Abstract
Micro-magnetic sensing and actuation have emerged as powerful tools for the diagnosis and monitoring of cancer. These technologies can be miniaturized and integrated onto compact, microfluidic platforms, enabling molecular diagnostics to be performed in practical clinical settings. Molecular targets tagged with magnetic nanoparticles can be detected with high sensitivity directly in unprocessed clinical samples (e.g. blood, sputum) due to the inherently negligible magnetic susceptibility of biological material. As a result, magnetic microchip-based diagnostics have been applied with great success to the isolation and detection of rare cells and the measurement of sparse soluble proteins. In this paper, we review recent advances in microchip-based detection of magnetically labeled biomarkers and their translation to clinical applications in cancer.
Keywords: Cancer, Diagnostics, Microfluidics, Magnetic nanoparticle
1. Introduction
In the last several decades, our ability to measure the molecular signals associated with cancer has advanced dramatically. Techniques such as genetic sequencing, high-throughput molecular screening, and flow cytometry have enabled sophisticated measurements that promise improvements in early diagnosis, personalized tailoring of treatment, and understanding of the underlying causes and mechanisms of cancer [1–3]. Unfortunately, the realization of tangible improvements in patient care from these molecular measurements has been constrained by significant engineering challenges in their translation to clinical applications. These challenges stem from the small concentration of cancer biomarkers in clinical samples, the heterogeneity of biomarker expression, and the extensive sample preparation that is often necessary prior to these measurement techniques [4,5]. Here, we review the use of magnetic actuation and sensing on microfluidic chips as a modality that is uniquely well suited to address these challenges.
The fundamental benefit of using magnetic fields to measure and control biological systems, rather than alternatives such as optical, acoustic, or electrical fields, is the negligible intrinsic magnetic susceptibility of biological systems. Magnetic sensing and sorting are based on the selective labeling of biological targets with magnetic nanoparticles (MNPs) conjugated with appropriate affinity ligands. The lack of magnetic background enables sensing and sorting to be performed on magnetically labeled cells in unprocessed clinical samples without interference from host cells or variations in pH, salinity, or turbidity [6,7]. By eliminating sample processing, magnetic detection minimizes the loss of precious sample and simplifies clinical use.
Magnetic sensors and particles can be scaled down to the micro- and nano-levels, enabling measurements to be made on biologically relevant length-scales, such as that of circulating tumor cells (~10 μm) [6], circulating microvesicles (~100 nm), and soluble proteins (~1 nm) [8]. The ability to measure clinical samples on these length-scales enables rare cells to be resolved and sparse molecular signals to be detected. Furthermore, magnetic sensing and sorting can be integrated onto microchips for automated, portable use in practical clinical settings [4].
The invention of new techniques to measure molecular biomarkers with magnetic microchips promises enormous impact on many applications in cancer diagnostics and monitoring. One example is the measurement of soluble blood-borne cancer biomarkers, which currently suffers from a lack of predictive value [9,10]. It is hypothesized that these diagnostics can be improved by increasing detector sensitivity and specificity, by expanding the number of proteins that are measured, and by measuring these biomarkers as a function of treatment progression [9,10]. Magnetic sensing, with its ability for ultra-sensitive, multiplexed detection on low-cost, portable microchips has proven uniquely well suited to meet these goals [11]. Another example where magnetic detection can address an important challenge in cancer diagnostics and monitoring, is the detection of circulating tumor cells (CTCs). Monitoring cancer progression with CTCs is an emerging technology that has shown great potential for observing the complex molecular state of a tumor, via a non-invasive blood test [12]. However, the low concentrations of CTCs versus the vast backgrounds of host cells make it challenging to efficiently isolate and profile these cells. The detection of magnetic nanoparticle labeled cells, with its inherent insensitivity to background and minimum sample processing, has been demonstrated as an effective tool to improve resolution of these rare cells [6].
In this paper, the relative utility of magnetic sensing and actuation is outlined, and recently reported technologies that harness these approaches for applications in cancer are reviewed. The review is organized by the three basic elements of magnetic detection: the labeling of molecular markers with magnetic nanoparticles (MNPs) (Fig. 1a) and the quantitative sensing (Fig. 1b) and the magnetic isolation (Fig. 1c) of these labeled biomarkers.
Fig. 1.
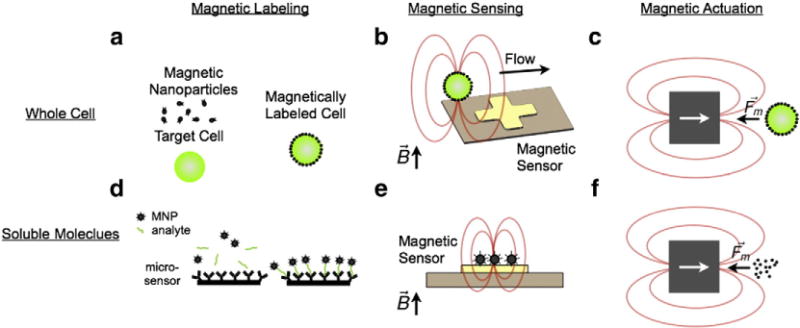
Magnetic sensing and actuation. a. Molecular markers of interest on cells can be labeled with magnetic nanoparticles (MNPs). A cell labeled with MNPs assumes a magnetic moment proportional to the expression of the targeted biomarker. b. Magnetic sensors can be used to quantitatively detect them. c. External magnetic field gradients can be used to apply forces to these cells and d. soluble biomarkers, such as proteinsor nucleic acid, can be captured onto magnetic beads for isolation or detection. For example, shown here a sandwich assay is used to capture an analyte onto a surface, and then label that analyte with MNPs. e. Magnetic sensors can be used to quantify soluble biomarkers labeled with MNPs. f. External magnetic field gradients can be used to isolate magnetic beads that have captured soluble biomarkers.
2. Recent developments in magnetic sensing and actuation
2.1. Magnetic nanoparticle labeling
Magnetic nanoparticles have physical properties than are qualitatively different than that of the bulk. These properties are controlled by the geometry of the particle and can be finely engineered for specific tasks [13]. The super-paramagnetic nature of very small (d < 20 nm) magnetic nanoparticles (MNPs) (Fig. 2a) has many advantages for biological sensing and actuation applications. At this size-scale, particles consisting of most magnetic materials (ferrites, iron) will contain only a single magnetic domain with an orientation defined by the magnetic anisotropy of the particle [5]. When these particles are suspended in fluid, thermal fluctuations at room temperature overcome this anisotropy barrier, causing the magnetic moment to spontaneously and randomly flip. An ensemble of MNPs displays a negligible net remnant magnetic moment in the absence of an external magnetic field, but become strongly magnetized in the presence of an applied field. As such, superparamagnetic particles may be described with a high magnetic susceptibility χ at fields lower than their saturation value Bs and, above Bs, by a constant magnetization Ms (Fig. 2b).
Fig. 2.
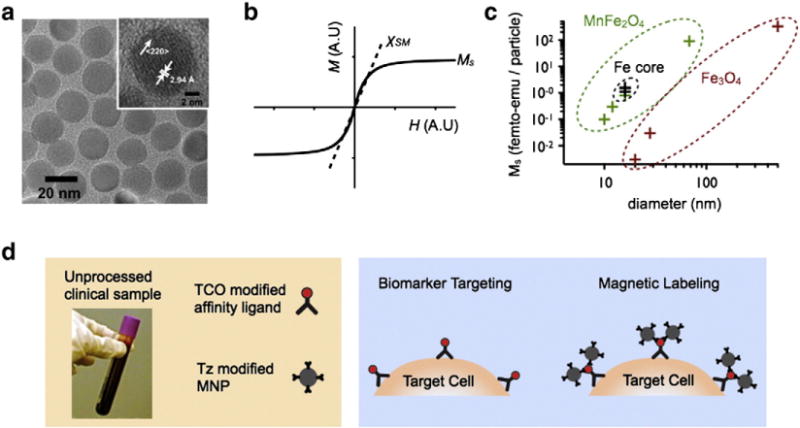
Magnetic nanoparticle labeling. a. A transmission electron micro-graph (TEM) images of manganese-doped ferrite nanoparticles (MnFe2O4) [26]. b. A graph demonstrating a typical magnetization curve for a suspension of superparamagnetic nanoparticles, with susceptibility χSM and saturation magnetization Ms. c. A graph summarizing the magnetization Ms and size of recently reported particles [5]. d. A summary of two-step magnetic labeling. First a clinical sample is mixed with a TCO modified affinity ligand (e.g. antibody), and then subsequently labeled with Tz modified MNPs.
These superparamagnetic MNPs offer several important advantages for diagnostic applications:
MNPs conjugated with the appropriate affinity ligands can be made to selectively bind to a molecular target of interest. Highly efficient two-step bio-orthogonal magnetic labeling strategies may be utilized which enable the use of generic nanoparticles, the efficient utilization of valuable affinity ligands, and amplified magnetic labeling (>105 MNP/cell) [5] (Fig. 2d).
MNPs facilitate molecular-specific mechanical actuation of intended targets. Because biological objects have negligible intrinsic magnetic moments, only magnetically-labeled targets will respond to external magnetic field gradients and experience a mechanical force [14]. Due to the superparamagnetic nature of MNPs, the magnetization of the MNPS vanishes when the external field gradient is removed, enabling stable long-term storage of these reagents.
Biological targets labeled with MNPs assume a magnetic moment proportional to their expression of a specific biomarker, enabling quantitative measurements of molecular signals.[7,15]
Ferrite particles are among the most widely utilized MNP. In particular, cross-linked iron oxide (CLIO) nanoparticles have found wide application due to their stability and biocompatibility [16]. CLIO nanoparticles contain a superparamagnetic iron oxide core (3–5 nm monocrystalline iron oxide) composed of ferrimagnetic magnetite (Fe3O4). The metallic core is coated with biocompatible dextran, cross-linked, and functionalized with primary amine, resulting in an average hydrodynamic diameter of 25–40nm [17].
Much work has been done to enhance the magnetization of MNPs. Highly magnetic particles become increasingly important when labeling cells with weakly expressing biomarkers[5] or when trying to detect small objects, such as a bacteria [18]. Doping of ferrite MNPs with elements such as manganese (Mn), cobalt (Co) or nickel (Ni) has been shown to improve the MNP magnetization (Fig. 2c) [19]. For even larger gains in magnetization, nanoparticles have been synthesized with ferromagnetic metals rather than their corresponding oxides [20] (Fig. 2c). To protect these highly reactive cores from oxidization and to further enhance magnetization, the particles have been coated with artificial ferrite shells [21]. Another strategy to enhance the magnetization of MNPs is to increase their size [22] (Fig. 2c). As the volume of the particle increases, there are more magnetic spins to contribute to the net magnetic moment. Additionally, surface effects that act to diminish the net moment of particles reduce as particles get larger due to the drop in their surface-to-volume ratio [5,13].
Polydispersity and aggregation are an impediment to the synthesis of increasingly large MNPs, as these issues become worse with increasing particle size (d > 50 nm) [21]. To avoid these problems, research groups have shown that multiple magnetic nanoparticles can be embedded into larger silica or polystyrene beads [21,23]. In addition to achieving large magnetization with these “multi-core particles”, these particles can be synthesized with multiple components (e.g. MNPs, fluorophores, quantum dots), creating multi-functional nanomaterials [23,24]. As particles get too large (d>100nm), issues arise with stability, optimal binding to biomarkers, and permeation into cells [25].
In addition to increasing the magnetization of MNPs, much work has been done to enhance the targeting of the MNPs to molecular targets of interest. One successful approach has been to break the magnetic labeling into two steps. In this method, molecular biomarkers are first targeted with affinity ligands modified with a molecular label that is not reactive with biological material. Subsequently, MNPs are introduced and are functionalized with a molecule that is selectively and highly reactive with the modified affinity ligand. Two step labeling enables the use of affinity ligands to be reduced by ~10×, generic MNPs to be used for a wide range of molecular targets, the magnetic signal to be amplified, and non-specific binding to be reduced [5]. Bioorthogonal cycloaddition between tetrazine (Tz) and trans-cyclooctene (TCO) is a fast and chemoselective reaction that does not require a catalyst, and has been used with great success for two-step magnetic labeling [26]. Recently, alternative schemes based on complementary oligonucleotide approaches[27] and cyclodextrin/adamantane chemistry [28] have been used to improve labeling and expand the scope of this methodology.
2.2. Magnetic sorting
The efficient separation and enrichment of targeted cells from a heterogeneous suspension are a critical task in the detection of biomarkers for cancer monitoring and diagnostics [29], as well as drug discovery [30], and stem cell research [31]. Magnetic sorting of MNP labeled cells has emerged as a promising technique. Highly selective sorting of targeted cells can be performed on unprocessed clinical samples due to the negligible intrinsic magnetic moments of biological material. Magnetic sorting can be miniaturized and integrated into microchip-based diagnostics. Moreover, magnetic sorting can rapidly sort through large numbers of cells, as it can process many cells in parallel unlike flow-cytometer based methods that process cells one at a time (<108 cells per hour) [32]. This feature is especially relevant for isolating rare cells, such as circulating tumor cells and endothelial cells which are suspended among vast backgrounds of host cells [2,33].
Magnetophoresis is the induced motion of a magnetically susceptible object in a nonuniform magnetic field B, with the force given by the expression:
| (1) |
The magnetophoretic force is a function of both the gradient and the strength of the magnetic field For a spherical object in a magnetic field B that is less than the saturation field B<Bs, the force is given by a simple analytic expression:
| (2) |
where a is the radius of the particle, χ is the effective magnetic susceptibility of the particle relative to the medium, and μ0 is the vacuum permeability. Note, the magnetic force scales with the volume of the particle (~a3), leading to a strong dependence of magnetic force on particle size.
Much work has been done to develop and improve magnetic separation through the application of microfabrication. Using lithography, magnetic field profiles can be engineered by patterning current carrying wires and magnetic materials. Lithographically patterned Ni, magnetized with an external permanent magnet, has been used to sort cells with high precision and accuracy [14,34]. Recently, a device was demonstrated with microfabricated permanent magnetic material (NdFeB), enabling strong magnetic forces without the need for a bulky external magnet [35].
There have been a number of creative approaches to magnetic sorting that achieve micro- and nano-patterned magnetic fields without the need for photolithography. Obviating photolithography enables devices to be fabricated that are larger, three-dimensional, inexpensive, and more practical for clinical use [36–38]. One recent device used three-dimensional columns of self-assembled super-paramagnetic beads that reach into a microfluidic channel to create an efficient sorting structure [37]. One recent work by the Lee group uses micrometer-scale grains of permanently magnetic material self-assembled to form alternating dipoles to efficiently trap cells in a microfluidic chip [36]. In another novel approach, a device was fabricated by depositing Nickel onto shrinkable polymer. Once the polymer was shrunk, the material buckled and wrinkled, forming many nanoscale magnetic traps [38].
One outstanding recent application of magnetic sorting to cancer diagnostics, is a recent work by Memhet Toner’s group at Massachusetts General Hospital. Their “iChip” [39] combines three microfluidic technologies in sequence on a single chip, for highly efficient sorting of rare circulating tumor cells from whole blood. In the first stage, red blood cells and platelets are removed from the blood using size-based deterministic lateral displacement through an array of microposts. In the second stage, the remaining cells are aligned to a narrow stream using inertial flow focusing. In the third stage, magnetically tagged cells are selectively deflected from this narrow stream into a collection channel for downstream analysis. In their paper, the iChip is used to isolate tumor cells using both positive selection based on antigen expression on the CTCs and negative selection based on antigen expression on the leukocytes. This capability allowed them to use this chip to isolate and analyze epithelial and nonepithelial cancers, including lung, prostate, pancreas, breast, and melanoma.
Much work has been done to integrate magnetic actuation onto microfluidic chips for compact diagnostics. Continuous sorting of devices has been presented that combine magnetic actuation with laminar flow microfluidics to achieve extremely high selectivity [14,40]. Recently, magnetic sorting has been integrated with droplet based microfluidics for single cell analysis [41] and digital electrowetting-based microfluidics for particle based assays [42]. Active magnetic sorting, using arrays of electronically controlled electromagnets has enabled the control of magnetically labeled objects that can be programmed for a wide array of tasks [43–45].
One outstanding example of integrating magnetic sorting with other functionalities for the diagnosis of disease is the Magnetic Integrated Microfluidic Electrochemical Detector (MIMED) from the Soh group at University of California, Santa Barbara (Fig. 3) [34]. This device integrates sample preparation and electrochemical sensors into a monolithic disposable device to detect RNA-based virus directly from throat swab samples. On this chip, viruses are first immunomagnetically isolated from throat swabs, subsequently RNA is extracted and amplified using reverse-transcriptase polymerase chain reaction (RT-PCR), and finally the products of the RT-PCR are read-out using sequence-specific electrochemical sensors. With this chip, influenza H1N1 is detected in throat swabs with four orders of magnitude with better sensitivity than clinical titer, all on a disposable, compact chip. While this chip was designed to detect H1N1 virus, this strategy generally enables sequence-specific genetic information to be efficiently collected from complex biological samples, and could find application in the genetic profiling of tumor cells for cancer diagnostics and monitoring.
Fig. 3.
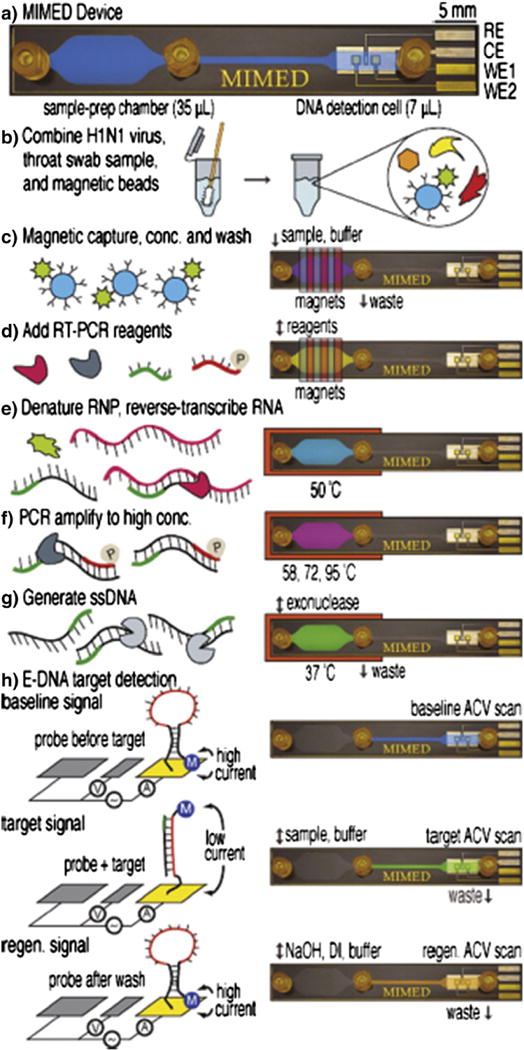
Magnetic trapping. Genetic analysis of H1N1 virus, on a microfluidic chip that integrates magnetic isolation of the virus from a throat swab, PCR amplification, and electrochemical detection. Reprinted from Ref. 35 with permission from the American Chemical Society.
2.3. Magnetic resonance based detection
Magnetic nanoparticle labeled targets can be detected with high sensitivity by harnessing nuclear magnetic resonance (NMR) as the detection mechanism. When placed in the large static magnetic fields used for NMR, MNPs create local magnetic fields that dephases the NMR signal. This dephasing leads to a measurable change in the magnetic resonance signal, manifested as a shortening of the longitudinal (T1) and transverse (T2) relaxation times (Fig. 4a). The sensitivity of this method arises from the inherent signal amplification, as each MNP affects many surrounding water molecules [46]. There are two main MNP based assays that have been utilized, 1. Cellular labeling, in which MNPs are targeted to surface biomarkers on cells and unbound MNPs are washed away [25,47] and 2. Clustering assays, in which target antigens cause dispersed MNPs to cluster and change their net relaxivity [46,48]. These techniques have been used to detect a wide range of biological targets including small molecules [49], proteins [17], nucleic acids [46], drugs (~1 pM) [50], bacteria [25], and tumor cells (1 cell) [47].
Fig. 4.
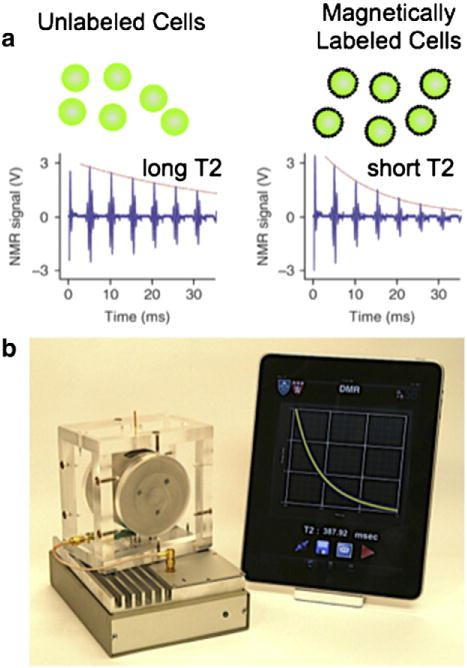
Magnetic resonance based detection. a. Principal of μNMR-based cell detection. When magnetic nanoparticles are bound to cells, they enhance the dephasing of surrounding water molecule’s NMR signal, causing the relaxation time (T2) to be reduced. b. A photograph of the DMR-III, a miniature magnetic resonance system for portable clinical use. The system consists of a permanent NdFeB magnet, micro-coil, and custom electronics. The system interfaces with a mobile device to facilitate system control and data sharing over wireless networks [56].
Technological advances in magnetic labeling are helping to extend this approach to additional applications. A targeting strategy was recently developed that uses oligonucleotide hybridization to achieve amplified and multiplexed labeling of multiple targets [27]. A bacteria labeling method was developed that is specific for gram-negativity [51].
One important recent development in the use of magnetic resonance, was a technique developed by Ralph Weissleder and Hakho Lee’s group at Massachusetts General Hospital to sensitively detect circulating microvesicles [52]. In the last several years, circulating microvesicles (CμVs) have been shown to contain a wealth of proteomic and genetic information that can guide the treatment of cancer [52]. Unfortunately, the utilization of this information to improve patient care has been limited by fundamental technical challenges that stem from the small size of CμVs (d<1μm) and the extensive sample preparation required prior to measurement. The Lee/Weissleder group developed a highly sensitive technique to detect circulating microvesicles in blood samples to diagnose glioblastoma. To this end, a microfluidic chip was developed upon which microvesicles were first labeled with target-specific magnetic nanoparticles and subsequently detected by nuclear magnetic resonance. Due to the high sensitivity of magnetic resonance measurements, the limit of detection was much lower than with conventional methods. On this chip, glioblastoma multiforme (GBM) microvesicles were successfully differentiated from microvesicles from healthy cells. Utilizing this technique, the Weissleder/Lee group demonstrated that mutations in a brain tumor, which could guide treatment decisions, could be detected through a simple blood test.
There have been a number of technological advances that are enabling NMR-based diagnostics to be miniaturized into portable systems that can be used in clinical environments. Most notably, Donhee Ham’s group at Harvard has developed an NMR system that is 1200× smaller than a benchtop NMR and 150× more sensitive [53]. The use of permanent magnets to create the NMR field, integrated circuit-based transceiver circuits to sensitively detect the weak NMR signal, surface microcoils for excitation and readout, and integrated microfluidics has enabled facile incorporation of NMR into diagnostic platforms [54]. Subsequent developments involved the integration of microfluidic components to incorporate sample preparation and concentration for enhanced sensitivity [52,55]. One major hurdle for bringing NMR to the point-of-care has been its sensitivity to temperature variation. As an alternative to costly and bulky mechanisms to control temperature, one recent paper utilized an automated feedback system to track and compensate for the temperature drift [56]. In a recent piece of work by the Han group at MIT, a new technique was developed to fabricate surface coils using multilayer liquid-metal microcoils integrated with a microfluidic network by lamination of dry adhesive sheets [57]. This new technique offers lower cost of use, as the detachable sample chamber can be disposed after each use and the microcoil can be reused without cross-contamination.
2.4. Micro-magnetic sensors
Extremely high sensitivity can be achieved by microfabricating magnetic sensors to have detection volumes similar to that of the objects they measure. A microfabricated10×10μm2 sensor has adetection volume of only ~1 pL, which is one million times smaller than is typically used in conventional diagnostics (~1 μL). These small sensors have been used to detect individual microbeads [58,59], magnetically labeled single cells [6,60], and extremely small concentrations of soluble molecules (50 aM) [8,7,62].
One important advantage of single cell detection is that it enables rare cells suspended among vast backgrounds of host cells to be detected. In bulk measurements, such as the magnetic resonance based detection described above, non-specific labeling of the background cells overwhelms the relatively small signal that comes from rare cells. The use of single cell resolution enables each individual cell to be detected one-by-one without the vast background of host cells present. This lack of background coupled with the enhanced sensitivity of single cell detectors, enables rare cells to be resolved directly in unprocessed clinical samples [6].
There are two main types of microfabricated magnetic field sensors, Hall and magnetoresistance. These techniques each have particular advantages that are useful for specific applications. For the detection of soluble molecular biomarkers, magnetoresistance-based sensors have been favored owing to the higher sensitivity at low-fields. In these detection schemes, the MNPs are magnetized in their low-field linear regime (Fig. 2b), and as such are able to harness lock-in detection on the induced magnetic moments for remarkably high sensitivity (~5 aM) [63].
For the detection of cells and microbeads, Hall sensors have been favored. Due to the short period of time that cells spend over the sensors in flow (~10μs), the lock-in techniques used with magnetoresistance-based assays cannot be utilized. Large magnetic fields (>0.1 T), which fully magnetize the MNPs can be applied to improve detection sensitivity without saturating the sensors. Owing to linearity in signal strength, cells with non- specifically bound MNPs can be accurately excluded by gating the measured signals above a particular threshold value. Furthermore, the fabrication of Hall sensors is fully compatible with standard semiconductor processing, enabling integration with auxiliary electronics for large arrays of sensors [59].
In one recent example of using micromagnetic sensors to measure soluble biomarkers, the Wang group at Stanford demonstrated an assay utilizing giant magnetoresistance (GMR) to detect sparse molecular targets in complex biological samples [7]. In this assay, the target antigen is detected using a sandwich assay between an antibody bound to the surface of the GMR sensor and an antibody attached to a superparamagnetic nanoparticle (Fig. 5a). The presence of a target antigen, causes MNPs to become bound to the sensor surface. The presence of the MNP on the sensor’s surface is detected by the GMR sensor using an applied magnetic field to magnetize the MNPs. Each chip contains an array of 64 GMR sensors, each of which could be utilized to detect a different type of protein. Multiplexed protein detection was demonstrated with a linear dynamic range of over six orders of magnitude, with a limit of detection of ~50 aM (Fig. 5b). Recently, the Wang group has combined this GMR detection scheme with a magnetic isolation technique, to detect and profile circulating tumor cells (CTCs) in whole blood [64].
Fig. 5.
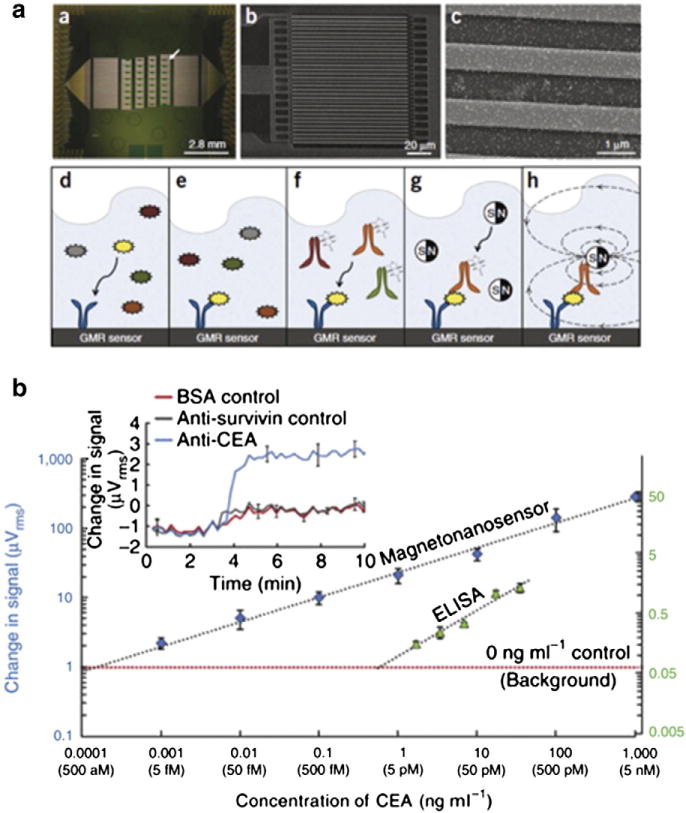
Giant magnetoresistance (GMR) based sensing of proteins by the Wang group at Stanford. a. The top figures show micrographs of the array of sixty four 100×100μm2 GMR sensors, below, a schematic of the magnetic nanoparticle based sandwich assay. b. Sensitivity and linear dynamic range of the GMR sensor and ELISA. Reprinted from Ref. 7 with permission from the Nature Publishing Group.
The utility of micro-Hall (μHall) sensors to detect magnetically labeled cells was recently demonstrated by Issadore et al. in the Weissleder/Lee group at Massachusetts General Hospital [6]. The ongoing challenge with the measurement of rare cells (e.g. cancer cells, stem cells) is that they often go undetected by conventional technologies, because current approaches require extensive sample purification and because many types of rare cells have limited half-lives outside of the body. The μHall sensor chip uses an array of eight 8×8μm2 Hall sensors to measure the magnetic moments of individual immunomagnetically tagged cells (Fig. 6a). Because the chip detects cells individually, it is able to ignore not only unbound MNPs but also other cells with inadvertently (non-specifically) attached MNPs. As a result, the μHall sensor can detect single cells even in the presence of vast numbers of blood cells and unbound reactants, and does not require any washing or purification steps. The micro-chip consists of a semiconductor substrate (GaAs) containing the μHall sensors with a PDMS microfluidic network placed on top. Hydrodynamic focusing is used to position cells into the middle and bottom of the channel towards the μHall sensors to maximize sensitivity (Fig. 6b). In a small trial of late stage ovarian cancer patients, this device was able to detect circulating cancer cells in all patients, even those that tested negative with current clinical standards (the CellSearch system) (Fig. 6c) [6]. Demonstrating the broad utility of this approach, the Weissleder/Lee group recently published a paper demonstrating that this same chip can be used to detect gram negative bacteria [65].
Fig. 6.
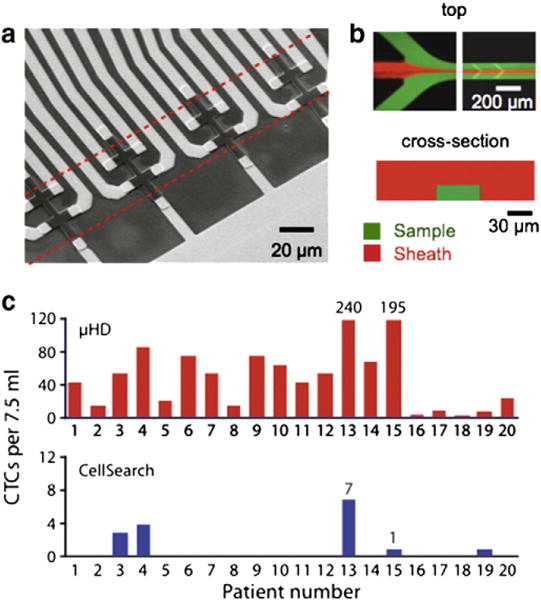
Micro-Hall detection of rare cells. a. A SEM micrograph of an array of eight 8×8μm2 Hall sensors. b. A fluorescence micrograph of a-hydrodynamic focusing structure, demonstrating that focusing can be controlled by utilizing a combination of vertical and lateral sheaths. c. Clinical applications of the μHall sensor. CTCs in patient blood samples with late stage ovarian cancer (n=20) were detected using the μHall sensor and with a clinical gold standard system, CellSearch (bottom). The μHall detected CTCs in all cancer patients, even those that were not detected using the gold standard [6].
3. Current challenges and future perspectives
An important next step for micromagnetic sensors is to extend the technology from the measurement of human cells (~10μm) to smaller objects, such as pathogens (~1μm) and microvesicles (~100 nm). This transition requires a number of fundamental developments in the technology. Most notably, the size of the sensors would need to be scaled to that of the individual object being measured. These sensors would be sub-micrometer, but well within the realm of modern integrated circuit (IC) foundries. The most recent Intel chip has feature sizes as small as 14 nm [66]. Secondly, smaller immunomagnetically tagged objects tend to have fewer surface biomarkers, and consequently weaker, more difficult to measure magnetic moments. The use of highly magnetic nanoparticles, described in Section 1, and bio-orthogonal magnetic labeling strategies to multiply the number of MNPs per biomarker, can be used to enhance this signal. Finally, it is difficult to design microfluidics to accurately position the flow of sub-micrometer objects to sub-micrometer sensors. One potential approach to solving this problem is utilizing many sub-micrometer sized sensors in parallel to reduce the required accuracy of the microfluidics.
The integration of large arrays (>1000 sensors) onto single, monolithic chips is possible using integrated circuit (IC) manufacturing [67,61]. On an IC, multiple layers of metallic wires are integrated with the semiconductor structure, allowing complex circuits that incorporate sensors, analog and digital electronics, as well as memory [43,45]. By incorporating a large number of sensors and their control circuits, an IC chip could provide enhanced throughput and obviate the need for microfluidics. With the beneficial features of low-cost electronics, μHall sensors are poised to offer clinician-friendly tools for molecular diagnostics at the point of care.
The simultaneous detection of multiple biomarkers in a sample, or on an individual cell, is essential for many biomedical applications. Unfortunately, magnetic sensing is not easily extended to multiplexed detection because the analytical signal arises from a single physical parameter, the magnetic moment m. By exploiting the distinct magnetization properties of different types of MNPs, several groups have shown that it is now possible to add “color” to the typically “gray-scale” modality of magnetic detection. Issadore et al. have demonstrated that different types of superparamagnetic MNPs (e.g. different sizes, different materials) can be distinguished by their unique non-linear magnetization curves [6]. By measuring individual cells at several different field strengths, the relative quantity of three different types of MNPs, each tagging a different biomarker, was calculated. This is analogous to the use of multiple fluorescent molecules in optical measurements, however the system currently has very low resolution and is not easily extended to more than 3–4 markers. Other prospective techniques such as detecting the harmonics generated by the non-linear magnetization curve of MNPs [67] and the detection of the particle’s ferromagnetic resonance (FMR) [68,69] offer potential ways forward.
Further integration of multiple microfluidic structures, sensors, and actuators onto single monolithic chips will enable the microchips to perform more complex, multi-step diagnostics, including sample preparation, molecular labeling, detection, and analysis. For commercialization of many of these approaches, these tools must be made less reliant on external instrumentation. The ultimate goal is to make “sample-to-answer” chips, with no external peripherals, in which an unprocessed clinical sample is the device’s input and digital, electronic data, presented in a useful format for clinicians, is the device’s output [4].
Overall, the work on magnetic microchip diagnostics described in this paper paints a hopeful picture. By enabling miniaturization of molecular diagnostics for caner, patients will gain access to more accurate, timelier, and far less expensive diagnosis and disease monitoring.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Cancer Nanotechnology”.
References
- 1.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: past studies and future opportunities. Lab Chip. 2007;7:41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 4.Issadore D, Westervelt RM. Point-of-Care Diagnostics on a Chip. Springer; 2012. [Google Scholar]
- 5.Shao H, Min C, Issadore D, Liong M, Yoon TJ, Weissleder R, Lee H. Magnetic nanoparticles and microNMR for diagnostic applications. Theranostics. 2012;2:55. doi: 10.7150/thno.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-Hall detector. Sci Transl Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall DA, Gaster RS, Makinwa KA, KA, Wang SX, Murmann B. IEEE Journal of Solid-State Circuits. 2013;48:1290–1301. doi: 10.1109/JSSC.2013.2245058. Chicago. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaster R, Xu L, Han SJ, Wilson R, Hall D, Osterfeld S, Yu H, Wang S. Quantification of protein interactions and solution transport using high-density GMR sensor arrays. Nat Nanotechnol. 2011;6:314–320. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 10.Bidart JM, Thuillier F, Augereau C, Chalas J, Daver A, Jacob N, Labrousse F, Voitot H. Kinetics of serum tumor marker concentrations and usefulness in clinical monitoring. Clin Chem. 1999;45:1695–1707. [PubMed] [Google Scholar]
- 11.Llandro J, Palfreyman JJ, Ionescu A, Barnes CHW. Magnetic biosensor technologies for medical applications: a review. Med Biol Eng Comput. 2010;48:977–998. doi: 10.1007/s11517-010-0649-3. [DOI] [PubMed] [Google Scholar]
- 12.Lang JM, Casavant BP, Beebe DJ. Circulating tumor cells: getting more from less. Sci Transl Med. 2012;4:141 ps13–141 ps13. doi: 10.1126/scitranslmed.3004261. [DOI] [PubMed] [Google Scholar]
- 13.Gubin PS. Magnetic Nanoparticles. Wiley; 2009. [Google Scholar]
- 14.Adams JD, Kim U, Soh HT. Multitarget magnetic activated cell sorter. Proc Natl Acad Sci U S A. 2008;105:18165–18170. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Sun E, Ham D, Weissleder R. Chip–NMR biosensor for detection and molecular analysis of cells. Nat med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Josephson L, Tung CH, Moore A, Weissleder R. High-efficiency intracellular magnetic labeling with novel superparamagnetic-Tat peptide conjugates. Bioconjug Chem. 1999;10:186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 17.Perez JM, Josephson L, O’Loughlin T, Högemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 18.Liong M, Fernandez-Suarez M, Issadore D, Min C, Tassa C, Reiner T, Fortune SM, Toner M, Lee H, Weissleder R. Specific pathogen detection using bioorthogonal chemistry and diagnostic magnetic resonance. Bioconjug Chem. 2011;22:2390–2394. doi: 10.1021/bc200490r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Huh YM, Jun YW, Seo JW, Jang JT, Song HT, Kim S, Cho EJ, Yoon HG, Suh JS, Cheon J. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 20.Peng S, Wang C, Xie J, Sun S. Synthesis and stabilization of monodisperse Fe nanoparticles. J Am Chem Soc. 2006;128:10676–10677. doi: 10.1021/ja063969h. [DOI] [PubMed] [Google Scholar]
- 21.Yoon TJ, Lee H, Shao H, Weissleder R. Highly magnetic core-shell nanoparticles with a unique magnetization mechanism. Angew Chem Int Ed Engl. 2011;50:4663–4666. doi: 10.1002/anie.201100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun YW, Huh YM, Choi JS, Lee JH, Song HT, Kim S, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. Nanoscale size effect of magnetic nanocrystals and their utilization for cancer diagnosis via magnetic resonance imaging. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Kim HS, Lee N, Kim T, Kim H, Yu T, Song IC, Moon WK, Hyeon T. Multifunctional uniform nanoparticles composed of a magnetite nanocrystal core and a mesoporous silica shell for magnetic resonance and fluorescence imaging and for drug delivery. Angew Chem Int Ed. 2008;47:8438–8441. doi: 10.1002/anie.200802469. [DOI] [PubMed] [Google Scholar]
- 24.Insin N, Tracy JB, Lee H, Zimmer JP, Westervelt RM, Bawendi MG. Incorporation of iron oxide nanoparticles and quantum dots into silica microspheres. ACS Nano. 2008;2:197–202. doi: 10.1021/nn700344x. [DOI] [PubMed] [Google Scholar]
- 25.Lee H, Yoon TJ, Weissleder R. Ultrasensitive detection of bacteria using core-shell nanoparticles and an NMR-filter system. Angew Chem Int Ed Engl. 2009;48:5657–5660. doi: 10.1002/anie.200901791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liong M, Tassa C, Shaw SY, Lee H, Weissleder R. Multiplexed magnetic labeling amplification using oligonucleotide hybridization. Adv Mater Weinheim. 2011;23:H254–H257. doi: 10.1002/adma.201101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agasti SS, Liong M, Tassa C, Chung HJ, Shaw SY, Lee H, Weissleder R. Supramolecular host–guest interaction for labeling and detection of cellular biomarkers. Angew Chem Int Ed. 2012;51:450–454. doi: 10.1002/anie.201105670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 30.Mattanovich D, Borth N. Applications of cell sorting in biotechnology. Microb Cell Factories. 2006;5:12. doi: 10.1186/1475-2859-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagasse E, Connors H, Al-Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman IL, Grompe M. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 32.Miltenyi S, Muller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 33.Meye A, Bilkenroth U, Schmidt U, Fussel S, Robel K, Melchior AM, Blumke K, Pinkert D, Bartel F, Linne C, et al. Isolation and enrichment of urologic tumor cells in blood samples by a semi-automated CD45 depletion autoMACS protocol. Int J Oncol. 2002;21:521–530. [PubMed] [Google Scholar]
- 34.Ferguson BS, Buchsbaum SF, Wu TT, Hsieh K, Xiao Y, Sun R, Soh HT. Genetic analysis of H1N1 influenza virus from throat swab samples in a microfluidic system for point-of-care diagnostics. J Am Chem Soc. 2011;133:9129–9135. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanini LF, Osman O, Frénéa-Robin M. Micromagnet structures for magnetic positioning and alignment. J Appl Phys. 2012;111:07B312–07B312. [Google Scholar]
- 36.Issadore D, Shao H, Chung J, Newton A, Pittet M, Weissleder R, Lee H. Self-assembled magnetic filter for highly efficient immunomagnetic separation. Lab Chip. 2011;11:147–151. doi: 10.1039/c0lc00149j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc Natl Acad Sci U S A. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawarathna D, Norouzi N, McLane J, Sharma H, Sharac N, Grant T, Chen A, Strayer S, Ragan R, Khine M. Shrink-induced sorting using integrated nanoscale magnetic traps. Appl Phys Lett. 2013;102:063504. doi: 10.1063/1.4790191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P-i, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47–179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia N, Hunt TP, Mayers BT, Alsberg E. Combined microfluidic–micromagnetic separation of living cells in continuous flow. Biomed Microdevices. 2006;8:299–308. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- 41.Chen A, Byvank T, Bharde A, Miller BL, Chalmers JJ, Sooryakumar R, Chang W-J, Bashir R. On-chip magnetic separation and cell encapsulation in droplets. Bull Am Phys Soc. 2012;57 doi: 10.1039/c2lc41201b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng AH, Choi K, Luoma RP, Robinson JM, Wheeler AR. Digital microfluidic magnetic separation for particle-based immunoassays. Anal Chem. 2012;84:8805–8812. doi: 10.1021/ac3020627. [DOI] [PubMed] [Google Scholar]
- 43.Issadore D, Franke T, Brown KA, Hunt TP, Westervelt RM. High-voltage dielectrophoretic and magnetophoretic hybrid integrated circuit/microfluidic chip. J Microelectromech Syst. 2009;18:1220–1225. doi: 10.1109/JMEMS.2009.2030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee H, Ham D, Westervelt RM. CMOS Biotechnology. Springer; 2007. [Google Scholar]
- 45.Issadore D, Franke T, Brown KA, Westervelt RM. A microfluidic microprocessor: controlling biomimetic containers and cells using hybrid integrated circuit/microfluidic chips. Lab Chip. 2010;10:2937–2943. doi: 10.1039/c0lc00092b. [DOI] [PubMed] [Google Scholar]
- 46.Josephson L, Perez JM, Weissleder R. Magnetic nanosensors for the detection of oligonucleotide sequences. Angew Chem. 2001;113:3304–3306. doi: 10.1002/1521-3773(20010903)40:17<3204::AID-ANIE3204>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 47.Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, Weissleder R. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3:71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown K, Vassiliou C, Issadore D, Berezovsky J, Cima M, Westervelt R. Scaling of transverse nuclear magnetic relaxation due to magnetic nanoparticle aggregation. J Magn Magn Mater. 2010;322:3122–3126. doi: 10.1016/j.jmmm.2010.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsourkas A, Hofstetter O, Hofstetter H, Weissleder R, Josephson L. Magnetic relaxation switch immunosensors detect enantiomeric impurities. Angew Chem Int Ed. 2004;43:2395–2399. doi: 10.1002/anie.200352998. [DOI] [PubMed] [Google Scholar]
- 50.Ullal AV, Reiner T, Yang KS, Gorbatov R, Min C, Issadore D, Lee H, Weissleder R. Nanoparticle-mediated measurement of target-drug binding in cancer cells. ACS Nano. 2011;5:9216–9224. doi: 10.1021/nn203450p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Budin G, Chung HJ, Lee H, Weissleder R. A magnetic gram stain for bacterial detection. Angew Chem Int Ed Engl. 2012;51:7752–7755. doi: 10.1002/anie.201202982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun N, Yoon T-J, Lee H, Andress W, Demas V, Prado P, Weissleder R, Ham D. Palm NMR and one-chip NMR Solid-State Circuits Conference Digest of Technical Papers (ISSCC) 2010 IEEE Int. 2010:488–489. [Google Scholar]
- 54.Lee H, Sun E, Ham D, Weissleder R. Chip–NMR biosensor for detection and molecular analysis of cells. Nat med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee H, Yoon T-J, Weissleder R. Ultrasensitive detection of bacteria using core–shell nanoparticles and an NMR-filter system. Angew Chem Int Ed. 2009;48:5657–5660. doi: 10.1002/anie.200901791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Issadore D, Min C, Liong M, Chung J, Weissleder R, Lee H. Miniature magnetic resonance system for point-of-care diagnostics. Lab Chip. 2011;11:2282–2287. doi: 10.1039/c1lc20177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong TF, Peng WK, Luong TD, Nguyen NT, Han J. Adhesive-based liquid metal radio-frequency microcoil for magnetic resonance relaxometry measurement. Lab Chip. 2012;12:287–294. doi: 10.1039/c1lc20853e. [DOI] [PubMed] [Google Scholar]
- 58.Li G, Joshi V, White RL, Wang SX, Kemp JT, Webb C, Davis RW, Sun S. Detection of single micron-sized magnetic bead and magnetic nanoparticles using spin valve sensors for biological applications. J Appl Phys. 2003;93:7557–7559. [Google Scholar]
- 59.Liu P, Skucha K, Duan Y, Megens M, Kim J, Izyumin I, Gambini S, Boser B. Magnetic relaxation detector for microbead labels. IEEE J Solid-State Circ. 2012;99:1–9. doi: 10.1109/jssc.2012.2185339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loureiro J, Andrade PZ, Cardoso S, da Silva CL, Cabral JM, Freitas PP. Magnetoresistive chip cytometer. Lab Chip. 2011;11:2255–2261. doi: 10.1039/c0lc00324g. [DOI] [PubMed] [Google Scholar]
- 61.Hall DA, Gaster RS, Makinwa K, Wang SX, Murmann B. A 256 pixel magnetoresistive biosensor microarray in 0.18um CMOS. J Solid State Circ. 2013;48:1290–1301. doi: 10.1109/JSSC.2013.2245058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu L, Yu H, Akhras MS, Han S-J, Osterfeld S, White RL, Pourmand N, Wang SX. Giant magnetoresistive biochip for DNA detection and HPV genotyping. Biosens Bioelectron. 2008;24:99–103. doi: 10.1016/j.bios.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaster R, Hall D, Wang S. nanoLAB: an ultraportable, handheld diagnostic laboratory for global health. Lab Chip. 2011;11:950–956. doi: 10.1039/c0lc00534g. [DOI] [PubMed] [Google Scholar]
- 64.Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014 doi: 10.1039/c3lc50580d. http://dx.doi.org/10.1039/C3LC50580D. [DOI] [PMC free article] [PubMed]
- 65.Issadore D, Chung H, Budin G, Weissleder R, Lee H. μHall chip for sensitive detection of bacteria. Adv healthc mater. 2013 doi: 10.1002/adhm.201200380. http://dx.doi.org/10.1002/adhm.201200380. [DOI] [PMC free article] [PubMed]
- 66.James D. Intel Ivy Bridge unveiled–he first commercial tri-gate, high-K, metal-gate CPU Custom Integrated Circuits Conference (CICC), 2012. IEEE. 2012:1–4. [Google Scholar]
- 67.Gambini S, Skucha K, Liu PP, Kim J, Krigel R. A 10 kPixel CMOS Hall Sensor Array With Baseline Suppression and Parallel Readout for Immunoassays. 2013;48:302–317. [Google Scholar]
- 68.Weaver JB, Rauwerdink AM, Sullivan CR, Baker I. Frequency distribution of the nanoparticle magnetization in the presence of a static as well as a harmonic magnetic field. Med phys. 2008;35:1988. doi: 10.1118/1.2903449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilhelm C, Gazeau F, Bacri J-C. Magnetophoresis and ferromagnetic resonance of magnetically labeled cells. Eur Biophys J. 2002;31:118–125. doi: 10.1007/s00249-001-0200-4. [DOI] [PubMed] [Google Scholar]


