE2-1-N7dG (12%), and m/z = 438, corresponding to the major depurinating adduct, 4-OHE2-1-N7Gua (48%). Purification of the adducts was conducted on preparative HPLC. The major adduct was compared with authentic depurinating adduct, 4-OHE2-1-N7Gua, and found to be identical, whereas the minor adduct, 4-OHE2-1-N7dG, was not isolable because the deoxyribose moiety was lost during the process of purification and drying. The loss of deoxyribose from 4-OHE2-1-N7dG was 80% complete after 10 h and 100% by 1 day (Table 1).
Table 1.
Comparison of adducts formed by reaction of E2-3,4-Q with dG, Guo, ds-DNA, ss-DNA, or t-RNA
| Adducts, µmol/mol DNA-P | ||||||||
|---|---|---|---|---|---|---|---|---|
| dGa | Guoa | ds-DNAb | ss-DNAb | t-RNAb | ||||
| Time | N7Gua | N7Gua | N3Ade | N7Gua | N3Ade | N7Gua | N3Ade | N7Gua |
| 10 h | 80 | 5 | 123 | 135 | 21.2 | 23.4 | <0.7 | <0.7 |
| 1 day | 100 | 14 | 131 | 138 | 19.8 | 20.7 | 2.9 | 2.5 |
| 2 days | 22 | 4.9 | 4.7 | |||||
| 5 days | 38 | 7.0 | 7.3 | |||||
| 10 days | 62 | 9.2 | 8.4 | |||||
Reaction at room temperature, pH 4.0.
Reaction at 37 °C, pH 7.0.
Reaction of E2-3,4-Q with Guo
E2-3,4-Q (8.7 mM) was reacted with Guo (43.6 mM) under the same conditions described for dG. HPLC analysis showed the 4-OHE2-1-N7Guo as the major adduct (60%) and the depurinating adduct, 4-OHE2-1-N7Gua, as a minor adduct (5%). These were further confirmed by mass spectrometry and after purification of the adducts by preparative HPLC. The loss of ribose to form 4-OHE2-1-N7Gua was only 5% complete after 10 h and 14% at 1 day. After 10 days, the loss of ribose was only 62% (Table 1).
1H NMR analysis of the major adduct showed the product to be a mixture of two conformational isomers attached to the C1 position of the estrogen A ring and the N7 position of Guo. Generation of two isomers had already been observed in the reaction between E2-3,4-Q and dG (17) and is due to a rotational barrier around the C1—N7 bond. Thus, in one isomer, the Guo moiety is located primarily on the α side of the estrogen ring system (below the plane of the estrogen), while the other isomer has the Guo moiety located on the β side of the estrogen ring system (above the plane). Rotation from the α isomer to the β isomer is hindered because the C8 proton of the Guo moiety would occupy the same van der Waals radius as the C11 protons of the estrogen C ring (Fig. 1). Based on similar arguments provided earlier (18), the major isomer (60%) was assigned as the α-isomer and the minor isomer (40%) was assigned as the β-isomer.
The characteristic ribose signals in the NMR spectrum indicated the presence of the ribose moiety in the adduct. This was further confirmed by FAB-MS analysis of the adduct, giving a molecular ion [M + 1]+ at m/z 570.2555 (calcd m/z 570.2564), corresponding to the molecular formula C28H36N5O8. Two sharp singlets, integrating to one proton at 11.78 and 11.73 ppm (α-NH- and β-NH-Guo), a broad singlet, integrating to two protons at 7.36 ppm (α- and β-NH2), and the C8 proton resonance at 9.66 and 9.58 ppm (both α- and β-isomers) indicated that these nucleophilic sites in the Guo moiety are not substituted, thereby corroborating substitution at N7. The C8 protons of the Guo moiety were significantly shifted downfield (1.68 ppm for the α-isomer and 1.76 ppm for the β-isomer) when compared with the C8 protons in 4-OHE2-1-N7Gua (18). This was due to a positive charge in the nearby nitrogen (i.e., the N7 of Guo), thus rendering the C8 proton of the Guo adduct deshielded. The deshielding effect of the positive charge was also observed on the resonances at NH-Guo, NH2-Guo, and the C2 proton at 6.78 (β-isomer), 6.74 (α -isomer) of the estrogen moiety. All other resonances in the NMR spectrum were the same as those described previously (18).
Rate of Loss of the Ribose Moiety from 4-OHE2-1-N7Guo
To begin to investigate the factors affecting depurination of adducts from RNA, loss of the ribose moiety from a model compound, 4-OHE2-1-N7Guo, was studied at three different pHs and temperatures (Fig. 2). At pH 1.2 (Fig. 2A), the reaction at 22 °C produced only 2% of the depurinating adduct, 4-OHE2-1-N7Gua, after 6 h, 10% after 12 h, and 18% after 24 h. At 37 °C, formation of 4-OHE2-1-N7Gua was 10% after 6 h, 19% after 12 h, and 36% after 24 h. Increasing the temperature to 80 °C at the same pH, we observed 70% formation of the depurinating adduct after 6 h, and almost all of the 4-OHE2-1-N7Guo was converted to the depurinating 4-OHE2-1-N7Gua adduct after 12 h at this temperature and pH.
Raising the pH to 4.0, the loss of the ribose moiety at 22 °C was observed to be 3, 12, and 20% after 6, 12, and 24 h (Fig. 2B). These values were not significantly different from those observed at pH 1.2. This means that the pH change did not affect the rate of loss of the ribose moiety from the adduct. At 37 °C at pH 4.0, loss of the ribose moiety was 12, 22, and 40% after 6, 12, and 24 h, respectively, again matching the values obtained at pH 1.2. Finally, increasing the temperature up to 80 °C yielded a loss of ribose of 97% after 6 h and almost 100% after 12 h. These findings indicate that the temperature has much more effect on the rate of loss of the ribose moiety than does the pH variation.
When the pH of the reaction mixture was raised to 7.0 (Fig. 2C), decomposition of the product was seen. At lower temperature (22 °C), the rate of loss of ribose was not different from those observed at pH 1.2 and 4.0. However, a slight rise in the temperature to 37 °C led to decomposition of the starting material, and after 24 h, all of the starting material was decomposed, with 12% formation of the depurinating adduct. At 80 °C, only 5% formation of depurinating adduct was seen, and after 12 h, all of the starting material and initially formed depurinating adduct had been decomposed (Fig. 2C).
Depurination of ds-DNA, ss-DNA, and t-RNA after Reaction with E2-3,4-Q
To investigate the factors involved in depurination of estrogen adducts, we studied depurination of adducts from DNA and RNA. E2-3,4-Q was reacted with ds-DNA under previously described conditions and the adducts were analyzed as described previously (15, 19). Approximately, equal amounts of the two depurinating adducts, 4-OHE2-1-N7Gua and 4-OHE2-1-N3Ade, were observed. In a time-course study, the depurination of the N3Ade adduct was found to be instantaneous, whereas the N7Gua adduct depurinates slowly, with a half life of ~3 h, as previously observed (15). Depurination of both the N3Ade and N7Gua adducts was complete in 10 h at 37 °C (15) (Table 1).
When E2-3,4-Q was reacted with ss-DNA, the levels of depurinating adducts, although in equal amount, were much lower than those observed with ds-DNA (Table 1). Efficient reaction of E2-3,4-Q with DNA relies on preliminary intercalation of the quinone into ds-DNA. The limited binding of E2-3,4-Q with ss-DNA occurs from random interactions. Reaction of E2-3,4-Q with t-RNA did not produce detectable levels of depurinating adducts in 10 h. We observed the depurinating adducts for RNA at later time points, but after 10 days, the levels were only 6.8% of those detected with ds-DNA and 39% of those with ss-DNA after 10 h.
Reaction of E2-3,4-Q with CdG
E2-3,4-Q was reacted with dG or CdG under the conditions described in the Experimental section. At selected timepoints over a period up to 27 days, an aliquot of each reaction mixture was monitored by LC-MS. After 1 h, the reaction of E2-3,4-Q with dG yielded an LC peak at 11.4 min (Fig. 3A) corresponding to 4-OHE2-1-N7dG, as identified by MS (Fig 3A-1). After 1 day (Fig. 3B), 4-OHE2-1-N7Gua is the only product detected by LC and identified as m/z 438 (Fig. 3B-1). Thus, the N7dG product has completely depurinated.
Figure 3.
HPLC chromatogram and LC-MS spectrum for the reaction mixtures of E2-3,4-Q with dG and CdG: (A) HPLC chromatogram of 1 h reaction of E2-3,4-Q with dG; (A-1) Mass spectrum of 1 h reaction peak at 11.4 min; (B) HPLC chromatogram of 1 day reaction of E2-3,4-Q with dG; (B-1) Mass spectrum of 1 day reaction peak at 14.2 min; (C) HPLC chromatogram of 1 h reaction of E2-3,4-Q with CdG; (C-1) Mass spectrum of 1 h reaction peak at 11.6 min; (D) HPLC chromatogram of 27 day reaction of E2-3,4-Q with CdG; (D-1) Mass spectrum of 27 day reaction peak at 14.2 min.
The reaction between E2-3,4-Q and CdG shows an LC peak at 12.6 min (Fig. 3C), which corresponds to 4-OHE2-1-N7CdG, with m/z 552. After 27 days, a second peak is observed on LC at 14.2 min (Fig. 3D) and its molecular weight of m/z 438 identifies it as 4-OHE2-1-N7Gua (Fig. 3D-1), identical to the product in Fig. 3B-1.
The rate of depurination of the adducts is depicted in Fig. 4. As observed, depurination of 4-OHE2-1-N7CdG is barely detected at 180 h and increases to 16% of product by 648 h. In contrast, depurination of 4-OHE2-1-N7dG is complete in 10 h.
Figure 4.
Rate of depurination of 4-OHE2-1-N7CdG adduct to form 4-OHE2-1-N7Gua.
DISCUSSION
Metabolic activation of PAH via radical cations or diol epoxides generally leads to the predominant formation of depurinating DNA adducts (2, 3, 20–23). This occurs when the electrophilic attack of PAH takes place at the N3 and N7 of Ade and at the N7 and sometimes at C8 of Gua. Analogous results are obtained by reaction of the electrophilic catechol estrogen quinones with DNA (4, 5, 15). Depurination occurs instantaneously after formation of the N3Ade, N7Ade, and N7Gua adducts of PAH. In contrast, the C8Gua adducts of BP and DB[a,l]P (11– 13) and the N7Gua adducts of the estrogen quinones (14, 15) depurinate more slowly. This is seen in the reaction of E2-3,4-Q with DNA (15).
The predominant formation of specific depurinating DNA adducts of the carcinogens BP and DB [a,l]P in mouse skin correlates with mutations induced in the H-ras oncogene in both papillomas (7) and preneoplastic skin (8). Treatment of mouse skin (9) or rat mammary gland (10) with E2-3,4-Q predominantly induces H-ras mutations at Ade residues, the sites of formation of rapidly depurinating N3Ade adducts. Thus, formation of depurinating adducts appears to play a significant role in the induction of tumor-initiating mutations.
Rate of Depurination
Cleavage of the glycosyl bond after reaction of E2-3,4-Q with dG takes place with a half-life of 3 h at room temperature (14), whereas the loss of ribose following reaction of E2-3,4-Q with Guo is extremely slow, with a half-life of about 8 days (Table 1).
Similar effects are observed when E2-3,4-Q is reacted with ds-DNA, ss-DNA, or t-RNA (Table 1). With ds-DNA and ss-DNA, depurination of both the N3Ade and N7Gua adducts is complete within 10 h, but the level of adduct formation is much lower with ss-DNA than with ds-DNA (Table 1).
Extent of Binding to ds-DNA, ss-DNA, and t-RNA
The relatively low binding of E2-3,4-Q to ss-DNA (17% of the amount with ds-DNA) is analogous to the low levels of binding of rat liver microsome-activated BP to ss-DNA (25% of ds-DNA) previously observed, but with BP, 77% of the adducts formed with ds-DNA were depurinating adducts and all of the adducts formed with ss-DNA were stable adducts (20). With DB[a,l]P activated by rat liver microsomes, 84% of the adducts formed with ds-DNA were depurinating adducts (23). With ss-DNA, the overall level of binding of DB[a,l]P was 30% of that observed with ds-DNA, and only depurinating adducts formed from DB[a,l]P diol epoxide were detected. These results are consistent with the importance of PAH intercalation between the stacking bases of ds-DNA prior to metabolic activation to the reactive intermediate that forms the DNA adduct. E2-3,4-Q or its precursor, 4-OHE2, also needs to intercalate in ds-DNA for efficiently producing a DNA adduct. This intercalation has been clearly demonstrated in the formation of DNA adducts by the human carcinogen diethylstilbestrol (24).
BP was found to bind even more poorly to RNA after activation by rat liver microsomes (20). The overall binding was only 19% of that with ds-DNA and no depurinating adducts were detected. The binding of E2-3,4-Q to RNA produced very low levels of N3Ade and N7Gua adducts that depurinated extremely slowly. In fact, the level of depurinating adducts detected after 10 days at 37 °C was less than half the levels found for ss-DNA after 10 h (Table 1).
Mechanism of Depurination
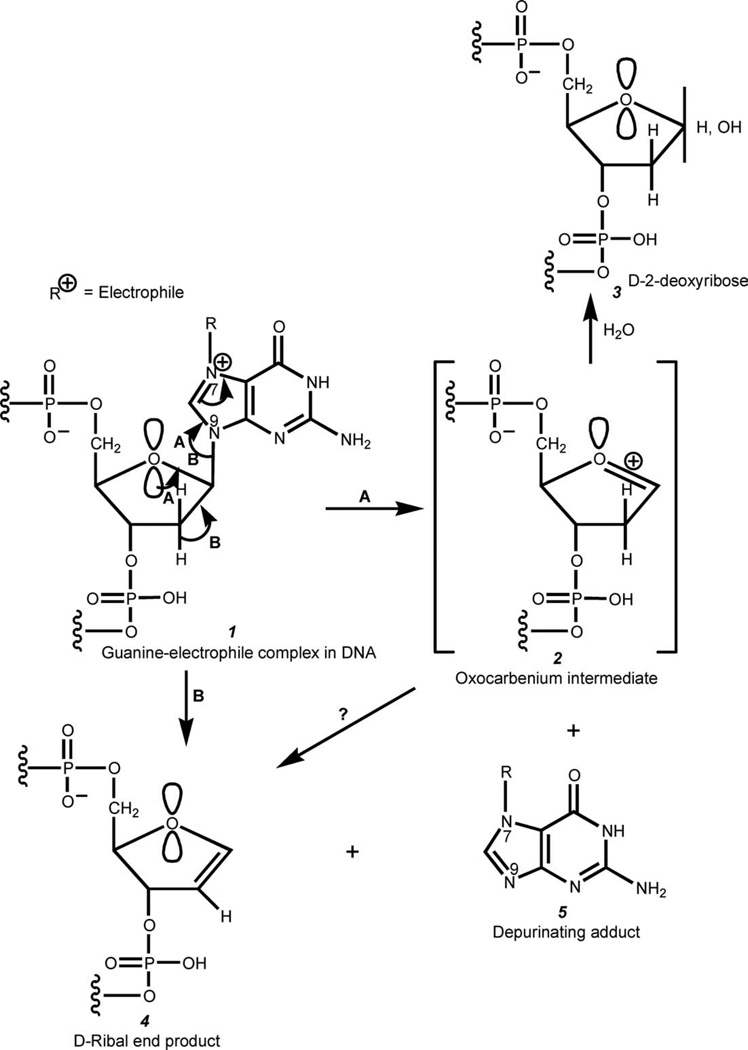
Substitution of an electrophile at the N7 of Gua in DNA gives rise to a positive charge on the N atom (Fig. 5). Depurination with cleavage of the glycosyl bond occurs to stabilize the positive charge in the adduct.
Figure 5.
Formation of depurinating DNA adducts and generation of apurinic sites. Path (A), currently accepted mechanism involving an intermediate oxocarbenium ion 2 with possible formation of a ribal intermediate 4. Path (B), mechanism in which the glycosyl bond cleavage is assisted by elimination of an anti-proton at C2′ in the deoxyribose ring with formation of a ribal intermediate 4.
The currently accepted mechanism of DNA depurination (25, 26) is illustrated in Fig. 5 (path A). The attack of an electrophile at the N7 of Gua results in the formation of a positively charged Gua-electrophile adduct, which can be stabilized by cleavage of the C1′—N9 glycosyl bond. Cleavage of the C—N glycosyl bond takes place most likely by participation of one pair of nonbonding electrons of the deoxyribose ring oxygen (the pair that is in the anti orientation to the C—N glycosyl bond due to the anomeric effect). Cleavage of the C—N glycosyl bond results in formation of the oxocarbenium ion intermediate and an electrically neutral Gua-electrophile adduct. The extremely reactive oxocarbenium ion adds a water molecule, giving deoxyribose as the end product. The loss of the nucleic acid base could theoretically also take place by elimination of the C2′ hydrogen that is anti-oriented to the C—N glycosyl bond, in which case D-ribal would be the end product (Fig. 5, path B). Finally, it should be mentioned that D-Ribal could also be formed from oxocarbenium ion 2 by elimination of the C2′ hydrogen.
Formation of the oxocarbenium ion intermediate in the process of depurination is supported by the strong resistance of RNA to depurination. The reason for this is the positive charge located on the C2′ carbon of ribose due to the presence of the strongly polarized C2′—O bond. Thus, formation of the oxocarbenium ion would require formation of a structure in which the two neighboring carbon atoms (C1′ and C2′) are positively charged, which is obviously unfavorable.
As we already said, due to the presence of the anomeric effect in nucleosides, depurination of DNA most likely occurs via formation of an oxocarbenium ion, with subsequent generation of an apurinic site (3 in Fig. 5, path A). However, it is not known whether elimination of the C2′ hydrogen that is trans to the glycosyl bond takes place concurrently with the formation of the oxocarbenium ion in the depurination process and if it does, how important is this pathway in the overall depurination process. To determine whether elimination of the C2′ hydrogen is a viable pathway for depurination of DNA, we examined whether CdG, in which the ring oxygen is replaced with a CH2 group, will be subject to depurination because now the formation of the oxocarbenium ion intermediate mechanism is excluded due to the lack of the ring oxygen. We found that the depurination product, 4-OHE2-1-N7Gua, is again formed, although at a very slow rate (Figs. 3 and 4). As in this case, depurination can take place only by elimination of the C2′ hydrogen that is in trans orientation to the nucleic acid base, we have concluded that the pathway that involves formation of the oxocarbenium ion is definitely the major pathway of depurination, although elimination of the C2′ hydrogen anti to the C—N glycosyl bond also plays a role.
Knowledge of the mechanism of depurination provides insight into cancer initiation, because formation of depurinating DNA adducts plays a major role in generating the critical mutations leading to cancer (2–5, 7–10). Paradoxically, if evolution had been able to create a double-stranded RNA instead of DNA, depurination would be a negligible event, and the chemically induced mutations leading to cancer would be extremely rare.
CONCLUSIONS
Formation of the covalent bond between chemical carcinogens and DNA is the first critical step in the initiation of cancer. The depurinating carcinogen-DNA adducts, which leave apurinic sites in the DNA, generate mutations that can initiate the series of events leading to cancer (2–5). The adducts most efficiently formed by the ultimate carcinogenic metabolites of estrogens, E2-3,4-Q, are the depurinating N3Ade and N7Gua adducts (98–99% of total adducts) (15). Formation of adducts becomes much less efficient when the E2-3,4-Q reacts with ss-DNA when compared with ds-DNA. This is because the preliminary intercalation of the estrogen metabolite in the ds-DNA structure provides a more efficient reaction with the strongest nucleophilic sites of DNA, namely the N3 of Ade and N7 of Gua. E2-3,4-Q binds inefficiently to RNA and the resulting adducts depurinate at a very slow rate from RNA. This explains why the depurinating adducts observed in humans and animals arise from DNA but not RNA.
We have demonstrated in this article that the mechanism of depurination of adducts from DNA mostly involves the accepted formation of an oxocarbenium ion and, to a very minor extent, cleavage of the C—N glycosyl bond by elimination of the C2′ hydrogen of the deoxyribose moiety that is anti to the C1′—N9 glycosyl bond. The extremely slow rate of depurination in RNA is due to the higher-energy transition state in the formation of the oxocarbenium ion. This is a consequence of the partial positive charge on the C2′ atom due to the presence of the strongly polarized C2′—O bond of the C2′ hydroxyl group in ribose. The oxocarbenium ion mechanism for depurination is widely accepted, and in this article, we have provided the first experimental evidence for it. The β-elimination mechanism is a new pathway and plays a minor role in depurination.
Demonstration of the mechanism of depurination enables us to explain the results obtained in a variety of studies of PAH and estrogen adducts formed with DNA and RNA (11–15, 19–23), including the depurinating estrogen-DNA adducts observed in humans (27–31). Understanding why the depurinating estrogen-DNA adducts found in humans come from DNA, and not RNA, underscores the critical role of these adducts in cancer initiation.
ACKNOWLEDGEMENTS
The authors thank Dr. John A. Secrist, Southern Research Institute, for providing the key compound carbocyclicdeoxyguanosine to demonstrate the mechanism of depurination. This work was supported by Grant P01 CA49210 from the National Cancer Institute and Grant DAMD17-03-10229 from the Department of Defense Breast Cancer Research Program. Core support at the Eppley Institute was provided by grant P30 CA36727 from the National Cancer Institute.
REFERENCES
- 1.Miller JA. Carcinogenesis by chemicals: an overview—G. H. A. Clowes memorial lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 2.Cavalieri EL, Rogan EG. The approach to understanding aromatic hydrocarbon carcinogenesis. The central role of radical cations in metabolic activation. Pharmacol. Ther. 1992;55:183–199. doi: 10.1016/0163-7258(92)90015-r. [DOI] [PubMed] [Google Scholar]
- 3.Cavalieri EL, Rogan EG. Mechanisms of tumor initiation by polycyclic aromatic hydrocarbons in mammals. In: Neilson AH, editor. The Handbook of Environmental Chemistry. Heidelberg, Germany: Springer-Verlag; 1998. pp. 81–117. [Google Scholar]
- 4.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, et al. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim. Biophys. Acta. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Cavalieri EL, Rogan EG. Depurinating estrogen-DNA adducts in the etiology and prevention of breast and other human cancers. Future Oncology. 2010;6:75–91. doi: 10.2217/fon.09.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pullman A, Pullman B. Molecular electrostatic potential of the nucleic acids. Q. Rev. Biophys. 1981;14:289–380. doi: 10.1017/s0033583500002341. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti D, Pelling JC, Cavalieri EL, Rogan EG. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc. Natl. Acad. Sci. USA. 1995;92:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravarti D, Mailander P, Cavalieri EL, Rogan EG. Evidence that error-prone DNA repair converts dibenzo[a,l]pyrene-induced depurinating lesions into mutations: formation, clonal proliferation and regression of initiated cells carrying H-ras oncogene mutations in early preneoplasia. Mutat. Res. 2000;456:17–32. doi: 10.1016/s0027-5107(00)00102-0. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti D, Mailander P, Li K-M, Higginbotham S, Zhang H, et al. Evidence that a burst of DNA depurination in SENCAR mouse skin induces error-prone repair and forms mutations in the H-ras gene. Oncogene. 2001;20:7945–7953. doi: 10.1038/sj.onc.1204969. [DOI] [PubMed] [Google Scholar]
- 10.Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T to G.C mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J. Steroid Biochem. Mol. Biol. 2006;101:204–215. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Rogan E, Cavalieri E, Tibbels S, Cremonesi P, Warner C, et al. Synthesis and identification of benzo[a]pyrene-guanine nucleoside adducts formed by electrochemical oxidation and horseradish peroxidase-catalyzed reaction of benzo[a]pyrene with DNA. J. Am. Chem. Soc. 1988;110:4023–4029. [Google Scholar]
- 12.Rama Krishna NV, Gao F, Padmavathi NS, Cavalieri EL, Rogan EG, et al. Model adducts of benzo[a]pyrene and nucleosides formed from its radical cation and diol epoxide. Chem. Res. Toxicol. 1992;5:293–302. doi: 10.1021/tx00026a023. [DOI] [PubMed] [Google Scholar]
- 13.Rama Krishna NV, Padmavathi NS, Cavalieri EL, Rogan EG, Cerny RL, et al. Synthesis and structure determination of the adducts formed by electrochemical oxidation of the potent carcinogen dibenzo[a,l]pyrene in the presence of nucleosides. Chem. Res. Toxicol. 1993;6:554–560. doi: 10.1021/tx00034a026. [DOI] [PubMed] [Google Scholar]
- 14.Saeed M, Zahid M, Gunselman SJ, Rogan E, Cavalieri E. Slow loss of deoxyribose from the N7deoxyguanosine adducts of estradiol-3,4-quinone and hexestrol-3′,4′-quinone. Implications for mutagenic activity. Steroids. 2005;70:29–35. doi: 10.1016/j.steroids.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem. Res. Toxicol. 2006;19:164–172. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 16.Zoltewicz JA, Clark DF, Sharpless TW, Grahe G. Kinetics and mechanism of the acid-catalyzed hydrolysis of some purine nucleosides. J. Am. Chem. Soc. 1970;92:1741–1749. doi: 10.1021/ja00709a055. [DOI] [PubMed] [Google Scholar]
- 17.Saeed M, Zahid M, Rogan E, Cavalieri E. Synthesis of the catechols of natural and synthetic estrogens by using 2-iodoxybenzoic acid (IBX) as the oxidizing agent. Steroids. 2005;70:173–178. doi: 10.1016/j.steroids.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Stack DE, Byun J, Gross ML, Rogan EG, Cavalieri EL. Molecular characteristics of catechol estrogen quinones in reactions with deoxyribonucleosides. Chem. Res. Toxicol. 1996;9:851–859. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- 19.Li KM, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, et al. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 20.Rogan EG, Devanesan PD, RamaKrishna NV, Higginbotham S, Padmavathi NS, et al. Identification and quantitation of benzo[a]pyrene-DNA adducts formed in mouse skin. Chem. Res. Toxicol. 1993;6:356–363. doi: 10.1021/tx00033a017. [DOI] [PubMed] [Google Scholar]
- 21.Cavalieri EL, Rogan EG, Li K-M, Todorovic R, Ariese F, et al. Identification and quantification of the depurinating DNA adducts formed in mouse skin treated with dibenzo[a,l]pyrene (DB[a,l)P), or its metabolites and in rat mammary gland treated with DB[a,l]P. Chem. Res. Toxicol. 2005;18:976–983. doi: 10.1021/tx049682k. [DOI] [PubMed] [Google Scholar]
- 22.Todorovic R, Devanesan P, Rogan E, Cavalieri E. Identification and quantification of stable DNA adducts of dibenzo[a,l]pyrene or its metabolites in vitro, and in mouse skin and rat mammary gland. Chem. Res. Toxicol. 2005;18:984–990. doi: 10.1021/tx049681s. [DOI] [PubMed] [Google Scholar]
- 23.Li KM, Todorovic R, Rogan EG, Cavalieri EL, Ariese F, et al. Identification and quantitation of dibenzo[a,l]pyrene--DNA adducts formed by rat liver microsomes in vitro: preponderance of depurinating adducts. Biochemistry. 1995;34:8043–8049. doi: 10.1021/bi00025a010. [DOI] [PubMed] [Google Scholar]
- 24.Saeed M, Rogan E, Cavalieri E. Mechanism of metabolic activation and DNA adduct formation by the human carcinogen diethylstilbestrol: the defining link to natural estrogens. Int. J. Cancer. 2009;124:1276–1284. doi: 10.1002/ijc.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, Ohsumi S, Makino K. Mechanistic studies on depurination and apurinic site chain breakage in oligodeoxyribonucleotides. Nucleic Acids Res. 1994;22:4997–5003. doi: 10.1093/nar/22.23.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gut IG. Depurination of DNA and matrix-assisted laser desorption/ionization mass spectrometry. Int. J. Mass Spectrom. Ion Process. 1997;169/170:313–322. [Google Scholar]
- 27.Gaikwad NW, Yang L, Muti P, Meza JL, Pruthi S, et al. The molecular etiology of breast cancer: evidence from biomarkers of risk. Int. J. Cancer. 2008;122:1949–1957. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaikwad NW, Yang L, Pruthi S, Ingle JN, Sandhu N, et al. Urine biomarkers of risk in the molecular etiology of breast cancer. Breast Cancer (Auckl) 2009;3:1–8. doi: 10.4137/bcbcr.s2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markushin Y, Gaikwad N, Zhang H, Kapke P, Rogan EG, et al. Potential biomarker for early risk assessment of prostate cancer. Prostate. 2006;66:1565–1571. doi: 10.1002/pros.20484. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Gaikwad N, Meza J, Cavalieri E, Muti P, et al. Novel biomarkers for risk of prostate cancer. Results from a case-control study. Prostate. 2009;69:41–48. doi: 10.1002/pros.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaikwad N, Yang L, Weisenburger DD, Vose J, Beseler C, et al. Urinary biomarkers suggest that estrogen-DNA adducts may play a role in the aetiology of non-Hodgkin lymphoma. Biomarkers. 2009;14:502–512. doi: 10.3109/13547500903121715. [DOI] [PMC free article] [PubMed] [Google Scholar]