Abstract
Background
The red turpentine beetle (RTB), Dendroctonus valens LeConte (Coleoptera: Curculionidae, Scolytinae), is a destructive invasive pest of conifers which has become the second most important forest pest nationwide in China. Dendroctonus valens is known to use host odors and aggregation pheromones, as well as non-host volatiles, in host location and mass-attack modulation, and thus antennal olfaction is of the utmost importance for the beetles’ survival and fitness. However, information on the genes underlying olfaction has been lacking in D. valens. Here, we report the antennal transcriptome of D. valens from next-generation sequencing, with the goal of identifying the olfaction gene repertoire that is involved in D. valens odor-processing.
Results
We obtained 51 million reads that were assembled into 61,889 genes, including 39,831 contigs and 22,058 unigenes. In total, we identified 68 novel putative odorant reception genes, including 21 transcripts encoding for putative odorant binding proteins (OBP), six chemosensory proteins (CSP), four sensory neuron membrane proteins (SNMP), 22 odorant receptors (OR), four gustatory receptors (GR), three ionotropic receptors (IR), and eight ionotropic glutamate receptors. We also identified 155 odorant/xenobiotic degradation enzymes from the antennal transcriptome, putatively identified to be involved in olfaction processes including cytochrome P450s, glutathione-S-transferases, and aldehyde dehydrogenase. Predicted protein sequences were compared with counterparts in Tribolium castaneum, Megacyllene caryae, Ips typographus, Dendroctonus ponderosae, and Agrilus planipennis.
Conclusion
The antennal transcriptome described here represents the first study of the repertoire of odor processing genes in D. valens. The genes reported here provide a significant addition to the pool of identified olfactory genes in Coleoptera, which might represent novel targets for insect management. The results from our study also will assist with evolutionary analyses of coleopteran olfaction.
Introduction
A sophisticated olfactory system is crucial to insects for survival and reproduction [1,2]. Antennae are the primary olfactory sensors of insects, where odorant messages (such as host volatiles, pheromones, or non-host volatiles) are perceived and subsequently translated into physiological signals that ultimately influence the insect’s behavior [3]. Antennae are accordingly well-equipped with a wide variety of sensilla. These sensilla are small sensory hair structures in which olfactory receptor neurons (ORNs) extend dendrites into the antennal lymph, where peripheral olfactory signal transduction events occur [4]. The ORNs act as biological transducers that convert ecologically relevant volatile signals into a sensory input [3]. The entire olfactory system is heavily dependent on the types of receptors expressed on peripheral ORNs [2]. The ability of the insect’s peripheral system to selectively detect and rapidly inactivate minute amounts of odorants once they have conveyed information is the cornerstone of a sophisticated olfactory system [2]. Diverse peripheral olfactory proteins have been reported to have roles in olfaction. These include the odorant binding proteins (OBPs), chemosensory proteins (CSPs), gustatory receptors (GRs), olfactory receptor proteins (ORs), sensory neuron membrane proteins (SNMPs), and ionotropic receptors (IRs), all of which are involved in different steps in the insect olfactory signal transduction pathway [4].
OBPs are small, hydrophilic proteins that are secreted by the accessory cells and accumulate in the sensillar lymph [2,3]. The soluble OBPs facilitate the transport of odorant molecules through the sensillar lymph and are the liaison between the external environment and Ors [2,4]. CSPs, like OBPs, are another class of small soluble proteins that are expressed at high levels in the sensillar lymph [3,5]. The exact role of CSPs in olfactory transduction remains largely unknown. However, the binding affinity of some CSPs for pheromone molecules supports their putative role in insect olfaction [6–9].
ORs have seven transmembrane domains with inverted membrane topology [10,11] and are embedded in the dendrites of ORNs. ORs are the key players in chemosensory signal transduction processes [4]. Two ORs are required in order to transduce odor-evoked signals, an olfactory receptor coreceptor (Orco) [12,13] and a specific OR, which varies according to ORN type [14]. Orco is both highly conserved across insect orders and widely expressed in the majority of ORNs, acting as an ion channel [15,16]. The individual ORs are associated with odorant-binding specificity [17]. ORs respond to a variety of volatile chemicals, including pheromones, and plant- and microbe-derived compounds [16,18,19].
IRs are relatives of ionotropic glutamate receptors (IGluRs) with atypical binding domains that are conserved across proteosome lineages; IRs are far more ancient than ORs [20]. IRs were recently discovered as another class of insect-specific gene products that are involved in odorant recognition in Drosophila [3]. The IR family contains a conserved subgroup, the antennal IRs, and a species-specific subgroup, the divergent IRs [20]. The antennal IRs are a novel group of chemosensory receptors and are expressed in sensory dendrites [4].
SNMPs were first found in the dendritic membrane of sex pheromone-sensing olfactory neurons (ORNs) of the wild silk moth, Antheraea polyphemus (Saturniidae), [21] and are proposed to play an important role in pheromone reception in insects [22–25]. SNMPs are homologous to the mammalian CD36 protein and have been shown to be required for detecting an aggregation pheromone in Drosophila [22]. SNMP1 and SNMP2 genes has been found in several different coleopteran insects [5,26]. Insects express SNMP1 and SNMP2 in pheromone-sensitive hairs, but in different locations: SNMP1 is specifically expressed in the dendritic membrane of a neuron of an olfactory sensillum, whereas SNMP2 is only found in supporting cells or the sensillar lymph of the antenna [27–29].
The red turpentine beetle (RTB), Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae), is a secondary pest of pines in its native range in North and Central America. Dendroctonus valens was introduced into China in the early 1980s, where it aggressively kills pine species native to China [30–32]. The behavior of D. valens in its native range is clearly different from that in China [31]. Since the outbreaks of D. valens in 1999, it has infected over 5,000,000 ha of pine forest, and more than 10 million Pinus tabuliformis Carr. as well as other pine species such as Pinus bungeana Zucc [31,33–35] and has resulted in unprecedented economic losses. At present, D. valens is the second most important forest pest nationwide in China [31] and its spread continues. As with most insect species, antennal olfaction is of the utmost importance in D. valens fitness because the beetle uses aggregation pheromones, as well as host and non-host volatiles in intra- and inter-specific communication [36–50]. However, there is little information on the molecular mechanisms underlying olfaction in D. valens. Investigating the repertoire of odor processing genes involved in D. valens olfaction could provide valuable insights into the molecular mechanisms of insect olfaction, and also identify possible molecular targets that could be manipulated for D. valens control.
Although the genome of Tribolium castaneum has been sequenced, our knowledge of the molecular basis of odorant reception in Coleoptera, the largest insect order, remains relatively limited. Recently, the advent of RNA-Seq approaches (next generation sequencing techniques) triggered an exponential growth in our knowledge of insect olfaction. Genes involved in odor processing in Coleoptera have been identified from insects whose genomes have not been sequenced, such as Ips typographus, Dendroctonus ponderosae [5], and Agrilus planipennis [26]. Additional beetle species need to be investigated to reach a better understanding of the molecular biology of coleopteran and insect olfaction.
Here, we employed a transcriptome approach based on next-generation sequencing of antennae of D. valens to identify the olfactory gene repertoire involved in D. valens’ odor-processing. In this study, we conducted a transcriptome analysis of antennae of adult beetles, and identified 68 putative chemosensory transcripts comprising 21 OBPs, six CSPs, four SNMPs, 22 ORs, four GRs, three IRs, and eight ionotropic glutamate receptors. We also obtained 155 odorant/xenobiotic degradation enzymes from the antennal transcriptome, putatively identified to be involved in olfaction processes including cytochrome P450s, glutathione-S-transferases, and aldehyde dehydrogenase.
Results
Transcriptome Sequencing and Assembly
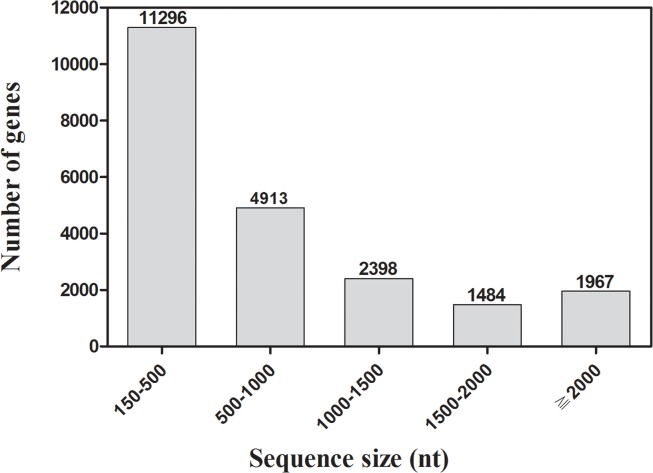
A total of approximately 51 million reads (4.86 Gb) were obtained from a D. valens antennaapp:addword:antenna sample by the Illumina HiSeq 2000 platform, which were assembled into 39,831 contigs with a mean length of 409 bp and an N50 length of 814 bp. After clustering and redundancy filtering, we finally acquired 22,058 unigenes (4,281 clusters and 17,777 singletons) with a mean length of 828 bp and a N50 length of 1360 bp. Of the 22,058 unigenes, those with a sequence length of more than 500 bp accounted for 48.79% of the transcriptome assembly (Fig 1). All the unigenes are referred to as transcripts hereafter and given a unique unigene ID.
Fig 1. Distribution of unigene size in the D. valens transcriptome assembly.
Homology Analysis and Gene Ontology (GO) Annotation
Among 22,058 transcripts, 15,531 (70.40%) were matched by the BlastX homology search to entries in the NCBI non-redundant (nr) protein database with a cut-off E-value of 10−5. The species distribution of the best match result for each sequence is shown in Fig 2. Overall, there is a strong match preference with T. castaneum genes, comprising 69.10%. Interestingly, there was only an 18.29% similarity of D. valens antennal transcripts to those of the congener D. ponderosae. However, this result does not mean that the identified transcripts in D. valens share more similarity with T. castaneum transcripts than D. ponderosae transcripts. Instead, it is likely due to the availability of more T. castaneum than D. ponderosae sequences in the NCBI protein database used in the homology analysis. The protein number of T. castaneum in the NCBI database is 36,478, 1.2 times higher than that of D. ponderosae (29,316). With more protein sequences annotated from D. ponderosae, we believe that more sequences in D. valens will align to those of D. ponderosae.
Fig 2. Percentage of homologous hits of the D. valens transcripts to other insect species.
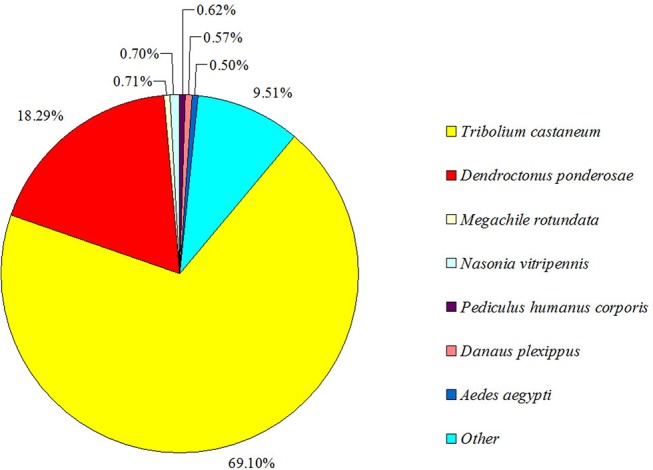
The D. valens transcripts were searched by BlastX against the non-redundancy protein database with a cutoff E-value of 10−5. Species which have more than 0.5% matching hits to the D. valens transcripts are shown.
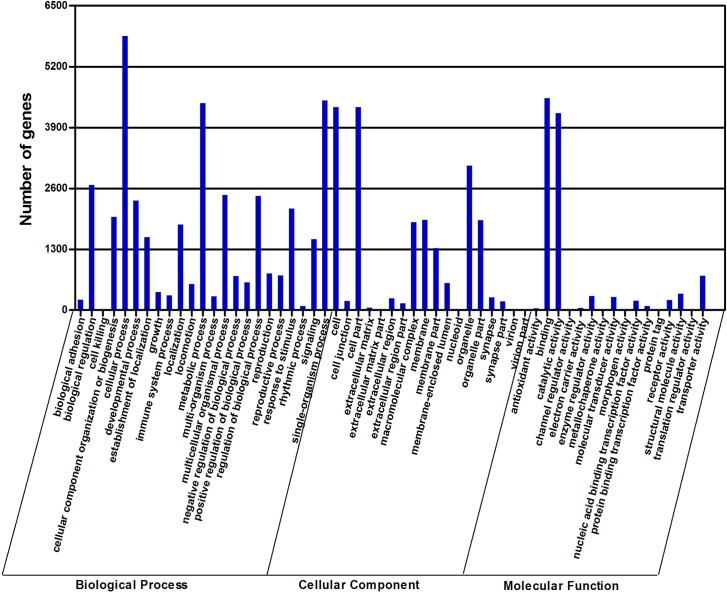
The gene functional annotation was first performed by GO annotation using Blast2GO. Of 22,058 transcripts, 8,743 (39.64%) could be annotated based on sequence homology. Because one transcript could align to more than one biological process, the 8,743 transcripts were assigned to the biological process category (38,416 alignments), cellular component category (20,508 alignments), and molecular function category (11,016 alignments). The major GO terms associated with molecular function were binding (41.09%) and catalytic activity (38.19%), which potentially reflects the metabolic processes of the antennal tissue (Fig 3). Cellular processes (15.23%), single-organism processes (11.66%), and metabolic processes (11.50%) were the main subcategories of biological processes, indicative of the important metabolic activities within D. valens antennae (Fig 3 and S1 Table). Under the category of cellular components, cell (21.11%) and cell parts (21.11%) were among the most highly represented subcategories (Fig 3 and S1 Table).
Fig 3. Gene ontology (GO) classification of the D. valens transcripts with Blast2GO program.
Identification of Putative Olfactory Genes
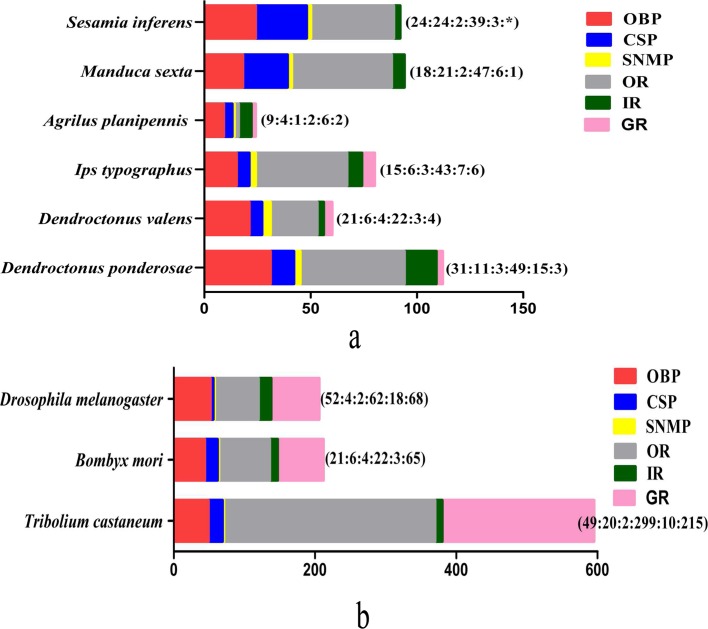
Further analysis on the transcriptome of D. valens focused on gene families associated with olfactory processing. By homology analysis, we identified 68 putative odor-reception genes, including 21 OBPs, six CSPs, four SNMPs, 22 ORs, four GRs, three IRs, and eight IGluRs. As well, 155 putative odorant/xenobiotic degradation enzymes were found (Table 1). The number of major olfactory genes obtained in the current study was larger than that of A. planipennis [26], but less than that of D. ponderosae and I. typographus (Fig 4A and 4B).
Table 1. Summary of candidate genes from the antennal transcriptome of Dendroctonus valens.
| Candidate genes | # in occurrence |
|---|---|
| Odor-reception | |
| Odor binding proteins | 21 |
| Odorant receptors | 22 |
| Ionotropic receptor | 3 |
| Ionotropic glutamate receptors | 8 |
| Gustatory receptors | 4 |
| Sensory neuron membrane proteins | 4 |
| Chemsensory proteins | 6 |
| Odor/xenobiotic degradation | |
| Cytochrome P450s | 49 |
| Glutathione S-transferases | 11 |
| Esterases | 66 |
| Aldehyde dehydrogenases | 14 |
| Epoxide hydrolases | 4 |
| Catalases | 3 |
| Superoxide dismutase | 5 |
| Glutathione peroxidase | 3 |
Fig 4. The number of chemosensory genes in different insect species, obtained from antenna transcriptome (4a) or genome (4b).
The digits by the histogram bars represent number of chemosensory genes in different subfamilies (OBP:CSP:SNMP:OR:IR). The data were obtained from the current study for D. valens and from references [7,20,24,91] for Drosophila melanogaster, [92] for Sesamia inferens,[20,24,62,93–95] for Bomby mori, [7,20,24,91] for Tribolium castaneum,[5] for I. typographus and D. ponderosae, [1] for Manduca sexta, [26] for Agrilus planipennis.
Identification of Putative Odorant-binding Proteins
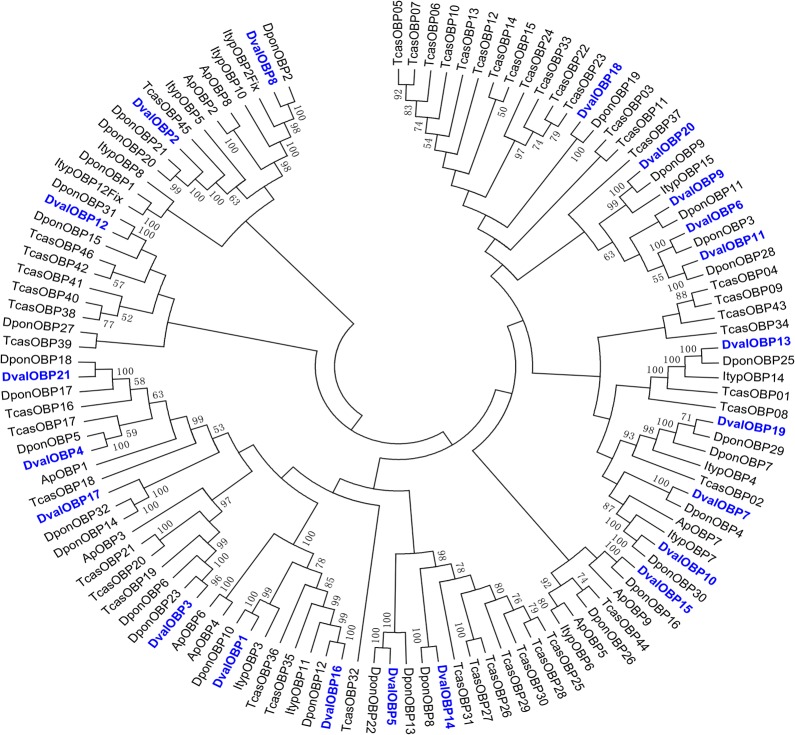
We identified 21 transcripts encoding putative OBPs in the D. valens antennal transcriptome. The number of putative OBP-coding genes in D. valens is less than that of D. ponderosae (31), but more than I. typographus (15). Sequence analysis identified 18 unigenes with a full length ORF (Open Reading Frame) with predicted signal peptide sequences (Table 2). Twenty of the 21 putative OBPs had high similarity to known coleopteran OBPs, (Table 2). DvalOBP3, 4, 7, 10, 14, 16, 18, and 21 had very high similarity with DponOBP6, 5, 4, 30, 8, 12, 19, and 18, respectively, with at least 95% identity (Table 2). A phylogenetic tree based on the neighbor-joining method is shown in Fig 5. Among the 21 putative OBPs, 19 sequences were clustered with at least one coleopteran orthologous gene (Fig 5). Not surprisingly, the identified DvalOBPs sequences were mostly clustered together with DponOBPs, and fourteen OBP orthologous pairs shared high amino acid identity (>85%) between the two species (Fig 5 and Table 2). The amino acid sequences of all OBPs are listed in S1 Fig.
Table 2. The Blastx match of D. valens putative OBPs,CSPs and SNMPs genes.
| Gene | Gene | ORF | Complete | Signal | Best Blastx Match | ||||
|---|---|---|---|---|---|---|---|---|---|
| Name | ID | Length(bp) | ORF | Peptide | Name | Acc.number | Species | E value | Identity(%) |
| Odorant Binding Protein(OBP) | |||||||||
| OBP1 | 455 | 438 | Yes | 1–23 | odorant-binding protein 10 | AFI45063.1 | [Dendroctonus ponderosae] | 6.00E-95 | 91 |
| OBP2 | 840 | 726 | Yes | 1–19 | odorant-binding protein 21 | AGI05159.1 | [Dendroctonus ponderosae] | 1.00E-120 | 87 |
| OBP3 | 849 | 417 | Yes | 1–20 | odorant-binding protein 6 | AGI05177.1 | [Dendroctonus ponderosae] | 1.00E-75 | 95 |
| OBP4 | 10282 | 408 | Yes | 1–19 | odorant-binding protein 5 | AFI45059.1 | [Dendroctonus ponderosae] | 2.00E-72 | 95 |
| OBP5 | 10284 | 405 | Yes | 1–23 | odorant-binding protein 22 | AGI05180.1 | [Dendroctonus ponderosae] | 5.00E-79 | 93 |
| OBP6 | 1456 | 426 | Yes | 1–20 | odorant-binding protein 3 | AGI05174.1 | [Dendroctonus ponderosae] | 7.00E-85 | 82 |
| OBP7 | 14597 | 222 | No | 0 | odorant-binding protein 4 | AGI05167.1 | [Dendroctonus ponderosae] | 3.00E-52 | 99 |
| OBP8 | 5689 | 522 | No | 0 | odorant-binding protein 2 | AGI05158.1 | [Dendroctonus ponderosae] | 2.00E-109 | 94 |
| OBP9 | 5818 | 395 | Yes | 1–17 | odorant-binding protein 28 | AGI05178.1 | [Dendroctonus ponderosae] | 3.00E-10 | 32 |
| OBP10 | 7393 | 402 | Yes | 1–18 | odorant-binding protein 30 | AGI05176.1 | [Dendroctonus ponderosae] | 4.00E-77 | 98 |
| OBP11 | 7769 | 507 | No | 0 | odorant-binding protein 28 | AGI05178.1 | [Dendroctonus ponderosae] | 9.00E-63 | 93 |
| OBP12 | 8450 | 387 | Yes | 1–24 | odorant-binding protein 31 | AGI05165.1 | [Dendroctonus ponderosae] | 3.00E-67 | 93 |
| OBP13 | 8460 | 447 | Yes | 1–19 | odorant binding protein | ADY17884.1 | [Spodoptera exigua] | 1.00E-16 | 46 |
| OBP14 | 9456 | 475 | Yes | 1–27 | odorant-binding protein 8 | AGI05175.1 | [Dendroctonus ponderosae] | 1.00E-120 | 96 |
| OBP15 | 9499 | 354 | Yes | 1–17 | odorant-binding protein 16 | AGI05186.1 | [Dendroctonus ponderosae] | 7.00E-72 | 91 |
| OBP16 | 9616 | 426 | Yes | 1–18 | odorant-binding protein 12 | AFI45058.1 | [Dendroctonus ponderosae] | 1.00E-93 | 95 |
| OBP17 | 9643 | 435 | Yes | 1–21 | odorant-binding protein 13 | AFI45057.1 | [Dendroctonus ponderosae] | 9.00E-83 | 88 |
| OBP18 | 9644 | 426 | Yes | 1–18 | odorant-binding protein 19 | AGI05183.1 | [Dendroctonus ponderosae] | 1.00E-65 | 95 |
| OBP19 | 9906 | 411 | Yes | 1–19 | odorant-binding protein 29 | AGI05182.1 | [Dendroctonus ponderosae] | 3.00E-61 | 94 |
| OBP20 | 9965 | 423 | Yes | 1–20 | odorant-binding protein 9 | AGI05185.1 | [Dendroctonus ponderosae] | 2.00E-80 | 90 |
| OBP21 | 9974 | 408 | Yes | 1–19 | odorant-binding protein 18 | AFI45062.1 | [Dendroctonus ponderosae] | 2.00E-71 | 95 |
| Chemosensory Protein(CSP) | |||||||||
| CSP1 | 10610 | 432 | Yes | 1–26 | chemosensory protein5 | AFI45003.1 | [Dendroctonus ponderosae] | 2.00E-81 | 94 |
| CSP2 | 8762 | 375 | Yes | 1–16 | chemosensory protein 1 | AGI05161.1 | [Dendroctonus ponderosae] | 3.00E-54 | 90 |
| CSP3 | 3805 | 102 | No | 0 | chemosensory protein 2 | AGI05172.1 | [Dendroctonus ponderosae] | 5.00E-33 | 89 |
| CSP4 | 168 | 822 | Yes | 1–18 | chemosensory protein 6 precursor | NP_001039288.1 | [Tribolium castaneum] | 1.00E-49 | 67 |
| CSP5 | 8684 | 423 | No | 0 | chemosensory protein 8 | AGI05164.1 | [Dendroctonus ponderosae] | 2.00E-82 | 96 |
| CSP6 | 11692 | 309 | Yes | 1–23 | chemosensory protein 11 | AGI05163.1 | [Dendroctonus ponderosae] | 3.00E-55 | 94 |
| Sensory Neuron Membrane Protein(SNMP) | |||||||||
| SNMP1 | 561 | 954 | No | No | sensory neuron membrane protein1 | AFI45066.1 | [Dendroctonus ponderosae] | 0 | 94 |
| SNMP1a | 10985 | 1674 | No | No | sensory neuron membrane protein 1a | AGI05171.1 | [Dendroctonus ponderosae] | 0 | 95 |
| SNMP2 | 7572 | 1338 | No | No | sensory neuron membrane protein 2, isoform B | NP_001036593.1 | [Drosophila melanogaster] | 2.00E-83 | 35 |
| SNMP | 11087 | 225 | No | No | sensory neuron membrane protein | AFI45067.1 | [Dendroctonus ponderosae] | 4.00E-39 | 85 |
Fig 5. Phylogenetic tree of putative OBPs from Dendroctonus valens (Dval), Ips typographus (Ityp), Dendroctonus ponderosae (Dpon), Tribolium castaneum (Tcas) and Agrilus planipennis (Ap).
The D. valens translated unigenes are shown in blue. Amino acid sequences are given in S1 Fig. The tree was constructed with MEGA5.0, using the neighbor-joining method. Values indicated at the nodes are bootstrap values based on 1000 replicates, and the bootstrap values below 50% are not shown.
In general, OBPs are divided into different subclasses. The classic OBPs are characterized by a pattern of six conserved cysteines. Multiple amino acid sequence alignment revealed the conserved and defined spacing of six cysteine residues (C1-X15-39-C2-X3-C3-X21-44-C4-X7-12-C5-X8-C6; Fig 6), which are considered to form disulfide bonds that stabilize the three-dimensional structure of the OBP [51].
Fig 6. Multiple sequences alignment of OBPs of Dendroctonus valens (Dval) with other insects OBPs.
Analyses included OBPs and pheromone binding proteins (PBPs). Amino acid sequences are given in S2 Fig.
The amino acid sequences used for multiple sequence alignment are listed in S2 Fig. The Plus-C class has more than 6 cysteines, whereas the Minus-C class has lost cysteine residues, generally C2 and C5. In our study, the members of the Minus-C class were DvalOBP2, 5, 6, 9, 10, 18, 19, and 20 and all of these are missing C2 and C5 cysteine residues, whereas only DvalOBP8 was classified as Plus-C.
Identification of Candidate Chemosensory Proteins
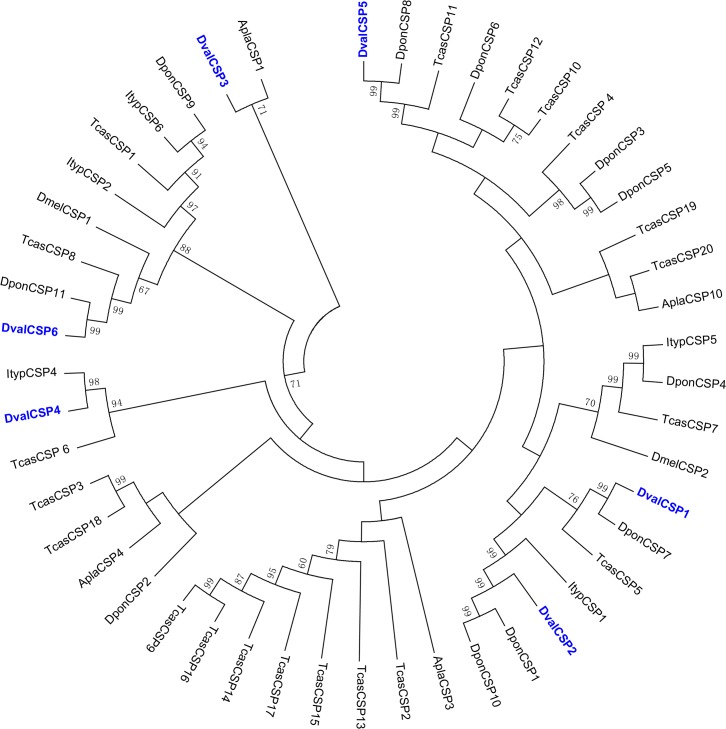
Bioinformatic analysis led to the identification of six different sequences encoding candidate CSPs in D. valens (Table 2), among which four sequences were predicted to be full length and all of them had a signal peptide. We compared all predicted CSPs of D. valens with 42 coleopteran CSPs and two dipteran CSPs in order to reveal the diversity of CSPs within the insect order (Fig 7). The CSPs were dispersed in different branches of the phylogenetic tree (Fig 7). Among the six candidate CSPs, five sequences were clustered together with bark beetles CSPs (Fig 7). The information of all the six CSPs is listed in Table 2. DvalCSP1, 5, and 6 had at least 94% similarity with the corresponding CSPs of D. ponderosae (DponCSP5, 8, and 11; Table 2). The amino acid sequences of all six CSPs are listed in S3 Fig.
Fig 7. Phylogenetic tree of putative CSPs from Dendroctonus valens (Dval), Ips typographus (Ityp), Dendroctonus ponderosae (Dpon), Tribolium castaneum (Tcas) and Drosophila melanogaster (Dmel).
The D. valens translated unigenes are shown in blue. Amino acid sequences are given in S3 Fig. The tree was constructed with MEGA5.0, using the neighbor-joining method. Values indicated at the nodes are bootstrap values based on 1000 replicates, and the bootstrap values below 50% are not shown.
Identification of Putative OR/GR Superfamily Members
Within the D. valens antennal transcriptome, 22 OR candidates were identified. Among these ORs, only three identified DvalOR sequences (Dval 7, 8 and 11) had a full length ORF (Table 3). As expected, 17 of the 22 putative DvalORs shared high amino acid identity with known coleopteran ORs, and six ORs in D. valens (DvalORs1, 3, 14, 15, 16, and 17) had at least 87% similarity with corresponding ORs of D. ponderosae (Table 3).
Table 3. The BlastX match of D. valens putative GRs, IRs and ORs genes.
| Gene | Gene | ORF | Complete | Signal | Best BlastX Match | ||||
|---|---|---|---|---|---|---|---|---|---|
| Name | ID | Length(bp) | ORF | Peptide | Name | Acc.number | Species | E value | Identity(%) |
| Gustatory Receptor(GR) | |||||||||
| GR1 | 14175 | 396 | No | 0 | gustatory receptor 2 | NP_001161916.1 | [Tribolium castaneum] | 2.00E-62 | 73 |
| GR2 | 16566 | 207 | No | 0 | Gustatory receptor 21a, putative | XP_001655150.1 | [Aedes aegypti] | 3.00E-26 | 74 |
| GR3 | 2217 | 375 | Yes | 1–22 | putative gustatory receptor candidate 59 | EHJ69979.1 | [Danaus plexippus] | 2.00E-04 | 74 |
| GR4 | 6503 | 576 | No | 0 | gustatory receptor 101 | EFA02934.1 | [Tribolium castaneum] | 5.00E-14 | 30 |
| Ionotropic Receptor(IR) | |||||||||
| IR1 | 11453 | 171 | No | 0 | ionotropic receptor 8a | AGI05169.1 | [Dendroctonus ponderosae] | 3.00E-31 | 87 |
| IR2 | 12733 | 264 | No | 0 | putative ionotropic receptor IR75, partial | AFC91756.1 | [Cydia pomonella] | 1.00E-10 | 36 |
| IR3 | 67 | 2367 | No | 0 | ionotropic receptor 8a | AGI05169.1 | [Dendroctonus ponderosae] | 0 | 97 |
| Odorant Receptor(OR) | |||||||||
| OR1 | 11271 | 780 | No | 0 | olfactory receptor | AEE62122.1 | [Dendroctonus ponderosae] | 0 | 97 |
| OR2 | 40 | 795 | No | 0 | odorant receptor 20 | AEE63423.1 | [Dendroctonus ponderosae] | 7.00E-173 | 77 |
| OR3 | 2158 | 705 | No | 0 | odorant receptor 18 | AEE62488.1 | [Dendroctonus ponderosae] | 1.00E-155 | 97 |
| OR4 | 5354 | 789 | No | 0 | odorant receptor | XP_001651754.1 | [Aedes aegypti] | 2.00E-13 | 29 |
| OR5 | 8369 | 528 | No | 0 | olfactory receptor | XP_001864544.1 | [Culex quinquefasciatus] | 2.00E-17 | 32 |
| OR6 | 5 | 723 | No | 0 | odorant receptor 2 | ACH69148.1 | [Anopheles stephensi] | 6.00E-06 | 27 |
| OR7 | 13471 | 279 | Yes | 1–20 | odorant receptor 10 | ACH69151.1 | [Anopheles stephensi] | 9.00E-10 | 41 |
| OR8 | 2313 | 300 | Yes | 1–17 | odorant receptor 14 | EFA09245.1 | [Tribolium castaneum] | 4.00E-05 | 42 |
| OR9 | 12394 | 273 | No | 0 | odorant receptor 23 | AGI05173.1 | [Dendroctonus ponderosae] | 8.00E-10 | 37 |
| OR10 | 2425 | 171 | No | 0 | odorant receptor 23 | AGI05173.1 | [Dendroctonus ponderosae] | 7.00E-10 | 44 |
| OR11 | 2712 | 241 | Yes | 1–19 | odorant receptor 23 | AGI05173.1 | [Dendroctonus ponderosae] | 5.00E-10 | 39 |
| OR12 | 4846 | 390 | No | 0 | odorant receptor 23 | AGI05173.1 | [Dendroctonus ponderosae] | 8.00E-22 | 40 |
| OR13 | 5024 | 179 | No | 0 | odorant receptor 23 | AGI05173.1 | [Dendroctonus ponderosae] | 1.00E-12 | 43 |
| OR14 | 2032 | 197 | No | 0 | odorant receptor 10 | AEE61404.1 | [Dendroctonus ponderosae] | 1.00E-45 | 99 |
| OR15 | 9058 | 1215 | No | 0 | odorant receptor 38 | AEE63155.1 | [Dendroctonus ponderosae] | 0.00E+00 | 92 |
| OR16 | 9393 | 378 | No | 0 | odorant receptor 23 | AGI05173.1 | [Dendroctonus ponderosae] | 3.00E-61 | 87 |
| OR17 | 7310 | 792 | No | 0 | odorant receptor 24 | AGI05166.1 | [Dendroctonus ponderosae] | 4.00E-148 | 88 |
| OR18 | 2992 | 267 | No | 0 | odorant receptor 73 | EFA05710.1 | [Tribolium castaneum] | 6.00E-06 | 35 |
| OR19 | 1231 | 195 | No | 0 | odorant receptor 46 | EFA02901.1 | [Tribolium castaneum] | 6.00E-11 | 46 |
| OR20 | 13762 | 705 | No | 0 | odorant receptor 59 | EEZ99171.1 | [Tribolium castaneum] | 2.00E-10 | 48 |
| OR21 | 7442 | 357 | No | 0 | olfactory receptor 60 | NP_001155301.1 | [Bombyx mori] | 9.00E-13 | 32 |
| OR22 | 313 | 798 | No | 0 | odorant receptor 64 | EFA10800.1 | [Tribolium castaneum] | 3.00E-112 | 59 |
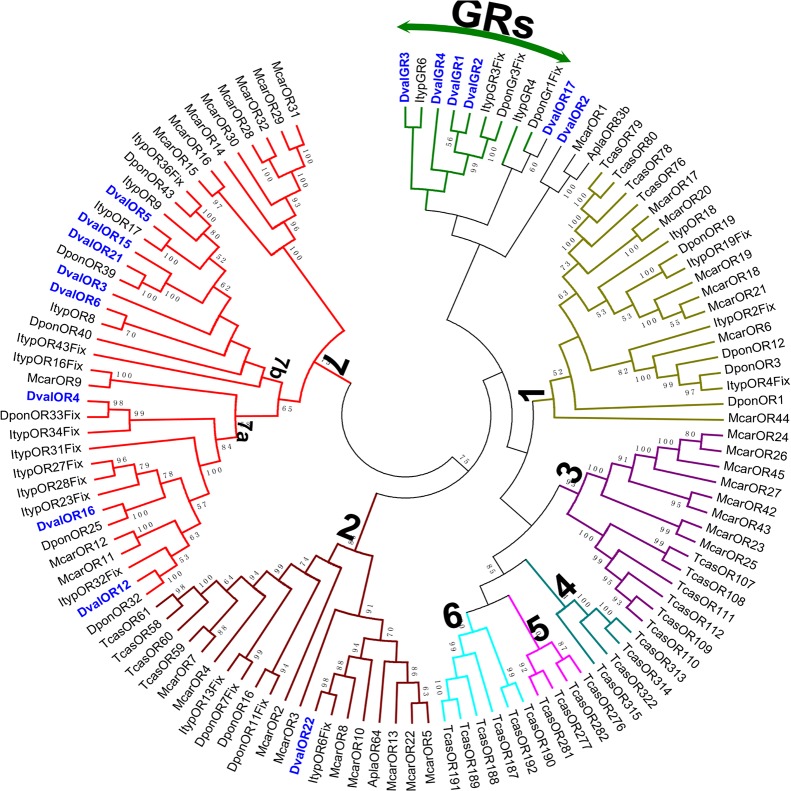
A phylogenetic tree of ORs based on the neighbor-joining method was constructed using protein sequences of ORs from D. valens, two other bark beetles (I. typographus, D. ponderosae), T. castaneum, Megacyllene caryae, and Agrilus planipennis. Eleven DvalORs were excluded from analysis due to insufficiently long amino acid sequences.
ORs are divided into several subgroups of various size and content in the phylogenetic tree (Fig 8). Almost all ORs of D. valens were clustered with at least one bark beetle orthologous gene in the phylogenetic tree except for two ORs (DvalOR2 and DvalOR17). Among them, five ORs from D. valens were clustered with one D. ponderosae orthologous gene.
Fig 8. Phylogenetic tree of putative ORs and GRs from Dendroctonus valens (Dval), Ips typographus (Ityp), Dendroctonus ponderosae (Dpon)and Tribolium castaneum (Tcas).
The D. valens translated unigenes are shown in blue. The branch containing GRs was used as outgroup to root the tree. The different subgroups (numbered 1–7 according to [35,53], and 7a-7b) are discussed in the main text. Originally, DponOR15Fix was found within group 2 [29], as indicated here by the numbers in brackets. Amino acid sequences are given in S4 Fig. The tree was constructed with MEGA5.0, using the neighbor-joining method. Values indicated at the nodes are bootstrap values based on 1000 replicates, and the bootstrap values below 50% are not shown.
The numbering of coleopteran OR subfamilies has been standardized in previous studies [5,51], and numbered from 1 to 7. Groups 1 and 2 included OR representatives from all six species. Receptor group 3 contained ORs only from T. castaneum and M. caryae. Receptor groups 4–6 contained ORs only from T. castaneum. Group 7 contained 11 ORs from M. caryae, but no ORs from T. castaneum or A. planipennis. The results showed eight of 11 ORs from D. valens were present within group 7. These eight DvalORs were further divided into two subgroups: group 7a and 7b. Subgroup 7b was exclusive to bark beetle species, including ORs from D. valens, I. typographus, and D. ponderosae. In three groups (1, 2, and 7), we found most of the candidate genes were grouped together with orthologs in D. ponderosae and I. typographus. The amino acid sequences of all the 11 ORs are listed in S4 Fig.
In the OR phylogenetic tree, DvalOR2 was clustered with MacrOR1 and AplaOR83b, which are Orco homologs. In addition, a similarity search for the DvalOR1 against the non-redundant nucleotide (nr) database at NCBI using BlastX revealed 97% identity with the Orco homolog of D. ponderosae (AEE62122.1,0; DponOR1; Table 3) at the nucleotide level.
Four GR candidates (GR1-4) were identified in D. valens, which is in agreement with several other recent antennal transcriptomic studies [3,5,26]. Only one of these likely represented a full-length gene (GR3). Because GRs and ORs belong to the same superfamily, both were included in the same dendrogram analysis, in which GRs formed a distinct clade (Fig 8). The information of all four GRs is listed in Table 3. The amino acid sequences of all four GRs are listed in S4 Fig.
Identification of Candidate Sensory Neuron Membrane Proteins
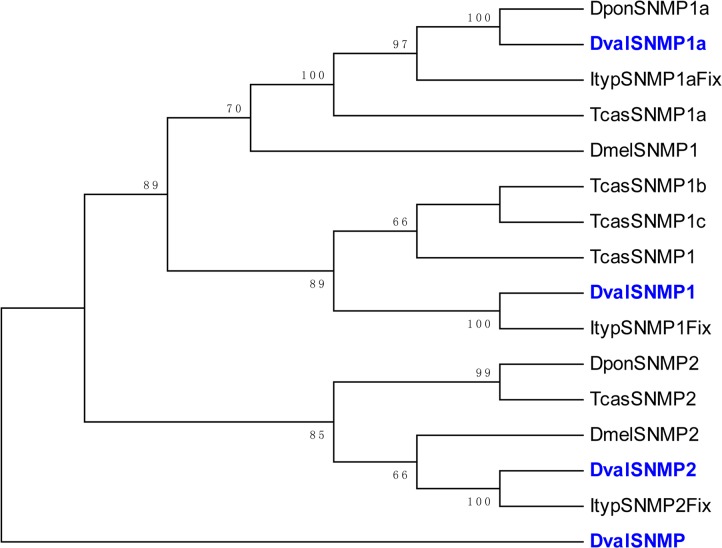
We also found four SNMPs (Table 2) in the antennal transcriptome of D. valens, none of which represented full-length genes. Among them, SNMP1 and SNMP1a are orthologs of SNMP1. DvalSNMP1 and DvalSNMP1a were very similar to the DponSNMP1 and DponSNMP1a published in GenBank, with 94% and 95% identity, respectively (Table 2). The genes grouped together with orthologs in T. castaneum, I. typographus, D. ponderosae, and A. planipennis. We found 1:1 orthologous candidate (SNMP1, SNMP1a, and SNMP2) relationships among the SNMPs (Fig 9). DvalSNMP was scattered in a monophyletic clade in the outer part of the tree, indicating an ancestral relationship to the rest of the tree. DvalSNMP was searched against the non-redundant nucleotide (nr) database at NCBI using BlastX (BLAST 2.2.23+, E-value < e-5), and showed 85% identity with a sensory neuron membrane protein of D. ponderosae (GenBank accession number: AFI45067.1). The information for all four SNMPs is listed in Table 2 with the amino acid sequences listed in S5 Fig.
Fig 9. Phylogenetic tree of putative SNMPs from Dendroctonus valens (Dval), Ips typographus (Ityp), Dendroctonus ponderosae (Dpon), Tribolium castaneum (Tcas) and Drosophila melanogaster (Dmel).
The D. valens translated unigenes are shown in blue. Accession numbers are given in S5 Fig. The tree was constructed with MEGA5.0, using the neighbor-joining method. Values indicated at the nodes are bootstrap values based on 1,000 replicates, and the bootstrap values below 50% are not shown.
Identification of Candidate Ionotropic Receptors
We discovered three antennal IRs from antennal transcriptome assembly in D. valens. IR1 and IR3 have high sequence similarity to the ionotropic receptor 8a of D. ponderosae (GenBank accession number: AGI05169.1) with amino acid identities of 87%, and 97%, respectively (Table 3), suggesting that IR1 and IR3 are homologs. We also identified eight ionotropic glutamate receptor genes (IGluR) (Table 3), likely as cell surface proteins which allow neurons to communicate with cells in the insect [20].
Identification of Candidate Odor/Xenobiotic Degradation Enzymes
A large number of odor/xenobiotic degradation genes were found within the D. valens antennal transcriptome (155). These genes are likely involved in olfaction processes. Among the odor/xenobiotics degradation genes, the number of esterase genes (66) was the highest, followed by cytochrome P450 genes (49). Cytochrome P450s are thought to participate in detoxification, odor processing, and neuro/development functions in insects [52–55]. Esterases are thought to function in metabolic resistance and odor degradation in several insect species [56–60]. The remaining genes identified in D. valens antennae are involved in odor/xenobiotic degradation, including glutathione S-transferases (GSTs, 11), aldehyde dehydrogenase (14), epoxide hydrolases (four), and several antioxidants (Table 1).
Discussion
The antennal transcriptome described here represents the first study on the repertoire of odor processing genes of the destructive and invasive red turpentine beetle, Dendroctonus valens. In this study, 21 OBPs, six CSPs, four SNMPs, 22 ORs, four GRs, three IRs, eight ionotropic glutamate receptors and 155 odorant/xenobiotic degradation enzymes were identified as putative olfactory system genes. These genes are not only involved in signal detection and transduction, but also in metabolic activity. It is well known that D. valens exploits host kairomones (such as (+)-3-carene), as well as non-host volatiles (e.g., 1-octen-3-ol, (Z)-3-hexen-1-ol, and (E)-2-hexen-1-ol) during host location, and uses aggregation pheromones and congeneric kairomones (trans-verbenol, myrtenol, myrtenal, and frontalin) in intra- and interspecific communication [36–50]. The odor reception and odor degradation genes identified here likely play important physiological roles in these aspects of D. valens’s biology and provide a foundation to further unravel the molecular mechanisms underlying olfaction in D. valens. Functional studies will be required to ascertain the specific role of these genes in D. valens olfaction. Furthermore, critical molecular targets of important semiochemicals such as (+)-3-carene, trans-verbenol, and frontalin could be identified via computational and molecular methods, such as homology modeling, molecular docking, and RNA interference (RNAi). By collating chemical and ecological knowledge with the putatively identified genes in this study, we can begin to elucidate the olfactory mechanisms of D. valens.
Prior to this study, coleopteran species’ odorant reception genes had been characterized from the genome of T. castaneum [51], the antennal transcriptomes of I. typographus and D. ponderosae [5], and the antennal transcriptome of A. plannipennis [26]. However coleopteran odorant degrading enzymes have received minimal study, except for the DponCYP345E2, an antenna-specific cytochrome P450 in D. ponderosae [61], and several odorant degrading enzymes in the antennal trasncriptome of A. plannipennis [26]. Odorant degrading enzymes are needed to quickly deactivate the odorants once the signal has been transduced so that the olfactory system can respond to continuing plumes of odor [2,62]. In the context of pest insect management, some of these genes or their products could be targets for development of specific inhibitors that interfere with the insect’s ability to respond appropriately to olfactory cues from mates or host plants[4,5,26]. The odorant degrading genes reported here provide a significant addition to the pool of identified olfactory genes in Coleoptera, and will advance our understanding of coleopteran olfactory molecular mechanisms.
In the D. valens transcriptome analyzed here, 70.41% of 22,058 transcripts had homologous matches in GenBank with the cutoff value of 10−5. Among these transcripts, 39.6% of the genes were annotated on one or more GO terms by GO analyses. More than 60% of the transcripts had no GO terms, similar to what was found with Manduca sexta [1], I. typographus and D. ponderosae [5], and A. planipennis [26] This result indicates high levels of unknown processes in antennal tissue and a large number of D. valens transcripts that may potentially represent novel genes.
OBPs play an important role in odor processing by insects, facilitating the transport of odorant molecules through the sensillar lymph and serving as the liaison between the external environment and ORs [2,4]. In this study, we identified 21 transcripts encoding putative OBPs in the D. valens antennal transcriptome. Even though the number of putative OBP-coding genes in D. valens is much lower than that of D. ponderosa (31), the majority of putative OBPs showed a high similarity to those of D. ponderosa, and most of the OBPs of D. valens clustered together with DponOBPs. CSPs are another more conserved class of small binding proteins which can bind pheromone compounds [6–9]. Here we identified six transcripts encoding putative CSPs in the D. valens antennal transcriptome. Among the six candidate CSPs, four sequences were clustered together with DponCSPs. Dendroctonus valens and D. ponderosae are sympatric in North America, and the sequence similarity of olfactory-related orthologous genes is very high between the two species, suggesting that the OBPs and CSPs of D. valens may have similar forms of expression and function as those of D. ponderosae. It was interesting that nearly one third of the transcripts which encoded putative OBPs and CSPs in D. ponderosae were identified only in non-sensory tissues rather than the antennal tissues [2,4], suggesting that these proteins might have physiological functions independent of olfaction.
SNMPs are membrane proteins observed to associate with chemosensory neurons in insects [24]. There are two representatives of insect SNMPs (SNMP1 and SNMP2). To date, the general mechanism of SNMP function is still poorly understood. Benton et al.[22] demonstrated that SNMP1 was essential for the detection of the pheromone (Z)-11-octadecenyl acetate (a volatile male-specific fatty-acid-derived pheromone) in Drosophila melanogaster and (Z)-11-hexadecenal (a lipid-derived pheromone) in Heliothis virescens, and suggested that SNMP acts in concert with odorant receptors to capture pheromone molecules on the surface of olfactory dendrites, particularly pheromones with hydrophobic tails [5]. In the current antennal transcriptome of D. valens, four SNMPs were identified. Because bark beetle pheromones lack the hydrophobic tails of D. melanogaster and Heliothis virescens pheromones, we assume that SNMPs may have an alternate functional role. The expression of antennal SNMPs in D. valens, similar to I. typographus and D. ponderosae, suggests that SNMPs may be involved in pheromone detection by bark beetles.
The IR family contains two major groups, the conserved antennal IRs involved in olfaction and species-specific divergent IRs that are expressed in other tissues including gustatory organs [63]. We identified three IRs from antennal transcriptome assembly in D. valens. The number of IR genes in D. valens is much lower than that of D. ponderosae (15). We could not exclude the possibility that some of transcripts were missed in our antennal transcriptome, because of the lower sequencing depth of D. valens than that of D. ponderosae. Sequence alignments showed that the putative D. valens IRs have high similarity with the known ionotropic receptor of D. ponderosae, DponIR8a. DponIR8a belongs to the group of IR8a, which is a broadly expressed co-receptor and necessary for odor responses[5]. Originally, IR8a was identified in the fruitfly D. melanogaster and it may form heteromers with another variant ionotropic receptor [64].
ORs belong to a large multigene family encoding seven transmembrane domain proteins [10,11], and are the key players in insect olfactory reception [4]. Twenty-two OR candidates were identified from the D. valens antennal transcriptome. This number is much lower than for two other bark beetles (I. typographus, 43, and D. ponderosae, 49). The lower number of receptors in D. valens may be due to technical reasons. Despite the number difference in the putative OR-coding genes, the majority of DvalORs grouped together with orthologs in D. ponderosae and I. typographus. In agreement with previous results, we found distinct expansions of OR lineages in bark beetles, and the subgroup 7b was exclusive to three bark beetle species in the OR phylogenetic tree [2,5]. In addition, DvalOR1 and DvalOR2 were identified as co-receptors in the D. valens antennal transcriptome. Further function studies on DvalOR1 and DvalOR2 may help in deciphering their role in D. valens olfaction.
GRs are mostly expressed in gustatory receptor neurons in taste organs and are involved in detection of sugars, bitter compounds, and contact pheromones [2,5]. In the antennal transcriptome of D. valens, four GR candidates (GR1-4) were identified. In the phylogenetic tree, Gr1, Gr 2, and Gr4 in D. valens grouped together with ItypGR3 and DponGR3, which may function as carbon dioxide receptors, and DvalGr3 was clustered with ItypGR6, tentatively assigned as a trehalose receptor [2,5]. So far, insect GRs have been identified as sugar receptors—BmorGR8 of B. mori [65] and HarmGR9 of Helicoverpa armigera [66]—heat sensors—DmelGR28B(D) of D. melanogaster [67]—and carbon dioxide and bitter receptors [68]. It is therefore likely that the D. valens GRs play important roles in the detection of carbon dioxide, sugars, or bitter compounds. Future studies on GR expression patterns of D. valens in chemosensory and non-chemosensory tissues (e.g. the head and maxillary palps of both sexes) will be helpful in understanding the actual roles of GRs in D. valens.
It is generally accepted that insect semiochemicals require specific odorant degrading enzymes based on the functional group(s) present on each semiochemical. In our study, a high number of odor/xenobiotic degradation genes were found in the D. valens antennal transcriptome. This is the first report of genes involved in odor degradation in D. valens. These genes, including esterases, cytochrome P450 genes, glutathione S-transferases, aldehyde dehydrogenase, epoxide hydrolases, and antioxidants, are likely involved in odor degradation. For insects, the olfactory system must not only precisely detect conspecific semiochemicals, but also rapidly inactivate the chemical signal once the message is conveyed to allow for continued reception of signals[2]. Mounting evidence suggests that various types of pheromones are degraded by antennae-specific esterases, aldehyde oxidases, aldehyde dehydrogenases, epoxide hydrolases, glutathione-S-transferases, and cytochrome P450s [2,57,61,69–78]. Given the diverse expression and functions of these odorant degrading enzymes, identification and characterization of the enzymes specializing in odorant degradation is challenging [62]. At present, in contrast to odorant reception genes, there have been relatively few studies of odor degradation genes in insects. The first odorant degrading enzyme identified was an antennal-specific esterase from A. polyphemus (ApolSE), which could effectively degrade the acetate component of the pheromone blend [59,60]. A glutathione-S-transferase in M. sexta (GST-msolf1) has been shown to play a key role in sex pheromone detection by inactivating the aldehyde component of the sex pheromone blend [79]. Cytochrome P450 is one of the most studied groups of detoxification enzymes linked to odorant degradation [62]. DponCYP345E2, an antenna-specific cytochrome P450 found in D. ponderosae, degrades several pine host monoterpenes [61]. This was the first P450 to be functionally characterized in insect olfaction in bark beetles. In another study, several antennal-specific cytochrome P450’s were found in D. valens, suggesting that these P450 genes may be involved in detoxification or odor degradation of these monoterpenes [80]. Here, we found 49 novel P450 genes in the D. valens antennal transcriptome, but their functional roles remain to be determined. Understanding the molecular mechanism(s) of signal inactivation is important in fundamental biology and may lead to novel molecular targets for insect pest control [2].
Conclusions
This study reports the first antennal transcriptome analysis in Dendroctonus valens. The genes reported here provide valuable insight into the molecular mechanisms of insect olfaction, and also represent possible novel targets for D. valens control or management. In addition, the results from our study provide the basis for a deeper understanding of coleopteran olfaction in the context of complex behavior, and information for comparative and functional genomic analyses of related species.
Recent studies have shown that the numbers of functional chemosensory receptor genes vary enormously among the genomes of different animal species [3–5]. Despite the number of chemosensory transcripts being highly variable within the Scolytinae, the putative olfactory genes in D. valens demonstrated close similarity in sequence alignments to those of two other two bark beetles. The phylogenetic trees showed that the majority of the odor processing genes of D. valens cluster with the analogous genes in D. ponderosae. In addition, we also found a distinct species-specific expansion of OR lineages in our results. Dendroctonus valens and D. ponderosae are sympatric in their host range in North America, and the two species live in similar coniferous habitats and share a number of biologically relevant compounds, including host compounds, pheromone compounds and non-host volatiles (S2 Table). The low degree of species-specific diversification in chemosensory genes can be explained by the shared semiochemical space of the three bark beetles. Ecological adaptation and life history parameters might play important roles in shaping the olfactory gene repertoire [81]. The patterns seen in the current study likely reflect the evolutionary and ecological relatedness of these species. Further functional evidence is required to support this hypothesis.
The insect olfactory gene repertoire may be larger than currently understood. For example, nearly one third of the transcripts encoding putative OBPs and CSPs in D. ponderosae were identified only in non-sensory tissues instead of from the antennal tissue [2,5], suggesting that these proteins might have physiological functions independent of or in addition to olfaction. Here, we analyzed the antennal chemosensory repertoire of D. valens of a specific life stage (newly emerged adults) and a specific organ (antennae). As the first step towards understanding their functions, a comprehensive examination of the chemosensory gene expression patterns in the context of different tissues and life stages in D. valens could provide important information on the functions of the chemosensory genes. Following that, functional studies on the chemosensory genes will help to depict the molecular mechanisms underlying D. valens olfactory detection from an evolutionary viewpoint.
Materials and Methods
Insect and RNA Extraction
Adults of Dendroctonus valens were collected by Lindgren funnel traps baited with a kairomone lure (3-carene) in P. tabuliformis plantations at the Tunlanchuan Forest Farm (N 37° 48′, E 111° 57′, average elevation 1,400 m), Gujiao, Shanxi Province, China. Forest Pest Control Station of Shanxi Province issued the permit for the field
collections (by the director, Zhenwang, Miao). About one thousand newly emerged adults (650 females and 350 males) were collected. All D. valens were identified using the morphological characteristics of front and elytral declivity [82] and sexed based on distinguishing characters on the seventh abdominal tergite [83]. In total, 2,000 antennae from both sexes were used for RNA extraction. The antennae were cut off and immediately frozen in liquid nitrogen, and stored at—70°C until RNA extraction. The samples were homogenized using a Tissue-tearor, and total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions.
cDNA Library Construction
cDNA library construction and Illumina sequencing of the samples were performed at Beijing Genomics Institute Shenzhen, China [84]. Briefly, the mRNA was purified from 20 mg of total RNA (a mixture of RNAs from the antennae) using oligo (dT) magnetic beads and fragmented into short sequences in the presence of divalent cations at 94°C for 5 min. Then, the first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by synthesis of the second-strand cDNA using RNaseHand DNA polymerase I. After end-repair and ligation of adaptors, the products were amplified by PCR and purified using the QIAquick PCR Purification Kit to create a cDNA library.
Transcriptome Sequencing and Assembly
The cDNA library constructed from the antennae of Dendroctonus valens were sequenced on the Illumina HiSeq 2000 platform. Transcriptome de novo assembly was carried out using the short reads assembling program Trinity-2014 [85]. Trinity-2014 first combines reads with a certain length of overlap to form longer fragments without N (N represents unknown sequence) to produce contigs. The reads are then mapped back to contigs, by using paired-end reads that enable identification of contigs from the same transcript and the distances between these contigs. Next, Trinity-2014 connects the contigs based on the paired-end reads for gap filling between each pair of contigs to build scaffold sequences with the least Ns. Such sequences are defined as unigenes.
Homology Analysis and Gene Ontology (GO) Annotation
The unigenes or contigs were matched by a BlastX homology search to the entries in the NCBI non-redundant (nr) protein database with a cut-off E-value of 10−5 to find similarities to the unigenes or contigs of other species. Unigenes larger than 150 bp were first aligned by BlastX to protein databases, including Nr, Swiss-Prot, KEGG and COG (e-value, 1025), retrieving proteins with the highest sequence similarity with the given unigenes along with their protein functional annotations. Then, we used the Blast2GO program [86] to get GO annotation of the unigenes, and got GO functional classification by using WEGO software [87]. The ORFs of the unigenes were predicted by using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The signal peptides of the protein sequences were predicted using SignalP 4.0 [88].
Sequence Alignment and Phylogenetic Analyses
The amino acid sequence alignment of the candidate OBPs, CSPs, ORs, GRs and SNMPs from D. valens and other insect species (S1–S5 Figs) were performed using ClustalX 2.0 [89]. The OBP data set contained 21 sequences from D. valens, 11 from Ips typographus, 31 from D. ponderosae, 45 from T. castaneum and nine from A. plannenis. The CSP data set contained six sequences from D. valens, four from I. typographus, seven from D. ponderosae, 20 from T. castaneum, four from A. planipennis, and two from D. melanogaster. The SNMP data set contained four sequences from D. valens, three from I. typographus, two from D. ponderosae, five from T. castaneum, one from A. planipennis, and two from D. melanogaster. The OR data set contained 11 sequences from D. valens, 18 from I. typographus, 13 from D. ponderosae, two sequences from A. planipennis, 36 sequences from M. caryae, and 28 from T. castaneum. The GR data set contained three sequences from D. valens, three from I. typographus, and two from D. ponderosae. The unrooted phylogenetic trees were constructed by MEGA5.0 [90] using the Neighbor-joining method with Poisson correction of distances. Node support was assessed using a bootstrap procedure base on 1,000 replicates.
Data Deposition
The sequences of 60 odorant reception genes (21 OBPs, six CSPs, four SNMPs, four GRs, three IRs and 22 ORs) of D. valens antennae were submitted to the GenBank database (accession number GenBank KP736107-KP736166).
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Professor Miao ZhenWang (Forest Pest Control Station of Shanxi Province) for help in collecting D. valens. The Tunlanchuan Forest Farm, Gujiao, Shanxi Province, provided logistical support in collecting insects. We thank Dr. Jocelyn Millar and R. Maxwell Collignon (University of California, Riverside), who kindly provided critical comments on early drafts. We also acknowledge two anonymous reviewers for useful comments on the submitted manuscript.
Data Availability
Relevant data are within the paper and supporting information files. The sequences of 60 odorant reception genes (21 OBPs, six CSPs, four SNMPs, four GRs, three IRs and 22 ORs) of D. valens antenna were submitted to the GenBank database (accession number GenBank KP736107-KP736166).
Funding Statement
This study was funded by the National Natural Science Foundation of China (31170616, 31000304). L-WZ received the funding and the founders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS (2011) Antennal transcriptome of Manduca sexta . Proc Natl Acad Sci U S A 108: 7449–7454. 10.1073/pnas.1017963108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leal WS (2013) Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol 58: 373–391. 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- 3. Nishimura O, Brillada C, Yazawa S, Maffei ME, Arimura G (2012) Transcriptome pyrosequencing of the parasitoid wasp Cotesia vestalis: genes involved in the antennal odorant-sensory system. PLoS One 7: e50664 10.1371/journal.pone.0050664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y, Gu S, Zhang Y, Guo Y, Wang G (2012) Candidate olfaction genes identified within the Helicoverpa armigera Antennal Transcriptome. PLoS One 7: e48260 10.1371/journal.pone.0048260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andersson MN, Grosse-Wilde E, Keeling CI, Bengtsson JM, Yuen MM, Li M, et al. (2013) Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics 14: 198 10.1186/1471-2164-14-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pelosi P, Zhou JJ, Ban LP, Calvello M (2006) Soluble proteins in insect chemical communication. Cell Mol Life Sci 63: 1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanchez-Gracia A, Vieira FG, Rozas J (2009) Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb) 103: 208–216. 10.1038/hdy.2009.55 [DOI] [PubMed] [Google Scholar]
- 8. Liu R, He X, Lehane S, Lehane M, Hertz-Fowler C, Berriman M, et al. (2012) Expression of chemosensory proteins in the tsetse fly Glossina morsitans is related to female host-seeking behaviour. Insect Mol Biol 21: 41–48. 10.1111/j.1365-2583.2011.01114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Briand L, Swasdipan N, Nespoulous C, Bezirard V, Blon F, Huet JC, et al. (2002) Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. European Journal of Biochemistry 269: 4586–4596. [DOI] [PubMed] [Google Scholar]
- 10. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR (1999) A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila . Neuron 22: 327–338. [DOI] [PubMed] [Google Scholar]
- 11. Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R (1999) A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96: 725–736. [DOI] [PubMed] [Google Scholar]
- 12. Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, Witzgall P, et al. (2012) Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS One 7: e31620 10.1371/journal.pone.0031620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, et al. (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452: 1007–1011. 10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- 14. Hallem EA, Ho MG, Carlson JR (2004) The molecular basis of odor coding in the Drosophila antenna. Cell 117: 965–979. [DOI] [PubMed] [Google Scholar]
- 15. Mitchell RF, Hughes DT, Luetje CW, Millar JG, Soriano-Agaton F, Hanks LM, et al. (2012) Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae . Insect Biochem Mol Biol 42: 499–505. 10.1016/j.ibmb.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR (2010) Odorant reception in the malaria mosquito Anopheles gambiae . Nature 464: 66–71. 10.1038/nature08834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hallem EA, Carlson JR (2006) Coding of odors by a receptor repertoire. Cell 125: 143–160. [DOI] [PubMed] [Google Scholar]
- 18. Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, et al. (2004) Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori . Proc Natl Acad Sci U S A 101: 16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stensmyr MC, Dweck HK, Farhan A, Ibba I, Strutz A, Mukunda L, et al. (2012) A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila . Cell 151: 1345–1357. 10.1016/j.cell.2012.09.046 [DOI] [PubMed] [Google Scholar]
- 20. Zhang S, Zhang Z, Wang H, Kong X (2014) Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (Lepidoptera: Lasiocampidae). Insect Biochem Mol Biol 52: 69–81. 10.1016/j.ibmb.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 21. Rogers ME, Sun M, Lerner MR, Vogt RG (1997) Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J Biol Chem 272: 14792–14799. [DOI] [PubMed] [Google Scholar]
- 22. Benton R, Vannice KS, Vosshall LB (2007) An essential role for a CD36-related receptor in pheromone detection in Drosophila . Nature 450: 289–293. [DOI] [PubMed] [Google Scholar]
- 23. Rogers ME, Krieger J, Vogt RG (2001) Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol 49: 47–61. [DOI] [PubMed] [Google Scholar]
- 24. Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, Staples J, et al. (2009) The insect SNMP gene family. Insect Biochem Mol Biol 39: 448–456. 10.1016/j.ibmb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 25. Jin X, Ha TS, Smith DP (2008) SNMP is a signaling component required for pheromone sensitivity in Drosophila . Proc Natl Acad Sci U S A 105: 10996–11001. 10.1073/pnas.0803309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mamidala P, Wijeratne AJ, Wijeratne S, Poland T, Qazi SS, Doucet D, et al. (2013) Identification of odor-processing genes in the emerald ash borer, Agrilus planipennis . PLoS One 8: e56555 10.1371/journal.pone.0056555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forstner M, Gohl T, Gondesen I, Raming K, Breer H, Krieger J (2008) Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem Senses 33: 291–299. 10.1093/chemse/bjm087 [DOI] [PubMed] [Google Scholar]
- 28. Gu SH, Yang RN, Guo MB, Wang GR, Wu KM, Guo YY, et al. (2013) Molecular identification and differential expression of sensory neuron membrane proteins in the antennae of the black cutworm moth Agrotis ipsilon . J Insect Physiol 59: 430–443. 10.1016/j.jinsphys.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 29. Liu C, Zhang J, Liu Y, Wang G, Dong S (2014) Expression of SNMP1 and SNMP2 genes in antennal sensilla of Spodoptera exigua (Hubner). Arch Insect Biochem Physiol 85: 114–126. 10.1002/arch.21150 [DOI] [PubMed] [Google Scholar]
- 30. Taerum SJ, Duong TA, de Beer ZW, Gillette N, Sun JH, Owen DR, et al. (2013) Large shift in symbiont assemblage in the invasive red turpentine beetle. PLoS One 8: e78126 10.1371/journal.pone.0078126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun JH, Lu M, Gillette NE, Wingfield MJ (2013) Red turpentine beetle: innocuous native becomes invasive tree killer in China. Annu Rev Entomol 58: 293–311. 10.1146/annurev-ento-120811-153624 [DOI] [PubMed] [Google Scholar]
- 32. Yan ZL, Sun JH, Don O, Zhang ZN (2005) The red turpentine beetle, Dendroctonus valens LeConte (Scolytidae): an exotic invasive pest of pine in China. Biodivers Conserv 14: 1735–1760. [Google Scholar]
- 33. Li J, CG, Song Y, Wang Y, Chang B (2001) Control project on red turpentine beetle (Dendroctonus valens). For Pest Dis 4: 41–44. [Google Scholar]
- 34. Legeai F, Malpel S, Montagne N, Monsempes C, Cousserans F, Merlin C, et al. (2011) An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics 12: 86 10.1186/1471-2164-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allen JE, Wanner KW (2011) Asian corn borer pheromone binding protein 3, a candidate for evolving specificity to the 12-tetradecenyl acetate sex pheromone. Insect Biochem Mol Biol 41: 141–149. 10.1016/j.ibmb.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 36. Hobson KR WD (1993) Stereospecific attraction of the red turpentine beetle to volatiles of host pines. Bulletin of the Ecological Society of America 74: 278–279. [Google Scholar]
- 37. Rappaport NG, Owen DR, Stein JD (2001) Interruption of semiochemical-mediated attraction of Dendroctonus valens (Coleoptera: Scolytidae) and selected nontarget insects by verbenone. Environ Entomol 30: 837–841. [Google Scholar]
- 38. Sun JH, Gillette NE, Miao ZW, Kang L, Zhang ZN, Owen DR, et al. (2003) Verbenone interrupts attraction to host volatiles and reduces attack on Pinus tabuliformis (Pinaceae) by Dendroctonus valens (Coleoptera: Scolytidae) in the People's Republic of China. Can Entomol 135: 721–732. [Google Scholar]
- 39. Fettig CJ, Borys RR, Cluck DR, Smith SL (2004) Field response of Dendroctonus valens (Coleoptera: scolytidae) and a major predator, Temnochila chlorodia (Coleoptera: Trogositidae), to host kairomones and a Dendroctonus spp. pheromone component. J Entomol Sci 39: 490–499. [Google Scholar]
- 40. Miao Z-W Zhang Z-N, Wang P-X, Guo Y-Y, Sun J-H (2004) Response of the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Scolytidae) to host semiochemicals and its implication in management. Acta Entomologica Sinica 47 47: 360–364. [Google Scholar]
- 41. Sun JH, Miao ZW, Zhang Z, Zhang ZN, Gillette NE (2004) Red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Scolytidae), response to host semiochemicals in China. Environ Entomol 33: 206–212. [Google Scholar]
- 42. Fettig CJ, Borys RR, Dabney CP, McKelvey SR, Cluck DR, Smith SL (2005) Disruption of red turpentine beetle attraction to baited traps by the addition of California fivespined ips pheromone components. Can Entomol 137: 748–752. [Google Scholar]
- 43. Zhang LW, Sun JH, Clarke SR (2006) Effects of verbenone dose and enantiomer on the interruption of response of the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Scolytidae), to its kariomones. Environ Entomol 35: 655–660. [Google Scholar]
- 44. Zhang LW, Sun JH (2006) Electrophysiological and behavioral responses of Dendroctonus valens (Coleoptera: Curculionidae: Scolytinae) to candidate pheromone components identified in hindgut extracts. Environ Entomol 35: 1232–1237. [DOI] [PubMed] [Google Scholar]
- 45. Erbilgin N, Mori SR, Sun JH, Stein JD, Owen DR, Merrill LD, et al. (2007) Response to host volatiles by native and introduced populations of Dendroctonus valens (Coleoptera: Curculionidae, Scolytinae) in North America and China. J Chem Ecol 33: 131–146. [DOI] [PubMed] [Google Scholar]
- 46. Fettig CJ, McKelvey SR, Dabney CP, Borys RR (2007) The response of Dendroctonus valens and Temnochila chlorodia to Ips paraconfusus pheromone components and verbenone. Can Entomol 139: 141–145. [Google Scholar]
- 47. Zhang LW, Gillette NE, Sun JH (2007) Electrophysiological and behavioral responses of Dendroctonus valens to non-host volatiles. Annals of Forest Science 64: 267–273. [Google Scholar]
- 48. Zhang LW, Ming L, Liu ZD, Sung JH (2007) Progress on invasion biology and chemical ecology of red turpentine beetle, Dendroctonus valens . Chinese Bulletin of Entomology 44 44: 171–178. [Google Scholar]
- 49. Koch SI, Groh K, Vogel H, Hansson BS, Kleineidam CJ, Grosse-Wilde E (2013) Caste-specific expression patterns of immune response and chemosensory related genes in the leaf-cutting ant, Atta vollenweideri . PLoS One 8: e81518 10.1371/journal.pone.0081518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Z, Xu B, Miao Z, Sun J (2013) The pheromone frontalin and its dual function in the invasive bark beetle Dendroctonus valens . Chem Senses 38: 485–495. 10.1093/chemse/bjt019 [DOI] [PubMed] [Google Scholar]
- 51. Engsontia P, Sanderson AP, Cobb M, Walden KK, Robertson HM, Brown S (2008) The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum . Insect Biochem Mol Biol 38: 387–397. 10.1016/j.ibmb.2007.10.005 [DOI] [PubMed] [Google Scholar]
- 52. Wojtasek H, Leal WS (1999) Degradation of an alkaloid pheromone from the pale-brown chafer, Phyllopertha diversa (Coleoptera: Scarabaeidae), by an insect olfactory cytochrome P450. FEBS Lett 458: 333–336. [DOI] [PubMed] [Google Scholar]
- 53. Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, et al. (2004) CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila . J Biol Chem 279: 35942–35949. [DOI] [PubMed] [Google Scholar]
- 54. Warren JT, Petryk A, Marques G, Parvy JP, Shinoda T, Fujimoto Y, et al. (2004) Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem Mol Biol 34: 991–1010. [DOI] [PubMed] [Google Scholar]
- 55. Petryk A, Warren JT, Marques G, Jarcho MP, Gilbert LI, Kahler J, et al. (2003) Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Natl Acad Sci U S A 100: 13773–13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gu SH, Wu KM, Guo YY, Pickett JA, Field LM, Zhou JJ, et al. (2013) Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics 14: 636 10.1186/1471-2164-14-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ishida Y, Leal WS (2002) Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus . Insect Biochem Mol Biol 32: 1775–1780. [DOI] [PubMed] [Google Scholar]
- 58. Ishida Y, Leal WS (2008) Chiral discrimination of the Japanese beetle sex pheromone and a behavioral antagonist by a pheromone-degrading enzyme. Proc Natl Acad Sci U S A 105: 9076–9080. 10.1073/pnas.0802610105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vogt RG, Riddiford LM, Prestwich GD (1985) Kinetic properties of a sex pheromone-degrading enzyme: the sensillar esterase of Antheraea polyphemus . Proc Natl Acad Sci U S A 82: 8827–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vogt RG, Riddiford LM (1981) Pheromone binding and inactivation by moth antennae. Nature 293: 161–163. [DOI] [PubMed] [Google Scholar]
- 61. Keeling CI, Henderson H, Li M, Dullat HK, Ohnishi T, Bohlmann J. (2013) CYP345E2, an antenna-specific cytochrome P450 from the mountain pine beetle, Dendroctonus ponderosae Hopkins, catalyses the oxidation of pine host monoterpene volatiles. Insect Biochem Mol Biol 43: 1142–1151. 10.1016/j.ibmb.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 62. Younus F, Chertemps T, Pearce SL, Pandey G, Bozzolan F, Coppin CW, et al. (2014) Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster . Insect Biochem Mol Biol 53: 30–43. 10.1016/j.ibmb.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 63. Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, et al. (2010) Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6: e1001064 10.1371/journal.pgen.1001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R (2011) Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69: 44–60. 10.1016/j.neuron.2010.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang HJ, Anderson AR, Trowell SC, Luo AR, Xiang ZH, Xia QY (2011) Topological and functional characterization of an insect gustatory receptor. PLoS One 6: e24111 10.1371/journal.pone.0024111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu W, Zhang HJ, Anderson A (2012) A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera . J Chem Ecol 38: 1513–1520. 10.1007/s10886-012-0221-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ni L, Bronk P, Chang EC, Lowell AM, Flam JO, Panzano VC, et al. (2013) A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila . Nature 500: 580–584. 10.1038/nature12390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Briscoe AD, Macias-Munoz A, Kozak KM, Walters JR, Yuan F, Jamie GA, et al. (2013) Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet 9: e1003620 10.1371/journal.pgen.1003620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. He P, Zhang YN, Li ZQ, Yang K, Zhu JY, Liu SJ, et al. (2014) An antennae-enriched carboxylesterase from Spodoptera exigua displays degradation activity in both plant volatiles and female sex pheromones. Insect Mol Biol 23: 475–486. 10.1111/imb.12095 [DOI] [PubMed] [Google Scholar]
- 70. He P, Zhang J, Li ZQ, Zhang YN, Yang K, Dong SL, et al. (2014) Functional characterization of an antennal esterase from the noctuid moth, Spodoptera exigua . Arch Insect Biochem Physiol 86: 85–99. 10.1002/arch.21164 [DOI] [PubMed] [Google Scholar]
- 71. He P, Li ZQ, Liu CC, Liu SJ, Dong SL (2014) Two esterases from the genus Spodoptera degrade sex pheromones and plant volatiles. Genome 57: 201–208. 10.1139/gen-2014-0041 [DOI] [PubMed] [Google Scholar]
- 72. Choo YM, Pelletier J, Atungulu E, Leal WS (2013) Identification and characterization of an antennae-specific aldehyde oxidase from the navel orangeworm. PLoS One 8: e67794 10.1371/journal.pone.0067794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pottier MA, Bozzolan F, Chertemps T, Jacquin-Joly E, Lalouette L, Siaussat D, et al. (2012) Cytochrome P450s and cytochrome P450 reductase in the olfactory organ of the cotton leafworm Spodoptera littoralis . Insect Mol Biol 21: 568–580. 10.1111/j.1365-2583.2012.01160.x [DOI] [PubMed] [Google Scholar]
- 74. Chertemps T, Francois A, Durand N, Rosell G, Dekker T, Lucas P, et al. (2012) A carboxylesterase, Esterase-6, modulates sensory physiological and behavioral response dynamics to pheromone in Drosophila . BMC Biol 10: 56 10.1186/1741-7007-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Durand N, Carot-Sans G, Bozzolan F, Rosell G, Siaussat D, Debernard S, et al. (2011) Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis . PLoS One 6: e29147 10.1371/journal.pone.0029147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Merlin C, Rosell G, Carot-Sans G, Francois MC, Bozzolan F, Pelletier J, et al. (2007) Antennal esterase cDNAs from two pest moths, Spodoptera littoralis and Sesamia nonagrioides, potentially involved in odourant degradation. Insect Mol Biol 16: 73–81. [DOI] [PubMed] [Google Scholar]
- 77. Merlin C, Francois MC, Bozzolan F, Pelletier J, Jacquin-Joly E, Maibeche-Coisne M, et al. (2005) A new aldehyde oxidase selectively expressed in chemosensory organs of insects. Biochem Biophys Res Commun 332: 4–10. [DOI] [PubMed] [Google Scholar]
- 78. Maida R, Ziegelberger G, Kaissling KE (1995) Esterase activity in the olfactory sensilla of the silkmoth Antheraea polyphemus . Neuroreport 6: 822–824. [DOI] [PubMed] [Google Scholar]
- 79. Rogers ME, Jani MK, Vogt RG (1999) An olfactory-specific glutathione-S-transferase in the sphinx moth Manduca sexta . J Exp Biol 202: 1625–1637. [DOI] [PubMed] [Google Scholar]
- 80. Lopez MF, Cano-Ramirez C, Cesar-Ayala AK, Ruiz EA, Zuniga G (2013) Diversity and expression of P450 genes from Dendroctonus valens LeConte (Curculionidae: Scolytinae) in response to different kairomones. Insect Biochem Mol Biol 43: 417–432. 10.1016/j.ibmb.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 81. Nei M, Niimura Y, Nozawa M (2008) The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet 9: 951–963. 10.1038/nrg2480 [DOI] [PubMed] [Google Scholar]
- 82. Yin HF (2000) The synopsis on morphological and biological characters of Dendroctonus valens LeConte. Aata Zootaxonomica Sinica 25: 120. [Google Scholar]
- 83. Lyon RL (1958) A Useful Secondary Sex Character in Dendroctonus Bark Beetles. Can Entomol 90: 582–584. [Google Scholar]
- 84. Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, et al. (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20: 646–654. 10.1101/gr.100677.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20: 265–272. 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gress JC, Robertson HM, Weaver DK, Dlakic M, Wanner KW (2013) Odorant receptors of a primitive hymenopteran pest, the wheat stem sawfly. Insect Mol Biol 22: 659–667. 10.1111/imb.12053 [DOI] [PubMed] [Google Scholar]
- 87. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. (2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 35: D5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 89. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 90. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen H, Lin L, Xie M, Zhang G, Su W (2014) De novo sequencing, assembly and characterization of antennal transcriptome of Anomala corpulenta Motschulsky (Coleoptera: Rutelidae). PLoS One 9: e114238 10.1371/journal.pone.0114238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang YN, Jin JY, Jin R, Xia YH, Zhou JJ, Deng JY, et al. (2013) Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS One 8: e69715 10.1371/journal.pone.0069715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Menuz K, Larter NK, Park J, Carlson JR (2014) An RNA-seq screen of the Drosophila antenna identifies a transporter necessary for ammonia detection. PLoS Genet 10: e1004810 10.1371/journal.pgen.1004810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chandler JC, Aizen J, Elizur A, Hollander-Cohen L, Battaglene S, Ventura T (2014) Discovery of a novel insulin-like peptide and insulin binding proteins in the Eastern rock lobster Sagmariasus verreauxi. Gen Comp Endocrinol. [DOI] [PubMed]
- 95. Cao D, Liu Y, Wei J, Liao X, Walker WB, Li J, et al. (2014) Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int J Biol Sci 10: 846–860. 10.7150/ijbs.9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Relevant data are within the paper and supporting information files. The sequences of 60 odorant reception genes (21 OBPs, six CSPs, four SNMPs, four GRs, three IRs and 22 ORs) of D. valens antenna were submitted to the GenBank database (accession number GenBank KP736107-KP736166).