Abstract
Recent advances in microfluidics to generate and control picoliter emulsions of water in oil have enabled ultra-sensitive assays for small molecules, proteins, nucleic acids, and cells. Unfortunately, the conventional fluorescence detection used to measure the outcome of these droplet-based assays has not proven suited to match the time and space multiplexing capabilities of microfluidic systems. To address this challenge, we developed an in-flow fluorescence detection platform that enables multiple streams of droplets to be monitored using only a single photodetector and no lenses. The key innovation of our technology is the amplitude modulation of the signal from fluorescent droplets using distinct micro-patterned masks for each channel. By taking advantage of the high bandwidth of electronics, our technique enables the velocity-independent recovery of weak fluorescent signals (SNR ≪ 1) using only simple hardware, obviating the need for lasers, bulky detectors, and complex fluid control. We demonstrated a handheld-sized device that simultaneously monitors four independent channels with the capability to be scaled-up to more than sixteen, limited primarily by the droplet density.
Introduction
Droplet-based assays, in which microscale emulsions are used as isolated compartments to run many independent chemical reactions, have gained popularity in recent years as a platform for a wide range of biomedical applications.1–6 Compared to the conventional laboratory approach of using millimeter-sized well plates to isolate fluids, micrometer-scale droplets contain only picoliters (10−12 L) of fluid, offering a 106× reduction in volume. Furthermore, compared to the hundreds of compartments available on a conventional well plate, microfluidics allow droplets to be created at rates as high as 106 per minute,7–9 offering a >104× increase in the number of compartments over conventional techniques. The enormous increase in sensitivity that comes from massively parallelized, ultra-small volume assays, has been harnessed to detect both single molecules of protein and nucleic acid,1,2 to monitor molecular concentrations as a function of time,3,4 to perform high-throughput screens for directed evolution,5 and to assay single cells.4,6
While the microfluidics to produce and process droplets can be miniaturized and integrated onto compact, monolithic chips,5,6,10–12 the read-out of droplet-based assays have been more difficult to miniaturize. Fluorescence-based sensing continues to be the favored detection modality due to three distinct advantages: 1. molecular beacons, which can turn on or off fluorescence based on binding events, obviate extra steps to wash away excess reagents,13–15 2. differently colored fluorophores allow for the detection of multiple targets in a single droplet,16,17 3. widely available fluorescence-based reagents ease assay development.18 Much previous work has been done to integrate fluorescence detection with droplet microfluidics, and related work has been done to miniaturize fluorescence detection of cells.11,19–26 Wide-field microscopy techniques have been developed that can take micrographs of static droplets, with an ability to resolve as many as 106 in a single-shot.22–24 Other groups have developed in-flow detection systems, which have the advantage of real-time sorting, down-stream processing, and an ability to measure a far greater numbers of droplets than possible with the static techniques, measuring as many as 104 droplets per second.10,11,27,28 However, because these techniques require complex optics and are not easily amenable to monitoring more than one channel, they cannot take full advantage of the multiplexing capability of microfluidics or its potential for portable point-of-care use.12
To address these challenges, we have developed a miniaturized in-flow droplet detection scheme that can simultaneously measure fluorescent droplets in multiple channels. Our chip uses only a single silicon photodetector, no lenses, and no sophisticated fluid control, making it well suited for easy integration into a microfluidic platform. The key innovation of our technology is the use of amplitude modulated encoding of the fluorescence signal from passing droplets, using a distinct micro-patterned (“barcoded”) mask for each of the channels (Fig. 1a). This patterning allows the one-dimensional signal from the photodetector to be decompressed into a set of vectors, each representing one of the individual channels. This time encoding serves two functions, 1. it allows the recovery of weak signals (SNR ≪ 1) using correlation-based signal recovery,29 and therefore enables a simple hardware implementation that does not include lenses, lasers, or highly sensitive detectors, 2. it allows for independent monitoring of multiple channels using only a single detector, enabling additional assays to be implemented on a single chip without having to add to the complexity of the hardware. In this study, we experimentally demonstrated a device with four channels and provide analysis to show that this particular design can be scaled to monitor many more channels (c > 16), primarily limited by the droplet density and the distribution of droplets in the channel. Furthermore, we coupled this detection platform with a fabrication strategy that enables multiple droplet makers to be integrated onto a single chip, demonstrating a compact, self-contained platform to generate and readout parallelized droplet based assays.
Fig. 1.
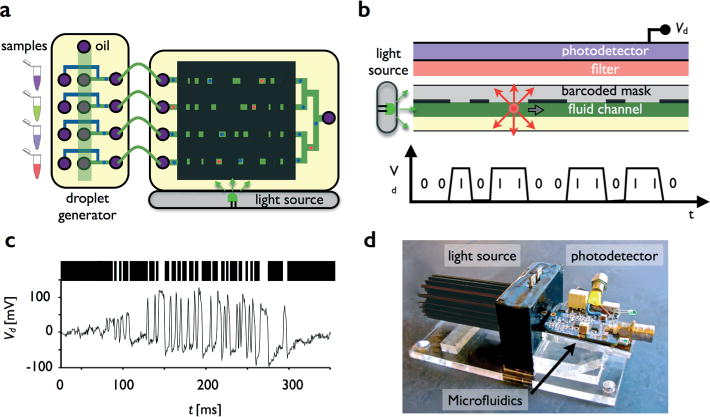
Design and implementation of a miniaturized, multichannel platform to read-out fluorescence droplet-based assays. a. The chip consists of four components, integrated droplet generators, a side-illuminator, a mask, and an optical detection system. Here, the top view is shown. b. A cross-sectional schematic of the chip. c. An example of a 105 bit long mask pattern, and the corresponding amplitude modulated signal Vd from a passing droplet. d. A photograph of the device, which is roughly the size of a cell phone (10 × 5 × 2 cm3).
Methods
Amplitude modulated encoding of the fluorescence signal
The chip consists of four modular components, shown in Fig. 1a: 1. the droplet generators to make monodispersed emulsions, 2. a side-illumination system to excite the fluorescent dyes within the droplets, 3. a mask to modulate the fluorescent signals of passing droplets, and 4. an optical detection system to measure and record the emitted light. The basic operating mechanism of the chip is shown in Fig. 1b. A side illuminator, consisting of a light emitting diode (LED) shines light, which is tuned to the excitation wavelength of the fluorophore λex, into the microfluidic chip. Anti-resonant coupling contains the light within the microchip and uniformly and intensely illuminates the fluid channel.30 A droplet containing a fluorescent dye, passing through the microchannel, will absorb the excitation light and fluoresce. The isotropically emitted fluorescence light passes through the patterned mask and into the Silicon photodetector. As the droplet moves down the channel, its emitted light is amplitude modulated by the mask (Fig. 1c). Between the mask and photodetector is a long pass optical filter to diminish background signal from scattered excitation light. The system is roughly the size of a cell phone (10 × 5 × 2 cm3) (Fig. 1d).
As the droplet passes under the mask, its emitted light transitions from being blocked to being transmitted, which results in a binary, amplitude modulated signal Vd(t) (Fig. 1b). We define the mask pattern mn, for each of the n channels, as a series of 1s and 0s, with 1 being transparent and 0 being opaque. Each mask pattern consists of 105 bits, with each bit being 80 μm long, resulting in an 8.4 mm long detection region. The photodetector, a 10 × 10 mm2 Silicon photodiode (Thor Labs, PDA100a) sits immediately on top of the mask (Fig. 1d). The fluid channel is 150 μm wide, 70 μm deep, and the droplets are approximately 60 μm in diameter. The mask, which consists of lithographically patterned metal on glass, also serves as the roof of the microfluidic channel. The close proximity of the mask to the droplets ensures that each bit in the mask pattern subtends the largest possible solid angle of light emitted (Fig. 1d) from the droplets and therefore ensures contrast between 1s and 0s.
Signal extraction
When processing the signal from the photodetector Vd(t), we aim to determine if a fluorescent droplet has passed through the detection region, and if so, to determine which channel it passed through. To this end, we project the signal onto a set of vectors Ψn, each representing the relative correlation of the signal from the passing droplet Vd(t) and each of the masks mn. We can calculate the cross-correlation Ψn(τ) = Vd(t)*mn(x/v) = ∫Vd(t)mn(x/v + τ)dt from −∞ to ∞, where mn(x) are each of the n mask patterns, scaled in time by the droplet velocity mn(x/v) (Fig. 2a).
Fig. 2.
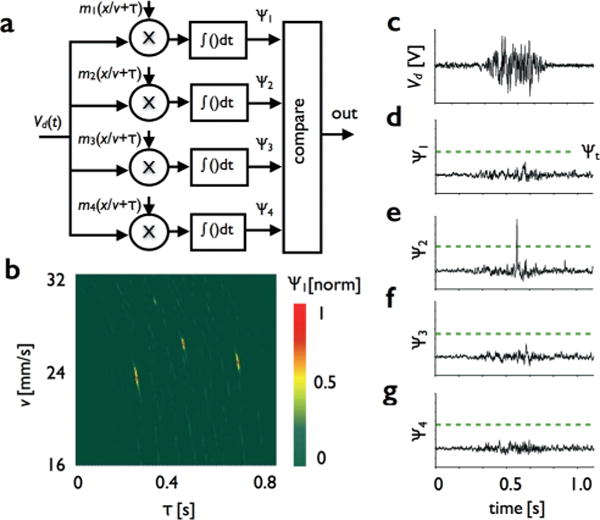
Amplitude modulated detection. a. The signal measured by the photodetector Vd(t) is projected into vectors Ψn(τ) = Vd(t)*mn(x/v) = ∫Vd(t)mn(x/v + τ)dt from −∞ to ∞, each representing the likelihood of the droplet having passed through an individual channel n and traveling at velocity v. b. To make the detection scheme robust against droplets traveling at different velocities, we performed this correlation for a range of velocities, leading to a two-dimensional correlation over a range of velocities. In the example shown, three droplets at three distinct velocities can be observed. c. An example of raw data Vd from a single passing droplet. A set of vectors are generated, Ψ1 (d), Ψ2 (e), Ψ3 (f), and Ψ4 (g). The droplet causes a large peak in Ψ2, allowing us to identify the droplet as having passed through that channel.
An example of raw data that comes from the photodetector as a droplet, loaded with 10 nM rhodamine B, passes through the detection region is shown in Fig. 2c. After the signal is passed through the bank of correlators (Fig. 2a), a set of vectors are generated, Ψ1 (Fig. 2d), Ψ2 (Fig. 2e), Ψ3 (Fig. 2f), and Ψ4 (Fig. 2g). The droplet causes a large peak in Ψ2, allowing us to identify the droplet as having passed through that channel. We define a gating threshold value Ψt that is applied to each of these vectors (shown as a green dashed line) above which we consider a fluorescent droplet detected.
Mask design
To generate the optimal mask patterns, we defined two design considerations. First, the masks should be minimally correlated with themselves, such that the energy E(ma*ma) is minimal. This constraint allows the droplets to be concurrently present in the detection region in the same channel and be distinguished from one another. Second, the masks should be minimally cross-correlated with one another, such that the energy E(ma*mb) is minimal for a ≠ b. This constraint ensures that the signals from different channels are maximally separable. To generate such masks, we referred back to a mathematical framework developed to generate maximally uncorrelated, pseudo-random sequences, for applications in radar and telecommunications.31
Pseudorandom vectors, known as maximum length sequences (MLS), can be generated using a feedback register.31 For a sequence of length L = 2M − 1, the shift register’s elements are defined by a primitive polynomial h(x) of degree M. By iterating this shift register, a series of 1s and 0s can be generated that are minimally autocorrelated.31 To create multiple channels, we generated a two-dimensional MLS by folding the one-dimensional MLS into a two-dimensional array as described by MacWilliams and Sloane, 1976.31
The length of the mask patterns for the device that we designed was 105 bits. The sequence was made as long as possible to minimize each channel’s autocorrelation and the cross-correlation between channels.31 Its length was constrained by the size of the photodetector. The Silicon photodetector (Thor Labs, PDA100A) that we used was 10 × 10 mm2, and since we did not use any lenses, this set the size of the detection region. The size of each pixel in the mask was determined by the size of the droplets. To ensure that a large fraction of the isotropically emitted light from each droplet was blocked by the mask pattern, the pixel size of the mask was matched to that of the droplets. We used 60 μm droplets and chose 80 μm pixels. The total number of bits per channel was thus constrained to be <125 (10 mm/80 μm).
To create a set of masks each 105 bits long, which matched our specifications for minimal auto and cross-correlation, we used the process in MacWilliams and Sloane to create a pseudo random matrix array from pseudo random maximum length sequences (MLS). The dimensions of the pseudo random matrix are certain permissible factorizations of the MLS sequence. Here we used 105 × 39 = 4095 = 212 − 1 (M = 12). We compared these sequences to randomly generated sequences of masks using MATLAB, and found the MLS generated masks to have, as expected,31 a significantly smaller autocorrelation and cross-correlation (P < 10−4, two-tailed t test). We also developed an alternative strategy to generate masks, in which we generated a large library of random masks, and then selected a subset of masks that had low autocorrelation. From that subset, we then selected a subset of pairs that had low cross-correlation, and from those pairs, sets of four that have low cross-correlation. The two procedures yielded similar results, but the MLS technique took only a few seconds of computation time whereas the selection technique took several hours, and became untenably slow for greater than four channels. Both implementations were performed in MATLAB.
Device fabrication
We fabricated the chip using a combination of soft lithography to make the fluidic channels and standard planar lithography to create the masks (Fig. 3a). The molded PDMS layer was fabricated using standard single-layer SU-8 lithography (SU-8 2025, MicroChem) with 80 μm thick features. The mask was patterned into Ni on glass, using standard planar photolithography. First, Ni was thermally evaporated onto glass slides (Kurt Lesker PVD 75, Wolf Nanofabrication Facility, University of Pennsylvania). Subsequently, the mask pattern was lithographically defined (Shipley 1813) using wet etching of the Ni (iron(III) Chloride). The Ni patterned glass slide was permanently bonded to the molded PDMS layer using PDMS-stamped bonding (Fig. 3b).7
Fig. 3.
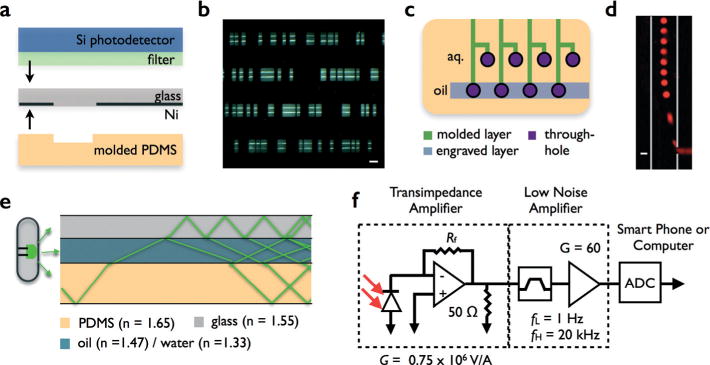
Prototype design. a. The device was fabricated using a combination of soft lithography to make molded PDMS fluidic channels and standard planar lithography of Ni on glass to create the masks. The Si photodetector sits immediately on top of the chip, a filter is used to block scattered excitation light. b. An optical micrograph of a portion of the device. The scale bar is 160 μm. c. A schematic of the integrated droplet generators. Four T-Junction droplet generators were integrated onto a single device, with only a single oil input. Hybrid soft-lithography/laser machining was used to fabricate this chip, in which soft lithography was used to fabricate a molded layer for the fluidic channels and direct laser micromachining was used to create vias, connecting these channels to deep laser-engraved channels that bring oil to the individual droplet makers. d. A fluorescence micrograph of one of the droplet makers. The drops were loaded with rhodamine. The scale bar is 60 μm. e. A schematic showing how antiresonant coupling was used to deliver light via side illumination into the chip. f. A schematic of the electronic readout of the Si photodetector.
Droplet design
Droplets were generated using T-Junctions (Fig. 3c).32 Four T-Junctions were integrated onto the device, with only a single oil input, using hybrid soft-lithography/laser machining.7 We fabricated the droplet generators using a combination of 1. soft lithography to make micro-scale droplet makers and fluidic channels in a layer of PDMS, and 2. direct laser micromachining to make vias to a second layer of PDMS that has much deeper (d = 200 μm) laser-engraved channels to deliver oil evenly to the individual droplet makers.7 The T junction’s aperture was 100 μm wide and 50 μm deep (Fig. 3c). For the continuous phase, we used mineral oil, with 5% V/V Span 80 and 1% V/V Tween (Fisher Scientific). Flow rates were 0.1 mL h−1 for the aqueous phase and 1 mL h−1 for the oil phase, and were controlled using syringe pumps (Braintree Scientific). The aqueous phase was prepared by creating a 10 nM Rhodamine B solution (Fisher Scientific) with DI water. Prior to use, the microfluidic channels were coated with Aquapel (PPG Industries) to ensure that the walls were preferentially wet by the mineral oil. The mean droplet size was 60 μm and the coefficient of variation (CV) was 5% (Fig. 3d). Using on-chip reservoirs for the four aqueous inputs, and connecting the two chips together using tygon tubing, a single syringe pump was capable of driving the device. Using tubing to connect two separate chip modules was done for convenient prototyping. The droplet generator and detection module, made using compatible fabrication strategies, were also seamlessly integrated onto the same chip.
Electronics design
The output current from the photodetector was amplified, digitized, and processed using custom electronics (Fig. 3c). The photodetector sensor had a responsivity of 200 mA W−1 at λ = 600 nm. The photodetector was connected to a G = 0.75 × 106 V A−1 transimpedance amplifier with a DC-20 kHz bandwidth (Thor PD100A). The output of the transimpedance amplifier was AC coupled to a pre-amplifier (PA) with a gain of 20, and a high-pass frequency of 1 Hz and a low pass frequency of 20 kHz (Ithaco). The output of the PA was connected to an analog to digital converter, (National Instruments, NI USB-6009) which digitized the signal at 40 kS s−1 before sending the signal to a computer or smart phone over USB for analysis. All analysis was done on a personal computer (MATLAB), but for a portable implementation this processing could be done using either a digital signal processing (DSP) chip or using cloud computing.12
Optics design
The optics in the chip were kept as small and inexpensive as possible, using commercial products that could be packaged into a small-footprint, portable device (Fig. 1d). The excitation light was provided by an ultra-bright LED (λex = 530 nm) (Luminus, CBT-90-G-C11-JK201) packaged into a custom laser machined (Universal Laser) acrylic box with heat sink (Wake-field Thermal Solutions, 19757-M-AB). The box was designed such that the chip could slide into it, similar to a slot connector, and the light would couple to the chip at an oblique angle for maximum anti-resonant coupling.30 A silicon photo-detector mounted on a printed circuit board, with integrated transimpedance amplification was used (Thor Labs, PDA 100a). A λ = 600 nm long pass filter (Edmund optics) was placed between the chip and detector to diminish (95% reflectance) the effect of scattered light at the excitation wavelength.
Results
Velocity independent detection
The detection scheme described above requires accurate knowledge of the droplet’s velocity v. This requirement poses a major challenge to this technique because controlling droplet velocity would necessitate the use of expensive syringe pumps, tight restrictions on fabrication tolerances, and greater control over droplet size and position within the microchannels, adding significantly to the cost and complexity of the platform.
Therefore, we decided that rather than control the droplet velocity, to instead use an algorithm that could adapt to droplets with dispersed velocities. To this end, rather than calculate a one-dimensional cross-correlation between the output signal Vd(t) and each mask pattern mn(x/v), we calculated the two-dimensional correlation mn (x/V)*Vd(t), where V is a 1-d matrix with a range of velocities [vmin : vmax]. This two-dimensional correlation was efficiently calculated using MATLAB. The output is a two-dimensional matrix (Fig. 2b) in velocity v and time t, from which peak positions (vp,tp) could accurately be found in the 2-d space. Due to the use of uncorrelated, pseudo-random sequences for the masks, there were no significant false correlations with other masks due to the dispersion of droplet velocities (Fig. S1†). Variations in velocity could be caused by dispersion in droplet size or by the increase in hydrodynamic resistance when a droplet enters a channel.33 An assumption of this technique is that the droplet velocity is constant for the duration of time (~50 ms) in the detection region, which we verified experimentally. Through simulation, we demonstrated that the device performance was unaffected by small linear accelerations (as much as 5% change in the initial velocity v0) of the droplets while they pass through the detection region (Fig. S2†).
Measuring fluorescent droplets below the noise floor
Our correlation-based detection scheme allowed extremely weak signals (SNR ≪ 1) to be recovered from below the noise floor. Our platform can efficiently separate signal from the noise because the signal correlates with the pattern of the mask, whereas the noise does not. The capability to recover weak signals is important for our design because it allows for lens free use making it well suited for miniaturization. To demonstrate this capability, we measured weakly fluorescent droplets with an SNR ~ 0.25, which could not be resolved in the raw data (Fig. 4a). The signal to noise ratio was calculated by dividing the energy in the signal from a passing droplet by the average energy of the noise over the same time interval. The primary source of noise was found to come from the Silicon photodetector, and not from ambient light or the excitation source, with a noise equivalent power NEP = 1.22 × 10−12 W √Hz−1. After correlating this data with the correct mask m2, the peak in its correlation vector Ψ was well above the noise floor (SNR > 10) in the correlation data and easily detected (Fig. 4b).
Fig. 4.
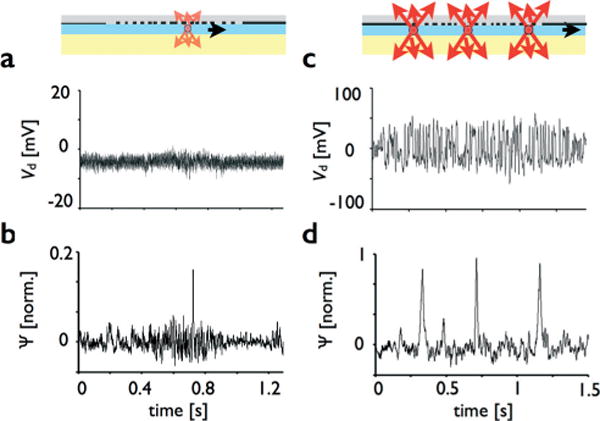
Features of amplitude modulated detection. a. Weak signals below the sensor’s noise floor (SNR ~ 0.25) in the raw data Vd can be readily resolved in Ψ after being correlated with the correct mask (b). c. Multiple droplets in the detection region concurrently, which are not resolvable in the raw data Vd, can be separated and resolved in Ψ after being correlated with the correct mask (d).
Measuring multiple droplets in the detection region simultaneously
The high sensitivity of our platform comes partially from the detection region’s large area (10 × 10 mm2), which collects many photons from fluorescing droplets as they pass. However, the trade-off for having a large detection region is that if we restrict ourselves to one droplet in the detection region at a time as in conventional cytometry, it severely limits the device’s throughput. To this end, we designed the masks to be capable of resolving multiple droplets within the detection region simultaneously. We chose mask patterns that have low autocorrelation and low cross-correlation with one another, such that our correlation-based detection strategy could resolve droplets in different positions along the same channel or in different channels concurrently.
To demonstrate this capability, we measured three droplets which passed through the detection region concurrently, all in the same channel. In the raw data, the three signals overlap and cannot be resolved (Fig. 4c). However, after correlating this data with the correct mask, the three peaks in the correlation vector Ψ become well separated and could be individually resolved (Fig. 4d).
Multichannel detection
A key feature of our platform is the ability to detect droplets in multiple channels simultaneously. To characterize this capability, we performed a set of experiments where we sent droplets through specific channels and compared the output of the device to the expected outcomes. This functionality is demonstrated in Fig. 5a, which shows two droplets passing through the detection region, one of the droplets passing in channel 1 and the other in channel 2. After correlating this data with the correct masks, we can see a clear peak in channel 2 corresponding to the first droplet and a clear peak in channel 1 corresponding to the second droplet, allowing each droplet’s correct channel to be readily identified (Fig. 5a – bottom).
Fig. 5.
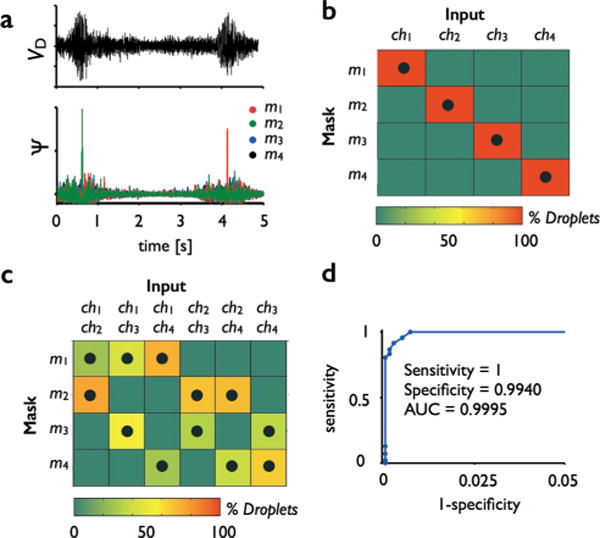
Multi-channel detection. Parallel streams of droplets can be monitored simultaneously by using the unique pattern of each channels’ mask to identify each passing droplet’s channel. a. Two droplets are shown passing in the raw data. After correlation with the set of four masks, the first droplet creates a peak in m2 and the second in m1, allowing their channels to be correctly identified. These curves are the maximum projection of correlations at varying velocity v. b. To test this platform, we passed droplets through each of the four channels and quantified the fraction of droplets measured in each channel in a heat map. Black dots show the channels where droplets were passed. c. We similarly tested the chip’s capability for measuring multiple channels simultaneously, by passing droplets through each of the six possible pairs of channels. d. The sensitivity and specificity was quantified, and summarized on a receiver operator characteristic (ROC) curve.
Quantification of device sensitivity and specificity
To characterize the tradeoff between sensitivity and specificity, we tested the device using a range of threshold values Ψt and generated a receiver operator characteristic curve (ROC). We defined the sensitivity = TP/P, where TP is the number of instances the detector successfully detected a passing droplet and correctly identified its channel and P is the total number of droplets. We defined the specificity = TN/N of the detector, where TN = N − FP is the true negatives and is defined by the total false positives FP and the total number of negatives N. The total number of negatives N = P*(c − 1) is defined by the total number of droplets P and the total number of channels c.
We first passed droplets through one of the four channels at a time and quantified the sensitivity and specificity of detection. We summarize the results of the experiments in a heat map (Fig. 5b). The device performed as expected; droplets were detected in the channels where droplets were passing, along the diagonal of the heat map, and not in the incorrect channels, off diagonal. For each of the tests, we passed ~400 droplets with an SNR ~ 1. For the threshold value Ψt chosen, the sensitivity was 1.0 and the specificity was 0.994.
To demonstrate the chip’s ability to simultaneously detect droplets in multiple channels, we next passed droplets through each of the six possible sets of channels (ch1 & ch2, ch1 & ch3, etc.…) We summarize the results of the experiments in a heat map (Fig. 5c). Droplets were detected in the correct sets of channels and not in the incorrect channels. For each of the tests, we passed ~800 droplets with an SNR ~ 1. For the threshold value Ψt chosen, the average sensitivity was 1.0 and the specificity was 0.993.
We characterized the tradeoff between sensitivity and specificity by generating an ROC curve that summarizes the results described above. For droplets with an approximate SNR ~ 1, we found an area under the curve, AUC = 0.9995, demonstrating the ability to robustly monitor parallel streams of droplets using only a single photodetector (Fig. 5d).
Characterizing the effect of design parameter choices on performance
To characterize and to aid in the design of the system, we supplemented our physical experiments with a model to simulate a wider range of parameters than was plausible with prototyping. The model was carried out using Matlab. The point in time tp that a droplet passes through a channel and the specific channel the droplet passes through n were generated stochastically using random number generators. The signal from the passing droplet was created using the mask pattern m(n,x/v − tp), scaled by the droplet velocity v and placed into the output signal Vd(t) at time point tp. N droplets were iteratively placed. Gaussian noise was added to the signal to obtain the appropriate signal to noise ratio (SNR). The model was verified by direct comparison to our experimental data. Using this model, we were able to determine the limits of our detection strategy, which sets the groundwork for future applications and development.
First, we investigated how the sensitivity and specificity of droplet detection were a function of the signal to noise ratio (SNR) of the photodetector. For the setup used in this study (L = 100 bits, c = 4 channels) we found that droplets could be detected with high sensitivity and accuracy (AUC = 0.9991) with an SNR as low as −10 dB (0.1) (Fig. 6a). Below an SNR of −13 dB (0.05) the sensitivity and specificity began to fall off rapidly. The ability to resolve droplets when the signal is only 5% of noise, enables weakly fluorescent droplets to be resolved using a simple, lens free photodetector.
Fig. 6.
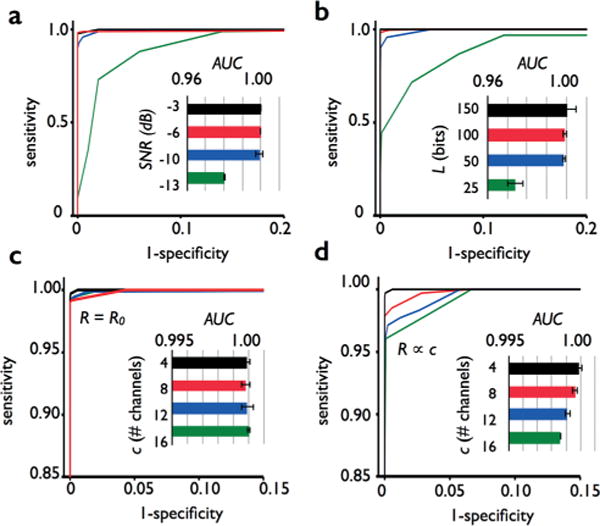
Characterizing the effect of design parameter choices on performance. The effect of various design parameters on sensitivity and specificity of droplet detection were characterized using a model system. The results were summarized by a receiver operator characteristic (ROC) curve and quantified by the curve’s area under the curve (AUC). a. The AUC remains unchanged as the signal to noise ratio (SNR) is dropped to as low as −10 dB (0.1), but then falls precipitously below −13 dB (0.05). b. The AUC remains unchanged as the number of bits in the mask (L) was dropped as low as L = 100, but then begins to fall off below L = 50. c. The AUC is unaffected by increasing the number of channels as high as c = 16, when the total droplet rate is held constant R = R0, leading to an average of one droplet in the detection region at a time. d. When the droplet rate is increased proportionally with the numbers of channels R ∝ c, the AUC begins to fall as the average number of droplets in the detection region becomes greater than two.
Next, we measured the effect of the number of bits in the mask L on performance. For a system matched to the one that we built (SNR = −6 dB, c = 4 channels) we found that we could reduce the mask length to as low as L = 100 bits, (AUC = 0.999) without a significant reduction in performance. Below L = 50 bits, sensitivity and specificity fell off rapidly. We attribute the increase in performance with bit length based on two factors, 1. As the number of bits is increased, the droplet is effectively measured more instances, leading to an increase in the effective SNR by signal averaging ~√L 2. As the number of bits is increased, the cross-correlation between the channels ma*mb≠a correspondingly decreases, which reduces the background signal in Ψa when a droplet passes through a channel b≠a.
Characterizing the limitations for increasing the number of channels
Finally, we used our model system to demonstrate the feasibility of adding more channels than the c = 4 demonstrated in our prototype. In our first experiment, we kept the total droplet rate constant R = R0, such that the average number of droplets in the detection region was ~1. For a device matched to the one that we built (L = 100 bits, SNR = −6 dB), we found that as the number of channels increased from c = 4 to 16 the sensitivity and specificity remained constant (Fig. 6c). We understand this result based on two factors, 1. the increase in correlation ma*mb≠a between channels as c is increased from 4 to 16 is insignificant, because the number of uncorrelated masks that can be generated from L = 100 bits is much larger than c.31 2. As the number of channels is increased, there are more correlation vectors Ψa per droplet. The peak-finding algorithm identifies peaks by comparing peak height to the local variance in the other vectors Ψb≠a. As we increase the number of channels c, there was increased averaging, which decreases the false positives and negatives.
We found that the ability of the chip to multiplex was ultimately limited by the droplet density, with performance falling off when the average number of droplets in the detection region increased beyond ~2 (Fig. 6d). Because in our model the position of the droplets in the channel followed Poisson statistics, as the average number of droplets in the detection region became greater than 2, it became increasingly likely to find >5 droplets in the detection region at any given instance, at which point that detector was no longer able to resolve individual events. To test this phenomenon, rather than keep the total droplet rate constant R = R0, we kept the droplet rate per channel constant R ∝ c. We chose the droplet rate such that for 4 channels, there was on average one droplet in the detection region at a time, and the rate increased proportionally as channels were added. The performance of the device fell off proportionally with each added channel (Fig. 6d, inset).
Discussion
We have developed a miniature fluorescence droplet detector with the ability to simultaneously monitor multiple streams of droplets. We demonstrated that on a cell phone sized platform, using only off the shelf optics, extremely high sensitivity and specificity could be achieved. Our chip used only a single silicon photodetector, no lenses, and no sophisticated fluid control making it well suited for low-cost, portable diagnostics. Amplitude modulation of the fluorescence signal using distinct micro-patterned masks for each of the channels, allowed multiple channels of droplets to be monitored. This technique also allowed weak signals (SNR as low as 0.05) to be recovered using correlation-based signal recovery. It also allowed for multiple droplets concurrently passing through the detection region to be resolved. By coupling this detection platform with a chip that integrates multiple droplet makers, we demonstrated a miniaturized platform to perform parallel, independent droplet-based assays.
This work builds upon previous work to encode optical signals from cells in the time-domain. Most notably, Kiesel et al. have developed a compact flow cytometer using spatial modulation of fluorescently labeled cells as they pass down a channel.19,29,34 By correlating the detected signal with the known pattern, high signal-to-noise was achieved using relatively simple optics. This technique has been implemented to monitor CD4+ cells in whole blood,34 and has been extended to multi-color fluorescence detection.19 In related work, Yu-Hwa Lo et al. have used time-domain encoding to measure weak side-scattering signals from unlabeled microbeads and cells.35,36 Similar approaches have also been developed to improve fluorescence imaging of cells, by encoding two dimensional images into a serial time-domain waveform for imaging dynamic processes37 and to reduce image blur of fluorescence imaging in microfluidic chips.38 Our work is most differentiated from prior work by its use of multiple uncorrelated masks in parallel to independently monitor multiple channels and its integration with droplet-based microfluidics.
Our platform’s capability to perform and read-out multiple parallel droplet based assays on an integrated chip is well suited for many applications. One key application is digital polymerase chain reaction (DPCR), where each stream of droplets can be mixed with a different primer to test for a different target DNA or RNA.39,40 Digital PCR can achieve sensitivity of a single molecule, and there are many applications where screening for many markers would allow for complex signatures of disease to be resolved.41 There has been great interest in recent years in designing PCR assays to quantify multiple targets in single reactions using differently colored dyes42 and different lengths of amplicons.43 Similarly, amplification strategies that enable single molecules of protein to be detected using droplet based assays have been recently reported.44 Furthermore, our chip’s compact, microfluidic format can be seamlessly integrated with front-end sample processing, such as immunomagnetic isolation of rare cells,45,46 or on-chip staining,47 enabling highly sensitive molecular analysis directly on clinical samples.
There are several aspects of the our platform that can be further developed to expand the functionality of the system. Currently, our system detects only a single color for each droplet. However, as has been previously shown by other groups on cell-based detection systems,25,48 multicolor detection can be integrated with masked systems. The data acquisition electronics, which currently fundamentally limits the overall throughput to 104 drops min−1, could be further improved (~50×) to exploit the high bandwidth of microelectronics. The maximum throughput demonstrated on our chip, 103 drops min−1, was set by the leakage of our device at high flow rates due to the viscosity of the mineral oil that was used. This throughput could be improved by switching to a less viscous continuous phase or increasing the cross sectional area of the fluid channel. Furthermore, the data processing that is currently done off-chip using MATLAB could be carried out in real-time using a digital signal processing chip,49 enabling real-time sorting to be integrated into this platform. Additionally, this correlation-based detection scheme can be extended to other modalities, including dielectric50 and magnetic sensing.50,51
Fully integrating this device with mobile technology will allow it to be used directly where molecular diagnostics are needed. By taking advantage of low-cost electronics and data processing, this chip-based platform is poised to offer a new tool for clinicians and researchers seeking rapid insight into molecular markers.
Supplementary Material
Acknowledgments
This work was supported by the Department of Bioengineering, University of Pennsylvania, a pilot grant from the University of Pennsylvania Nano/Bio Interface Center, National Science Foundation DMR 08-32802, and a pilot grant from the University of Pennsylvania Center for AIDS Research (AI 045008).
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc00819g
References
- 1.Gorris HH, Walt DR. Angew Chem, Int Ed. 2010;49:3880–3895. doi: 10.1002/anie.200906417. [DOI] [PubMed] [Google Scholar]
- 2.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 3.Chen D, Du W, Liu Y, Liu W, Kuznetsov A, Mendez FE, Phipipson LH, Ismagilov RF. Proc Natl Acad Sci U S A. 2008;105:16843–16848. doi: 10.1073/pnas.0807916105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtois F, Olguin LF, Whyte G, Bratton D, Huck WTS, Abell C, Hollfelder F. ChemBioChem. 2008;9:439–446. doi: 10.1002/cbic.200700536. [DOI] [PubMed] [Google Scholar]
- 5.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Proc Natl Acad Sci U S A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, Frenz L, Blouwolff J, Humphry KJ, Koster S, Duan H, et al. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Muluneh M, Issadore D. Lab Chip. 2013;13:4750–4754. doi: 10.1039/c3lc50979f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nisisako T, Torii T. Lab Chip. 2008;8:287–293. doi: 10.1039/b713141k. [DOI] [PubMed] [Google Scholar]
- 9.Romanowsky MB, Abate AR, Rotem A, Holtze C, Weitz DA. Lab Chip. 2012;12:802–807. doi: 10.1039/c2lc21033a. [DOI] [PubMed] [Google Scholar]
- 10.Guo MT, Rotem A, Heyman JA, Weitz DA. Lab Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 11.Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, et al. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 12.Issadore D, Westervelt RM. Point-of-care Diagnostics on a Chip. Springer; 2013. [Google Scholar]
- 13.Rane TD, Puleo CM, Liu KJ, Zhang Y, Lee AP, Wang TH. Lab Chip. 2010;10:161–164. doi: 10.1039/b917503b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh ATH, Pan PJ, Lee AP. Anal Bioanal Chem. 2014;406:3059–3067. doi: 10.1007/s00216-014-7721-z. [DOI] [PubMed] [Google Scholar]
- 15.Guillaume G, Saint-Pierre C, Gasparutto D. Anal Chim Acta. 2014;812:168–175. doi: 10.1016/j.aca.2013.12.038. [DOI] [PubMed] [Google Scholar]
- 16.Zhong Q, Bhattacharya S, Kotsopoulos S, Olson J, Taly V, Griffiths AD, Link DR, Larson JW. Lab Chip. 2011;11:2167–2174. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
- 17.Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, Millot F, El Harrak A, Hutchison JB, Larson JW, Link DR, Laurent-Puig P, Griffiths AD, Taly V. Lab Chip. 2011;11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 18.Joensson HN, Svahn HA. Angew Chem, Int Ed. 2012;51:12176–12192. doi: 10.1002/anie.201200460. [DOI] [PubMed] [Google Scholar]
- 19.Martini J, Recht MI, Huck M, Bern MW, Johnson NM, Kiesel P. Lab Chip. 2012;12:5057–5062. doi: 10.1039/c2lc40515f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna BK, Evans JG, Cheung MC, Ehrlich DJ. Nat Methods. 2011;8:401–403. doi: 10.1038/nmeth.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhagat AAS, Kuntaegowdanahalli SS, Kaval N, Seliskar CJ, Papautsky I. Biomed Microdevices. 2010;12:187–195. doi: 10.1007/s10544-009-9374-9. [DOI] [PubMed] [Google Scholar]
- 22.Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, Yang DL, Lee AP. Lab Chip. 2011;11:3838–3845. doi: 10.1039/c1lc20561g. [DOI] [PubMed] [Google Scholar]
- 23.Lim J, Gruner P, Konrad M, Baret JC. Lab Chip. 2013;13:1472–1475. doi: 10.1039/c3lc41329b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coskun AF, Sencan I, Su TW, Ozcan A. Opt Express. 2010;18:10510–10523. doi: 10.1364/OE.18.010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SH, Qiao W, Tsai FS, Yamashita K, Lo YH. Appl Phys Lett. 2010;97:093704. doi: 10.1063/1.3481695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ateya DA, Erickson JS, Howell J, Peter B, Hilliard LR, Golden JP, Ligler FS. Anal Bioanal Chem. 2008;391:1485–1498. doi: 10.1007/s00216-007-1827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi JW, Kang DK, Park H, deMello AJ, Chang SI. Anal Chem. 2012;84:3849–3854. doi: 10.1021/ac300414g. [DOI] [PubMed] [Google Scholar]
- 28.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc Natl Acad Sci U S A. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiesel P, Bassler M, Beck M, Johnson N. Appl Phys Lett. 2009;94:041107. [Google Scholar]
- 30.Schmidt O, Bassler M, Kiesel P, Knollenberg C, Johnson N. Lab Chip. 2007;7:626–629. doi: 10.1039/b618879f. [DOI] [PubMed] [Google Scholar]
- 31.MacWilliams FJ, Sloan NJA. Proc IEEE. 1976;64:1715–1729. [Google Scholar]
- 32.Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Lab Chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- 33.Vanapalli SA, Banpurkar AG, van den Ende D, Duits MH, Mugele F. Lab Chip. 2009;9:982–990. doi: 10.1039/b815002h. [DOI] [PubMed] [Google Scholar]
- 34.Kiesel P, Beck M, Johnson N. Cytometry, Part A. 2011;79:317–324. doi: 10.1002/cyto.a.21042. [DOI] [PubMed] [Google Scholar]
- 35.Mei Z, Wu T, Pion-Tonachini L, Qiao W, Zhao C, Liu Z, Lo Y. Biomicrofluidics. 2011;5:034116–034116. doi: 10.1063/1.3624740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu T, Mei Z, Lo Y. Lab Chip. 2012;12:3791–3797. doi: 10.1039/c2lc40560a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goda K, Tsia KK, Jalali B. Nature. 2009;458:1145–1149. doi: 10.1038/nature07980. [DOI] [PubMed] [Google Scholar]
- 38.Gorthi SS, Schaak D, Schonbrun E. Opt Express. 2013;21:5164–5170. doi: 10.1364/OE.21.005164. [DOI] [PubMed] [Google Scholar]
- 39.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beer NR, Wheeler EK, Lee-Houghton L, Watkins N, Nasarabadi S, Hebert N, Leung P, Arnold DW, Bailey CG, Colston BW. Anal Chem. 2008;80:1854–1858. doi: 10.1021/ac800048k. [DOI] [PubMed] [Google Scholar]
- 41.Park S, Zhang Y, Lin S, Wang TH, Yang S. Biotechnol Adv. 2011;29:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong Q, et al. Lab Chip. 2011;11:2167–2174. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
- 43.McDermott GP, et al. Anal Chem. 2013;85:11619–11627. doi: 10.1021/ac403061n. [DOI] [PubMed] [Google Scholar]
- 44.Walt DR. Anal Chem. 2012;85:1258–1263. doi: 10.1021/ac3027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muluneh M, Shang W, Issadore D. Adv Healthcare Mater. 2014;3:1078–1085. doi: 10.1002/adhm.201300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen A, Byvank T, Chang WJ, Bharde A, Vieira G, Miller BL, Chalmers JJ, Bashir R, Sooryakumar R. Lab Chip. 2013;13:1172–1181. doi: 10.1039/c2lc41201b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung J, Issadore D, Ullal A, Lee K, Weissleder R, Lee H. Biomicrofluidics. 2013;7:54107. doi: 10.1063/1.4821923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiesel P, Martini J, Recht MI, Bern MW, Johnson NM, Huck M. Point of Care Diangostics on a Chip. Springer; 2013. pp. 47–69. [Google Scholar]
- 49.Fields AP, Cohen AE. Proc Natl Acad Sci U S A. 2011;108:8937–8942. doi: 10.1073/pnas.1103554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood DK, Oh SH, Lee SH, Soh HT, Cleland AN. Appl Phys Lett. 2005;87:184106. [Google Scholar]
- 51.Issadore D, Chung HJ, Chung J, Budin G, Weissleder R, Lee H. Adv Healthcare Mater. 2013;2:1224–1228. doi: 10.1002/adhm.201200380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


