Abstract
Magnetic resonance imaging (MRI) of autism populations is confounded by the inherent heterogeneity in the individuals’ genetics and environment, two factors difficult to control for. Imaging genetic animal models that recapitulate a mutation associated with autism quantify the impact of genetics on brain morphology and mitigate the confounding factors in human studies. Here, we used MRI to image three genetic mouse models with single mutations implicated in autism: Neuroligin-3 R451C knock-in, Methyl-CpG binding protein-2 (MECP2) 308-truncation and integrin β3 homozygous knockout. This study identified the morphological differences specific to the cerebellum, a structure repeatedly linked to autism in human neuroimaging and postmortem studies. To accomplish a comparative analysis, a segmented cerebellum template was created and used to segment each study image. This template delineated 39 different cerebellar structures. For Neuroligin-3 R451C male mutants, the gray (effect size (ES) = 1.94, FDR q = 0.03) and white (ES = 1.84, q = 0.037) matter of crus II lobule and the gray matter of the paraflocculus (ES = 1.45, q = 0.045) were larger in volume. The MECP2 mutant mice had cerebellar volume changes that increased in scope depending on the genotype: hemizygous males to homozygous females. The integrin β3 mutant mouse had a drastically smaller cerebellum than controls with 28 out of 39 cerebellar structures smaller. These imaging results are discussed in relation to repetitive behaviors, sociability, and learning in the context of autism. This work further illuminates the cerebellum’s role in autism.
Keywords: animal models, neuroimaging, neuroanatomy, structural MRI, genetics
Introduction
Recent progress has provided evidence for a genetic basis to the etiology of autism spectrum disorders (ASD). This evidence includes a concordance rate of > 70% in identical twins, a rate higher than observed in fraternal twins and siblings [Bailey et al., 1995; Hallmayer et al., 2011]. Furthermore, from genome-wide association studies (GWA), over 250 genes have been associated with ASD [Banerjee-Basu & Packer, 2010]; although no gene appears to contribute to more than 2% of ASD cases [Abrahams & Geschwind, 2010]. While the neurobiological underpinnings of ASD have not yet been fully established, neuroimaging and histopathology have consistently implicated the cerebellum [Fatemi et al., 2012]. Here, we propose to study the cerebellum in ASD by applying neuroimaging to animal models that recapitulate the single-nucleotide polymorphisms (SNP) found in human GWA. Furthermore, the mouse models that recapitulate ASD genetic lesions have behavior changes thought to reflect some of the behavior seen in ASD patients [Ey, Leblond, & Bourgeron, 2011; Robertson & Feng, 2011; Silverman, Yang, Lord, & Crawley, 2010]. Also shown recently with Fragile X, an ASD-related disorder, was that a mouse model of disorder and Fragile X patients do respond similarly to pharmacological treatments [Berry-Kravis et al., 2012; Henderson et al., 2012]. Neuroimaging therefore provides a starting point to look at shared and unshared biological traits of the brain, and specifically the cerebellum, across genetic models. Future work can use shared traits as a stepping-stone to develop and validate new treatments.
How the cerebellum is implicated in ASD can be understood through its anatomy and functional circuitry. The cerebellum is comprised of three main functional units, the spinocerebellum, cerebrocerebellum, and vestibular cerebellum [Sillitoe, Fu, & Watson, 2011]. The spino-cerebellum functional unit comprises the vermis and the intermediate hemispheres. This unit projects from the cerebellar cortical areas to the fastigial and interposed cerebellar nuclei, and then to brain stem nuclei and the spinal cord. The spinocerebellum functional unit is involved in movement coordination, proprioception, and other motor and premotor functions. The cerebrocerebellum creates a thalamic–cortex–pontine nucleus loop with the lateral hemispheres, posterior vermis, and dentate nuclei, which functions together in planning, initiation, and execution of movement. An example of the cerebellum’s function in this loop, shown using functional Magnetic Resonance Imaging (fMRI), revealed that crus I and II of the cerebellar hemispheres are a part of the executive control network [Habas et al., 2009]. Additionally, several mouse models of ASD have repetitive behaviors and social deficits, and have shown neuroanatomical abnormalities in the cerebrocerebellum loop constituents (e.g., [Carter et al., 2011; Horev et al., 2011]). The last functional unit is the vestibular cerebellum. It is made up of vermis lobules IX and X, the flocculus, and the paraflocculus [Sillitoe et al., 2011; Voogd, Gerrits, & Ruigrok, 1996]. It functions in balance, posture, and eye movement. Within these three functional circuits, there is evidence for a cerebellar role in complex cognitive functions as well as in its traditional role of motor control.
Almost since researchers started imaging the brain of autism patients, cerebellar abnormalities have been reported [Allen & Courchesne, 2003; Courchesne, 1995; Courchesne, Yeung-Courchesne, Press, Hesselink, & Jernigan, 1988; Hashimoto et al., 1995; Piven, Saliba, Bailey, & Arndt, 1997]. The first report, in 1988, found decreased volumes in vermis lobules VI and VII [Courchesne et al., 1988]. Later experiments, for example Piven et al. [1997], found no abnormalities. To better describe the brain of ASD patients, further neuroimaging studies were carried out and culminated in several recent reviews, identifying the cerebellum as a structure associated with ASD [Amaral, Schumann, & Nordahl, 2008; Anagnostou & Taylor, 2011; Stanfield et al., 2008]. However, disagreement remains in the type of cerebellar changes and in understanding which particular cerebellar regions are affected. This disagreement arises partially from the genetic heterogeneity intrinsic to the often-underpowered human imaging studies. In addition to neuroimaging, researchers investigate the neurobiology of autism through postmortem tissue samples. This effort has consistently found a decrease in the number of Purkinje cells (PC) within the hemispheres of the cerebellum [Bauman & Kemper, 1985; Fatemi et al., 2002; Ritvo et al., 1986]. Neuroimaging and postmortem studies have been unable to say what regions of the cerebellum are affected by specific genetic mutations associated with the disorder and whether specific regions of the cerebellum are more consistently affected than others. This shortfall of current human neuroimaging and postmortem studies includes the discrepancy between the two methods in which cerebellar regions are structurally abnormal: neuroimaging reporting vermis abnormalities and postmortem studies finding hemispheric abnormalities. Although it should be mentioned that suggestions have been made as to why this discrepancy exists. One line of reasoning is from Dr. Amaral et al. [Amaral et al., 2008] who has pointed out that neuroimaging studies typically use high-functioning ASD patients, whereas postmortem studies are not so selective, and both typically suffer from small and not statistically powerful sample sizes.
The largest problem with neuroimaging and postmortem studies may be the diversity in genetic and environmental factors contained within the sampled populations, which likely confound our understanding of the cerebellum in ASD. An alternative approach, where MRI determines the neuroanatomical phenotype of genetic ASD mouse models, allows for a direct link between brain morphology and ASD genetics. To perform such mouse neuroimaging studies in a high-throughput automated fashion, MRI atlases have been developed [Dorr, Lerch, Spring, Kabani, & Henkelman, 2008; Ullmann et al., 2012]. Since ASD has been reported as both a white matter and gray matter disorder [Amaral et al., 2008; Anagnostou & Taylor, 2011], atlases to segment the cerebellum defects should evaluate both tissues. Until recently, the majority of MR mouse atlases defined the cerebellum as the cerebellar cortex and arbor vita, a region containing the cerebellar white matter and deep nuclei. This definition overlooked the finer cerebellum regions known to be distinct in function and connectivity. Then in 2012, Ullmann et al. [2012] partitioned the cerebellar cortex and deep cerebellar nuclei at the necessary finer resolution; however, they did not parse the white matter. This left the white matter insufficiently parcellated, preventing a comprehensive analysis of the cerebellum in ASD mouse models.
This article constructs a cerebellar MRI atlas with 39 unique structures for cerebellar nuclei and white and gray matter. This MRI atlas is applied to three genetic mouse models of ASD. The models investigated are: (a) Neuroligin-3 R451C knock-in (NL3 KI), an ASD-associated SNP; (b) methyl-CpG binding protein-2 (MECP2) truncated at the 308th amino acid and associated with an ASD-related disorder, Rett Syndrome; and (c) integrin β3 (ITGB3), an ASD-associated mutation closely tied to the serotonin system. These three cases were chosen to represent a diverse set of genetic ASD associations. Neuroligin-3 (NL3) is located on the X chromosome, codes for a synapse adhesion molecule and the knock-in model results in decreased expression of the neuroligin-3 protein [Tabuchi et al., 2007]. The MECP2 mutation causes a transient ASD-associated syndrome and affects the expression of many proteins as a transcription factor [Shahbazian et al., 2002]. ITGB3 is associated with ASD susceptibility, whole brain serotonin levels, and the serotonin transporter SLC6A4 [Weiss, Kosova et al., 2006; Weiss, Ober, & Cook, 2006]. The link of ITGB3 to the serotonin system is important because this neurotransmitter system is involved in behaviors such as mood, cognition, and learning and memory [Sodhi & Sanders-Bush, 2004], and high levels of serotonin is a consistently replicated finding in ASD patients [Chugani, 2004; Coutinho et al., 2007]. Together, the three models come from three important groups associated with ASD genetics: ASD-related syndromes, synapse surface proteins, and serotonin system-associated proteins. Therefore, the purpose of this study is to illustrate the power of MRI structural analysis in the cerebellum, understand the individual cerebellar phenotypes of three mouse models, and collectively assess how their neuroanatomy may relate to ASD-associated behavior.
Methods
Sample Preparation
For NL3 KI, 16 male mice were purchased from Jackson Laboratory comprised of eight NL3 KI (Jackson Laboratory #008475) and eight non-littermate wild-type (WT; Jackson Laboratory #101045) mice. Perfusion was performed on postnatal day 108 [Ellegood, Lerch, & Henkelman, 2011]. The MECP2 group was derived from Huda Zoghbi’s line [Shahbazian et al., 2002] (Jackson Laboratory #005439). This group included 12 homozygous females (MECP2 308 −/−), 12 heterozygous females (MECP2 308 −/+), 10 hemizygous males (MECP2 308 −/Y), and 20 non-littermate WT (nine males, Jackson Laboratory #000664) from a C57BL/6 background. The mice were perfused at approximately postnatal day 250. For MECP2, both the WT and MECP 308-truncation mice were retired breeders. For ITGB3, 24 male mice were purchased from Jackson Laboratory comprised of 12 ITGB3 knock-out mice (Jackson Laboratory #004669) and 12 non-littermate WT (Jackson Laboratory #101045) mice. The 24 ITGB3 cohort mice were perfused on postnatal day 60 [Ellegood, Henkelman, & Lerch, 2012]. All mice were perfused using a solution containing the gadolinium contrast agent ProHance (Bracco Diagnostics Inc., Princeton, NJ, USA) and followed the procedure as described in Cahill and colleagues [2012], Lerch and colleagues [2011], and Spring and colleagues [2007].
Magnetic Resonance Imaging
A 7.0 Tesla MRI scanner (Varian Inc., Palo Alto, CA, USA) with a 40-cm inner bore diameter was used. To increase throughput, a custom-built 16-coil solenoid array imaged 16 mice in parallel [Bock, Konyer, & Henkelman, 2003]. A T2-weighted three-dimensional (3D) fast spin echo (FSE) sequence was used and optimized for gray–white matter contrast [Nieman et al., 2005] with repetition time TR = 2000 ms, echo train length = 6, TEeff = 42 ms, field of view (FOV) 25 mm × 28 mm × 14 mm, and matrix size of 450 × 504 × 250, which provided an isotropic resolution of 56 μm. Total scan time was 11.7 hours. This sequence was used for the ITGB3 mouse model. For the MECP2 and NL3 KI mouse models, a 32 μm isotropic T2 weighted 3D FSE sequence was used with a TR of 325 ms and a TE of 10 ms with 6 echos. The FOV was 14 mm × 14 mm × 25 mm with a matrix size of 432 × 432 × 780. These images were then resampled to 56 μm with isotropic voxels for analysis.
Segmentation of the Mouse Cerebellum
Using the average MR image atlas created from 40 mouse brains (20 females), the cerebellum was segmented [Dorr et al., 2008] after resampling to a resolution of 56 μm with isotropic voxels. In Dorr and colleagues’ atlas, the cerebellum was defined as cerebellar cortex and arbor vita, with a separate designation for the right and left sides. This is not sufficient detail for studying the specific anatomy of the cerebellum. To segment the gray matter, the anatomical definitions of Larsell [1952] were used, guided by two stereotactic atlases [Franklin & Paxinos, 2007; Hof, Young, Bloom, Belichenko, & Celio, 2000].
To segment the white matter, anatomical definitions were based on the gray matter lobule proximity along with the developmental trajectory and connectivity of the cerebellum. For example, specific incoming white matter fibers, such as climbing fibers from the inferior olivary complex, connect with precise PC anterior–posterior strips [Voogd, 2012]. Furthermore, anchor regions form early in development and are a crucial event in folia development [Sudarov & Joyner, 2007]. These anchor regions help demarcate folia and begin to segregate spatially specific white matter fibers. This supports an anterior–posterior differentiation of white matter based on connectivity and spatial separation in the cerebellar cortex. In the mediolateral direction, it has been shown that PCs are born at different time points depending on final migration location [Hashimoto & Mikoshiba, 2003]. Lateral hemisphere PCs differentiate before those that migrate to the vermis, which is located midline. Additionally, the lateral cerebellar cortex connects to the dentate nucleus of the deep cerebellar nuclei, while the paravermis connects to the interposed nucleus, and the vermis connects to the fastigial nucleus. This mediolateral specific connectivity with the cerebellar nuclei is sufficient evidence that white matter tracts from cerebellar cortical regions are distinct until they join together in the trunk of the arbor vita.
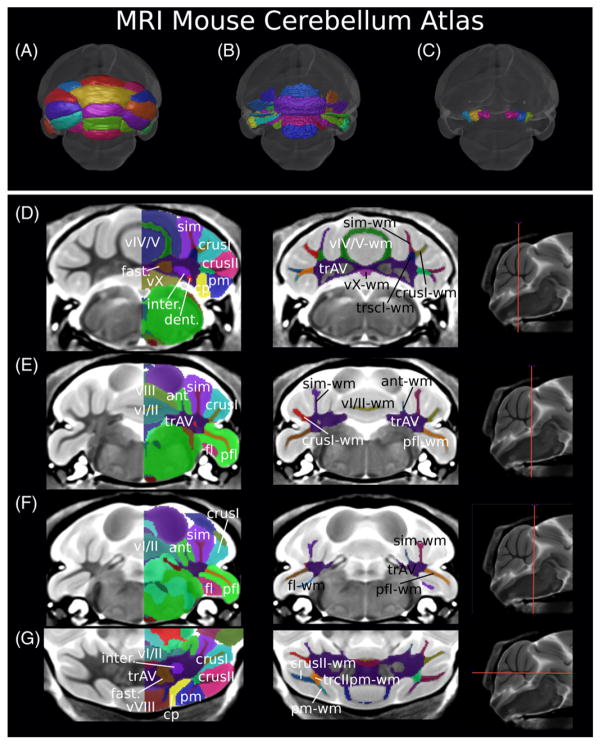
This reasoning guided segmentation (Fig. 1) using coronal slices and was performed using the software package display (http://www.bic.mni.mcgill.ca/ServicesSoftwareVisualization/Display, Montreal Neurological Institute, Montreal, Canada). Segmentations were subsequently refined using the transverse and sagittal orientations, ensuring smooth 3D volumes. The cerebellar cortex was segmented first, allowing segmentation of the larger lobules to help delineate the smaller ones, then the deep cerebellar nuclei, and finally the white matter components. A list of the structures segmented can be found in Table 1, and several two-dimensional representations of the atlas can be found in Figure 1. In total, 60 unique labels for 39 structures with 21 of those having a left and right side definition were introduced. This expanded Dorr et al.’s atlas [2008] from 62 to 99 structures after removing the two labels previously used for the cerebellum. Lobule boundaries were determined using the fissures as done by Ullmann [Ullmann et al., 2012], whereas white matter boundaries were determined using reasoning around connectivity and spatial orientation.
Figure 1.
The magnetic resonance imaging MRI mouse cerebellar atlas. A, B, and C represent surface renderings illustrating the cortical surface, white matter surface, and deep cerebellar nuclei, respectively. D, E, and F are coronal slices with labeled structures, gray matter structures labeled on the left column, and white matter structures labeled on the right (with the exception of the arbor vita trunk, labeled in both columns). G is a transverse slice with gray matter structures labeled in the left image and white matter structures in the right. The colors are distinct for each label; therefore, structures with a right and left label will be colored differently depending on the side of the brain. For the left column, only one side is labeled and colored so the underlying MRI contrast can be seen.
Table 1.
Cerebellar Structures Segmented in the MRI Cerebellar Atlas Including Their Volume
| Structure | Abbreviation | Hierarchy | Mean Volume ± SEM (mm3) |
|---|---|---|---|
| Lobules I–II | vI/II | Vermis | 1.83 ± 0.03 |
| Lobule III | vIII | Vermis | 1.97 ± 0.04 |
| Lobule IV–V | vIV/V | Vermis | 4.37 ± 0.06 |
| Lobule VI | vVI | Vermis | 2.70 ± 0.05 |
| Lobule VII | vVII | Vermis | 0.86 ± 0.02 |
| Lobule VIII | vVIII | Vermis | 1.47 ± 0.03 |
| Lobule IX | vIX | Vermis | 2.92 ± 0.05 |
| Lobule X | vX | Vermis | 1.47 ± 0.02 |
| Anterior lobule (lobules 4–5) | ant | Paravermis | 1.81 ± 0.02 |
| Simple lobule (lobule 6) | sim | Hemisphere | 5.20 ± 0.07 |
| Crus I (lobule 6) | crusI | Hemisphere | 4.50 ± 0.06 |
| Crus II (lobule 7) | crusII | Hemisphere | 3.47 ± 0.05 |
| Paramedian lobule (lobule 7) | pm | Hemisphere | 4.00 ± 0.04 |
| Copula of the pyramis (lobule 8) | cp | Hemisphere | 2.65 ± 0.04 |
| Flocculus | fl | Flocculonodular lobe | 1.02 ± 0.02 |
| Paraflocculus | pfl | Flocculonodular lobe | 4.00 ± 0.09 |
| Trunk of arbor vita | trAV | White matter | 4.30 ± 0.05 |
| Lobule I–II white matter | vI/II-wm | Vermis white matter | 0.089 ± 0.002 |
| Lobule III white matter | vIII-wm | Vermis white matter | 0.226 ± 0.004 |
| Trunk of lobules I–III white matter | trI-III-wm | Vermis white matter | 0.144 ± 0.002 |
| Lobules IV–V white matter | vIV/V-wm | Vermis white matter | 0.71 ± 0.01 |
| Lobules VI–VII white matter | vVI/VII-wm | Vermis white matter | 0.79 ± 0.01 |
| Lobule VIII white matter | vVIII-wm | Vermis white matter | 0.154 ± 0.004 |
| Trunk of lobules VI–VIII white matter | trVI-VIII-wm | Vermis white matter | 0.118 ± 0.001 |
| Lobule IX white matter | vIX-wm | Vermis white matter | 0.343 ± 0.006 |
| Lobule X white matter | vX-wm | Vermis white matter | 0.103 ± 0.001 |
| Anterior lobule white matter | ant-wm | Paravermis white matter | 0.102 ± 0.002 |
| Simple lobule white matter | sim-wm | White matter | 0.52 ± 0.01 |
| Crus I white matter | crusI-wm | White matter | 0.474 ± 0.007 |
| Trunk of simple and crus I white matter | trscI-wm | White matter | 0.188 ± 0.003 |
| Crus II white matter | crusII-wm | White matter | 0.278 ± 0.0405 |
| Paramedian lobule | pm-wm | White matter | 0.172 ± 0.003 |
| Trunk of crus II and paramedian white matter | trcIIpm-wm | White matter | 0.371 ± 0.005 |
| Copula white matter | cp-wm | White matter | 0.094 ± 0.001 |
| Paraflocculus white matter | pfl-wm | White matter | 0.343 ± 0.006 |
| Flocculus white matter | fl-wm | White matter | 0.077 ± 0.001 |
| Dentate nucleus | dent | Deep nuclei | 0.307 ± 0.005 |
| Nucleus interpositus | inter | Deep nuclei | 0.402 ± 0.007 |
| Fastigial nucleus | fast | Deep nuclei | 0.442 ± 0.006 |
SEM, standard error of the mean.
White matter boundaries were determined moving outward from the trunk of the arbor vita to the cerebellar cortex. The trunk, encompassing the separately labeled deep cerebellar nuclei, comprised the innermost cerebellar white matter label. Moving out toward the cerebellar cortex, if a lobule’s white matter tract did not differentiate into two tracts more outwardly, then the white matter component was labeled and named for the lobule it was in. An example of this is the paraflocculus (Fig. 1E–F). If the white matter lead to several lobules, it was labeled as the trunk of the lobules it eventually split into. Moving out from these multilobule trunks, the white matter was labeled for each lobule once the trunk portion divided (Fig. 1D–G). An example of this is hemisphere simple and crus I lobules, which white matter is shared as a trunk initially and eventually separates as one moves mediolaterally.
Image Registration and Analysis
To compare neuroanatomy between groups, each model and its corresponding WT were aligned linearly first, using translations and rotations only. From this linear alignment, we followed the MAGeT procedure outlined in [Chakravarty et al., 2013], wherein multiple templates are created and used to improve the segmentation accuracy. Registration was performed using the toolkit Advanced Normalization Tools [Avants, Epstein, Grossman, & Gee, 2008; Avants et al., 2011]. This toolkit performs cross-correlation to compare the two images being registered for similarity. The toolkit uses symmetric normalization diffeomorphic registration to ensure differences between images arise from anatomical differences only [Avants, Epstein, Grossman, & Gee, 2008; Avants et al., 2011]. From the final voted segmentation, volume changes were calculated and expressed in absolute (mm3) or relative (percentage of total brain) volumes. Results are also reported as Glass’s effect sizes (ES) where μ is the mean volume of either the ASD mouse model or the WT group, and σ is the standard deviation of the WT volumes:
To control for multiple comparisons, we used the false discovery rate (FDR; [Benjamini & Hochberg, 1995; Genovese, Lazar, & Nichols, 2002]). The FDR threshold was determined using all 99 brain structures in the mouse brain atlas, a combination of the structures segmented here and in Dorr colleagues’ atlas [2008].
Results
Neuroligin-3 R451C
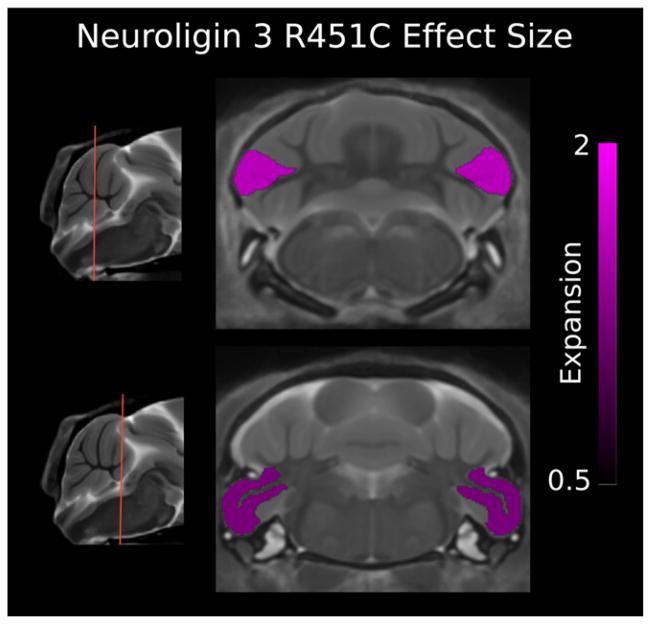
Previous work showed the brain volume of NL3 KI to be smaller than WT control mice [Ellegood et al., 2011]. Relative volumes, as a percent of total brain volume, are discussed here to minimize differences in total brain volume. The NL3 KI mouse model showed very specific cerebellar changes with three of the 39 structures significantly larger (FDR < 5%). The larger structures included crus II of the ansiform lobule, in both the gray and white matter, and the paraflocculus, with mean ES 1.94, 1.84, and 1.45, and percent volume difference 10%, 12%, and 12%, respectively (Fig. 2; Table 2).
Figure 2.
Two coronal slices of the Neuroligin-3 R451C (NL3 KI) mouse model compared with wild type (WT) showing the effect size of each significant false discovery rate < 5%) cerebellum structure. The paraflocculus gray matter and crus II gray and white matter are bilaterally larger in volume. A positive effect size represents an expansion of the region in the NL3 R451C mouse relative to controls (volumes have been normalized by total brain volume).
Table 2.
Volume Differences in the NL3 KI, MECP2 308 and ITGB3 Mouse Models
| Structure | Volume (mean ± SD) absolute (mm3)
|
Relative volume % difference | Effect size | q-value | |
|---|---|---|---|---|---|
| WT | ASD Model | ||||
| NL3 KI | |||||
| Total brain volume | 477 ± 19 | 438 ± 18 | −8a | −2.06 | 0.02 |
| Total cerebellum volume | 61 ± 2 | 58 ± 4 | −5a | −1.32 | 0.2 |
| Crus II: ansiform lobule (lobule 7) | 3.9 ± 0.2 | 4.0 ± 0.2 | 10 | 1.94 | 3 × 10−2 |
| Paraflocculus (PFL) | 4.4 ± 0.4 | 4.5 ± 0.4 | 12 | 1.45 | 5 × 10−2 |
| Crus II white matter | 0.34 ± 0.03 | 0.35 ± 0.03 | 11 | 1.84 | 4 × 10−2 |
| MECP2 hemizygous | |||||
| Total brain volume | 478 ± 11 | 465 ± 11 | −3a | −1.22 | 0.1 |
| Total cerebellum volume | 54 ± 1 | 55 ± 3 | 1a | 0.57 | 0.6 |
| Lobule IX | 2.8 ± 0.2 | 3.0 ± 0.3 | 11 | 1.78 | 2 × 10−2 |
| Lobule X | 1.29 ± 0.04 | 1.4 ± 0.1 | 11 | 3.95 | 3 × 10−3 |
| Lobule IX white matter | 0.38 ± 0.02 | 0.41 ± 0.04 | 12 | 2.30 | 1 × 10−2 |
| MECP2 heterozygous | |||||
| Total brain volume | 478 ± 11 | 464 ± 21 | −31 | −1.35 | 0.03 |
| Total cerebellum volume | 54 ± 1 | 55 ± 4 | 21 | 0.79 | 0.4 |
| Lobule VI | 2.51 ± 0.08 | 2.6 ± 0.2 | 7 | 1.72 | 6 × 10−4 |
| Lobule VIII | 1.54 ± 0.08 | 1.6 ± 0.2 | 8 | 1.54 | 5 × 10−2 |
| Lobule X | 1.29 ± 0.04 | 1.4 ± 0.1 | 12 | 4.28 | 2 × 10−2 |
| Anterior lobule (lobules 4–5) | 1.8 ± 0.1 | 1.8 ± 0.1 | 6 | 1.20 | 2 × 10−2 |
| Simple lobule (lobule 6) | 4.8 ± 0.1 | 4.9 ± 0.4 | 5 | 1.80 | 3 × 10−2 |
| Crus I: ansiform lobule (lobule 6) | 4.4 ± 0.1 | 4.6 ± 0.4 | 6 | 1.87 | 2 × 10−2 |
| Crus II: ansiform lobule (lobule 7) | 4.0 ± 0.2 | 4.2 ± 0.4 | 6 | 1.64 | 2 × 10−2 |
| Copula white matter | 0.09 ± 0.02 | 0.10 ± 0.01 | 9 | 1.33 | 2 × 10−2 |
| MECP2 homozygous | |||||
| Total brain volume | 478 ± 11 | 468 ± 12 | −2a | −0.93 | 9 × 10−2 |
| Total cerebellum volume | 54 ± 1 | 57 ± 1 | 5a | 2.41 | 6 × 10−3 |
| Lobule III | 1.9 ± 0.2 | 2.0 ± 0.2 | 8 | 1.01 | 4 × 10−2 |
| Lobules IV–V | 4.1 ± 0.3 | 4.3 ± 0.2 | 5 | 0.89 | 4 × 10−2 |
| Lobule VI | 2.51 ± 0.08 | 2.64 ± 0.08 | 7 | 1.72 | 4 × 10−4 |
| Lobule VII | 0.88 ± 0.05 | 0.93 ± 0.09 | 8 | 1.36 | 2 × 10−2 |
| Lobule VIII | 1.54 ± 0.08 | 1.7 ± 0.1 | 10 | 1.96 | 1 × 10−3 |
| Lobule IX | 2.8 ± 0.2 | 3.2 ± 0.2 | 16 | 2.50 | 8 × 10−5 |
| Lobule X | 1.29 ± 0.04 | 1.5 ± 0.1 | 14 | 5.05 | 8 × 10−6 |
| Anterior lobule (lobules 4–5) | 1.8 ± 0.1 | 1.85 ± 0.07 | 8 | 1.65 | 1 × 10−3 |
| Simple lobule (lobule 6) | 4.8 ± 0.1 | 5.0 ± 0.2 | 6 | 2.41 | 3 × 10−3 |
| Crus I: ansiform lobule (lobule 6) | 4.4 ± 0.1 | 4.7 ± 0.2 | 8 | 2.49 | 1 × 10−3 |
| Crus II: ansiform lobule (lobule 7) | 4.0 ± 0.2 | 4.3 ± 0.2 | 9 | 2.25 | 7 × 10−4 |
| Copula: pyramis (lobule 8) | 2.4 ± 0.1 | 2.5 ± 0.1 | 6 | 1.09 | 6 × 10−2 |
| Paraflocculus (PFL) | 3.9 ± 0.3 | 4.3 ± 0.4 | 13 | 1.94 | 2 × 10−3 |
| Lobule VIII white matter | 0.19 ± 0.01 | 0.20 ± 0.01 | 7 | 0.91 | 4 × 10−2 |
| Lobule IX white matter | 0.38 ± 0.02 | 0.42 ± 0.03 | 13 | 2.48 | 4 × 10−4 |
| Anterior lobule white matter | 0.09 ± 0.01 | 0.10 ± 0.01 | 8 | 1.27 | 2 × 10−2 |
| Crus I white matter | 0.45 ± 0.02 | 0.46 ± 0.02 | 6 | 1.47 | 4 × 10−2 |
| Copula white matter | 0.09 ± 0.02 | 0.10 ± 0.01 | 13 | 1.88 | 7 × 10−4 |
| Paraflocculus white matter | 0.36 ± 0.02 | 0.38 ± 0.02 | 7 | 1.25 | 6 × 10−3 |
| ITGB3 | |||||
| Total brain volume | 483 ± 21 | 430 ± 12 | −11a | −2.52 | 2 × 10−5 |
| Total cerebellum volume | 62 ± 4 | 49 ± 2 | −20a | −3.04 | 3 × 10−3 |
| Lobules I–II | 2.1 ± 0.3 | 1.4 ± 0.2 | −26 | −2.35 | 2 × 10−4 |
| Lobule III | 2.4 ± 0.3 | 1.9 ± 0.1 | −12 | −1.31 | 6 × 10−4 |
| Lobules IV–V | 4.9 ± 0.5 | 3.8 ± 0.2 | −12 | −1.76 | 4 × 10−3 |
| Lobule VI | 2.9 ± 0.3 | 2.24 ± 0.09 | −11 | −1.52 | 1 × 10−4 |
| Llobule VII | 0.9 ± 0.1 | 0.71 ± 0.07 | −14 | −1.37 | 2 × 10−4 |
| Lobule VIII | 1.6 ± 0.1 | 1.23 ± 0.09 | −15 | −2.28 | 5 × 10−4 |
| Lobule IX | 3.1 ± 0.3 | 2.3 ± 0.1 | −15 | −1.74 | 2 × 10−5 |
| Lobule X | 1.5 ± 0.2 | 1.17 ± 0.08 | −14 | −1.64 | 3 × 10−3 |
| Simple lobule (lobule 6) | 6.0 ± 0.4 | 4.6 ± 0.2 | −14 | −2.85 | 8 × 10−7 |
| Crus I: ansiform lobule (lobule 6) | 5.4 ± 0.4 | 4.3 ± 0.2 | −11 | −1.87 | 1 × 10−4 |
| Copula: pyramis (lobule 8) | 2.6 ± 0.2 | 2.1 ± 0.1 | −9 | −1.04 | 1 × 10−2 |
| Trunk of arbor vita | 4.4 ± 0.3 | 3.4 ± 0.1 | −12 | −3.29 | 1 × 10−7 |
| Lobule I–II white matter | 0.09 ± 0.01 | 0.05 ± 0.01 | −42 | −4.21 | 1 × 10−7 |
| Lobule III white matter | 0.29 ± 0.04 | 0.19 ± 0.02 | −27 | −2.20 | 2 × 10−5 |
| Trunk of lobules I–III white matter | 0.16 ± 0.02 | 0.11 ± 0.01 | −25 | −3.06 | 5 × 10−7 |
| Lobules IV–V white matter | 0.81 ± 0.09 | 0.62 ± 0.04 | −13 | −1.61 | 8 × 10−4 |
| Lobules VI–VII white matter | 0.88 ± 0.09 | 0.69 ± 0.04 | −11 | −1.55 | 2 × 10−3 |
| Lobule VIII white matter | 0.19 ± 0.02 | 0.14 ± 0.01 | −20 | −2.28 | 2 × 10−5 |
| Trunk of lobules VI–VIII white matter | 0.13 ± 0.01 | 0.10 ± 0.01 | −10 | −1.70 | 2 × 10−3 |
| Lobule IX white matter | 0.40 ± 0.03 | 0.28 ± 0.01 | −20 | −3.16 | 3 × 10−7 |
| Lobule X white matter | 0.11 ± 0.02 | 0.08 ± 0.01 | −21 | −1.52 | 5 × 10−4 |
| Anterior lobule white matter | 0.10 ± 0.01 | 0.09 ± 0.01 | −8 | −1.01 | 2 × 10−2 |
| Simple lobule white matter | 0.54 ± 0.04 | 0.35 ± 0.05 | −27 | −5.37 | 2 × 10−7 |
| Crus I white matter | 0.51 ± 0.04 | 0.33 ± 0.04 | −27 | −3.97 | 8 × 10−7 |
| Trunk of simple and crus I white matter | 0.19 ± 0.01 | 0.18 ± 0.01 | 8 | 1.38 | 1 × 10−2 |
| Copula white matter | 0.10 ± 0.01 | 0.08 ± 0.01 | −10 | −1.32 | 6 × 10−3 |
| Dentate nucleus | 0.34 ± 0.02 | 0.26 ± 0.01 | −14 | −2.53 | 3 × 10−6 |
| Nucleus interpositus | 0.43 ± 0.04 | 0.32 ± 0.02 | −15 | −2.12 | 3 × 10−5 |
| Fastigial nucleus | 0.43 ± 0.04 | 0.32 ± 0.01 | −15 | −2.64 | 2 × 10−6 |
Percent difference of absolute volume, not relative volume.
ASD, autism spectrum disorder; ITGB3, integrin β3; NL3 KI, neuroligin-3 R451C knock-in; MECP2, methyl-CpG binding protein-2; SD, standard deviation; WT, wild type.
MECP2 308 Truncation
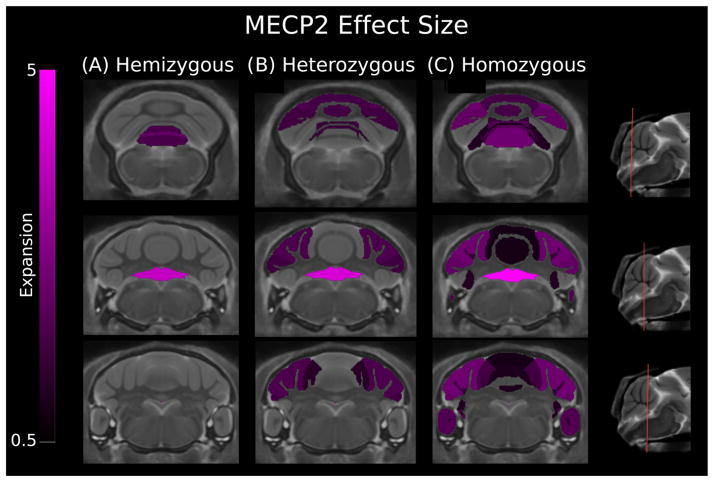
For total brain volume, the hemizygous genotype brain volume differed by −3% (t(50) = −2.44), heterozygous at −3% (t(50) = −2.85), and homozygous by −2% (t(50) =−1.98). Relative volumes, as a percent of total brain volume, are reported here. Each genotype was compared with a WT group comprised of males and females. We tested for and found no sex–genotype interaction. The hemizygous genotype had three structures out of 39 significantly larger (FDR < 5%), all confined to the vermis (Fig. 3; Table 2). Vermis lobule IX was smaller for both gray and white matter with mean ES of 1.78 and 2.30, respectively. Vermis lobule X, the nodulus, was the strongest change with an ES of 3.95. The heterozygous genotype, with the largest total brain volume difference, had eight structures smaller (FDR < 5%; Fig. 3; Table 2). Structures were confined to the posterior lobe of the cerebellum, with the vermis lobules VI, VII, and X affected with mean ES of 1.72, 1.54 and 4.58, respectively (FDR < 5%). For the hemispheres, the anterior lobule was the only anterior lobe portion with a volume larger than the WT and had a mean ES of 1.20. The lateral extensions of lobule VI, the simple lobule and crus I of the ansiform lobule, were larger in volume along with crus II, the second component of the ansiform lobule. The mean ES were 1.80, 1.87, and 1.64 for the simple lobule, crus I and crus II, respectively. The MECP2 308 homozygous model showed the largest and most comprehensive volume changes across the three MECP2 genotypes with 19 of the 39 structures in the cerebellum bigger (Fig. 3; Table 2). In the anterior lobe, vermis lobules II and IV–V along with the hemisphere extension of IV–V, the anterior lobule, were larger in volume as compared with the WT. The posterior lobe overall is larger in volume, which includes vermis lobules VI–X. The simple lobule, crus I, crus II, copula, and paraflocculus were enlarged in the homozygous group as well. Across all three MECP2 models, the vermis lobule X had the greatest or second largest mean ES.
Figure 3.
Three coronal slices of the Methyl-CpG binding protein-2 (MECP2) mouse models compared with wild type showing effect size for cerebellum structures. The hemizygous model showed increases in the gray and white matter of vermis lobule IX and gray matter of vermis lobule X. The heterozygous model showed expansion of vermis lobule VI, VIII, and X. In the hemispheres, the anterior, simple, crus I, and crus II gray matter are larger in volume. The homozygous model had expansion of vermis lobules III–X, the anterior lobule, the paraflocculus, and simple, crus I, and crus II hemisphere lobules. A positive effect size represents an expansion of the region in the MECP2 mouse relative to controls (volumes have been normalized to total brain volume). These changes have been corrected for multiple comparisons using a false discovery rate threshold of 5%. Genotypes: hemizygous: −/y, heterozygous: −/+, homozygous: −/−.
Integrin Beta-3
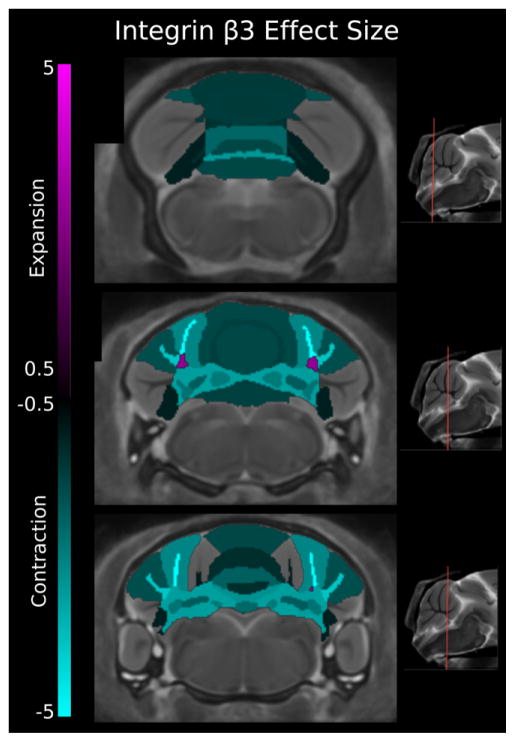
The total brain volume of ITGB3 KO mouse has been reported to be 11% smaller [Ellegood et al., 2012]. To assess cerebellar regions, volumes were normalized to brain volume, controlling for total brain volume differences between WT and ITGB3. Of the 39 structures in the cerebellum, 28 were significantly smaller at an FDR < 5% (Fig. 4), this held for absolute volumes as well (see Table 2). With respect to the vermis, its entirety was found to be smaller in both white and gray matter. The deep cerebellar nuclei were bilaterally smaller with a mean ES of −2.53, −2.11, and −2.64, respectively for the dentate, interposed, and fastigial nuclei. In the hemispheres, the two lateral extensions of lobule VI, the simple lobule and crus I were found to be smaller in both the white and gray matter. Lobule VIII’s lateral extension, the copula, was significantly smaller in both white and gray matter, mean ES of −1.32 and −1.04, respectively.
Figure 4.
Three coronal slices of the integrin β3 (ITGB3) mouse model compared with wild type showing effect size. Smaller volumes (cyan) were found in all vermis lobules (gray and white matter), the trunk of the arbor vita, and the gray and white matter of the simple, crus I, crus II, and copula hemisphere lobules. However, the trunk of the simple and crus I lobules was found to be larger in volume (magenta). All three deep cerebellar nuclei showed bilateral decreases in volume. A positive effect size represents an expansion of the region in the ITGB3 mouse relative to controls, and a negative value represents a contraction in that region of the ITGB3 mouse relative to controls (volumes have been normalized to total brain volume). These changes have been corrected for multiple comparisons using a false discovery rate threshold of 5%.
Discussion
The MRI cerebellum atlas represents a powerful and efficient method to study cerebellar anatomy. The data reported in our current study illustrate the ability of MRI followed by image processing and statistical analysis to discern volumetric changes within distinct anatomical units of the cerebellum. This has been demonstrated on three genetic mouse models of ASD. With some anatomical continuity across the three mouse models, we find specific cerebellar changes that relate closely to known behavioral traits (Fig. 5; Table 2). Furthermore, the ability to characterize the cerebellum across these ASD mouse models marks an improvement over current atlas-based analysis methods.
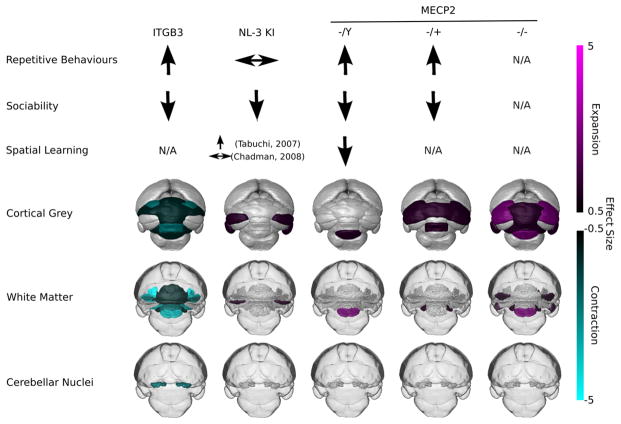
Figure 5.
Summary of behavioral and neuroanatomical findings for integrin β3 (ITGB3) knock out, Neuroligin-3 R451C knock in (NL3 KI), and methyl-CpG binding protein-2 (MECP2) knock out in hemizygous, heterozygous, and homozygous genotypes. Cortical gray matter images are surface renderings of the cortex of the cerebellum; white matter images (shown in darker gray) are surface renderings of the white matter aspects with a transparent outline of the brain to aid in orientation; and cerebellar nuclei images are the deep cerebellar nuclei surface renderings, again with a transparent outline of the brain to aid in orientation. These renderings illustrate volume changes across the cerebellum. Effect size is shown with cyan representing contraction and magenta being expansion. N/A; Not reported in the literature.
In the NL3 KI model, we show volumetric changes specific to crus II and the paraflocculus, providing additional volumetric data to our previously reported diffusion imaging results [Ellegood et al., 2011]. We also show, in ITGB3, distinct regions of the cerebellar gray and white matter with reduced volume (Table 2), building upon work reported in 2012 that found volume changes in a region encompassing the arbor vita and cerebellar cortex [Ellegood et al., 2012]. The MECP2 mouse model’s neuroanatomy has not previously been studied with MRI in this detail. We report relative volume increases in all three genotypes (Fig. 3).
Among the studied ASD mouse models, our volumetric results reveal several similarities and differences in cerebellar anatomy. For similarities, all three volumetric results have significant differences from the WT mice in the cerebellum. This shows that at least, these three ASD-associated genes either have a direct or indirect effect on cerebellum morphology. It also appears that the posterior vermis is morphologically altered in models with reported repetitive behaviors (Fig. 5, see further discussion below). For differences, the regions of the cerebellum with volumetric differences are not all the same across the three ASD mouse models. In addition to differences in cerebellar regions, some regions affected in multiple mouse models studied differ in magnitude and whether they are smaller or larger in volume. For example, the crus II gray matter is larger in volume for the NL3 and MECP2 heterozygous and homozygous models. But crus II gray matter is not significantly different in volume from WT mice for the ITGB3 mouse model and the MECP2 hemizygous mouse. This reveals that the ASD-associated genes studied here have effects that can be similar to other genes or entirely different in terms of morphology.
From human neuroimaging studies, the cerebellum appears to have an altered morphology [Anagnostou & Taylor, 2011; Stanfield et al., 2008], which is in agreement with our study. However, the Stanfield systematic review found the effect of the total cerebellum volume to be larger than controls. This result differs from the results of the NL3 and ITGB3 models studied but is in agreement with the MECP2 homozygous genotype volumetric results. This may explain why human neuroimaging studies, where samples are typically heterogeneous in ASD-associated genetic lesions, have had trouble forming a consensus. Several neuroimaging studies have reported no volumetric differences between controls in the cerebellum, which we find for NL3 and ITGB3. However, we are now able to look at the cerebellum’s substructures. This added level of detail localizes the volumetric changes, something not yet performed in humans and shows where morphological changes may be occurring at the regional level within the cerebellum.
The results reported here on the neuroanatomical phenotype are important because anatomy is closely connected to behavior. This connection is clear in Nieman and colleagues’ study, where 17 of 19 genetic mouse models with behavior or motor neural abnormalities were found with atypical neuroanatomy [2007]. The rest of the discussion focuses on how the reported results in the cerebellum may contribute the behavioral traits of these mice and seen in ASD. Although it should be clear that the behaviors observed in ASD are likely rooted in a network of brain structures, it remains important to understand how the cerebellum may contribute to those networks and their function.
The Vermis and Repetitive Behavior Traits
Repetitive behavior represents one of the three core features of autism. These behaviors are consistently observed in many transgenic mouse models of ASD [Silverman et al., 2010]. By using a mouse model that lost PS during the postnatal period and crossing it with WT mice, Martin, Goldowitz, and Mittleman [2010] were able to generate mice displaying a range of PC loss. These mice were found to have greater repetitive behaviors when the cerebellar PC population size was smaller. Additionally, PC population size in the cerebellar cortex is consistently found to be smaller in postmortem studies of ASD patients [Bauman & Kemper, 2005]. These findings, when taken together, connect the PC to the repetitive behavior observed in ASD. Furthermore, in a group of ASD patients, repetitive behavior severity was found to negatively correlate with the area of vermis VI–VII [Pierce & Courchesne, 2001], two lobules in the posterior vermis.
Across the three ASD models investigated here, two have known repetitive behaviors: ITGB3 [Carter et al., 2011] and MECP2 [Samaco et al., 2012; Shahbazian et al., 2002]. The third mouse model, NL3 KI, does not show increased repetitive self-grooming [Chadman et al., 2008]. We report only finding posterior vermis volumetric abnormalities in the two models with repetitive behavior. The ITGB3 mouse model has a smaller volume of 11% (ES = −1.52) and 14% (ES = −1.37) for vermis lobules VI and VII, respectively (Table 2). The MECP2 heterozygous model shows a 7% (ES = 1.72) larger vermis lobule VI and the homozygous model a 7% (ES = 1.72) bigger vermis VI and 8% (ES = 1.36) bigger vermis VII. The MECP hemizygous group shows strong changes in the vermis lobule IX with an 11% (ES = 1.78) and 12% (ES = 2.30) larger gray and white matter, respectively. From these results, we speculate that these regions have a role in the repetitive behaviors exhibited by these ASD mouse models. Future work should include these regions when examining other motor areas for links between repetitive behaviors and brain morphology.
Of note is the difference in direction of the changes across the two models with repetitive behavior phenotypes. The ITGB3 posterior vermis is smaller in volume, while the MECP2 is larger. Furthermore, the ITGB3 mice have both gray and white matter changes, whereas for the MECP2 mice, changes are contained within the gray matter. One possible reason could be that the PC have decreased in number [Martin et al., 2010] or altered in cellular morphology, similar to the increased spine density in fragile X syndrome [Koekkoek et al., 2005]. Both would have an effect on the cerebellar circuits that may increase repetitive behaviors and either grow or shrink the cerebellum structure depending on the cellular change. Future work should investigate these possibilities and how the cerebellum may contribute to repetitive behaviors in ASD in conjunction with the rest of the brain.
Lateral Hemisphere and Social Avoidance
A second core feature of ASD is social deficits. In 2012, Tsai and colleagues [2012] published results of an ASD mouse model that knocked out the TSC1 gene within the cerebellar PC only and then demonstrated social deficits in these mice. Further work on cerebellar function gives evidence for sociability being associated with the cerebellum through demands sociability places on the executive control network. The cerebellar contribution to the executive control network includes crus I and II of the cerebellar hemispheres [Stoodley & Schmahmann, 2009].
From the three models in this study, two have reported social avoidance behaviors and the third deficits for social novelty [Carter et al., 2011; Chadman et al., 2008; Etherton et al., 2011; Moretti, Bouwknecht, Teague, Paylor, & Zoghbi, 2005; Pearson et al., 2012; Pobbe, Pearson, Blanchard, & Blanchard, 2012; Samaco et al., 2012; Tabuchi et al., 2007]. In addition to reported social deficits, these mice have crus I and II morphological abnormalities (Table 2). These results suggest regions of interest in the cerebellum that may contribute to networks involved in sociability.
Lateral Hemispheres and Learning
The cerebellum plays an important role in the learning of motor tasks and a suggested role in higher order cognitive functions [D’Angelo & Casali, 2012; Fatemi et al., 2012]. A common example of cerebellum learning is the eye air puff classical conditioning, where the circuitry for this process in the cerebellum involves the simple lobule, crus I and posterior vermis in the cortex, and the interposed nuclei [Manto et al., 2012]. Deficits in the air puff classical conditioning have been reported in ASD population for acquisition [Sears, Finn, & Steinmetz, 1994]. In addition to this paradigm, a recent paper looking at brain plasticity in spatial learning of mice showed cerebellar changes within the simple lobule, crus I and posterior vermis [Lerch, Yiu, et al., 2011]. In the three models studied here, cerebellar hemispheric regions are altered in volume (Table 2). The anatomical results presented here contribute to the literature that these cerebellar regions may participate in networks essential to learning.
Limitations
The design of this study provides some limitations in interpreting and extending these results to ASD genetics and behaviors. Better study of the cerebellar abnormalities using littermate controls and assessing the behavioral phenotype in the same mice would provide a link between ASD behavior and the cerebellum. A recent study showed no morphological change in the cerebellum over the age range of mice in our study [Woodruff-Pak et al., 2010]. However, over time morphology changes; therefore, future work should investigate developmental trajectories of brain morphology and include more mouse models of ASD.
Isolating genetic factors in ASD and studying them with animal models provides valuable insight into the disorder. However, the human condition has considerable heterogeneity for which mouse studies may not entirely be able to explain. Mouse and human studies on ASD need to continue in conjunction to help understand the disorder and develop new treatments.
Conclusion
We used MRI and statistical image analysis to study three genetic mouse models of ASD. Harnessing the power of high-throughput MRI and statistical analysis techniques allowed for detailed investigation of neuroanatomy. To study specific regions of anatomy, an MRI cerebellar atlas was created for detailed volume analysis in the cerebellum. This allowed us to investigate genetic effects on cerebellar structure in a quantitative manner, where structural differences may explain known behavioral characteristics. We have highlighted potential roles of MECP2, ITGB3, and NL3 KI within the cerebellum on repetitive and social behaviors and learning. The analysis performed here assesses the contribution of genetics to the neuroanatomy of the cerebellum in autism and can easily be extended to additional models of disease. Imaging and analyzing the entire brain in additional genetic models of ASD and their littermate controls will strengthen the link between neuroanatomy and genetics.
Acknowledgments
Grant sponsor CIHR; Grant number: MOP-106418, Grant sponsor OBI; Grant number: IDS-11-02.
The authors would like to thank Dr. James Ellis for contributing the MECP2 mice used in this study. We would also like to thank the Canadian Foundation for Innovation, the Ontario Research Fund, the Canada Research Chairs, the Natural Sciences and Engineering Research Council of Canada, and the Faculty of Medicine at University of Toronto. This research was conducted with the support of the Canadian Institute for Health Research (CIHR) and the Ontario Brain Institute (OBI). OBI is an independent nonprofit corporation, funded partially by the Ontario government. The opinions, results, and conclusions are those of the authors and no endorsement by the OBI is intended or should be inferred.
References
- Abrahams BS, Geschwind DH. Connecting genes to brain in the autism spectrum disorders. Archives of Neurology. 2010;67:395–399. doi: 10.1001/archneurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G, Courchesne E. Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: An fMRI study of autism. The American Journal of Psychiatry. 2003;160:262–273. doi: 10.1176/appi.ajp.160.2.262. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends in Neurosciences. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Anagnostou E, Taylor MJ. Review of neuroimaging in autism spectrum disorders: What have we learned and where we go from here. Molecular autism. 2011;2:1–4. doi: 10.1186/2040-2392-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychological Medicine. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu S, Packer A. SFARI Gene: An evolving database for the autism research community. Disease Models Mechanisms. 2010;3:133–135. doi: 10.1242/dmm.005439. [DOI] [PubMed] [Google Scholar]
- Bauman M, Kemper TL. Histoanatomic observations of the brain in early infantile autism. Neurology. 1985;35:866–874. doi: 10.1212/wnl.35.6.866. [DOI] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: A review and future directions. International Journal of Developmental Neuroscience. 2005;23:183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Berry-Kravis EM, Hessl D, Rathmell B, Zarevics P, Cherubini M, et al. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: A randomized, controlled, phase 2 trial. Science Translational Medicine. 2012;4:1–7. doi: 10.1126/scitranslmed.3004214. [DOI] [PubMed] [Google Scholar]
- Bock NA, Konyer NB, Henkelman RM. Multiple-mouse MRI. Magnetic Resonance in Medicine. 2003;49:158–167. doi: 10.1002/mrm.10326. [DOI] [PubMed] [Google Scholar]
- Cahill LS, Laliberté CL, Ellegood J, Spring S, Gleave JA, et al. Preparation of fixed mouse brains for MRI. Neuroimage. 2012;60:933–939. doi: 10.1016/j.neuroimage.2012.01.100. [DOI] [PubMed] [Google Scholar]
- Carter MD, Shah CR, Muller CL, Crawley JN, Carneiro AMD, Veenstra-VanderWeele J. Absence of preference for social novelty and increased grooming in integrin β3 knockout mice: Initial studies and future directions. Autism Research. 2011;4:57–67. doi: 10.1002/aur.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadman KK, Gong S, Scattoni ML, Boltuck SE, Gandhy SU, et al. Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Research. 2008;1:147–158. doi: 10.1002/aur.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty MM, Steadman P, van Eede MC, Calcott RD, Gu V, et al. Performing label-fusion-based segmentation using multiple automatically generated templates. Human Brain Mapping. 2013;34:2635–2654. doi: 10.1002/hbm.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani DC. Serotonin in autism and pediatric epilepsies. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10:112–116. doi: 10.1002/mrdd.20021. [DOI] [PubMed] [Google Scholar]
- Courchesne E. New evidence of cerebellar and brainstem hypoplasia in autistic infants, children and adolescents: The MR imaging study by Hashimoto and colleagues. Journal of Autism and Developmental Disorders. 1995;25:19–22. doi: 10.1007/BF02178164. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL. Hypoplasia of cerebellar vermal lobules VI and VII in autism. New England Journal of Medicine. 1988;318:1349–1354. doi: 10.1056/NEJM198805263182102. [DOI] [PubMed] [Google Scholar]
- Coutinho AM, Sousa I, Martins M, Correia C, Morgadinho T, et al. Evidence for epistasis between SLC6A4 and ITGB3 in autism etiology and in the determination of platelet serotonin levels. Human Genetics. 2007;121:243–256. doi: 10.1007/s00439-006-0301-3. [DOI] [PubMed] [Google Scholar]
- D’Angelo E, Casali S. Seeking a unified framework for cerebellar function and dysfunction: From circuit operations to cognition. Frontiers in Neural Circuits. 2012;6:116. doi: 10.3389/fncir.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Ellegood J, Henkelman RM, Lerch JP. Neuroanatomical assessment of the integrin β3 mouse model related to autism and the serotonin system using high resolution MRI. Frontiers in Psychiatry. 2012;3:1–9. doi: 10.3389/fpsyt.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J, Lerch JP, Henkelman RM. Brain abnormalities in a Neuroligin3 R451C knockin mouse model associated with autism. Autism Research. 2011;4:368–376. doi: 10.1002/aur.215. [DOI] [PubMed] [Google Scholar]
- Etherton M, Földy C, Sharma M, Tabuchi K, Liu X, et al. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proceedings of the National Academy of Sciences. 2011;108:13764–13769. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ey E, Leblond CS, Bourgeron T. Behavioral profiles of mouse models for autism spectrum disorders. Autism Research. 2011;4:5–16. doi: 10.1002/aur.175. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CD, et al. Consensus paper: Pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Realmuto G, Earle J, Kist DA, et al. Purkinje cell size is reduced in cerebellum of patients with autism. Cellular and Molecular Neurobiology. 2002;22:171–175. doi: 10.1023/A:1019861721160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. 3. San Diego: Academic Press; 2007. [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Habas C, Kamdar N, Nguyen D, Prater K, Beckmann CF, et al. Distinct cerebellar contributions to intrinsic connectivity networks. Journal of Neuroscience. 2009;29:8586–8594. doi: 10.1523/JNEUROSCI.1868-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of General Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Mikoshiba K. Mediolateral compartmentalization of the cerebellum is determined on the “birth date” of Purkinje cells. Journal of Neuroscience. 2003;23:11342–11351. doi: 10.1523/JNEUROSCI.23-36-11342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Tayama M, Murakawa K, Yoshimoto T, Miyazaki M, et al. Development of the brainstem and cerebellum in autistic patients. Journal of Autism and Developmental Disorders. 1995;25:1–18. doi: 10.1007/BF02178163. [DOI] [PubMed] [Google Scholar]
- Henderson C, Wijetunge L, Kinoshita MN, Shumway M, Hammond RS, et al. Reversal of disease-related pathologies in the fragile X mouse model by selective activation of GABAB receptors with arbaclofen. Science Translational Medicine. 2012;4:1–11. doi: 10.1126/scitranslmed.3004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Young WG, Bloom FE, Belichenko PV, Celio MR. Comparative cytoarchitectonic atlas of the C57BL/6 and 129/Sv mouse brains. Amsterdam, The Netherlands: Elsevier; 2000. [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son YEE, Muthuswamy L, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koekkoek SKE, Yamaguchi K, Milojkovic BA, Dortland BR, Ruigrok TJH, et al. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Larsell O. The morphogenesis and adult pattern of the lobules and fissures of the cerebellum of the white rat. The Journal of Comparative Neurology. 1952;97:281–356. doi: 10.1002/cne.900970204. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods in Molecular Biology. 2011a;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Yiu AP, Martinez-Canabal A, Pekar T, Bohbot VD, et al. Maze training in mice induces MRI-detectable brain shape changes specific to the type of learning. Neuroimage. 2011b;54:2086–2095. doi: 10.1016/j.neuroimage.2010.09.086. [DOI] [PubMed] [Google Scholar]
- Manto M, Bower JM, Conforto AB, Delgado-García JM, da Guarda SNF, et al. Consensus paper: Roles of the cerebellum in motor control —the diversity of ideas on cerebellar involvement in movement. Cerebellum. 2012;11:457–487. doi: 10.1007/s12311-011-0331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LA, Goldowitz D, Mittleman G. Repetitive behavior and increased activity in mice with Purkinje cell loss: A model for understanding the role of cerebellar pathology in autism. The European Journal of Neuroscience. 2010;31:544–555. doi: 10.1111/j.1460-9568.2009.07073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Human Molecular Genetics. 2005;14:205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, Bock NA, Bishop J, Sled JG, Josette Chen X, Mark Henkelman R. Fast spin-echo for multiple mouse magnetic resonance phenotyping. Magnetic Resonance in Medicine. 2005;54:532–537. doi: 10.1002/mrm.20590. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, Lerch JP, Bock NA, Chen XJ, Sled JG, Henkelman RM. Mouse behavioral mutants have neuroimaging abnormalities. Human Brain Mapping. 2007;28:567–575. doi: 10.1002/hbm.20408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BL, Defensor EB, Pobbe RLH, Yamamoto LHL, Bolivar VJ, et al. Mecp2 truncation in male mice promotes affiliative social behavior. Behavior Genetics. 2012;42:299–312. doi: 10.1007/s10519-011-9501-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce K, Courchesne E. Evidence for a cerebellar role in reduced exploration and stereotyped behavior in autism. Biological Psychiatry. 2001;49:655–664. doi: 10.1016/s0006-3223(00)01008-8. [DOI] [PubMed] [Google Scholar]
- Piven J, Saliba K, Bailey J, Arndt S. An MRI study of autism: The cerebellum revisited. Neurology. 1997;49:546–551. doi: 10.1212/wnl.49.2.546. [DOI] [PubMed] [Google Scholar]
- Pobbe RLH, Pearson BL, Blanchard DC, Blanchard RJ. Oxytocin receptor and Mecp2(308/Y) knockout mice exhibit altered expression of autism-related social behaviors. Physiology & Behavior. 2012;107:641–648. doi: 10.1016/j.physbeh.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, et al. Lower Purkinje cell counts in the cerebella of four autistic subjects: Initial findings of the UCLA-NSAC Autopsy Research Report. The American Journal of Psychiatry. 1986;143:862–866. doi: 10.1176/ajp.143.7.862. [DOI] [PubMed] [Google Scholar]
- Robertson HR, Feng G. Annual Research Review: Transgenic mouse models of childhood-onset psychiatric disorders. Journal of Child Psychology and Psychiatry, and Allied Disciplines. 2011;52:442–475. doi: 10.1111/j.1469-7610.2011.02380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, McGraw CM, Ward CS, Sun Y, Neul JL, Zoghbi HY. Female Mecp2+/− mice display robust behavioral deficits on two different genetic backgrounds providing a framework for pre-clinical studies. Human Molecular Genetics. 2012;22:96–109. doi: 10.1093/hmg/dds406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears LL, Finn PR, Steinmetz JE. Abnormal classical eye-blink conditioning in autism. Journal of Autism and Developmental Disorders. 1994;24:737–751. doi: 10.1007/BF02172283. [DOI] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Sillitoe RV, Fu Y, Watson C. The mouse nervous system. Chapter 11. Amsterdam: Elsevier; 2011. Cerebellum; pp. 360–397. [DOI] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nature Reviews Neuroscience. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi MSK, Sanders-Bush E. Serotonin and brain development. International Review of Neurobiology. 2004;59:111–174. doi: 10.1016/S0074-7742(04)59006-2. [DOI] [PubMed] [Google Scholar]
- Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage. 2007;35:1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Stanfield AC, McIntosh AM, Spencer MD, Philip R, Gaur S, Lawrie SM. Towards a neuroanatomy of autism: A systematic review and meta-analysis of structural magnetic resonance imaging studies. European Psychiatry. 2008;23:289–299. doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Stoodley CJ, Schmahmann JD. Functional topography in the human cerebellum: A meta-analysis of neuroimaging studies. Neuroimage. 2009;44:489–501. doi: 10.1016/j.neuroimage.2008.08.039. [DOI] [PubMed] [Google Scholar]
- Sudarov A, Joyner AL. Cerebellum morphogenesis: The foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Development. 2007;2:26. doi: 10.1186/1749-8104-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann JFP, Keller MD, Watson C, Janke AL, Kurniawan ND, et al. Segmentation of the C57BL/6J mouse cerebellum in magnetic resonance images. Neuroimage. 2012;62:1408–1414. doi: 10.1016/j.neuroimage.2012.05.061. [DOI] [PubMed] [Google Scholar]
- Voogd J. A note on the definition and the development of cerebellar Purkinje cell zones. Cerebellum. 2012;11:422–425. doi: 10.1007/s12311-012-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd J, Gerrits NM, Ruigrok TJ. Organization of the vestibulocerebellum. Annals of the New York Academy of Sciences. 1996;781:553–579. doi: 10.1111/j.1749-6632.1996.tb15728.x. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Kosova G, Delahanty RJ, Jiang L, Cook EH, et al. Variation in ITGB3 is associated with whole-blood serotonin level and autism susceptibility. European Journal of Human Genetics. 2006a;14:923–931. doi: 10.1038/sj.ejhg.5201644. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Ober C, Cook EH. ITGB3 shows genetic and expression interaction with SLC6A4. Human Genetics. 2006b;120:93–100. doi: 10.1007/s00439-006-0196-z. [DOI] [PubMed] [Google Scholar]
- Woodruff-Pak DS, Foy MR, Akopian GG, Lee KH, Zach J, et al. Differential effects and rates of normal aging in cerebellum and hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1624–1629. doi: 10.1073/pnas.0914207107. [DOI] [PMC free article] [PubMed] [Google Scholar]