Significance
Decades of research have not deciphered the mechanism by which methyl-CpG binding protein 2 (MeCP2) regulates transcription and why Rett symptoms manifest 1 to 2 y after birth. We hypothesized that the temporal dynamics of MeCP2 binding might provide an answer. We developed mice with an EGFP-tagged MeCP2 allele to identify high-resolution MeCP2 binding profiles in the adult mouse brain. Using genomic binding profiles, methylation maps, and mRNA deep-sequencing data, we found MeCP2 binds to non-CG methylation (mCH, not mCG) to regulate expression of genes altered in mouse models of MeCP2 disorders. These data and the parallel timing of mCH and MeCP2 postnatal accumulation suggest MeCP2 binds mCH as neurons mature to regulate gene expression, offering an explanation for the delayed onset of Rett.
Keywords: MeCP2, transcriptional regulation, Non-CpG methylation, Rett syndrome, mCH
Abstract
Epigenetic mechanisms, such as DNA methylation, regulate transcriptional programs to afford the genome flexibility in responding to developmental and environmental cues in health and disease. A prime example involving epigenetic dysfunction is the postnatal neurodevelopmental disorder Rett syndrome (RTT), which is caused by mutations in the gene encoding methyl-CpG binding protein 2 (MeCP2). Despite decades of research, it remains unclear how MeCP2 regulates transcription or why RTT features appear 6–18 months after birth. Here we report integrated analyses of genomic binding of MeCP2, gene-expression data, and patterns of DNA methylation. In addition to the expected high-affinity binding to methylated cytosine in the CG context (mCG), we find a distinct epigenetic pattern of substantial MeCP2 binding to methylated cytosine in the non-CG context (mCH, where H = A, C, or T) in the adult brain. Unexpectedly, we discovered that genes that acquire elevated mCH after birth become preferentially misregulated in mouse models of MeCP2 disorders, suggesting that MeCP2 binding at mCH loci is key for regulating neuronal gene expression in vivo. This pattern is unique to the maturing and adult nervous system, as it requires the increase in mCH after birth to guide differential MeCP2 binding among mCG, mCH, and nonmethylated DNA elements. Notably, MeCP2 binds mCH with higher affinity than nonmethylated identical DNA sequences to influence the level of Bdnf, a gene implicated in the pathophysiology of RTT. This study thus provides insight into the molecular mechanism governing MeCP2 targeting and sheds light on the delayed onset of RTT symptoms.
Rett syndrome (RTT) is a debilitating postnatal neurological disorder characterized by an initial period of normal development followed by progressive neurological dysfunction and developmental regression. Although initially doubted to be a genetic disorder, we now know that loss of function of methyl-CpG binding protein 2 (MeCP2) is the root cause of RTT (1, 2).
Mouse models representing either a loss (3) or a gain of MeCP2 function (4) have proved invaluable toward the understanding of MeCP2-related disorders and revealed numerous transcriptional changes, establishing MeCP2 as an epigenetic regulator of transcription (5). The molecular details of MeCP2 transcriptional regulation have proven to be more complex than initially assumed, and little is known about how the broad binding pattern of MeCP2 regulates transcription or how the subsequent transcriptional changes contribute to the pathogenesis of RTT. The fact that doubling or tripling the levels of MeCP2 results in progressive neurological syndromes with increasing severity (MECP2 duplication and triplication syndromes, respectively) (6, 7) also raises questions about the binding patterns of MeCP2 and their functional consequences. Early studies found that MeCP2 interacts specifically with methylated cytosine in the CG context (mCG) (8–10). Recent in vivo studies in the adult mouse brain confirm the preference of MeCP2 for mCG over nonmethylated CG, although MeCP2 clearly binds widely across the entire genome (11). Other studies have observed that MeCP2 binds to nonspecific DNA elements via its A/T hooks and basic and disordered protein domains, as well as to mCH, although the functional relevance of these interactions has not been explored (12–16). Given that MeCP2 levels rise in mammalian neurons for some time after birth (11, 17) and approach those of the histone-octamer in the adult mouse brain (11), its differential binding to other DNA elements besides mCG is expected but has not yet been defined experimentally at high resolution. Moreover, the functional relevance of MeCP2 binding to non-CG methylated DNA has not been addressed.
Here we use a transgenic mouse line that expresses a fully functional EGFP-tagged MeCP2 that is the sole MeCP2 protein source when put on a MeCP2-null background (18). Using this model we overcome challenges of conventional MeCP2 ChIP to report a high-resolution genomic binding profile for MeCP2 in the hypothalamus of adult mice. We also performed mRNA deep-sequencing and analyzed hypothalamic gene-expression data from wild-type versus mouse models of RTT (3) and MeCP2 duplication syndrome (4, 5). Combining the MeCP2 binding data and the gene-expression data with reported genome-wide methylation patterns (19), we find MeCP2 preferentially binds to loci with marked mCH in adult mouse brain, and that this association between mCH and MeCP2 binding is particularly evident in genes with reciprocally misregulated expression in disease models of MeCP2 duplication and RTT. These data suggest that MeCP2 regulates gene expression via mCH, indicating the functional relevance for this interaction. Furthermore, the timing of regulation, demonstrated here for disease-relevant gene brain-derived neurotropic factor (Bdnf), provides a hypothesis for why RTT patients display delayed onset of symptoms.
Results
ChIP Using EGFP-Tagged MeCP2 Reveals High-Resolution Chromatin Binding Profiles.
To investigate the genomic binding of MeCP2 in the adult mouse brain, we used a transgenic mouse line that expresses an EGFP-tagged MeCP2 at levels similar to those of the endogenous protein (18). The transgene is fully functional in vivo when put on a MeCP2-null background (Mecp2-/y; MECP2-EGFP) and allows us to perform highly efficient and specific ChIP with an anti-GFP antibody followed by high-throughput sequencing (Fig. 1). In total, we obtained ∼230 million reads, achieving much more comprehensive MeCP2 binding profiles across the entire mouse genome than was possible in previous studies (11, 16). The profiles showed genome-wide MeCP2 binding with peaks at regions with high mCG density, in agreement with previous studies (11, 16) (Fig. 2A): in fact, the two profiles of MeCP2 binding from the adult mouse brain showed striking similarity at low resolution (r = 0.99) (Fig. 2B) (11). The correlation between our adult mouse brain profile and previously reported MeCP2 occupancy in embryonic stem cells (ES) was lower (r = 0.86) (Fig. 2B).
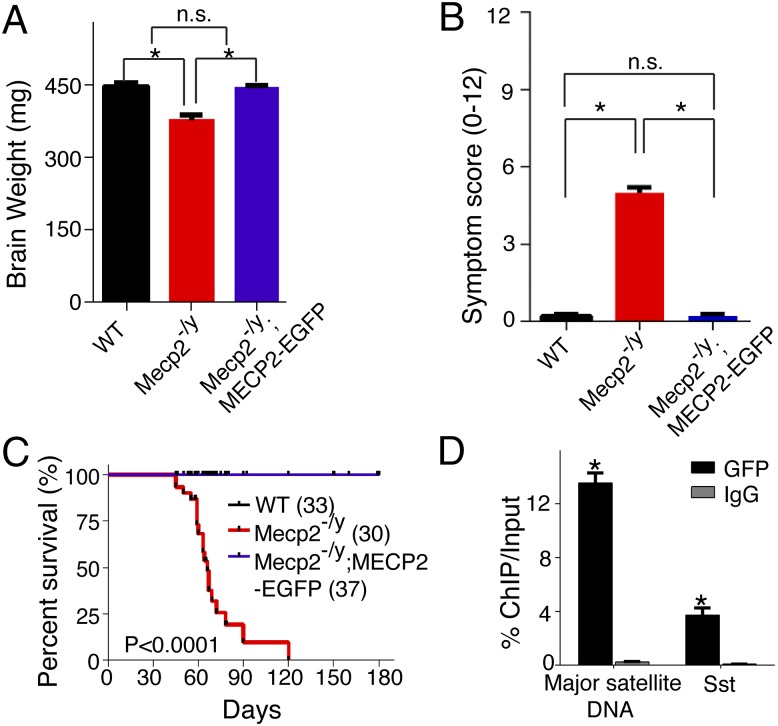
Fig. 1.
Characterization of a functional MECP2-EGFP transgenic mouse line that allows for highly efficient and specific ChIP. (A and B) MeCP2-EGFP rescued brain weight and disease symptoms of Mecp2 knockout mouse. Each bar represents SEM; n = 20–25 per genotype; *P < 0.01; n.s., not significant by paired, one-tailed Student’s t test. Symptom scoring was performed as described previously (29, 30). (C) MeCP2-EGFP rescued early lethality of Mecp2 knockout mouse. P < 0.0001 by Log-rank test. (D) MeCP2-EGFP binds to previously characterized DNA elements (13). In ChIP-PCR experiments, anti-GFP antibody displayed high efficiency (∼13% ChIP/Input for mouse major satellite DNA) and specificity (more than 50-fold enrichment over ChIP using control IgG antibody for mouse major satellite DNA and a promoter region of Sst). Each bar represents SEM; n = 3–4; *P < 0.05 by paired, one-tailed Student’s t test when comparing ChIP with GFP and control IgG antibodies.
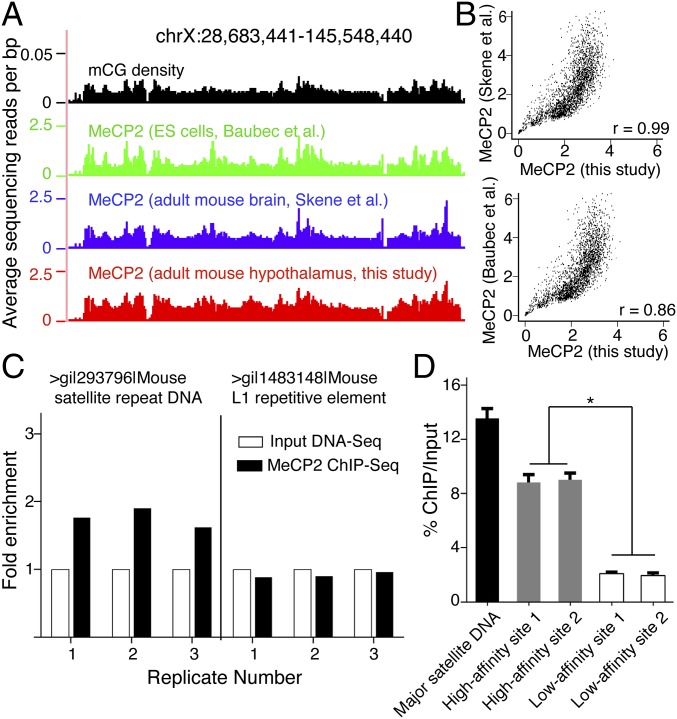
Fig. 2.
MeCP2 binds the entire genome with differential affinity in the adult mouse brain. (A) Browser representation of mCG density and normalized MeCP2 binding spanning example chromosomal region (Top, representing the x axis). Shown are mCG density in the adult mouse brain (black), previously reported MeCP2 binding in ES cells (green), adult mouse brain (blue), and results from the current study in the adult mouse hypothalamus (red). (B) Correlation between MeCP2 binding profiles from our high-resolution data and previous data from the adult mouse brain (Upper) and ES cells (Lower). Raw reads were binned per 1 Mb across the entire mouse genome. r denotes Pearson’s correlation. (C) Enrichment of repetitive DNA elements from three biological replicates, showing significant increase of mouse major satellite DNA, but not L1 repetitive element after MeCP2 ChIP-Seq relative to Input DNA-Seq. Data were normalized and represented as the fold-change relative to each Input DNA-Seq control. (D) RT-PCR analysis of isolated DNA from ChIP using primers for two representative high-affinity sites (gray) and two low-affinity sites (white) in the mouse genome. Mouse major satellite DNA (black) was used as a positive control for the high-affinity site. Each bar represents SEM; n = 3 or 4; *P < 0.05 by one-way ANOVA followed by Student’s t test.
MeCP2’s genome-wide binding renders conventional peak calling methods ineffectual, necessitating a different approach to identifying subtle differences in its binding. We sequenced the input mouse genomic DNA to the same depth as our MeCP2 ChIP-Seq and directly compared the read differences between them. This approach allowed us to uncover reliable differential MeCP2 binding patterns. As expected, previously known high-affinity sites for MeCP2 binding, including the mouse major satellite DNA that makes up about 3.6% of the mouse genome (20), were enriched in MeCP2 ChIP-Seq relative to input (Fig. 2C). Importantly, we were able to discover quantitative differences in MeCP2 binding across the entire mouse genome, which we further verified by ChIP followed by real-time PCR (RT-PCR) (Fig. 2D). These data provide a high-resolution profile of MeCP2 occupancy in the adult mouse brain and demonstrate that MeCP2 binds throughout the genome but with different affinities.
MeCP2 Binds to mCH in the Mature Brain to Regulate Transcription of Disease Relevant Genes.
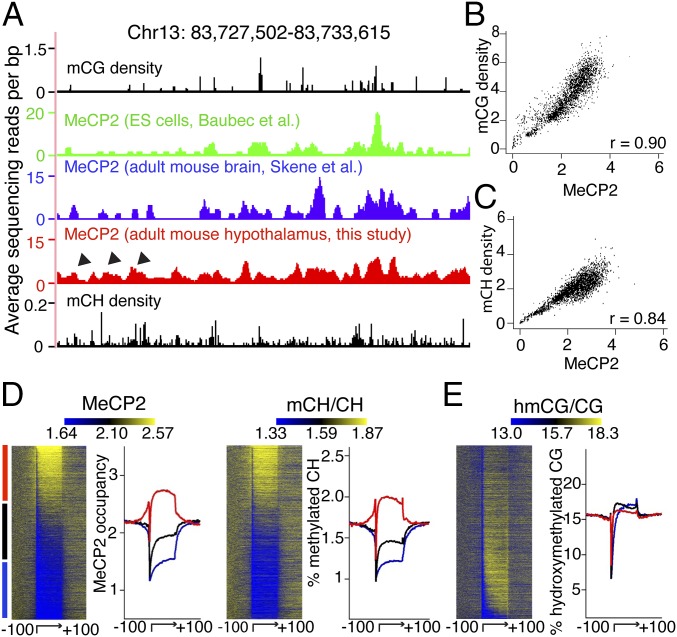
We next investigated the relationship between our high-resolution MeCP2 occupancy and DNA methylation, using a recent DNA methylation map of adult mouse brain (19). Inspection of representative chromosomal regions confirmed that MeCP2 occupancy coincides with mCG density, both globally and locally (Figs. 2A and 3A). For a more quantitative test, we calculated MeCP2 binding as a function of mCG density and found the two directly correlate (r = 0.90) (Fig. 3B), in agreement with prior studies (11, 16).
Fig. 3.
mCH is a distinct determinant of MeCP2 binding in the adult mouse brain. (A) Browser representation of MeCP2 binding spanning an example chromosomal region (Top, representing the x axis). Shown are mCG density in adult mouse brain (black) and MeCP2 binding in ES cells (green), whole adult mouse brains (blue), and adult mouse hypothalamus (red). Arrowhead denotes additional MeCP2 binding sites in the whole brain and hypothalamus that lack mCG density but coincide with mCH density peaks in the adult mouse brain (black). (B and C) Whole-genome representation of MeCP2 binding as a function of mCG density and mCH density in the adult mouse brain. r denotes Spearman’s correlation. (D) Heatmap representation of MeCP2 binding and mCH levels in the gene body and the flanking 100 kb for all mouse genes in the adult mouse brain, ranked by MeCP2 occupancy in the gene body. The graph to the right of each heatmap shows normalized MeCP2 and mCH profiles divided into levels of MeCP2 occupancy: highest third (red), intermediate (black), and lowest third (blue). (E) Heatmap representation and hmCG profiles for all mouse genes in the adult mouse brain ranked by the same method and depicted in the same color scheme as in D. Signal values in heatmap and average plot are normalized so that the mean signal value in flanking regions of each gene will be the same in D and E.
We reasoned that there might be binding cues for MeCP2 besides mCG in the mature nervous system when MeCP2 levels are much higher (11, 17). Indeed, unique peaks of MeCP2 binding were observed in the adult mouse brain in areas of lower mCG that correspond to elevated local mCH density (Fig. 3A and Fig. S1). A recent report demonstrated a conserved, genome-wide increase of mCH and 5-hydroxymethyl CG (5hmCG) in the face of relatively unchanging levels of mCG during postnatal brain development (19). We therefore assessed the relationship between MeCP2 binding and these two epigenetic marks genome-wide in the adult mouse brain. We found that MeCP2 binding correlated with mCH density (r = 0.84) (Fig. 3C). To further probe for distinct genomic elements of MeCP2 binding in the mCH context, we then plotted the levels of MeCP2 binding within each annotated gene body and flanking regions according to a previously described method (19). MeCP2 occupancy was greater in gene bodies with higher mCH levels (Fig. 3D). In contrast, there was no apparent correlation between MeCP2 binding and hmCG levels (Fig. 3E) at this resolution. These data suggest that, in addition to the expected binding to mCG, MeCP2 displays substantial binding to mCH sites that accumulate after birth and plateau when brain development is complete (19).
Interestingly, nucleosome occupancy positively correlates with mCG levels (21), and recent data suggest that mCH is spaced similarly to nucleosomes (15, 19). We mapped nucleosome positioning in adult mouse hypothalamus (Fig. S2) and found that the level of CH methylation was higher on nucleosomal DNA than flanking regions (Fig. S3A). As expected, MeCP2 binding, which tracks mCG and mCH at a genomic level (Fig. 3), was higher at nucleosomes in vivo (Fig. S3B). Our observations are consistent with a report that MeCP2 preferentially binds to methylated nucleosomes in an in vitro reconstitution assay (22) and further strengthens the binding principles of MeCP2 to methylated cytosine.
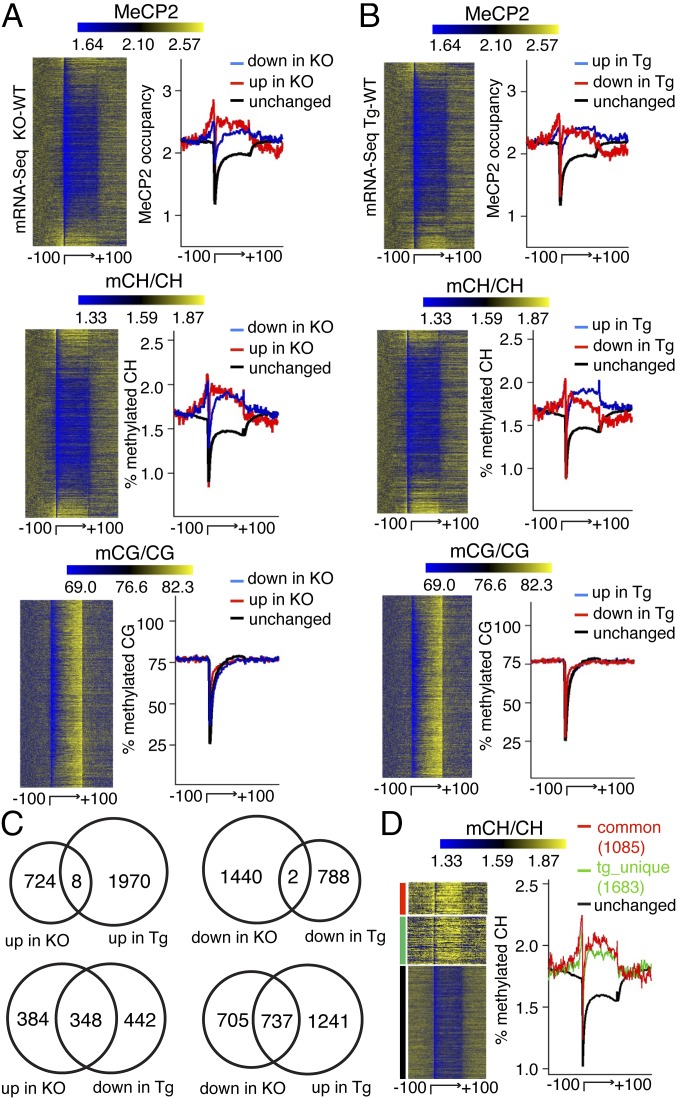
DNA methylation has been implicated in the regulation of gene expression (15, 19, 23, 24). To explore how MeCP2 binding to methylated DNA is involved in transcriptional regulation, we performed transcriptional profiling (mRNA-Seq) in adult MeCP2-null, MeCP2-overexpressing, and wild-type mouse hypothalami. We found numerous transcriptional changes caused by MeCP2 loss- (2,174 genes) or gain- (2,768 genes) of-function using negative binomial model in edgeR with a false-discovery cut-off of 1 × 10−5. Of interest is the reproducible finding that many (1,085, Fisher’s exact test P value < 1 × 10−300) genes exhibited inverse misexpression between the two mutants (Fig. 4C and Fig. S4A), with misregulated genes showing enrichment for neuronal processes (Fig. S4B and Dataset S1). Additionally, ranking all genes by mRNA transcript abundance revealed an inverse relationship between DNA methylation and transcriptional activity (Fig. S5), consistent with previous observations (15, 19).
Fig. 4.
MeCP2 binds mCH to influence transcription in the adult mouse brain. (A) Heatmap representation of MeCP2 binding, mCH and mCG in the gene body, and the flanking 100 kb for all mouse genes, ranked by mRNA expression changes in the adult MeCP2 knockout (KO) brain compared with wild-type (WT) and presented from top to bottom in ascending order (from most down-regulated to most up-regulated). Right panel of each heatmap shows quantitative profiles. Chart depicts up-regulated genes (red), down-regulated genes (blue), and unchanged genes (black) in MeCP2 KO brain. (B) Same as in A except that the data are ranked by mRNA expression changes in the adult MeCP2 transgenic (Tg) mouse brain compared with wild-type and presented from top to bottom in ascending order (from most down-regulated to most up-regulated). Chart depicts up-regulated genes (blue), down-regulated genes (red), and unchanged genes (black) in MeCP2 Tg adult mouse brain. (C) Transcriptional changes in MeCP2 KO and Tg adult mouse brain compared with their corresponding WT. (D) Heatmap representation of flank-normalized mCH in genes with inverse expression changes in both MeCP2 mutants (red); a subgroup of uniquely misregulated genes in MeCP2 Tg animals (green); and those genes that are unchanged (black). Right panel shows quantitative profiles. Signal values in heatmap and average plot are normalized so that the mean signal value in flanking regions of each gene will be the same in A, B, and D.
Given the broad localization of MeCP2 but the narrower extent of transcriptional changes in MeCP2 mutants, we next investigated the relationship between MeCP2 binding, DNA methylation, and gene-expression changes. Analysis of transcriptional changes in MeCP2-null brain compared with wild-type revealed that misregulated genes have higher MeCP2 binding and mCH levels in the gene body than unaffected genes. However, our data revealed no overall correlation between the misregulated genes and mCG levels in the adult mouse brain (Fig. 4A). Moreover, misregulated genes in MeCP2-overexpressing animals correlated with mCH, but not mCG levels in adult mouse brain (Fig. 4B). Taken together, these data suggest that mCG, although correlating strongly with MeCP2 binding at a global level (Fig. 3B), has little effect in directing the gene-expression changes that result from MeCP2 duplication or loss of function.
We hypothesized that, in transgenic animals that overexpress MeCP2 at levels exceeding those of the endogenous protein, MeCP2 might occupy certain sites with intermediate mCH levels and thus influence their expression. Indeed, comparing the transcriptional profiles revealed a broader spectrum of gene misregulation in the MeCP2-overexpressing mice (Fig. 4C). Genes with inverse gene-expression changes in the two MeCP2 mutant lines displayed the highest mCH levels in their gene bodies. The subgroup of uniquely misregulated genes found only in transgenic animals showed intermediate mCH levels (Fig. 4D), which would be consistent with an increase in MeCP2 levels enabling localization at lower mCH/affinity sites to influence transcription as a gain-of-function.
MeCP2 Binding and Misregulation of mCH-Marked, Disease-Modifying Gene, Bdnf, Occurs Uniquely in Adult Mice.
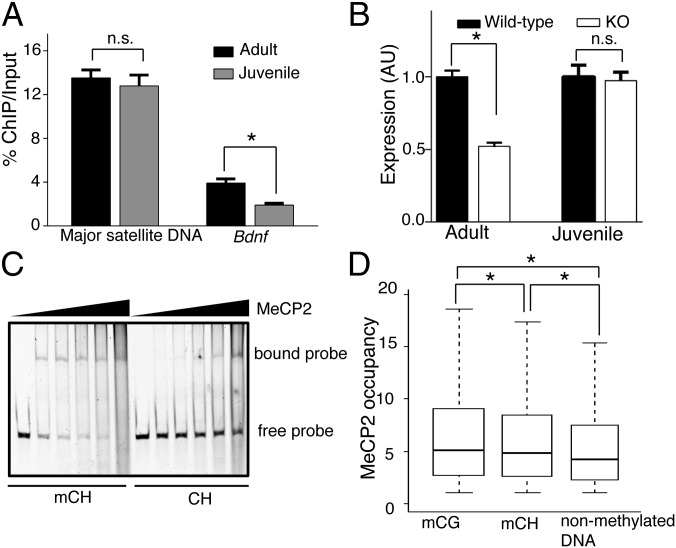
To determine whether MeCP2 recognizes mCH with higher affinity than nonmethylated DNA at otherwise identical genomic sequences over time within the brain, we analyzed MeCP2 binding at the Bdnf locus, known to be positively regulated by MeCP2 (5, 25). In line with this, enhancing the levels of BDNF in mouse models of Rett improves behavioral phenotypes and survival (25). We performed ChIP-qPCR for the Bdnf locus on adult (6-wk-old) and juvenile (3-wk-old) mouse hypothalamus. We chose these two ages because this locus shows dynamic mCH changes with brain maturation (19). MeCP2 binding at a site known to be mCH marked overdevelopment within the Bdnf locus (chr2:109,550,474–109,550,582), and was significantly higher in the adult mouse brains than in the brains of younger animals. In contrast, MeCP2 binding of mouse major satellite DNA, which exhibits methylation at eight CG sites (8), was not significantly affected by age (Fig. 5A). Interestingly, Bdnf was significantly down-regulated in adult but not juvenile MeCP2-null brains (Fig. 5B). To further evaluate the binding affinity of MeCP2 for methylated and nonmethylated DNA elements in the CH context, we purified recombinant full-length MeCP2 to homogeneity (Fig. S6) and tested its binding to a portion of the Bdnf locus by EMSA. We found that MeCP2 binding increased at the locus with only two cytosines methylated during brain development (19) (Fig. 5C). Collectively, our data demonstrated that substantial binding to mCH sites is necessary for MeCP2 to regulate the proper expression of Bdnf, a modulator of RTT-like phenotypes (25).
Fig. 5.
Molecular determinants of genomic targeting of MeCP2. (A) RT-PCR analysis of isolated DNA from MeCP2-ChIP in adult and juvenile mouse hypothalamus using primers for mouse major satellite DNA and Bdnf. Each bar represents SEM; n = 3 or 4; *P < 0.05; n.s., not significant by paired, one-tailed Student’s t test. (B) RT-PCR analysis of Bdnf transcript levels from adult and juvenile mouse hypothalamus. Data were normalized to Gapdh and represented as the fold-expression relative to its age-matched wild-type. Each bar represents SEM; n = 3; *P < 0.05; n.s., not significant by paired, one-tailed Student’s t test. (C) EMSA analysis for recombinant full-length MeCP2 binding to methylated and nonmethylated Bdnf nucleotide sequence in vitro, showing increased binding to methylated substrate. (D) Normalized MeCP2 binding to mCG, mCH, and nonmethylated DNA at the genomic level. Raw reads were binned per 10 base pairs across the entire mouse genome. *P < 10−300 by Kolmogorov–Smirnov test for each pair-wise comparison.
Discussion
Our in vivo analyses suggested that MeCP2 binds early on to mCG and then mCH to influence transcription (Figs. 3 and 4) as the latter accumulates during brain maturation (19). As noted above, MeCP2 binding is genome-wide in the adult brain, so it is necessary to sequence the entire genome several-fold to discover differential binding at high-resolution. Our MeCP2 binding profile thus allows us to compare the binding affinities of MeCP2 to various DNA elements. Whole-genome analysis using base-pair DNA methylation and MeCP2 binding profiles indicate that MeCP2 shows the highest binding affinity for mCG, followed by mCH, and then significantly lower affinity for nonmethylated DNA elements (Fig. 5D), in agreement with a report using synthesized nucleotide sequences in vitro (15). Such a targeting principle ensures proper spatiotemporal MeCP2 binding and likely contributes to transcriptional regulation. In the adult brain, when MeCP2 levels are significantly increased (11, 17), we observe a distinct trend toward increased binding to misregulated genes distinguished by mCH marks but not to those unaffected in MeCP2 mutants. Notably, while our manuscript was under review, a new study showed that MeCP2 binding coincides with mCH (narrowed down to mCA) at genes up-regulated upon MeCP2 loss that are enriched in neuronal functions (26), in agreement with data presented here. Our findings show that MeCP2 binding sites also correspond to mCH sites in genes down-regulated upon MeCP2 loss, and that such genes are also enriched for neuronal function (Fig. S4B and Dataset S1), suggesting that the mode of MeCP2 function at mCH is more complex than proposed (26).
Because the pattern of mCH becomes fully established only in the adult mouse brain, it is interesting to note that the establishment of mCH after birth requires de novo DNA methyltransferase 3A (Dnmt3A) (15, 19, 26). Mice with central nervous system deletion of Dnmt3A display several features characteristic of RTT mouse models, including hypoactivity, decreased grip strength, motor deficits, and premature death (27), underscoring the importance of this mark for neurological function. However, given the lack of specificity of such phenotypes, future studies aimed at comparing the cell-specific phenotypic consequences of loss of Dnmt3A (behavior, physiology, mCH distribution, and gene expression) to those of loss of MeCP2 in the same cell type are necessary to uncover the true functional overlap of these two epigenetic factors.
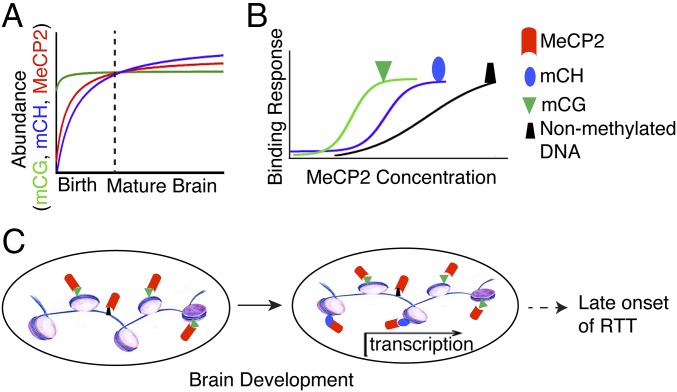
In conclusion, we show that MeCP2 adopts different roles over time in recognizing methylated DNA to influence transcription in the nervous system (Fig. 6). Before birth, MeCP2 tracks mCG and its expression levels are relatively low. This binding is largely at repetitive elements and does not seem to contribute to direct regulation of gene expression, consistent with the previous model (11). During brain development, mCH marks build up in a subgroup of genes (e.g., Bdnf), whereas mCG remains relatively unchanged. As MeCP2 accumulates during neuronal maturation, substantial amounts of MeCP2 occupy those mCH sites to influence transcription. We propose that this dynamic spatiotemporal relationship with methylated DNA in the CH context plays a central role in MeCP2’s epigenetic regulation of gene expression and in part might contribute to the late onset of RTT symptoms. Notably, although our model suggests the mechanism of differential transcriptional regulation across the genome is in part because of the intrinsic affinity of MeCP2 for different DNA modifications, further investigation is needed to assess the influence of local binding partners, as well as the total density and context of mC sequences to further elucidate the rules for functional MeCP2 binding. Although future studies are needed to determine precisely how the rise in MeCP2 and mCH levels leads to alterations in gene transcription, we now have a broad mechanistic framework in which to understand the modes of MeCP2 function in the mature nervous system.
Fig. 6.
Proposed model of MeCP2 genomic targeting to influence transcription. (A) During postnatal brain development the levels of MeCP2 (red) and mCH (blue) increase as neurons mature, whereas mCG (green) remains relatively stable. When DNA methylation patterns are fully developed together with higher levels of MeCP2 in the adult brain, differential binding affinity of MeCP2 for mCG (green), mCH (blue) and nonmethylated DNA (black) becomes apparent as illustrated in B. (C) In this context, MeCP2 can now bind available mCH sites to influence transcription. Thus, changes to MeCP2 levels or function, which lead to disease related phenotypes, only become apparent after the brain has fully matured.
Materials and Methods
For further details, see SI Materials and Methods.
Mice.
Mecp2 heterozygous mice, carrying Mecp2tm1.1Bird allele (3) were backcrossed and maintained on a pure 129SvEvTac background. MECP2 Tg3 mice in pure FVB background were previously described (4, 5). Wild-type male mice were crossed to Mecp2 heterozygous and transgenic female mice and the resulting F1 males were used for preparing total RNA and MNase-digested nucleosomes. Transgenic MECP2-EGFP mice on a pure FVB background (18) were crossed to Mecp2 heterozygous females resulting in the FVBx129SvEvTac F1 males and genotyped using the following primers detecting GFP (forward 5′-cagcaggaccatgtgatcgc-3′, reverse 5′-gtgaagttcgaggtgcgacac-3′). Mecp2-/y; MECP2-EGFP male mice were used for all MeCP2 ChIP experiments. All mouse studies were approved by the Institutional Animal Care and Use Committee for Baylor College of Medicine and were conducted under the guidelines of the Center for Comparative Medicine at Baylor College of Medicine.
ChIP and ChIP-Seq.
ChIP was performed as described previously (13). Libraries from input and ChIP-ed DNA were prepared by the Genomic and RNA Profiling Core at Baylor College of Medicine following the standard manufacture’s protocol (Illumina) and sequenced using an Illumina Solexa HiSeq system. For further details see SI Materials and Methods.
mRNA-Seq.
Mice (7-wk) were killed under anesthesia and the hypothalamus tissues were quickly dissected over ice. Tissue samples were homogenized in PureZOL Reagent (Bio-Rad) through a Polytron disruption method and purified using an Aurum Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad) following the manufacturer’s instructions. Isolated RNA was eluted in RNase-free water and submitted to the Genomic and RNA Profiling Core at Baylor College of Medicine. RNAs were polyA-selected, amplified, and sequenced using an Illumina Solexa HiSeq system following the manufacturer’s instructions. For details of mRNA-Seq and gene ontology analysis, see SI Materials and Methods.
MNase-Seq.
Mice (7-wk) were killed under anesthesia and the hypothalamus tissues were quickly dissected over ice. MNase digestion of chromatin was prepared as described previously (28). Briefly, samples washed in cold PBS and resuspended in RSB buffer (10 mM Tris⋅HCl, 10 mM NaCl, 3 mM MgCl2, 0.2% Nonidet P-40, protease inhibitors). Nuclei pellets were digested with Micrococcal Nuclease in the presence of 1 mM CaCl2. Reactions were terminated by adding 10 mM EDTA. The chromatin preparations were then purified using a Qiagen PCR Purification Kit. Libraries from purified mononucleosomes were prepared by the Genomic and RNA Profiling Core at Baylor College of Medicine following the standard manufacture’s protocols (Illumina) and sequenced using an Illumina Solexa HiSeq system. For details of MNase-Seq data analysis, see SI Materials and Methods.
EMSA.
Full-length MeCP2 was expressed and purified as described previously (13). The strands of the DNA probe (Sigma Life Science) were annealed by boiling both strands for 5 min and cooling down to room temperature over the course of 3 h. The sequences were as follows: forward 5′- ctcagatctaagagttagCaCacattcttcccttagggaaacaaaatgcaactgcattcatctt-3′, reverse 5′-aagatgaatgcagttgcattttgtttccctaagggaagaatgtgtgctaactcttagatctgag-3′. Two methylated cytosines in the mCH probe were capitalized and synthesized using 5-Me dC as substrates instead of dC.
EMSA was performed as previously described (13). Briefly, increasing amounts of MeCP2 protein (0 µg, 0.25 µg, 0.5 µg, 1 µg, 2 µg, and 4 µg) were incubated with 0.1 mg/mL BSA, 30 nM DNA probe in EMSA buffer [10 mM Tris HCl, 50 mM KCl, 0.5 mM MgCl2, 0.1 mM EDTA, 5% (vol/vol) glycerol] at room temperature for 30 min. DNA loading dye was added and the mixture of samples was separated on a 6% (vol/vol) acrylamide gel in 0.5× TBE. Gels were poststained with ethidium bromide and imaged under UV light to detect DNA.
RT-PCR Assay.
For RT-PCR analysis of gene expression, isolated RNA was reverse-transcribed using M-MLV (Invitrogen) and random hexamer primers. The resulting cDNA was diluted and stored at −20 °C until needed. Quantitation of gene expression was carried out in in triplicate reactions using iTaq SYBER Green Supermix (Bio-Rad) and a CFX96 Real-Time System (Bio-Rad) per the manufacturer’s instructions. Results were averaged for each sample and normalized to Gapdh. For RT-PCR analysis following ChIP experiments, the input and ChIP-ed DNA were diluted and stored at −20 °C until needed. Reactions were carried out in in triplicate reactions using iTaq SYBER Green Supermix (Bio-Rad) and a CFX96 Real-Time System (Bio-Rad) per the manufacturer’s instructions. Data were normalized to input and shown as percentage of ChIP/Input. Statistical analyses of RT-PCR data were indicated in figure legends. Data were shown as mean ± SEM, with significance set to P < 0.05. For detailed primer information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the H.Y.Z. laboratory and the W.L. laboratory for discussions and input on the manuscript; Ronald Richman for help with the purification of methyl-CpG binding protein 2; and Dr. Peng Yu for initial data analyses that did not make it into the current manuscript. This work was supported in part by the Genomic and RNA Profiling Core at Baylor College of Medicine and National Institutes of Health Grant P30HD024064 (Intellectual and Developmental Disabilities Research Center) to generate the datasets; National Institutes of Health Grant 5R01NS057819 (to H.Y.Z.); National Institutes of Health Grant HG007538 (to W.L.); and Cancer Prevention Research Institute of Texas Grants RP110471 and RP150292 (to W.L.). H.Y.Z. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE66871).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505909112/-/DCSupplemental.
References
- 1.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Lyst MJ, Bird A. 2015. Rett syndrome: A complex disorder with simple roots. Nat Rev Genet, 10.1038/nrg3897.
- 3.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 4.Collins AL, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13(21):2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- 5.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Esch H, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77(3):442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Gaudio D, et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet Med. 2006;8(12):784–792. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- 8.Lewis JD, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 9.Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16(1):414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 11.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37(4):457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J Biol Chem. 2007;282(20):15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- 13.Baker SA, et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152(5):984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo JU, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17(2):215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baubec T, Ivánek R, Lienert F, Schübeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153(2):480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11(2):115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 18.Heckman LD, Chahrour MH, Zoghbi HY. Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. eLife. 2014;3:02676. doi: 10.7554/eLife.02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterston RH, et al. Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 21.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143(3):470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 24.Weber M, Schübeler D. Genomic patterns of DNA methylation: Targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19(3):273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49(3):341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 26.Gabel HW, et al. 2015. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 10.1038/nature14319.
- 27.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236(6):1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123(7):1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315(5815):1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGraw CM, Samaco RC, Zoghbi HY. Adult neural function requires MeCP2. Science. 2011;333(6039):186. doi: 10.1126/science.1206593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.