Abstract
Formaldehyde is a reactive aldehyde that has been classified as a class I human carcinogen by the International Agency for Cancer Research. There are growing concerns over the possible adverse health effects related to the occupational and environmental human exposures to formaldehyde. Although formaldehyde-induced DNA and protein adducts have been identified, the genomic instability mechanisms and the cellular tolerance pathways associated with formaldehyde exposure are not fully characterized. This study specifically examines the role of a genome stability protein, Bloom (BLM) in limiting formaldehyde-induced cellular and genetic abnormalities. Here, we show that in the absence of BLM protein, formaldehyde-treated cells exhibited increased cellular sensitivity, an immediate cell cycle arrest, and an accumulation of chromosome radial structures. In addition, live-cell imaging experiments demonstrated that formaldehyde-treated cells are dependent on BLM for timely segregation of daughter cells. Both wild-type and BLM-deficient formaldehyde-treated cells showed an accumulation of 53BP1 and γH2AX foci indicative of DNA double-strand breaks (DSBs); however, relative to wild-type cells, the BLM-deficient cells exhibited delayed repair. In response to formaldehyde exposure, we observed co-localization of 53BP1 and BLM foci at the DSB repair site, where ATM-dependent accumulation of formaldehyde-induced BLM foci occurred after the recruitment of 53BP1. Together, these findings highlight the significance of functional interactions among ATM, 53BP1, and BLM proteins as responders associated with the repair and tolerance mechanisms induced by formaldehyde.
Keywords: ATM, BLM, 53BP1, Double-strand breaks, Formaldehyde
1. Introduction
Formaldehyde (HCHO) is a ubiquitous environmental and occupational pollutant. Both chronic and acute exposures to formaldehyde have been associated with adverse effects on human health including eye, nose, throat, and skin irritation, allergic contact dermatitis, altered lung functions and immune responses, occupational asthma, and cancer [1,2]. Based on epidemiological evidence associating formaldehyde exposure with nasopharyngeal cancer and myeloid leukemia [3-5], formaldehyde was classified as a class I human carcinogen by the International Agency on Cancer Research (IARC) in 2006. However, there exists some controversy regarding the nature, magnitude, and persistence of the adverse health effects related to formaldehyde exposure.
The primary DNA lesions resulting from formaldehyde exposure that contribute to its genotoxic and mutagenic potential are considered to be DNA-protein crosslinks (DPCs) [6-10], though the molecular mechanisms that lead to formaldehyde-induced carcinogenesis remain elusive. Formation and persistence of DPCs may pose a formidable challenge to genome stability by interfering with biological processes such as replication, recombination, and transcription. Thus, elucidating the roles of specific DNA repair and tolerance factors involved in DPC processing is important for understanding the mechanisms of formaldehyde-induced carcinogenesis. In this regard, several studies have shown that loss of specific DNA repair factors can promote formaldehyde-induced cellular and cytogenetic abnormalities [11-14].
In order to identify pathways involved in limiting formaldehyde-induced cell death, a formaldehyde cytotoxicity screen of the Saccharomyces cerevisiae gene deletion library was previously performed in our lab and multiple pathways were identified that are important for cell survival following formaldehyde exposure. Under low dose, chronic exposure conditions, homologous recombination was the primary pathway that conferred resistance to formaldehyde-induced lesions; while following acute, high dose exposure, the nucleotide excision repair (NER) pathway was critical for cell survival [15]. Interestingly, this investigation showed that a sgs1 (a member of the RecQ superfamily) deletion mutant exhibited increased cellular sensitivity to both chronic and acute formaldehyde exposures [15]. Consistent with the yeast study, an E. coli RecQ mutant was sensitive to formaldehyde treatment [12]. However, both E. coli and yeast studies were limited to measurements of cell viability and did not further investigate the molecular mechanisms mediated by RecQ helicases in limiting formaldehyde-induced cytotoxicity.
Members of the RecQ superfamily are important for maintaining genomic integrity and thus, are referred to as guardians of the genome. Although, there exists only one RecQ family member in bacteria and yeast, in humans, the RecQ superfamily of helicases is comprised of 5 known family members: BLM, WRN, RecQL1, RecQL4, and RecQ5. Among these, BLM was the first to be linked to a hereditary disease known as Bloom syndrome (BS) [16]. BS is a rare autosomal recessive disorder characterized by multiple abnormalities, including immunodeficiency, pre- and post-natal growth retardation, and a high incidence of cancer [17]. Biochemical and cellular studies have demonstrated that the BLM protein is a 3′–5′ helicase that participates in critical steps associated with replication, recombination and repair [18]. BLM protein has also been shown to be required for faithful chromosome segregation during mitotic cell division [19,20]. BS cells exhibit chromosomal instability and hypersensitivity in response to several genotoxic agents, including replication stressors, topoisomerase inhibitors, and DNA crosslinking agents [21]; however, the role of BLM following exposure to DNA-protein crosslinking agents remains to be elucidated.
Germane to our interest in formaldehyde-induced genomic instability, this study investigated the potential importance of human BLM in both the DNA damage response and maintenance of genomic integrity following formaldehyde exposure. Herein, we show that BLM rescues formaldehyde-treated cells from G2/M arrest by facilitating the repair of DSBs and regulating normal mitotic progression. Additionally, our studies demonstrate a co-localization of 53BP1 and BLM proteins at sites of formaldehyde-induced DNA damage where the recruitment of BLM protein to the damage sites was found to be ATM-dependent. Overall, our findings suggest an interplay between ATM, 53BP1 and BLM proteins that is critical for mitigating formaldehyde-induced genotoxic and cytotoxic effects.
2. Materials and methods
2.1. Cells and culture conditions
Patient derived BLM-deficient (GM08505) and ATM-deficient (GM05849) SV40-transformed fibroblast cells used in this study were purchased from Coriell Cell Repositories. Wild-type (GM639, also known as GM00639) SV40-transformed cells were a kind gift from Dr. Robb E. Moses (OHSU). Cells were grown in DMEM supplemented with 10% fetal bovine serum and antibiotics (ampicillin and streptomycin, Gibco) at 37 °C in a 5% CO2 incubator. For all experiments, sub-confluent cultures were treated for 4 h with various concentrations of formaldehyde (Fisher Scientific) and harvested at the indicated recovery times. For Click-iT EdU (5-ethynyl-2′-deoxyuridine) assays, cells were labeled with EdU (10 μM for 1 h) and processed for imaging following the manufacturer’s protocol (Invitrogen, C10377).
2.2. Colony forming assays
For colony forming assays, 300–1800 cells were seeded in 100 mm or 6-well plates and incubated overnight at 37 °C prior to acute formaldehyde treatment for 4 h at the indicated concentrations. Following a 4 h treatment, formaldehyde was removed, cells were washed with PBS, and fresh media was added. After 10–15 days, colonies (>30 cells) were fixed, stained with methylene blue diluted in methanol (4 g/L), and counted.
2.3. siRNA transfection
Wild-type cells were transfected with 100 nM scramble (Dharmacon, D001810-01-05) or BLM siRNA cocktail (Dharmacon, M-007287-02-0005) according to the manufacturer’s instructions using HiPerFect transfection reagent (QIAGEN). Briefly, HiPerFect (25 μL) was diluted into siRNA- or scramble-containing Opti-MEM (GIBCO) and incubated at room temperature for 10 min to allow the formation of lipid-siRNA complexes. A suspension of GM639 cells (0.2 × 106 cells in 600 μL DMEM) was added to the preformed lipid-siRNA complexes (400 μL) and incubated at room temperature for an additional 10 min. Transfected cells were diluted in DMEM and then seeded in a 12-well plate (0.1 × 106 cells per well). After 24 h incubation, transfected cells were subjected to acute formaldehyde treatment (0–400 μM) for 4 h. Formaldehyde-treated cells were washed with PBS twice and re-transfected with scramble or BLM siRNA. After 24 h of second transfection, cells were trypsinized and seeded in 6-well plates for colony forming assays. Following 10 days of growth, plates were stained with methylene blue and colonies were counted. In parallel, cells were plated in 60 mm plates and harvested at indicated times for Western blot analyses.
2.4. Cell cycle analyses
Cells were arrested at the G1/S phase boundary by treatment with a replication elongation inhibitor, aphidicolin (1 μg/mL) for 24 h. Prior to formaldehyde treatment, aphidicolin-synchronized cells were allowed a 2 h recovery to facilitate the progression of cells into S phase. Cells were harvested at the indicated times, fixed in ice-cold 70% ethanol, and stained with propidium iodide (PI) (Invitrogen). DNA content was measured using a FACSCalibur instrument (Becton Dickinson) (Flow Cytometry Core, OHSU). Aggregated cells were excluded from the PI-stained cell suspensions by passing them through strainer-capped tubes (BD Falcon) prior to analyses. The results were analyzed using FlowJo software (Tree Star V.7.5).
2.5. Live-cell imaging
For live-cell imaging, wild-type and BLM-deficient cells were plated (1–3 × 104 per plate), grown on a glass bottom microwell petridish (P35G-1.5-14-C, Mattek), and treated with formaldehyde (100 μM) for 4 h. Cells were then washed with PBS, and released into fresh media prior to imaging using a VivaView™ FL incubator microscope. For synchronization in S phase, plated cells were treated with aphidicolin (1 μg/mL) overnight, released into fresh media for 2 h prior to treatment with formaldehyde. Four random fields were selected per sample and images were captured every 30 min for 6 days. Image brightness and contrast enhancement, and conversion to QuickTime movies were performed with ImageJ software.
2.6. Immunofluorescence studies
Cells were fixed in 4% paraformaldehyde at room temperature for 10 min, washed with PBS buffer, permeabilized with PBS-T (PBS with 0.25% Triton-X100), and blocked with 5% non-fat milk in PBS. Cells were then incubated with the following antibodies individually or in combination: mouse monoclonal anti-53BP1 (BD Transduction Laboratories 612522; 1:1000 dilution) for 1 h; rabbit polyclonal anti-BLM (Abcam ab2179; 1:250 dilution) for 1 h; or rabbit polyclonal anti-γH2AX (Milipore Cat. 05-636, 1:1000 dilution) for 2 h. Appropriate fluorochrome-conjugated secondary antibodies (Alexa-Fluor 594-conjugated goat anti-rabbit (Invitrogen, 1:500 dilution) and Alexa-Fluor 488-conjugated anti-mouse (Invitrogen, 1:500 dilution)) were used for 45–60 min. Nuclei were stained with DAPI for 10 min prior to mounting the slides using Fluoro-Gel with Tris Buffer (EMS). Cells were visualized on an Axioskop 2 microscope (Zeiss) and images were processed using Axiovision Software (Zeiss).
2.7. Western blot analyses
For Western blot analyses, 4–9 × 106 cells were seeded in 100 mm plates and incubated overnight at 37 °C prior to a 4 h formaldehyde treatment (300 μM). For Western blot analyses of BLM protein, cells were harvested at indicated time points and disrupted in a lysis buffer (50 mM Tris–HCl, pH 8.0, 0.5% Nonidet P-40, 5 mM EDTA, 150 mM NaCl, 1 mM pepstatin A, and 1 mM PMSF). For detection of BLM and ATM proteins, aliquots of total cell lysate (35 or 50 μg, respectively) were run on a 3–8% NuPAGE® Tris-Acetate Gel (Life Technologies), followed by transfer onto PVDF membranes. The membranes were immunoblotted independently with the following primary antibodies: 3 h incubation with mouse monoclonal anti-ATM (GeneTex 2C1, 1:1000 dilution), overnight incubation with rabbit polyclonal anti-BLM (Santa Cruz H-300, 1:1000 dilution), and 1 h incubation with mouse monoclonal anti-α-tublin (Sigma clone B-5-1-2, 1:10,000 dilution). The membranes were then incubated for 1 h with appropriate secondary antibodies conjugated to horseradish peroxidase, and proteins were detected by the enhanced chemiluminescence detection system (Western LightningTM Plus-ECL from Perkin Elmer or Clarity Western ECL Substrate from Bio-Rad).
2.8. Cytogenetic analyses
Cells were harvested, treated with hypotonic solution (75 mM KCl, 5% fetal bovine serum) for 10 min, and fixed with 3:1 methanol:acetic acid. Cells were dropped onto slides for metaphase spreads and stained with Wright’s stain (Fisher Scientific). For each sample, fifty metaphases were analyzed for radial formation on a Zeiss Axioskop Brightfield microscope. Representative photographs were taken using CytoVision software (Applied Imaging). These experiments were performed at the Cytogenetics Research Facility, OHSU.
3. Results
3.1. BLM-deficient cells exhibit increased sensitivity to formaldehyde compared to BLM-proficient cells
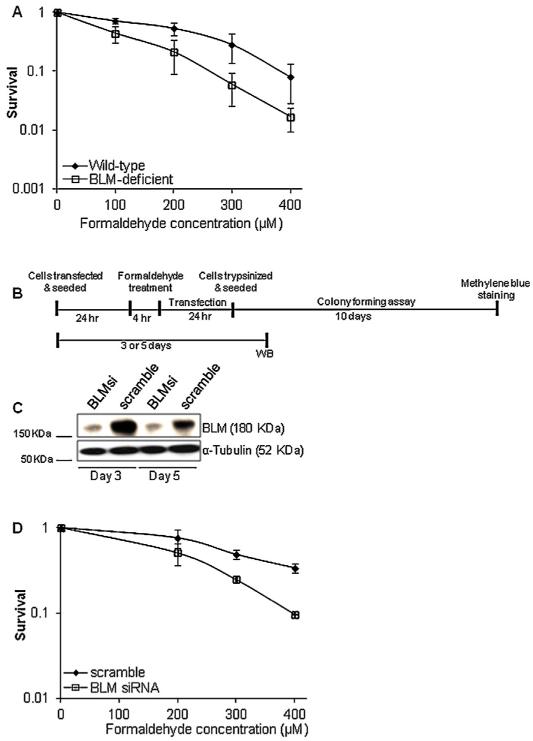
A screen of the yeast gene deletion library revealed that the sgs1 deletion strain was hypersensitive to chronic and moderately sensitive to acute treatments of formaldehyde [15]. Given the conserved nature of RecQ helicases from bacteria to humans, we hypothesized that loss of BLM function would sensitize human cells to formaldehyde. Formaldehyde-induced cytotoxicity was compared for wild-type BLM-proficient (GM639) and BLM-deficient (GM08505) fibroblast cells. Cells were subjected to acute treatment of formaldehyde (0–400 μM) for 4 h and cell viability was determined by colony forming assays. BLM-deficient fibroblast cells were observed to have reduced viability following formaldehyde treatment compared to wild-type cells (Fig. 1A).
Fig. 1.
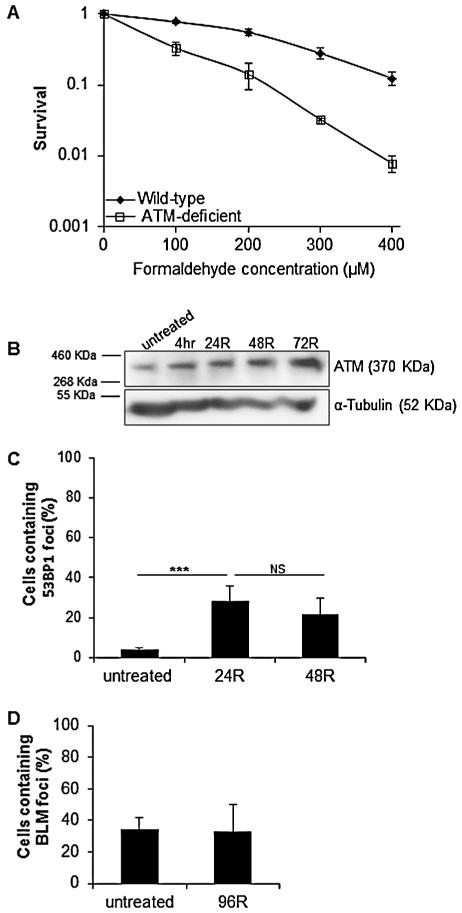
BLM protein deficiency leads to increased sensitivity to formaldehyde exposure. (A) Survival of wild-type (GM639) and BLM-deficient (GM08505) cells was determined by colony forming assay. Cells were treated with the indicated concentrations of formaldehyde for 4 h. (B) Experimental scheme employed for the colony forming assay with the scramble or BLM siRNA transfected cells. Freshly seeded wild-type cells were incubated overnight and transfected with scramble or BLM siRNA cocktail. Following a 24 h transfection, cells were treated with formaldehyde for 4 h, washed with PBS twice, and re-transfected with scramble or BLM siRNA. WB, Western blot. (C) Immunoblot analysis showing the depletion of BLM protein following transfection with BLM siRNA. Whole-cell extracts of scramble or BLM siRNA-treated cells were isolated on day 3 and day 5 following transfection (i.e., day 2 and day 4 following formaldehyde treatment as indicated in the scheme above). The expression of α-tubulin was used as a loading control for the Western blot. (D) Following formaldehyde treatment at the indicated concentrations, colony forming assay was used to measure the cell viability of scramble or BLM siRNA-treated cells. For A and D, the mean and standard deviation from three or more independent experiments are shown.
Although the normal human SV40-transformed fibroblast cell line, GM639 has been previously used as a wild-type BLM-proficient cell line as compared to GM08505 cells [22], it could be argued that the differential sensitivity to formaldehyde observed in the two cell lines was attributed to their non-isogenic backgrounds. To address this concern and further validate that the BLM protein has a role in limiting formaldehyde-induced cytotoxicity, we used a siRNA approach to knockdown BLM in wild-type cells (Fig. 1B, C). Following scramble or BLM siRNA treatment for 24 h, cells were exposed to acute treatment of formaldehyde at the indicated concentrations for 4 h. Cells were re-transfected with scramble or BLM siRNA for 24 h and seeded for colony forming assays. In parallel, the effectiveness of the siRNA knockdown was measured by Western blot analyses at 3 and 5 days post initial transfection (Fig. 1B and C). Depletion of BLM protein by siRNA in GM639 cells resulted in >85% depletion of BLM protein levels. Consistent with the survival assays in Fig. 1A, BLM siRNA-treated cells exhibited increased sensitivity to formaldehyde treatment than the scramble-treated cells (Fig. 1D). Overall, results from the above experiments suggest that BLM protein has a role in limiting formaldehyde-induced cytotoxicity.
3.2. Exposure to formaldehyde leads to an immediate and pronounced cell cycle arrest of BLM-deficient cells
BLM helicase is known to participate in the resolution and repair of certain DNA structures (such as Holliday Junctions, D-loops, G-quadruplexes, etc.) formed at replication forks [23]. Thus, loss of BLM function causes an S phase delay of the cell cycle presumably due to inhibition of these replication-dependent processes [24]. Given that formaldehyde induces a G2/M phase arrest of the cell cycle in Chinese Hamster Ovary cells (CHO) [11]; we hypothesized that the formaldehyde-induced G2/M arrest is a result of replication-associated DNA damage and furthermore, that BLM protein is required to overcome formaldehyde-induced replication stress.
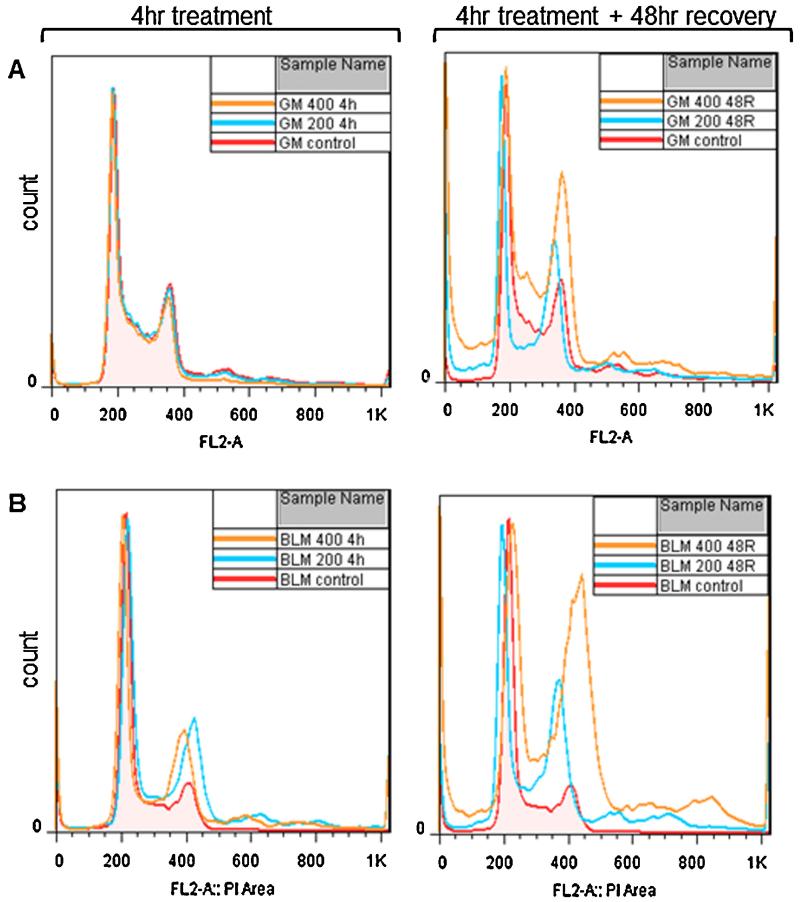
In order to examine whether formaldehyde-induced G2/M arrest occurred in a BLM-dependent manner, we studied the effects of formaldehyde treatment on the cell cycle profiles of wild-type and BLM-deficient cells. Since comparable trends for survival had been observed for BLM siRNA-treated and BLM-deficient cells in response to formaldehyde, further characterization of BLM’s function was carried out using wild-type and BLM-deficient cells rather than siRNA-treated cells. Both cell lines were treated with formaldehyde at 200 μM or 400 μM for 4 h and cells were harvested either immediately after the treatment or following a 48 h recovery. Modulation of cell cycle progression was analyzed by flow cytometry. The cell cycle analyses showed no change in the cell cycle profile of the wild-type cells immediately after the 4 h formaldehyde treatment (Fig. 2A, left panel). However, following a 48 h recovery, formaldehyde-treated wild-type cells showed a dose-dependent accumulation of cells in late S-G2/M phase (Fig. 2A, right panel). In contrast, a significant portion of the BLM-deficient cells harvested at 4 h post formaldehyde treatment accumulated in late S-G2/M phase (Fig. 2B, left panel). The magnitude of this effect increased following the 48 h recovery period (Fig. 2B, right panel). At 48 h post recovery, although both cell lines showed increased apoptosis and ploidy changes as indicated by sub-G1 and post-G2 populations, respectively, a higher percentage of cells with >4 N DNA were observed in the BLM-deficient population than the wild-type. These results suggest that BLM function is most likely associated with the G2/M checkpoint in response to formaldehyde-induced DNA damage; thus, loss of a normal G2/M checkpoint results in cells with abnormal ploidy levels, possibly as a consequence of nuclear and chromosomal segregation defects or exacerbated cytokinesis failure.
Fig. 2.
Formaldehyde induces a severe G2/M arrest in BLM-deficient cells. Wild-type (A) and BLM-deficient (B) cells were treated with formaldehyde at the indicated concentrations (200 μM and 400 μM) and harvested either immediately after a 4 h (4 h) treatment (left side panels) or following a 48 h recovery post treatment (48R) (right side panels). The DNA content of the cells was measured by flow cytometry. GM and BLM are abbreviations for wild-type and BLM-deficient cells, respectively (inset right corner of each panel). The flow cytometry results are summarized by generating overlay images representing all samples harvested at each time point.
Next, to examine whether formaldehyde-induced G2/M arrest was a replication-dependent process, we synchronized wild-type cells in S phase following aphidicolin treatment, and then monitored the changes in the molecular events following the release from S phase cell cycle block. Consistent with our prior data with Chinese Hamster Ovary (CHO) cells [11], we observed that the cell cycle effects were more pronounced when wild-type human cells were synchronized in S phase and then treated with formaldehyde (Fig. S1A). Furthermore, when synchronized cultures of wild-type cells were treated with formaldehyde, they displayed a delayed recovery from cell cycle arrest, accompanied by increased apoptosis (Fig. S1A) as indicated by the increased sub-G1 population. These data confirmed that formaldehyde-induced cell cycle perturbations occur in a replication-dependent manner.
To further investigate if formaldehyde has a direct effect on DNA replication and if this effect is BLM-dependent, wild-type and BLM-deficient cells were subjected to formaldehyde treatment and pulse-labeled with EdU prior to harvesting cells. It was observed that both wild-type and BLM-deficient cells showed a complete inhibition of DNA synthesis in the presence of formaldehyde; however, DNA synthesis resumed in both cell lines 24 h post recovery following formaldehyde exposure (Fig. S1B, C). These results demonstrate that formaldehyde-induced inhibition of DNA synthesis and the subsequent resumption of DNA synthesis occur in a BLM-independent manner.
3.3. Absence of BLM causes a severe delay in mitotic timing of formaldehyde-treated cells
Our cell cycle analyses (Fig. 2) revealed that following formaldehyde treatment, cells with higher ploidy (>4n) levels accumulate in both wild-type and BLM-deficient cells. These data are consistent with our karyotypic data which demonstrated that formaldehyde exposure causes an increase in aneuploidy and polyploidy in CHO cells [11]. Changes in ploidy levels are frequently associated with chromosomal instability resulting from defects in mitotic segregation of chromosomes [25].
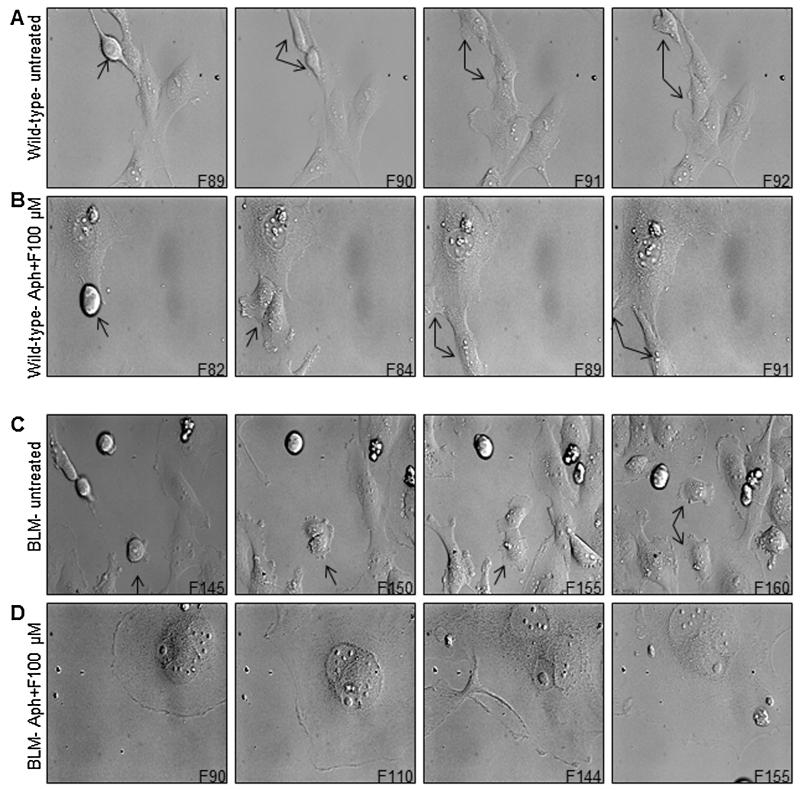
To examine the effects of formaldehyde treatment on mitotic cell division as a source of abnormal ploidy levels, we performed live-cell imaging on formaldehyde-treated wild-type and BLM-deficient cells for 6 days and studied the progression of the mitotic cells. In parallel, to study the replication-dependent effects of formaldehyde, cells were synchronized in S phase and then treated with formaldehyde. Four random fields for each sample were selected for imaging and the representative still images from movies for four samples (A. wild-type untreated, B. wild-type synchronized and formaldehyde-treated, C. BLM-deficient untreated, and D. BLM-deficient synchronized and formaldehyde-treated) are shown in Fig. 3 (Movies S1A, S1D, S1E, and S1H, respectively). Still images were extracted from the time-lapse movies and the time taken to complete cell division was estimated by multiplying the number of the frames a mitotic cell took to complete cell division × 30 min (the time difference between each frame). Most of the wild-type cells completed their cell division in 1.5–3.5 h (Fig. 3A, Movie S1A, n = 15 cells). Synchronization with aphidicolin alone did not result in any delay of cell division in wild-type cells (Movies S1B); however, several formaldehyde-treated wild-type cells (both unsynchronized and synchronized) exhibited a mitotic delay in completing their cell division, where the length of mitosis varied between 1.5 and 5 h (n = 11 and 13 cells, respectively) (Fig. 3B, Movies S1C and S1D), suggesting that formaldehyde-induced lesions hinder the process of cell division. Compared to the strong replication-dependency observed for the synchronized formaldehyde-treated wild-type cells in cell cycle analyses (Fig. S1), live-cell imaging observations with the wild-type cells under similar conditions could not be associated with replication. We believe this difference is largely due to the lower formaldehyde concentration used for the live-cell imaging experiments as compared to cell cycle analyses (100 μM versus 300 μM). It was necessary to lower the formaldehyde concentration for the live-cell imaging experiments in order to avoid excessive apoptosis of BLM-deficient cells following formaldehyde treatment.
Fig. 3.
Snapshot images from a time-lapse experiment showing mitotic cell division defects in formaldehyde-treated BLM-deficient cells. Untreated wild-type or BLM-deficient (A, C) cells are compared to S phase synchronized and formaldehyde-treated cells (B, D). Synchronized cultures were released from aphidicolin (24 h treatment), and 2 h later, treated with formaldehyde (100 μM for 4 h). Images were captured at 30 min intervals for 6 days. “F” followed by a number in the right corner of each image represents the frame number from the time-lapse movie. Arrows indicate dividing cells.
Compared to wild-type, untreated unsynchronized BLM-deficient cells took approximately 1.5–7 h (n = 12 cells) to complete a round of cell division (Fig. 3C and Movie S1E). Aphidicolin-synchronized BLM-deficient cells did not exhibit any obvious delay of cell division (Movie S1F) relative to unsynchronized BLM-deficient cells. However, both synchronized and unsynchronized formaldehyde-treated BLM-deficient cells exhibited delayed cell division, multi-nucleation and enlargement of nuclei (Fig. 3D and Movies S1G-H). In particular, synchronized formaldehyde-treated BLM-deficient cells had a remarkably extended mitosis, during which none of the cells were observed to complete the mitotic division during a 6-day period (Fig. 3D and Movie S1H). For this population, cells either underwent apoptosis or remained as giant cells. To rule out the possibility that these giant cells were undergoing senescence, independent experiments were carried out with cells stained for the proliferation marker, Ki-67. All giant cells were found to be positively labeled for Ki-67 (data not shown). Collectively, although wild-type synchronized populations failed to demonstrate replication-dependency, live-cell imaging results with BLM-deficient synchronous populations confirmed that formaldehyde-induced effects are strongly dependent on replication. Furthermore, the impaired exit from mitosis in BLM-deficient cells is suggestive of a possible role of BLM protein in accurate segregation of daughter nuclei.
3.4. BLM-deficient cells exhibit delayed DNA DSB repair following formaldehyde exposure
It is well established that DSBs are highly cytotoxic lesions and in response to DSB generation, numerous DNA damage-responsive proteins (such as H2AX, 53BP1, etc.) rapidly redistribute as microscopically visible foci at the sites of damage. Our lab has previously shown that formaldehyde exposure results in an accumulation of γH2AX (phosphorylated form of histone variant H2AX) foci [11]. In the current study, we examined whether BLM is involved in the generation or repair process of formaldehyde-induced DSBs. In order to address this question, DSB generation and repair capacity of wild-type and BLM-deficient cells were compared by studying the enrichment and disappearance of 53BP1 foci following formaldehyde treatment.
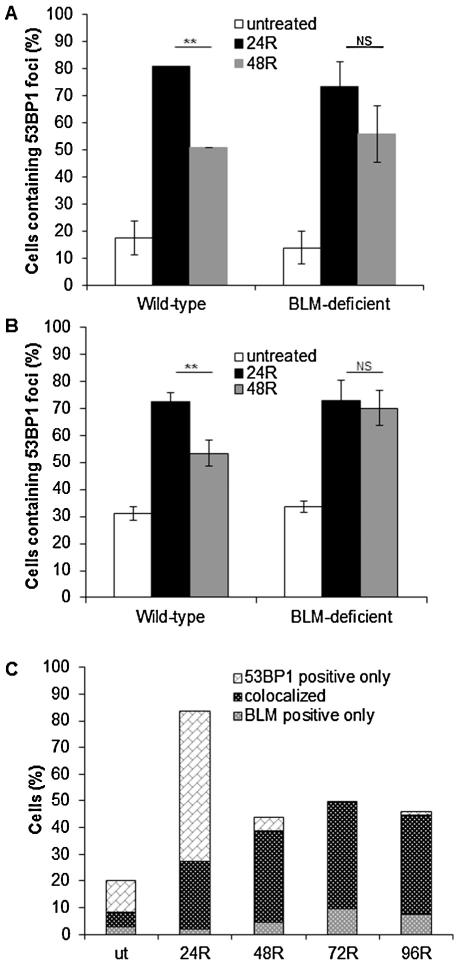
53BP1 (also known as TP53BP1) protein is a DSB marker and an important factor required for their effective repair [26-28]. Its recruitment to the sites of DSBs is critical for determining DSB repair pathway choice [29,30]. To study 53BP1 dynamics in response to formaldehyde-induced stress, cells were subjected to acute formaldehyde treatment (300 μM for 4 h) and scored for 53BP1 foci formation following a recovery of 24 or 48 h (Fig. 4). Nuclei containing >10 53BP1 foci were counted as 53BP1 positive. No induction of 53BP1 foci was observed after a 4 h formaldehyde treatment (data not shown) and this is an expected result given that the primary lesions induced by formaldehyde are not DSBs. We believe that formaldehyde-induced DSBs are generated as a result of processing of DPCs (i.e., the primary lesions induced by formaldehyde). After a 24 h recovery period, a 5-fold increase in the number of 53BP1 foci-containing cells (P ≤ 0.001) was observed in formaldehyde-treated wild-type and BLM-deficient cells compared to their respective untreated populations. In order to study the repair kinetics of these DSBs, 53BP1 foci accumulation was studied after 48 h recovery post formaldehyde treatment. Although both cell lines exhibited similar levels of accumulation of DSBs following formaldehyde treatment, their repair kinetics were notably different. After 48 h recovery, the number of wild-type cells with 53BP1 foci declined significantly (P ≤ 0.01; Figs. S2A and 4A), while 53BP1 positive BLM-deficient cells did not show a statistically significant reduction (P > 0.05; compare 24R and 48R in Fig. 4A). This observation was further validated by studying the repair kinetics of γH2AX foci formation following formaldehyde treatment. Similar to 53BP1 induction at 24 h recovery, both the cell lines had a comparable number (~70%) of γH2AX positive (>10 foci/cell) cells following a 24 h recovery after formaldehyde treatment (Fig. 4B). At 48 h recovery, wild-type cells showed ~a 20% drop in γH2AX positive cells; however, the number of BLM-deficient cells positive for γH2AX foci remained nearly unchanged at the 48R time-point (Fig. 4B). Together, these 53BP1 and γH2AX results suggest that BLM is not required for formaldehyde-induced DSB formation; however, loss of BLM results in a delayed DSB repair response.
Fig. 4.
Accumulation and co-localization of 53BP1 and BLM foci following formaldehyde treatment. Kinetics of 53BP1 (A) and H2AX (B) foci accumulation in wild-type and BLM-deficient cells following formaldehyde treatment. Cells were treated with formaldehyde (300 μM for 4 h), fixed following a 24 and 48 h recovery (24R and 48R, respectively), and stained with anti-53BP1 or γH2AX antibody. The mean and standard deviation from three or more independent experiments are shown. Significant difference (Student’s t-test, **P ≤ 0.01); NS, not significant. (C) Co-localization of 53BP1 and BLM in wild-type cells following formaldehyde exposure. Under the same treatment conditions as used for 53BP1 staining, cells were co-stained with anti-BLM and anti-53BP1 antibodies to follow the co-localization of the two proteins over a 4 day recovery period (R). For each sample, 100 cells were counted in every experiment. The mean of three or more independent experiments are shown. “ut” denotes untreated cells.
To further study the involvement of BLM protein in formaldehyde-induced DSB repair, we measured the kinetics of BLM foci accumulation in formaldehyde-treated wild-type cells. Unlike the trend observed for 53BP1 enrichment, BLM foci appeared much later during recovery from formaldehyde-induced DNA damage (Figs. S2B and 4C). Specifically, although wild-type cells containing 53BP1 foci declined by the 48 h recovery time, BLM foci continued to amass throughout the first 72 h post recovery time point and dropped only slightly by 96 h post recovery.
In addition, we investigated the focal co-localization of 53BP1 and BLM proteins in response to formaldehyde-induced DNA damage. In untreated cells, 53BP1 and BLM foci rarely co-localized. However, as the recovery progressed, a time-dependent enhancement of 53BP1 and BLM co-localized foci was observed, with the maximal co-localization seen at 72 h post formaldehyde treatment (Fig. 4C). It was also noted that while the appearance of the BLM foci remained unchanged throughout the recovery, 53BP1 foci appeared in various forms corresponding to different time points. At the 24 h recovery time point, 53BP1 foci observed were small and numerous, often exceeding more than 100 foci per nuclei. As the recovery process continued, the small foci gave way to gradually larger, less abundant foci that co-localized with BLM protein (Fig. S2C). Together, these data establish a time-dependent enrichment of both 53BP1 and BLM foci in response to formaldehyde exposure where 53BP1 focal accumulation precedes the recruitment of BLM foci at the sites of DSBs.
3.5. Accumulation of BLM, but not 53BP1, foci occurs in an ATM-dependent manner
It has previously been shown that ATM regulates the accumulation and phosphorylation of both 53BP1 and BLM proteins in response to genotoxic stress [31,32]. In order to determine whether formaldehyde-induced stress signaling was transmitted through ATM kinase, the survival response of ATM-proficient (GM639) and ATM-deficient (GM05849) cells to formaldehyde treatment was examined. Cells were treated with the indicated concentrations of formaldehyde for 4 h and cell viability was determined by colony forming assays. ATM-deficient cells displayed sensitivity to formaldehyde (Fig. 5A) that was comparable to the formaldehyde sensitivity of BLM-deficient cells. In addition, a time-dependent increase in total ATM protein levels was observed following formaldehyde treatment in wild-type cells (Fig. 5B). These findings are in accordance with previously published data suggesting a role for ATM in response to formaldehyde/DPC-inducing agents [14]; however, the prior study did not provide any direct evidence supporting the role of ATM protein in formaldehyde-induced damage response [14].
Fig. 5.
Recruitment of 53BP1 and BLM foci at the sites of formaldehyde-induced DNA damage in ATM-deficient cells. (A) Survival of wild-type and ATM-deficient cells was determined by colony forming assay. Cells were treated with the indicated concentrations of formaldehyde for 4 h. (B) Western blot analysis of ATM protein levels in wild-type cells treated with formaldehyde (300 μM for 4 h), and allowed to recover (R) for the indicated time period. (C) Accumulation of 53BP1 foci in formaldehyde-treated ATM-deficient cells. Cells were treated with formaldehyde (300 μM for 4 h), fixed following a 24 and 48 h recovery (24R and 48R, respectively), and stained with anti-53BP1 antibody. Significant difference (Student’s t-test, ***P ≤ 0.001); NS, not significant. (D) Quantification of BLM foci in ATM-deficient cells with or without formaldehyde exposure. Cells were treated with formaldehyde (300 μM for 4 h) and fixed following a 96 h recovery (96R). For each sample, 200–300 cells were counted in every experiment. The mean and standard deviation from three or more independent experiments are shown.
Thus, we next examined if ATM protein is an upstream effector regulating 53BP1 and BLM localization to the formaldehyde-induced DSB sites. In order to address if the recruitment of 53BP1 and BLM foci following formaldehyde treatment is regulated by ATM, immunofluorescence experiments were performed with ATM-deficient cells to visualize the formation/induction of 53BP1 and BLM foci following formaldehyde exposure. ATM-deficient cells were treated with formaldehyde for 4 h, followed by a 24 and 48 h recovery period. Similar to wild-type and BLM-deficient cells that showed a five-fold induction of 53BP1 foci 48 h following formaldehyde exposure, ATM-deficient cells, compared to the untreated population, exhibited a seven-fold increase in 53BP1 foci accumulation under the same conditions (Fig. 5C). Analogous to BLM-deficient cells, ATM-deficient cells exhibited no significant repair of formaldehyde-induced DNA DSBs as measured by the persistence of 53BP1 foci (P > 0.05; compare 24R and 48R in Fig. 5C). In parallel, γH2AX foci induction was examined in ATM-deficient cells following formaldehyde treatment. Similar to the trend observed for 53BP1, an induction of γH2AX foci was observed to be peaking after 24 h recovery following a 4 h formaldehyde treatment, implying that accumulation of γH2AX foci following formaldehyde exposure is not an ATM-dependent process (Fig. S3). Thus, it was concluded that formaldehyde-induced accumulation of 53BP1 and γH2AX foci is not regulated by ATM. These findings raise two possibilities. Either ATM works together with 53BP1 and/or γH2AX in the formaldehyde-induced DNA damage response pathway where 53BP1/γH2AX act upstream of ATM. This scenario would be analogous to the IR-induced damage response in which 53BP1 functions as an upstream activator of ATM [33]. Alternatively, formaldehyde-induced accumulation of 53BP1 and/or γH2AX foci is regulated by another kinase. In this regard, we cannot exclude the possibility that 53BP1/γH2AX may be recruited to the sites of DNA DSBs via an ATR-dependent process, as has been observed for UV-induced DNA damage [31].
Next, to determine whether the BLM protein is a downstream effector of ATM in the formaldehyde-induced damage response pathway, immunofluorescence experiments were performed to study the formation of BLM foci in formaldehyde-treated ATM-deficient cells following a 48 and 96 h recovery. Untreated ATM-deficient cells exhibit high endogenous levels of BLM foci compared to untreated wild-type cells, suggesting that replication-associated genomic instability in these cells might be promoting localization of BLM to the sites of damage. However, formaldehyde-treated ATM-deficient cells failed to show any induction of BLM foci formation above the basal levels. Similar trends were observed following 48 h (data not shown) and 96 h recovery (Fig. 5D). These data suggest that a functional ATM protein is required for the recruitment of BLM protein to the sites of formaldehyde-induced DNA DSBs.
3.6. BLM-deficient cells have elevated levels of radials following formaldehyde treatment
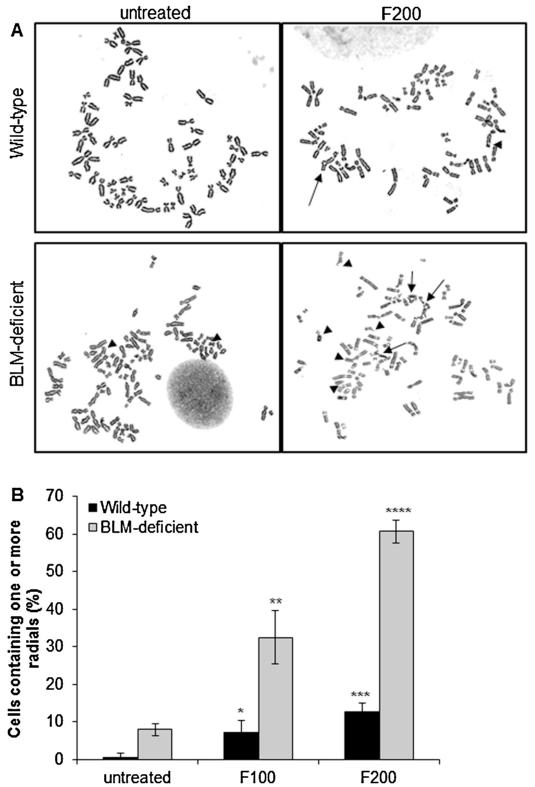
Alterations in S and G2/M phases of the cell cycle are an indication of perturbed replication and chromosome segregation defects that may be accompanied by DNA breaks and chromosomal aberrations, such as radials. Additionally, the appearance of larger 53BP1 foci in response to formaldehyde treatment is consistent with the IR-induced 53BP1 foci that have been shown to represent clustered DNA lesions [34]. These clustered DNA lesions, if left unrepaired, result in chromosomal breaks and radial formation. Although the precise mechanism leading to the formation of radials remains unknown, they are believed to appear secondarily to DSB formation [35,36]. In order to determine if formaldehyde-induced DSB generation was accompanied by radial formation, wild-type and BLM-deficient cells were treated with formaldehyde and examined for radial formation. Untreated wild-type cells exhibited a very low level of radial formation (<1%) whereas, approximately 8% of untreated BLM-deficient cells contained radials, indicating that loss of BLM function alone results in increased genome instability (Fig. 6). Following formaldehyde treatment, radials appeared in a dose-dependent manner in both wild-type and BLM-deficient cells. However, relative to wild-type, BLM-deficient cells exhibited dramatically elevated levels of formaldehyde-induced radials (15% versus 60% at 200 μM formaldehyde concentration, Fig. 6). These observations suggest that following formaldehyde-induced DNA damage, BLM activity is essential for maintaining genomic integrity by suppressing the formation of aberrant chromosomal intermediates, possibly resulting from inaccurate rejoining of DSBs.
Fig. 6.
Formaldehyde-induced radial formation in wild-type and BLM-deficient cells. (A) Cells were assessed for radial formation at the indicated concentrations following a 4 h formaldehyde treatment and a 48 h recovery period. Representative images of radials and chromosomal breaks are shown. Arrowheads point to chromosomal breaks, while arrows indicate radials. (B) Quantitation of cells containing radials. The mean and standard deviation from three or more independent experiments are shown. Significant difference between treated versus untreated populations (Student’s t-test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001). Abbreviations used: 53BP1, p53 binding protein; ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related; BLM, bloom protein; CHO, Chinese Hamster Ovary; DSB, double-strand break; DPC, DNA-protein crosslink.
4. Discussion
The cellular responses associated with formaldehyde-induced DNA damage are emerging as complex, multi-pathway processes (reviewed in [37]). Given the multifunctional nature of DNA repair proteins, elucidating their specific contributions and associations with other proteins is needed to delineate the different pathways related to formaldehyde-induced damage and repair responses. In this study, we demonstrate a role for the BLM helicase in mitigating formaldehyde-induced genomic instability. Though reactive aldehydes, including formaldehyde, share the ability to induce DNA-protein crosslinks, the protective role of BLM is not common to all the aldehydes, as Langevin et al. [38] have shown that BLM-deficient DT40 cells are not sensitive to acetaldehyde. The differential sensitivity of BLM-deficient cells to aldehydes could be attributed to either (1) the formation of different species of DNA-protein crosslinks in response to different aldehydes, or (2) the aldehydes induce different primary or secondary lesions that also contribute to cellular cytotoxicity.
This work provides strong evidence to support a BLM-mediated DNA damage response invoked by formaldehyde for (1) formaldehyde-induced DSB repair and suppression of radial formation and (2) accurate and timely progression through mitosis. Interestingly, cells deficient in the Werner helicase (WRN), also a member of the recQ family, show increased sensitivity to formaldehyde exposure but the mechanism of cytotoxicity appears to be different from that in the BLM-deficient cells (manuscript in preparation).
A collaborative role for BLM and FANCC has been shown during mitosis to prevent micro-nucleation and chromosome abnormalities [38]. As would be anticipated if BLM and FANCC were functioning together in the formaldehyde-initiated DNA damage response, FANCC-deficient human cells are hypersensitive to levels of formaldehyde detected in human plasma [13]. Thus, it is possible that a BLM/FANCC-mediated pathway plays a key role in prevention of formaldehyde-induced chromosome instability and ploidy changes that may otherwise represent an early step toward development of cancer.
Consistent with our results with human BLM-deficient cells, we have previously demonstrated that mammalian XPF-deficient cells exhibit an enhanced formaldehyde-induced cell cycle arrest, increased radials, and higher ploidy levels [11]. Similar to the contribution of BLM in formaldehyde-induced DSB repair, the function of XPF endonuclease was not required for the formation of formaldehyde-induced DSBs; however, the repair of these DSBs appeared to be XPF-dependent. Considering similar consequences of formaldehyde exposure are observed in XPF- and BLM-deficient cells, it remains to be determined if these two proteins participate in the same repair pathway. It is well known that BLM and XPF proteins are two key players of the ICL repair pathway where, the XPF-ERCC1 heterodimer is required for the incision step, while BLM is believed to be responsible for down-regulating homologous recombination [39] and/or facilitating nuclease events by unwinding damaged DNA substrates [40]. Thus, even though the primary lesions generated by an ICL-inducing agent (DNA-DNA crosslinks) versus formaldehyde (DNA-protein crosslinks) are different; there exists a possibility that the downstream events involved in the ICL- and formaldehyde-induced DNA damage and repair responses share common intermediates.
In the cellular response to DNA damage, BLM physically interacts with several DNA repair proteins (ATM, NBS, BRCA1, MSH6, Rad50, RFC, and MLH1) forming a super-complex called BRCA1-associated genome surveillance complex (BASC) [41]. In response to recognition of DSBs by γH2AX and 53BP1, it is well known that BLM acts as an important responder protein that can be regulated by either ATM or ATR sensor kinases via the signal transmitted by transducer proteins. The current work demonstrates that the recruitment of formaldehyde-induced BLM foci requires a functional ATM gene. This observation is consistent with previous reports showing that treatments with ionizing radiation and replication inhibitors result in phosphorylation of BLM in an ATM-dependent manner [32,42,43]. Thus, despite the differences between the types of DNA lesions generated by the different DNA damaging agents, a point of convergence appears to be in the recruitment of the BLM protein to the sites of DSBs in an ATM-dependent manner. However, ATM-dependent targeting of BLM in a formaldehyde-specific cellular response does not exclude a possible role for ATR kinase. Specifically, the G2/M cell cycle checkpoint proteins and the downstream substrates of ATM and ATR, Chk1 and Chk2 have been shown to be phosphorylated [14,44], followed by an activation of the downstream effector, cyclin B1 in response to formaldehyde treatment [11].
In conclusion, this study demonstrates a critical role for BLM in mitigating formaldehyde-induced genomic instability. Additionally, we provide evidence for collaboration between BLM and other players, ATM and 53BP1, that are involved in a formaldehyde-induced stress response. Further studies are required to address the specific roles and sequential recruitment of these factors in the formaldehyde-induced damage response pathway.
Supplementary Material
Acknowledgements
We thank R. Stephen Lloyd and Robb E. Moses for invaluable discussions and critical reading of the manuscript. We thank Adrienne Lloyd, Amy Hanlon Newell, and Susan Olson for their assistance with the flow cytometry and cytogenetic experiments, respectively. We are also thankful to Timothy Murphy (Olympus America Inc.) for his help with live-cell imaging experiments. This work was supported by NCI grant R01 CA106858.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2015.02.010.
References
- [1].Final Report on Carcinogens Background Document for Formaldehyde. Rep Carcinog Backgr Doc. 2010:i–512. [PubMed] [Google Scholar]
- [2].Formaldehyde. Rep Carcinog. 2011:195–205. [PubMed] [Google Scholar]
- [3].Hauptmann M, Lubin JH, Stewart PA, Hayes RB, Blair A. Mortality from solid cancers among workers in formaldehyde industries. Am. J. Epidemiol. 2004;159:1117–1130. doi: 10.1093/aje/kwh174. [DOI] [PubMed] [Google Scholar]
- [4].Beane Freeman LE, Blair A, Lubin JH, Stewart PA, Hayes RB, Hoover RN, Hauptmann M. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the National Cancer Institute Cohort. J. Natl. Cancer Inst. 2009;101:751–761. doi: 10.1093/jnci/djp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang L, Steinmaus C, Eastmond DA, Xin XK, Smith MT. Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutat. Res. 2009;681:150–168. doi: 10.1016/j.mrrev.2008.07.002. [DOI] [PubMed] [Google Scholar]
- [6].Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 2010;120(1):S130–S145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Craft TR, Bermudez E, Skopek TR. Formaldehyde mutagenesis and formation of DNA-protein crosslinks in human lymphoblasts in vitro. Mutat. Res. 1987;176:147–155. doi: 10.1016/0027-5107(87)90262-4. [DOI] [PubMed] [Google Scholar]
- [8].Crosby RM, Richardson KK, Craft TR, Benforado KB, Liber HL, Skopek TR. Molecular analysis of formaldehyde-induced mutations in human lymphoblasts and E. coli. Environ. Mol. Mutagen. 1988;12:155–166. doi: 10.1002/em.2860120202. [DOI] [PubMed] [Google Scholar]
- [9].Casanova M, Morgan KT, Gross EA, Moss OR, Heck HA. DNA-protein crosslinks and cell replication at specific sites in the nose of F344 rats exposed subchronically to formaldehyde. Fundam. Appl. Toxicol. 1994;23:525–536. doi: 10.1006/faat.1994.1137. [DOI] [PubMed] [Google Scholar]
- [10].Casanova M, Morgan KT, Steinhagen WH, Everitt JI, Popp JA, Heck HD. Covalent binding of inhaled formaldehyde to DNA in the respiratory tract of rhesus monkeys: pharmacokinetics, rat-to-monkey interspecies scaling, and extrapolation to man. Fundam. Appl. Toxicol. 1991;17:409–428. doi: 10.1016/0272-0590(91)90230-2. [DOI] [PubMed] [Google Scholar]
- [11].Kumari A, Lim YX, Newell AH, Olson SB, McCullough AK. Formaldehyde-induced genome instability is suppressed by an XPF-dependent pathway. DNA Repair (Amst.) 2012;11:236–246. doi: 10.1016/j.dnarep.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, Ide H. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- [13].Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- [14].Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst.) 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- [17].German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- [18].Tikoo S, Sengupta S. Time to bloom. Genome Integr. 2010;1:14. doi: 10.1186/2041-9414-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- [21].Hickson ID, Davies SL, Li JL, Levitt NC, Mohaghegh P, North PS, Wu L. Role of the Bloom’s syndrome helicase in maintenance of genome stability. Biochem. Soc. Trans. 2001;29:201–204. doi: 10.1042/0300-5127:0290201. [DOI] [PubMed] [Google Scholar]
- [22].Hemphill AW, Akkari Y, Newell AH, Schultz RA, Grompe M, North PS, Hickson ID, Jakobs PM, Rennie S, Pauw D, Hejna J, Olson SB, Moses RE. Topo IIIalpha and BLM act within the Fanconi anemia pathway in response to DNA-crosslinking agents. Cytogenet. Genome Res. 2009;125:165–175. doi: 10.1159/000230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mohaghegh P, Karow JK, Brosh RM, Jr., Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rassool FV, North PS, Mufti GJ, Hickson ID. Constitutive DNA damage is linked to DNA replication abnormalities in Bloom’s syndrome cells. Oncogene. 2003;22:8749–8757. doi: 10.1038/sj.onc.1206970. [DOI] [PubMed] [Google Scholar]
- [25].Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- [26].Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell. Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell. Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br. J. Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- [31].Jowsey P, Morrice NA, Hastie CJ, McLauchlan H, Toth R, Rouse J. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst.) 2007;6:1536–1544. doi: 10.1016/j.dnarep.2007.04.011. [DOI] [PubMed] [Google Scholar]
- [32].Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19:5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- [33].Mochan TA, Venere M, DiTullio RA, Jr., Halazonetis TD. 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst.) 2004;3:945–952. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]
- [34].Asaithamby A, Chen DJ. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat. Res. 2010;711:87–99. doi: 10.1016/j.mrfmmm.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].McCabe KM, Olson SB, Moses RE. DNA interstrand crosslink repair in mammalian cells. J. Cell. Physiol. 2009;220:569–573. doi: 10.1002/jcp.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- [37].Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AM. Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 2011;711:113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- [38].Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- [39].Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM., Jr. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 2011;30:692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- [42].Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF. Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangiectasia-mutated protein, ATM. J. Biol. Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- [43].Ababou M, Dumaire V, Lecluse Y, Amor-Gueret M. Bloom’s syndrome protein response to ultraviolet-C radiation and hydroxyurea-mediated DNA synthesis inhibition. Oncogene. 2002;21:2079–2088. doi: 10.1038/sj.onc.1205246. [DOI] [PubMed] [Google Scholar]
- [44].Wong VC, Cash HL, Morse JL, Lu S, Zhitkovich A. S-phase sensing of DNA-protein crosslinks triggers TopBP1-independent ATR activation and p53-mediated cell death by formaldehyde. Cell Cycle. 2012;11:2526–2537. doi: 10.4161/cc.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.