Significance
Alternative splicing is a key mechanism for gene regulation that is regulated in response to developmental and antigen signaling in T cells. However, the extent and mechanisms of regulated splicing, particularly during T-cell development, have not been well characterized. Here we demonstrate that expression of the RNA binding protein CELF2 (CUGBP, Elav-like family member 2) is increased in response to T-cell signaling through the combined regulation of transcription and mRNA stability. This increase in CELF2 expression drives widespread changes in mRNA splicing in cultured T cells and correlates with changes in mRNA splicing during T-cell development. These results provide unprecedented insight into the regulation of splicing during thymic development, and reveal an important biologic role of CELF2 in human T cells.
Keywords: alternative splicing, CELF2, thymic development, T cells, mRNA stability
Abstract
Studies in several cell types have highlighted dramatic and diverse changes in mRNA processing that occur upon cellular stimulation. However, the mechanisms and pathways that lead to regulated changes in mRNA processing remain poorly understood. Here we demonstrate that expression of the splicing factor CELF2 (CUGBP, Elav-like family member 2) is regulated in response to T-cell signaling through combined increases in transcription and mRNA stability. Transcriptional induction occurs within 6 h of stimulation and is dependent on activation of NF-κB. Subsequently, there is an increase in the stability of the CELF2 mRNA that correlates with a change in CELF2 3′UTR length and contributes to the total signal-induced enhancement of CELF2 expression. Importantly, we uncover dozens of splicing events in cultured T cells whose changes upon stimulation are dependent on CELF2 expression, and provide evidence that CELF2 controls a similar proportion of splicing events during human thymic T-cell development. Taken together, these findings expand the physiologic impact of CELF2 beyond that previously documented in developing neuronal and muscle cells to T-cell development and function, identify unappreciated instances of alternative splicing in the human thymus, and uncover novel mechanisms for CELF2 regulation that may broadly impact CELF2 expression across diverse cell types.
To survive and carry out their physiological function, human cells must express thousands of gene isoforms at the appropriate level in response to changing developmental and environmental cues. Transcription, pre-mRNA splicing, mRNA stability, translation, and protein stability have all been shown to contribute to the expression levels of individual genes and ultimately to cellular function (1–3). Importantly, these processes do not occur in isolation; rather, one mechanism may alter the expression of a protein that regulates many other gene-expression events via additional mechanisms (3, 4). Such gene regulation cascades serve to amplify the functional consequences of an initial trigger.
One important example of gene-regulation cascades is the coordinated alternative splicing of functionally related genes through the altered activity of a common regulatory protein (4). The inclusion of exons in a final mRNA message is generally determined by transacting factors that bind throughout a pre-mRNA and guide the activity of the spliceosome (5). In most described cases, alternative splicing of a given gene is controlled by the combinatorial activity of multiple regulatory proteins, and individual regulatory proteins bind to a large number of distinct transcripts (5). Therefore, regulation of the level of a particular splicing regulatory protein can impact the expression pattern of hundreds of genes, as had been demonstrated in numerous knock-down and knockout studies (4, 6, 7). Changes in the splicing of broad sets of genes have also been observed during developmental transitions or in response to signaling pathways (8–12). In a few cases, such sets of developmentally coregulated splicing events have been causally linked to the change in expression of a particular splicing factor (6, 7, 13–15). However, the mechanisms by which such coregulation is initiated remain largely unknown.
Previously, we have shown that the RNA binding protein CELF2 (CUGBP, Elav-like family member 2) controls the splicing of the gene encoding the transcription factor LEF1 (lymphoid enhancer binding factor 1) in a Jurkat T-cell line (16). Importantly, CELF2 expression is increased in response to stimulation of Jurkat cells, and this increase in expression leads to an increase in the binding of CELF2 to regulatory sequences within the LEF1 gene and a CELF2-dependent increase in the inclusion of a cassette exon within the LEF1 transcript (16). The biologic importance of this regulation is underscored by the fact that the CELF2-enchanced isoform of LEF1 is preferentially active in promoting transcription of the T-cell receptor (TCR)-α chain. Strikingly, we also found an increase in CELF2 expression—and a corresponding change in LEF1 splicing—during the double-negative (DN) to double-positive (DP) transition in thymic T-cell development, which is the stage at which transcription of the TCR-α chain is first induced (16).
The regulation of CELF2 expression was initially characterized during heart development through the activity of miRNAs (11). This developmentally regulated decrease in CELF2 expression leads to a transition in the splicing pattern of a large family of genes important for cardiac function (11, 15). By analogy, we hypothesized that regulation of CELF2 expression in developing T cells is also critical to establish an appropriate program of splicing for T-cell functions. However, the mechanism by which CELF2 expression is increased upon stimulation of Jurkat cells or pre-TCR signaling remained entirely unknown. Moreover, there have been no studies investigating the breadth of CELF2-dependent alternative splicing in developing or mature T cells.
Here we show that both transcription and stability of CELF2 mRNA is increased upon stimulation of Jurkat cells, resulting in the overall observed induction of CELF2 expression. The transcription factor NF-κB is required for the induction of CELF2 transcription shortly after the initial stimulation, whereas the stability of CELF2 mRNA increases on a longer time scale to promote prolonged and enhanced CELF2 expression. Unexpectedly, we show that this change in mRNA stability is dependent on the mRNA’s 3′UTR; however, miRNAs are not involved in controlling CELF2 expression in Jurkat cells. Instead, increased stability correlates with the retention of a 3′UTR intron, and perhaps a shift in the site of polyadenylation. Finally, we identify ∼70 alternative exons, in addition to the known alternative exon in LEF1, that exhibit significant CELF2-dependent changes in splicing upon phorbol myristate acetate (PMA) stimulation of Jurkat cells. These represent approximately one-third of the PMA-responsive exons that were interrogated, implying that induced expression of CELF2 plays a major role in shaping the isoform expression of stimulated T cells. Remarkably, we find a similar enrichment of CELF2-dependent exons among exons that undergo alternative splicing during thymocyte development. Taken together, our data reveal novel mechanisms of CELF2 regulation and demonstrate a broad role of CELF2 in shaping the transcriptome in developing and stimulated T cells.
Results
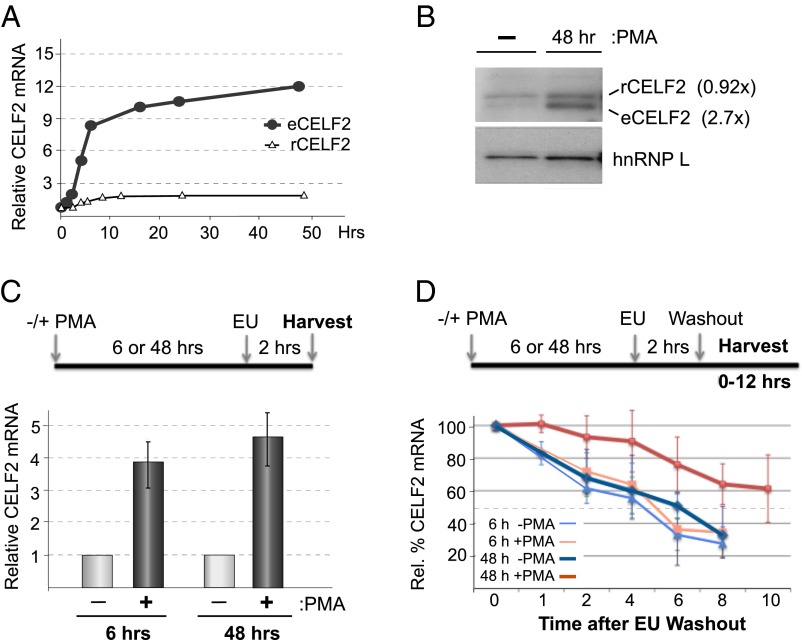
We have previously shown that expression of the splicing regulatory protein CELF2 increases following PMA stimulation of JSL1 Jurkat cells to drive alternative splicing of the gene encoding the LEF1 transcription factor (16) (Fig. 1 A and B and Fig. S1, eCELF2). Importantly, this PMA-induced change in CELF2 expression and LEF1 splicing in Jurkat cells mimics that observed during the pre-TCR signaling-dependent maturation of DN to DP thymocytes (16). Given the functional relevance of this stimulus-induced expression of CELF2 for appropriate expression of LEF1, as well as for other splicing events (see below), we sought to understand the mechanisms driving activation-induced expression of CELF2. Because thymocytes are both highly heterogeneous and difficult to manipulate, we focused on using the Jurkat system as an experimentally tractable model for T-cell development.
Fig. 1.
Stimulation-induced increase in CELF2 mRNA is a result of both increased transcription and mRNA stability. (A) Quantification of total mRNA of endogenous CELF2 or stably integrated Flag-tagged CELF2 cDNA in JSL1 cells following stimulation with 20 ng/mL PMA. Values are determined from RT-PCR analysis of at least three independent experiments and normalized to actin. (Fig. S1). (B) Western blot of endogenous (eCELF2) and cDNA-expressed (rCELF2) CELF2 in JSL1 cells incubated with or without PMA for the time indicated. hnRNP L is used as a loading control. The average difference in CELF2 protein between stimulated and resting (n > 3), relative to the hnRNP L loading control, is given in parentheses. (C) Quantification of newly synthesized endogenous CELF2 mRNA in JSL1 cells grown for 6 or 48 h in the absence or presence of PMA. Nascent transcripts were labeled and isolated by addition of EU at the indicated time (see Materials and Methods) and CELF2 expression was quantified by both RT-PCR and quantitative PCR relative to actin. Values shown are an average of at least three independent experiments and an average of both RT-PCR and quantitative PCR values. (D) Quantification of the stability of nascent CELF2 transcripts. Analysis was identical to that in C, except that cells were incubated for additional time indicated following wash-out of EU label.
Interestingly, whereas both the mRNA and protein of the endogenous CELF2 increase upon stimulation of the Jurkat cells, we observe no analogous change in the RNA or protein derived from a tagged recombinant CELF2 cDNA expression construct stably integrated into the same cells (Fig. 1 A and B and Fig. S1, rCELF2). Notably, the recombinant (r) CELF2 mRNA is driven by a constitutive heterologous promoter and lacks all of the 3′UTR sequences of the native endogenous CELF2 gene. Therefore, the differential regulation of the endogenous CELF2 compared with the rCELF2 suggests that the endogenous promoter and 3′UTR are responsible for the PMA-induced expression of both CELF2 mRNA and protein.
The expression of CELF2 in developing cardiomyocytes has been shown to be strongly regulated by miRNAs, as depletion of the miRNA processing factor Dicer results in a significant up-regulation of CELF2 expression in these cells (11). In contrast, we observe no effect of Dicer depletion on CELF2 expression in Jurkat cells, even under conditions in which known miRNA target genes in Jurkat cells are impacted (Fig. S2). Although we cannot fully exclude the possibility that we would see a change in CELF2 expression if greater than 50% depletion of Dicer could be achieved, we note that in cardiomyocytes a ∼66% reduction in Dicer was sufficient to yield a 10× increase in CELF2 protein (11). Thus, regulation of miRNA function is unlikely to play a primary role in controlling CELF2 up-regulation in activated T cells.
Given the requirement for the endogenous promoter and 3′UTR for CELF2 induction, we next investigated whether PMA alters the transcription or stability of endogenous CELF2 mRNA. Using ethynyl uridine (EU) labeling of nascent transcripts, we observe a ∼fourfold increase in transcription of the endogenous CELF2 mRNA 6 h after PMA stimulation, and continuing at least through 48 h poststimulation (Fig. 1C). By following the loss of nascent transcripts after wash-out of EU, we also find that the stability of CELF2 mRNA is increased upon stimulation, as indicated by a change in the half-life of CELF2 mRNA from ∼5 h to greater than 10 h by 48 h posttreatment with PMA (Fig. 1D). Notably, we do not observe an increase in the stability of CELF2 message when assayed only 6 h after the cells are stimulated. Therefore, we conclude that the PMA-induced increase in CELF2 mRNA and protein expression is driven initially by an increase in transcription, followed by an increase in message stability. Importantly, the four- to fivefold increase in transcription combined with the subsequent two- to threefold change in mRNA stability is sufficient to yield the ∼12-fold overall increase in message level that we observe without a need to invoke any additional layers of mRNA regulation (Fig. S3).
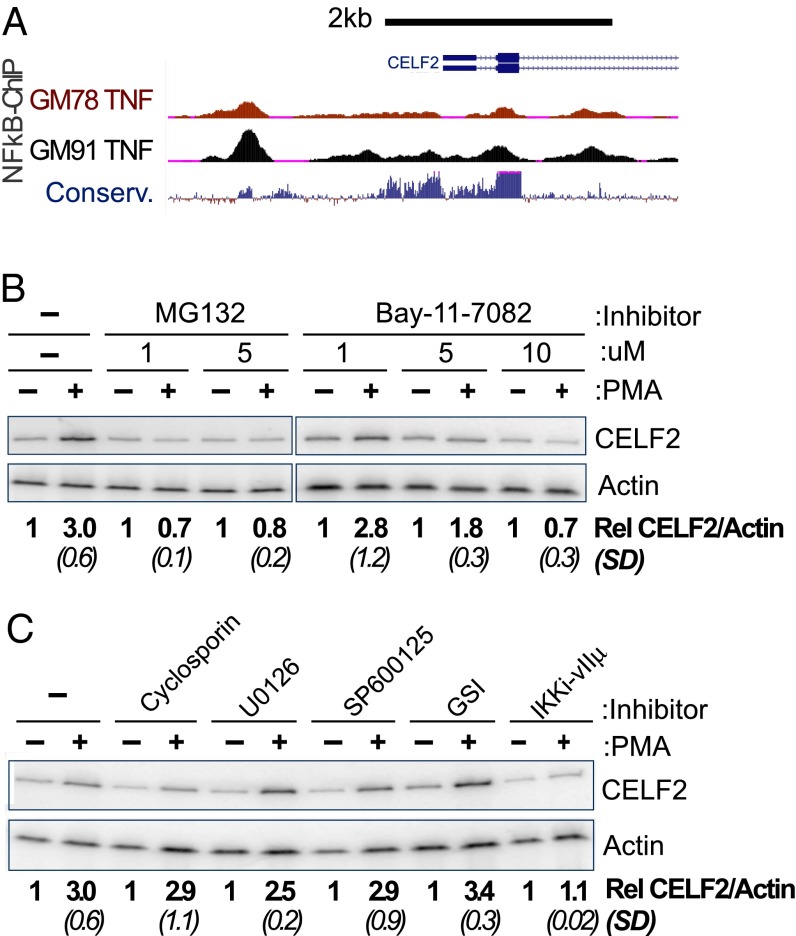
As increased transcription is the initial driver of enhanced CELF2 expression, we first investigated the mechanism of this transcriptional activation. Several transcription factors are known to be activated upon PMA stimulation of T cells, including NF-κB, nuclear factor of activated T cells (NFAT), and activator protein 1 (AP-1) (1, 17–19). Importantly, publically available ChIP-Seq data reveal multiple NF-κB–RelA binding sites around the primary transcription start site for CELF2 in at least two different cell types (Fig. 2A). Many of these NF-κB ChIP sites reside in highly conserved genomic regions, suggesting selective pressure to maintain NF-κB binding (Fig. 2A). Strikingly, when we treat cells with any of three well-documented inhibitors of NF-κB activity—MG132, BAY11-7082, or the VIIμ inhibitor of IKK (20–23)—we entirely abrogate the increase in mRNA expression normally observed after 6 h of PMA stimulation (Fig. 2 B and C). In contrast, treatment of cells with inhibitors of NFAT [cyclosporine (24)], the ERK pathway [U0126 (25)], AP-1 [SB600125 (26)], and Notch [γ-secretase inhibitor XXI (GSI) (27)] has minimal or no effect on PMA-induction of CELF2 expression (Fig. 2C). We conclude that activation of NF-κB promotes the transcription of CELF2 and is necessary for at least the initial induction of transcription in response to PMA stimulation.
Fig. 2.
Inhibition of NF-κB abrogates the rapid transcriptional induction of CELF2. (A) University of California Santa Cruz Genome Browser shot of 5′ end of the CELF2 locus, showing ChIP signal for NF-κB (RelA) at regions of high conservation upstream of CELF2 transcription start site in TNF-α–induced GM12878 and GM12891 lymphoblastoid cell lines (genome.ucsc.edu/) (68). (B and C) RT-PCR analysis, as in Fig. 1A, showing loss of CELF2 induction upon inhibition of NF-KB (B and C) or other PMA-induced transcription factors (C). Inhibitors are as follows: MG132 (5 μM, inhibits proteosome degradation of IkB), BAY11-702 (10 μM, inhibitor of IkB-a), cyclosporin A (10 μM, inhibits NFAT), U0126 (20 μM, inhibits MEK pathway), SP600125 (50 μM, inhibits JNK activation of AP-1), GSI (1 μM, inhibits Notch activation), IKK inhibitor VII (10 μM, inhibits IKK-mediated degradation of IkB). Induction of CELF2 mRNA is calculated as the fold-increase in CELF2 in PMA treated sample relative to unstimulated control, normalized to actin as a loading control. Values shown are the average of at least three independent experiments. SD are shown in parentheses.
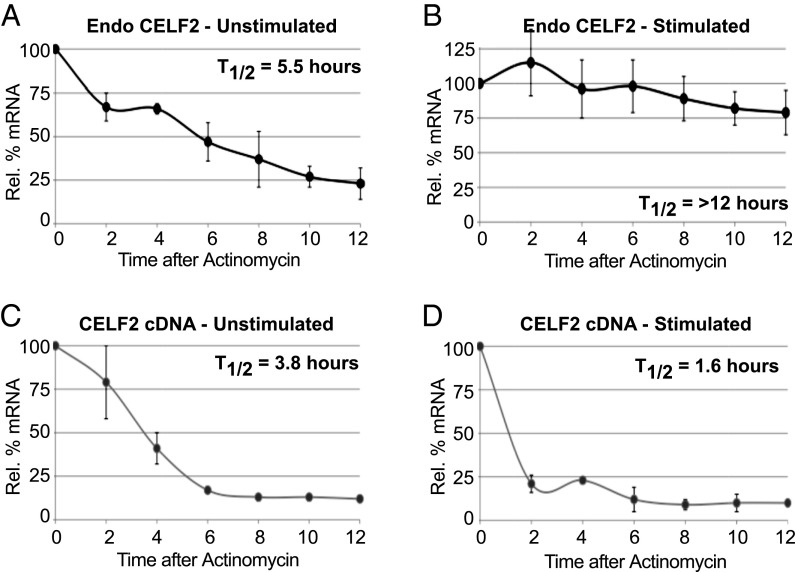
We next investigated the role and regulation of stimulation-induced CELF2 mRNA stability. To complement the EU pulse-chase analysis of CELF2 stability (Fig. 1D) with an orthogonal assay, we used the transcription-blocker actinomycin. Consistent with the EU experiment, upon inhibition of transcription we observe a two- to threefold increase in the half-life of CELF2 message in cells stimulated for 48 h with PMA compared with unstimulated cells (Fig. 3 A and B). Strikingly, mRNA derived from the integrated CELF2 cDNA vector exhibits a similar half-life as the endogenous message in resting cells (Fig. 3 A and C), but is not stabilized upon PMA-stimulation (Fig. 3D). In fact, the stability of the cDNA-derived CELF2 mRNA appears to be somewhat less than the endogenous message in resting cells and even shows a further decrease upon stimulation (Fig. 3 C and D). As the 3′UTR is the only distinction between vector-encoded and endogenous mRNAs, the differential half-lives of these messages strongly suggest that PMA-induced stabilization is conferred through the endogenous CELF2 3′UTR.
Fig. 3.
Stimulation-induced CELF2 mRNA stability is not an inherent feature of the cDNA. Quantification of mRNA from the endogenous CELF2 gene (A and B) or stably integrated cDNA expression construct (C and D) following actinomycin-induced transcriptional inhibition in unstimulated (A and C) or PMA-stimulated (B and D) JSL1 cells. Quantification of mRNA as in Fig. 1A, normalized to actin, n ≥ 3.
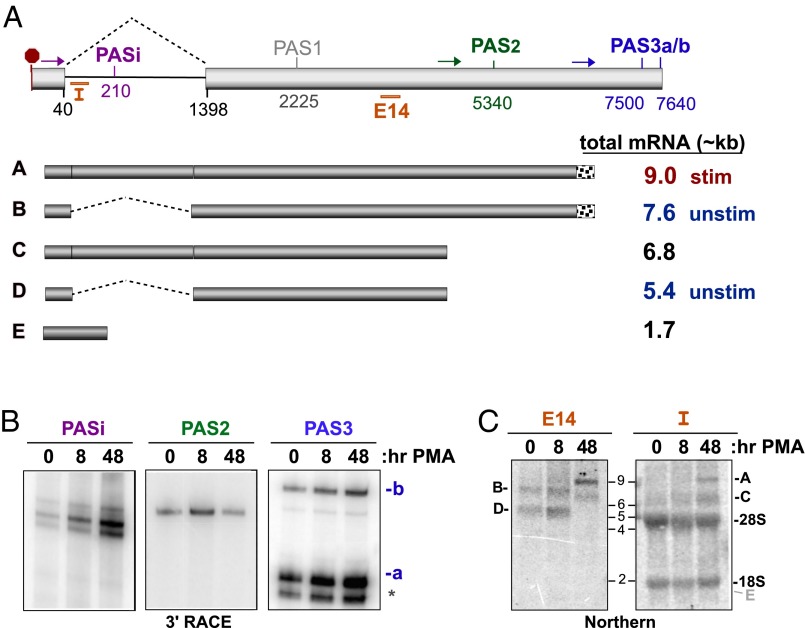
The 3′UTR of CELF2 contains an intron and multiple potential sites of cleavage and polyadenylation (Fig. 4A and Fig. S4) (www.ensembl.org/index.html). Therefore, our first step toward understanding the mechanism of CELF2 mRNA stability is to determine what polyadenylation sites are used in Jurkat T cells before and after stimulation with PMA. Using 3′ RACE, we find products corresponding to use of polyadenylation sites (PAS)i, PAS2, and PAS3 in Jurkat cells (Fig. 4 A and B and Fig. S5A), whereas evidence for other putative PAS sites is either absent or ambiguous (Fig. S5A). The emergence of PASi upon PMA treatment (Fig. 4B) correlates with an increase in intron retention as observed by RT-PCR (Fig. S5B). Use of PAS3 also increases throughout the duration of the PMA-stimulation, whereas 3′ RACE indicates an initial increase in PAS2-processed mRNA followed by a sharp decline in PAS2 use upon prolonged stimulation (Fig. 4B).
Fig. 4.
Alterations in CELF2 3′UTR expression correlate with changing mRNA stability. (A) Schematic of the exon 13–14 region that comprise the 3′UTR of CELF2. (Upper) Light gray boxes are exons, line is intron. Numbers indicate position of splice sites or Ref-Seq described putative PAS relative to the translation stop codon (red octogon). Colored arrows indicate position of primers used in 3′ RACE to detect the four PAS sites observed in JSL1 cells (PASi, PAS2, PAS3a, PAS3b). Orange bars indicate position of Intron (“I”) or exon 14 (“E14”) probes used for Northern blots. (Lower) Schematic of the five 3′UTRs for which there is experimental evidence in JSL1 cells and approximate size of the corresponding complete mRNA. Predominant forms in unstimulated and stimulated Jurkat cells are indicated. Note that mRNAs using PAS3a and PAS3b are grouped together as these sites are in close proximity and are not easily distinguished by Northern blot. (B) 3′ RACE analysis of PAS utilization in the endogenous CELF2 gene in unstimulated (0 h PMA) or stimulated (8 or 48 h PMA) JSL1 cells. Oligo-dT reverse transcription extension was amplified using the primers shown in A (see Materials and Methods). Primers immediately upstream of PASi, PAS2, and PAS3 result in products shown. Identity of PAS sites was confirmed by sequencing. (C) Northern blots of total RNA from JSL1 cells grown under indicated conditions were hybridized to oligonucleotides probes specific for a region of exon 14 upstream of both PAS2 and -3 (Left, E14, detects 3′UTRs A–D), or the 5′ end of intron 13 (right, I, detects 3′UTRs A, C, and E). Position where mRNA E would be detected is marked in gray.
To further quantify the relative use of PASi, PAS2, and PAS3, we interrogated the 3′UTR identity by Northern blot. Consistent with the 3′ RACE and RT-PCR results, only two mRNA species are detected in unstimulated cells with a probe that hybridizes to exon 14 (Fig. 4C, E14). These species correspond in size to 3′UTRs in which intron 13–14 is spliced out and PAS2 or PAS3 is used. Consistently, no mRNA species from unstimulated cells hybridize to a probe complementary to intron 13–14 (Fig. 4C, I). Similar results are observed when CELF2 mRNA is assayed 8 h after stimulation (Fig. 4C). However, following 48 h of stimulation there is a dramatic shift in mRNA size by >1 kb, and these larger mRNAs hybridize with both the E14 and intron probes (Fig. 4C), consistent with efficient intron retention 48 h after treatment with PMA (Fig. S5B). Interestingly, no significant mRNA population corresponding to PASi is detected at any of the time points tested, regardless of the intron probe used (Fig. 4C). Therefore, we conclude that PAS2 and PAS3 are the primary sites of polyadenylation in the CELF2 mRNA in Jurkat cells, with PASi accounting for only a minor population. Furthermore, our data show a clear shift toward use of PAS3 and retention of the 3′UTR intron as a consequence of prolonged stimulation (Fig. 4A). These results clearly demonstrate that the sequence of the 3′ UTR of CELF2 is regulated in response to T-cell stimulation and correlates with a change in mRNA abundance and stability.
We considered two possible mechanisms by which intron retention might impact mRNA stability. First, splicing of the 3′UTR intron may trigger nonsense mediated decay (NMD) that is avoided when the intron is retained. Notably, the stop codon within exon 13 is 43 nucleotides upstream of the intron, close to the typical threshold for NMD (28). To test if intron 13 splicing induced degradation by NMD, we inhibited the NMD process by either knock-down of Upf1 or treatment with cycloheximide (Fig. S6). Both conditions resulted in a marked increase in an isoform of Tra2B, previously shown to be sensitive to NMD (29). In contrast, neither expression of the intron 13-spliced nor the intron 13-retained versions of CELF2 were altered by either method of NMD inhibition (Fig. S6 C and D), thus arguing against a model of NMD regulation of CELF2.
In a second potential mechanism, intron 13 retention could lead to nuclear retention of the resulting mRNA, which in turn is sometimes manifest in transcript accumulation (30–32). To address the possibility that intron retention in the 3′UTR alters the nucleo-cytoplasmic localization of CELF2 mRNA, we fractionated cells and assayed for RNA abundance in the nuclear and cytoplasmic pools (Fig. S7). Importantly, incompletely spliced β-actin mRNA and the ncRNA MALAT1 were both observed almost exclusively in the nuclear fraction, consistent with previous reports (30). In contrast, approximately one-third of total CELF2 mRNA is present in the cytoplasm, regardless of intron retention, overall expression level, or stimulation-state of the cells (Fig. S7B). Therefore, we conclude that stability of CELF2 message upon stimulation is not a result of either escape from NMD or altered nucleo-cytoplasmic localization.
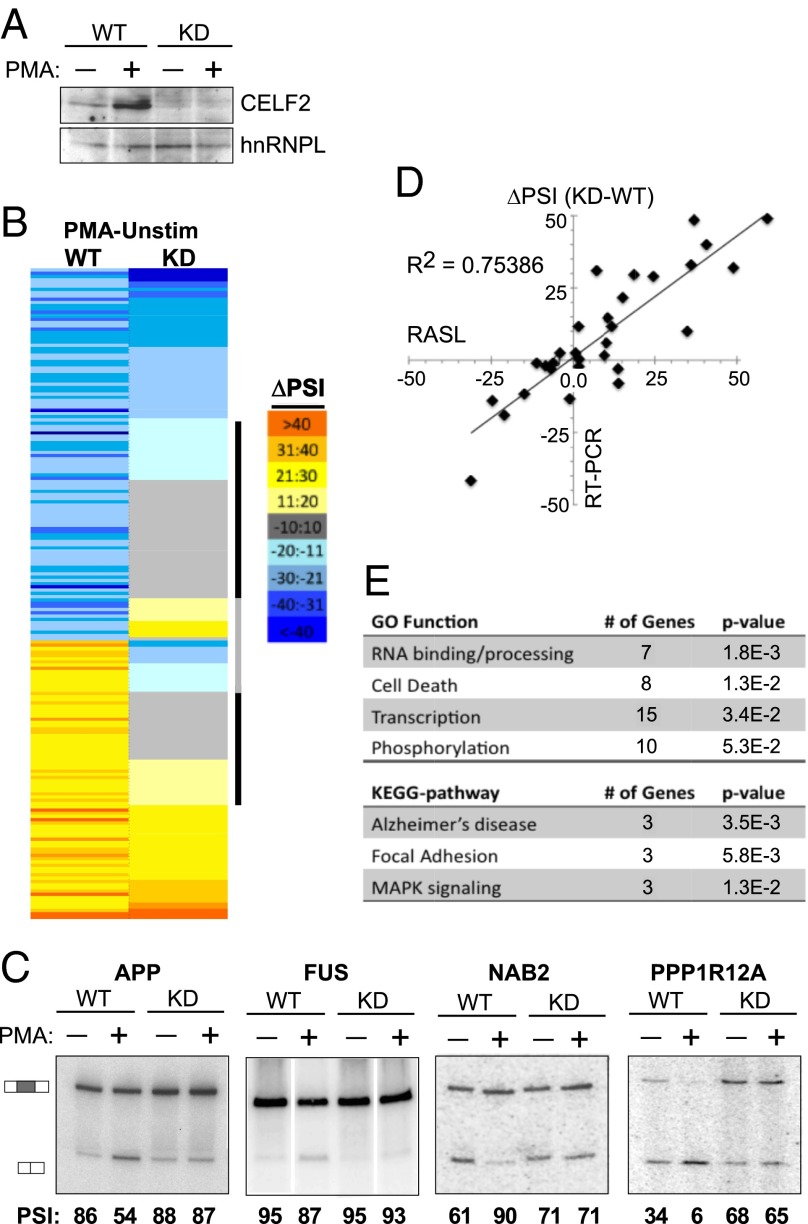
Taken together, the above experiments demonstrate that CELF2 mRNA is increased upon activation of Jurkat cells through a combination of NF-κB–mediated transcription and a change in 3′UTR isoforms that correlate with increased mRNA stability. Although the mechanism of the latter is not yet fully understood, it is clear that the increase in CELF2 mRNA expression leads to an increase in CELF2 protein expression (Fig. 1B) (16). We have previously shown that activation-induced CELF2 expression is critical for the alternative splicing of the at least one gene relevant to T-cell function, namely LEF1 (16). To determine if induced expression of CELF2 contributes more broadly to regulating alternative splicing in response to T-cell signaling, we compared PMA-induced changes in wild-type versus CELF2-depleted JSL1 cells using RNA annealing, selection, and ligation sequencing (RASL-Seq). RASL-Seq quantifies the inclusion of exons by hybridizing primers to sequences immediately at alternative splice junctions, followed by ligation of juxtaposed primers, PCR amplification, and next-generation sequencing (12, 33). For this study, we used a set of primers designed for ∼5,000 alternative splicing events, of which ∼3,000 yielded >10 reads average for both the short and long isoforms across all conditions and biologic replicates assayed (Dataset S1). For each of these events we calculated a percent inclusion of the variable exon in each of four cell conditions: comparing (i) wild-type JSL1 Jurkats to (ii) cells depleted of CELF2 by expression of a shRNA, and comparing (iii) cells grown under unstimulated conditions versus (iv) stimulated for 60 h with PMA (Fig. 5A). The percent inclusion is indicated as percent spliced in (PSI; also note that for events that are alternative splice sites, instead of cassette exons, PSI is designated as the longer isoform), and in each case the PSI value reported is the average of three biologic replicates.
Fig. 5.
Increased CELF2 expression drives a network of activation-induced alternative splicing events in Jurkat cells. (A) Western blot showing CELF2 depletion in JSL1 cells expressing a shRNA targeting CELF2, relative to wild-type JSL1 cells. Expression of CELF2 was assayed in both unstimulated (−PMA) and stimulated (+PMA) conditions. hnRNP L is used as a loading control. (B) Heat map of the ∼200 exons that exhibit a change in inclusion of >10 (P < 0.05) upon PMA stimulation in wild-type JSL1 cells, and the corresponding change in PSI upon PMA stimulation in the CELF2-depleted cells. Color scale is indicated to the right. Exons were sorted first by the directionality of PMA-induced change in wild-type cells and then each category (enhanced or repressed exons) was sorted for the extent of PMA-induced splicing change in the CELF2-depleted cells. Black bar indicates those exons for which CELF2 depletion has a significant effect on PMA-induced splicing regulation; gray bar indicates those exons for which CELF2 depletion switches the directionality of the PMA response in splicing. (C) Representative RT-PCR validations of CELF2-dependent stimulation-induced splicing events identified by the RASL-Seq data. Percent inclusion values shown are the average of at least three independent experiments. (D) Graphic comparison of ∼30 RASL-calculated changes in exon inclusion upon PMA stimulation (x axis) versus change in exon inclusion confirmed by RT-PCR. (E) Enriched Gene Ontology and Kyoto Encyclopedia of Genes and Genomes categories within the CELF2-dependent genes compared with the total ∼200 genes regulated by PMA. Analysis was done using DAVID (david.abcc.ncifcrf.gov).
We first identified 200 exons that exhibit a significant change in inclusion of more than 10 percentage points upon stimulation of wild-type cells with PMA (PSIWT−PMA – PSIWT+PMA >|10| and P < 0.05) (Fig. 5 B–D and Dataset S2). The identification of these ∼200 PMA-responsive exons among the 3,000 exons surveyed is consistent with our previous estimate that ∼10% of alternative exons are regulated in response to T-cell activation (8). We next investigated the effect of CELF2 depletion on the PMA-responsiveness of these alternative exons. Strikingly, for about one-third of the stimulation-responsive exons (72 of 200), CELF2 depletion reduced the PMA-induced change in inclusion by over 60% (Fig. 5 A–C and Table 1), indicating that the ability of PMA to regulate splicing of these exons is dependent on the expression of CELF2. Moreover, for 42 of these 72 exons, CELF2 depletion has only a minimal effect (<10%) on inclusion in resting cells (Table 1 and Dataset S2), suggesting that the low level of expression of CELF2 in resting cells is not sufficient to alter splicing of these genes but that the PMA-induced expression of CELF2 is a primary driver of their signal-responsive splicing.
Table 1.
Genes dependent on CELF2 expression for PMA-responsiveness
| Gene name | ΔPSI-WT | ΔPSI- KD | % CELF2 dependence |
| GK | −55.2 | −8.0 | 85.5 |
| PTS | −48.8 | −11.2 | 77.2 |
| CUGBP2 | −48.8 | 4.2 | 108.7 |
| APP | −36.7 | −1.3 | 96.3 |
| MATR3 | −36.1 | −5.0 | 86.1 |
| PKN2 | −34.1 | −1.4 | 95.8 |
| RAB3IP | −30.6 | −6.6 | 78.4 |
| BAZ2B | −28.1 | −5.0 | 82.2 |
| ADNP | −27.7 | 3.0 | 110.8 |
| BRD8 | −25.9 | −3.7 | 85.9 |
| RIMS2 | −25.4 | −1.2 | 95.3 |
| ZFP64 | −24.4 | 0.0 | 100.1 |
| HNRNPH3 | −23.7 | −7.4 | 68.8 |
| PRDM1 | −23.5 | 3.5 | 115.0 |
| OPA1 | −23.5 | −6.9 | 70.5 |
| FAM179B | −22.5 | 3.9 | 117.5 |
| SH3KBP1 | −22.2 | 4.9 | 122.0 |
| AKT2 | −20.8 | −7.4 | 64.7 |
| SRPK2 | −20.6 | −3.1 | 85.2 |
| UGCGL1 | −20.2 | 2.6 | 112.8 |
| NUB1 | −20.1 | −1.1 | 94.5 |
| MTHFD2 | −19.7 | 4.6 | 123.3 |
| PPP1R12A | −19.5 | −1.0 | 95.1 |
| NAPB | −19.0 | 3.2 | 116.9 |
| EPB41 | −16.4 | −1.6 | 90.3 |
| NCOR1 | −14.7 | −2.6 | 82.1 |
| RNF38 | −14.4 | 1.3 | 109.1 |
| MTRR | −13.7 | −4.4 | 67.7 |
| MTRR | −13.7 | −4.4 | 67.7 |
| dJ1198O21.1 | −13.7 | 3.6 | 126.2 |
| EIF4H | −13.1 | 1.0 | 107.6 |
| FUS | −13.0 | −4.6 | 64.7 |
| LOC643837 | −13.0 | 4.9 | 137.7 |
| PRC1 | −12.9 | −2.9 | 77.8 |
| TLK2 | −12.3 | 0.2 | 102.0 |
| NEO1 | −12.2 | −1.7 | 86.0 |
| C20orf199 | −11.9 | −2.2 | 81.1 |
| C10orf28 | −11.7 | 2.1 | 117.5 |
| SLTM | −11.3 | 2.1 | 118.5 |
| DKFZp762G094 | −11.1 | −3.1 | 72.2 |
| Chk2 | −11.0 | −0.9 | 91.7 |
| TSC22D2 | −11.0 | −2.2 | 80.0 |
| R3HDM2 | −11.0 | −3.2 | 70.8 |
| DKFZp547E092 | −10.4 | −1.7 | 83.5 |
| CCNT1 | 10.0 | −1.6 | 115.8 |
| LCK | 10.1 | 2.4 | 76.3 |
| NT5C3 | 10.4 | −3.5 | 133.4 |
| MAPKAPK2 | 10.7 | 2.9 | 72.8 |
| GGCT | 11.3 | −2.2 | 119.5 |
| MCCB | 11.8 | −3.0 | 125.4 |
| MRPL42 | 11.9 | 4.1 | 65.4 |
| PRMT1 | 12.2 | 3.7 | 69.4 |
| PIP5K1A | 13.1 | 5.2 | 60.7 |
| SLC7A6 | 14.0 | 3.1 | 78.2 |
| SPPL2A | 14.6 | 5.0 | 65.8 |
| USF2 | 14.8 | 5.0 | 66.0 |
| PRPF3 | 17.1 | 1.3 | 92.2 |
| RPLP0 | 17.5 | 3.7 | 79.0 |
| PPIL5 | 18.3 | 4.8 | 73.7 |
| CASP8 | 18.6 | 1.0 | 94.9 |
| FLNB | 18.6 | −2.1 | 111.1 |
| FLNB | 18.6 | −2.1 | 111.1 |
| C14orf135 | 19.3 | 7.5 | 61.2 |
| SCLY | 21.1 | −2.9 | 113.6 |
| BBX | 21.2 | 1.8 | 91.3 |
| SLC3A2 | 21.9 | −4.3 | 119.9 |
| CBFB | 22.3 | 5.8 | 74.3 |
| LETMD1 | 24.9 | 6.0 | 76.0 |
| DIS3 | 26.1 | 1.5 | 94.1 |
| CCNL1 | 34.1 | −0.8 | 102.3 |
| NDUFV3 | 42.1 | 10.5 | 75.0 |
| SRSF3 | 44.8 | 11.0 | 75.5 |
Seventy-two genes found by RASL-Seq to require CELF2 for at least 60% of their PMA-induced change in splicing. CELF2 dependence is calculated as the percentage of the ΔPSI in WT cells that is lost when cells are depleted of CELF2 [100 −(ΔPSICELF2KD/ΔPSIWT)]. Genes in bold are not altered by CELF2 depletion in unstimulated cells (see Dataset S2 for further details).
Importantly, we confirmed 26 of 32 (>80%) RASL-identified effects of CELF2-depletion by RT-PCR (Fig. 5 C and D), with a strong correlation between the extent of splicing change induced by CELF-depletion determined by both methods (Fig. 5D) (R2 = 0.75 for all data). Therefore, we conclude that induction of CELF2 expression contributes broadly to the changes in alternative splicing observed upon T-cell activation. We also note that CELF2-regulated exons are enriched in genes involved in RNA processing, cell death, transcription, and phosphorylation (Fig. 5E), all processes critical to shaping T-cell development and antigen-induced effector functions. Although the P value of the enrichment is relatively modest, approximately half of the 72 CELF2 targets identified fall into one of the above functional categories. We also observe enrichment of genes implicated in Alzheimer’s disease, including the amyloid protein APP and the RNA-binding protein FUS (Fig. 5 C and E).
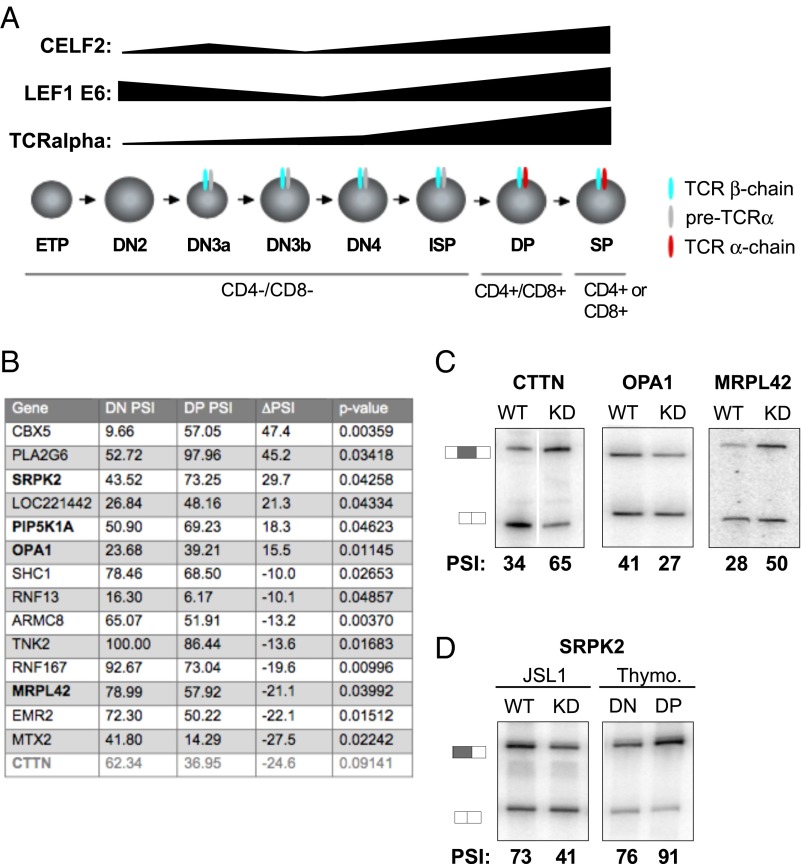
Finally, we sought to determine if the CELF2-dependent splicing events we observe in Jurkat cells are reflective of alternative splicing during T-cell development. We have previously shown that CELF2 expression is increased during the transition from DN to DP (CD4/CD8-expressing) thymocytes, and that CELF2-dependent alternative splicing of LEF1 also occurs during this DN to DP transition (16) (Fig. 6A). To identify additional changes in splicing during T-cell development, we performed RASL-Seq using RNA from FACS-isolated DN and DP cells from human thymocytes (Fig. S8). Given limited thymic material available and inherent donor variability, the RASL-Seq analysis only identified 14 exons that display >10% PSI changes that are statistically significant across the three donors surveyed (Fig. 6B and Dataset S3). Remarkably, however, 4 of these 14 events (29%) are among the 72 CELF2-dependent signal-responsive exons identified in the Jurkat cells (Fig. 6B) (P < 0.00036, two-tailed Fisher’s exact test). An additional CELF2-dependent event (CTTN) was also observed to change dramatically in the DN vs. DP cells, albeit with a modest P value (0.09). We validated the CELF2 dependence of four of these genes in JSL1 cells, and further confirmed the change in splicing between DN and DP cells for SRPK2 (Fig. 6 C and D). We were unable to obtain clear RT-PCR data on PIP5K5A given the presence of highly homologous intronless genes. Although none of CTTN, OPA1, or MPRL42 were expressed to sufficient levels in the minimal thymic RNA available for quantification by RT-PCR, depletion of CELF2 in the Jurkat cells altered splicing of these genes in a pattern opposite to that observed upon the transition from DN to DP (Fig. 6C), consistent with the increased CELF2 expression in DP cells contributing to the observed developmentally induced change in splicing of these genes. This correlation between CELF2-expression and thymic splicing is most strikingly shown in the case of SRPK2, where we can directly validate both the thymic regulation and CELF2 dependence identified by RASL-Seq (Fig. 6D). Taken together, our data indicate that signal-induced changes in CELF2 expression not only control splicing upon activation of cultured T cells, but also play an unappreciated and widespread role in shaping gene expression during T-cell development.
Fig. 6.
Alternative splicing events in thymocytes are enriched for CELF2 target genes. (A) Schematic of T-cell development showing DN and DP stage of thymic development and TCR-α expression and the corresponding changes in CELF2 mRNA expression and inclusion of LEF1 Exon 6 from ref. 16. (B) Table of significant changes in splicing between DN and DP stages of development, as determined by RASL-Seq. Last gene in gray has a P value > 0.05. (C) Validation of CELF2-dependence of genes overlapping in the thymocytes and CELF2-KD RASL experiments in JSL1 cells depleted of CELF2 as in Fig 5C. Percent inclusion values shown are the average of at least three independent experiments. (D) Same as C but including also RNA from FACS purified DN and DP thymocytes. PSI values for thymocytes are the average from at least three independent donors.
Discussion
Studies in several cell types have identified dramatic and diverse changes in mRNA processing that occur upon cellular stimulation (2, 9, 10, 12). However, there is limited understanding of the mechanisms through which individual splicing factors are controlled to mediate splicing changes, particularly with regard to the development and activation of immune system lineages (34). Here we demonstrate that expression of the CELF2 splicing factor is increased in response to T-cell signaling through the combined effects of NF-κB–induced transcription and increased mRNA stability. Furthermore, building upon the documented impact that increased CELF2 expression has on LEF1 splicing (16), we uncover dozens of splicing events whose changes upon T-cell maturation and activation require CELF2. These results provide a unique mechanistic link between cell signaling and alternative splicing and highlight a critical role for CELF2 in T-cell development and function.
CELF2 mRNA Regulation in T Cells Differs from That in Other Systems.
The regulation of CELF2 transcription and stability that we describe here add to a growing list of mechanisms by which expression of CELF2 is tightly regulated. In developing cardiac myocytes, the expression of CELF2 protein is regulated primarily through miRNAs (11). Multiple miRNAs that have complementarity to the CELF2 3′UTR have been identified and expression of these miRNAs exhibit a reciprocal pattern to CELF2 protein in the developing heart. In fetal myocytes miRNAs expression is low and CELF2 expression is high, whereas the converse is true in adult tissue. Importantly, depletion of Dicer in mature cardiac muscle leads to a restoration of fetal-levels of CELF2 expression and a return to an embryonic pattern of splicing of CELF2 target genes (11). CELF2 protein expression has also been shown to be controlled by an auto-inhibitory splicing event (35). CELF2 protein represses inclusion of exon 6 of the CELF2 pre-mRNA by competing with U2AF for binding to the 3′ splice site. Skipping of exon 6 results in a premature stop codon, thereby preventing additional expression of full-length protein. It is this negative feedback loop that limits the increase of CELF2 protein to ∼threefold in response to T-cell signaling, despite the 10- to 12-fold increase in CELF2 mRNA (16) (Table 1). We note that this auto-regulation is not unique to CELF2; rather, the expression of many splicing factors is self-limiting through auto-inhibitory splicing regulation (36–38). The frequency of auto-regulation of splicing factors, in addition to other mechanisms through which a protein such as CELF2 is regulated, suggests that maintaining appropriate expression of splicing regulatory factors is essential for cellular viability. Furthermore, such extensive layering of control points implies a high degree of sensitivity of splicing programs to even moderate alterations in the expression of regulatory proteins.
NF-κB Transcription in T-Cell Activation and Thymic Development.
NF-κB has been widely studied in T-cell activation (39). In unstimulated cells NF-κB proteins are present but inhibited from binding to DNA targets through the activity of IkB (inhibitor of NF-κB). Signaling through the T-cell receptor, or stimulation of cells with PMA, leads to phosphorylation and proteosome-mediated degradation of IkB, thus releasing NF-κB (39). Importantly, we show here that inhibition of either the phosphorylation or degradation of IkB blocks the initial PMA-mediated induction of CELF2 mRNA expression in JSL1 Jurkat cells, whereas inhibition of other common T-cell transcription factors has little to no effect on CELF2 expression. A critical role for NF-κB in the activation of CELF2 transcription is also consistent with the rapidity with this transcription is induced (Fig. 1), as NF-κB is activated within minutes of T-cell signaling (39). To our knowledge, our identification of NF-κB is the first transcription factor implicated in driving CELF2 expression. Although we cannot directly test the requirement of NF-κB for CELF2 expression in human thymocytes, NF-κB is known to be activated in response to pre-TCR signaling in thymocytes, and this activation correlates with the thymic developmental stages at which we observe increased CELF2 expression (40, 41). Therefore, we conclude NF-κB likely regulates CELF2 expression in thymic development as it does in our cell line model. Furthermore, despite the initial identification of NF-κB in lymphocytes, this transcription factor is present in a wide range of cell types (39). Therefore, NF-κB may be important for the expression of CELF2 in other tissues in addition to thymocytes. However, we emphasize that our results do not preclude the possible involvement of other transcription factors in regulating the expression of CELF2, particularly in nonlymphoid cell types.
Regulated mRNA Stability in T Cells.
Importantly, the transcriptional induction of CELF2 mRNA is not sufficient to account for the total increase in CELF2 expression upon PMA treatment. Previous studies have demonstrated that regulation of mRNA stability is as important to the overall regulation of gene expression upon T-cell activation as is transcription (42). Consistently, we observe that prolonged T-cell signaling results in a two- to threefold increase in CELF2 mRNA stability, and that this increased stability is essential for the full induction of CELF2 expression.
mRNA stability can be regulated by several different mechanisms (2, 43). MicroRNAs typically bind to complementary sequences in the 3′UTR, and can destabilize bound mRNAs in addition to inhibiting translation (44). In fact, microRNAs in the miR-181 family are known to influence T-cell development through mRNA destabilization (45). Despite this, we demonstrate here that Dicer-mediated microRNA mechanisms play a minimal role in regulating CELF2 mRNA in developing thymocytes. Alternative mechanisms for regulating mRNA stability include NMD and nuclear mRNA retention, both of which have also been implicated as important to various aspects of immune cell physiology (32, 46, 47). However, we also show here that neither NMD nor nuclear retention play a significant role in the stability or expression of any of the CELF2 isoforms. Therefore, we propose that the differential stability of CELF2 upon stimulation is most likely conferred through the activity of RNA binding proteins. For example, the ubiquitously expressed protein HuR is known to bind to AU-rich elements in 3′UTRs and ultimately increase the stability of target mRNA, whereas binding of the proteins TTP or KSRP to similar AU-rich elements typically destabilizes transcripts (48). The variable inclusion of intron 13–14 and shift in PAS use that we observe in the 3′UTR of CELF2, coincident with increased message stability 48 h after stimulation, suggests that the differential stability of CELF2 mRNA may be caused by altering the presence of binding sites for stabilizing or destabilizing proteins. Future work to identify the proteins, sequences, and mechanisms controlling CELF2 mRNA stability will be important to understanding the details of this regulation.
Impact of CELF2 Regulation on T-Cell Development and Function.
As discussed above, the extensive mechanisms that exist to regulate CELF2 expression suggest that tight control of CELF2 is essential for cellular function. Previous studies have indeed shown that misregulation of CELF2 expression in heart and neuronal cells results in widespread changes in splicing patterns and cellular pathology (15, 49, 50). Here we extend the spectrum of CELF2-mediated regulation to include the immune system, by showing that increased CELF2 expression is required for appropriate regulation of splicing in response to antigen or developmentally induced signaling pathways in T cells. Specifically, we find that among ∼200 splicing events that are substantially altered upon stimulation of cultured T cells, approximately one-third are dependent on CELF2. Importantly, analysis of splicing events that are regulated in the transition from DN to DP thymocytes suggests that enhanced expression of CELF2 in DP cells regulates a similar proportion of signal-induced splicing events during thymic development.
The role of alternative splicing during thymic development is largely unstudied. Our data herein provide some of the first insights into the breadth and impact of splicing in thymocytes. Interestingly, the CELF2-dependent splicing events that we have uncovered in this study are enriched for activities with known importance in T-cell development and function (Fig. 5E). Programmed cell death is essential for negative selection of auto-reactive thymocytes and for attenuation of an immune response following initial antigen challenge to ensure homeostasis of the immune system (51). Similarly, regulation of transcription and phosphorylation is essential for shaping effector functions of activated T cells and for propagating thymocytes through development (1, 52). Thus, CELF2-dependent regulation of genes affecting cell death, transcription, and phosphorylation is predicted to have direct consequences on T-cell function.
The presence of genes encoding RNA processing activities among CELF2-regulated splicing events is also notable. Cross-regulation of splicing factors has been described in a number of studies (14, 37, 53, 54), suggesting that splicing factors represent a tightly knit network of positive and negative regulation that finely shapes overall transcriptome processing. The results we present here suggest that CELF2 may be a key hub in this regulatory network, and that CELF2’s impact on global mRNA processing may include indirect as well as direct effects. We are particularly interested in the regulation of SRPK2 by CELF2, as this gene encodes a kinase that phosphorylates the SR family of RNA binding proteins, as well as other proteins involved in RNA processing (55, 56). The change in splicing induced by CELF2, and promoted during the transition from DN to DP thymocytes, alters the N terminus of the encoded SRPK2 protein, reducing expression of a form with a highly basic N terminus and favoring expression of a form with serine-rich N terminus. Although the functional consequence of this isoform change is unknown, given the role of SR proteins in all aspects of RNA biogenesis and translation and the role of phosphorylation in the regulation of SR protein function (57), the regulation of SRPK2 is likely to have a general impact on cellular protein expression.
Finally, several of the genes we validated as CELF2 targets in this study that are not part of an enriched functional group are still of known relevance for T-cell biology and human disease. For example, CTTN encodes cortactin, an actin-regulatory protein that is directly relevant to lymphocyte migration (58, 59). Overexpression of cortactin has also been causally implicated with invasiveness of a wide range of metastatic cancers (60). The OPA1 gene, which encodes a mitochondrial GTPase, was initially identified as the gene mutated in an inherited form of blindness, although this gene has subsequently also been linked to diabetes and “multiple system disease” (61). Finally, both the amyloid precursor protein APP and the RNA-binding protein FUS are well documented to be involved in multiple neuropathologies, including Alzheimer’s disease and amyotrophic lateral sclerosis (62, 63). Although further work remains to define the precise phenotypic effects of CELF2-regulated alternative splicing events on thymocyte development, the data we present here clearly demonstrate the biologic importance of CELF2 outside of the neuromuscular system, and identify new targets and mechanisms of CELF2 regulation that likely have broad impact across all tissues in which this ubiquitous protein is expressed.
Materials and Methods
Cell Culture.
JSL1 Jurkat cells were maintained and stimulated with 20 ng/mL PMA, as described previously (64). Transcription inhibition was achieved by incubation of cells with actinomycin D at a final concentration of 5 μg/mL. Pulse-labeling of transcripts was done as described previously (65) by a 2-h incubation in 200 μM EU, followed by capture of EU-labeled RNAs by reaction with Biotin-Azide and isolation on streptavidin beads (Click-iT Nascent RNA Capture Kit, Invitrogen C10365). Inhibitors of transcription pathways were as follows: MG132 (5 μM; Calbiochem, 474790), BAY11-702 (10 μM; InvivoGen, tiri-b82), cyclosporin A (10 μM; EnzoLife Sciences, BML-A195), U0126 (20 μM; Invitrogen, PHZ1283), SP600125 (50 μM; Millipore, 420119), GSI (1 μM; Millipore, 565790), and IKK inhibitor VII (10 μM; Calbiochem, 401486).
Human Thymocytes.
De-identified human thymus samples were generated as discards from pediatric cardiac surgery and obtained through the Cooperative Human Tissue Network in accordance with IRB #811028. Single-cell thymocytes suspensions were incubated with RBC lysis buffer to deplete for RBCs then stained with anti–CD8-APC (Becton-Dickinson #555369), anti–CD4-FITC (Becton-Dickinson #555346), and a mixture of antibodies to deplete nonthymocytes (human T-cell depletion antibody mix, Invitrogen #113.44D) that were labeled with PE via a secondary antibody (Becton-Dickinson rat anti-mouse PE #349073). DN and DP cells were isolated in the by gating for PE− viable cells, then collecting the most intense APC/FITC double stained (DP) or lowest University of Pennsylvania biohazard cell sorting facility 2–3% staining (DN).
CELF2 Depletion and cDNA Expression.
Expression of Flag-tagged CELF2 was done by stable integration of a cDNA expression construct into JSL1 cells with neomycin as a selectable marker, as described previously (66). The expression construct was generated by cloning of cDNA encoding the 489-aa full-length isoform of CELF2 (CCDS4188) downstream of an N-terminal Flag tag in the pEF-nFlag vector (67). Depletion of CELF2 was done by lentivirus encoding hairpins that target CELF2 cDNA encoding residues 57–63 or 322–330.
RT-PCR.
Low-cycle RT-PCR to quantify mRNA expression and splicing was done as previously described (66) using 32P-labeled primers listed in Table S1.
Quantitative PCR.
Quantitative PCR was performed using Applied Biosystems SYBR Green PCR Master Mix according to standard PCR conditions in an Applied Biosystems 7500 Fast Real-Time PCR System and an Applied Biosystems 7900HT Fast Real-Time PCR System.
Abundance was normalized to β-actin and calculated as 2(CtActin-CtCELF2). Primer sequences are listed in Table S1.
3′ RACE.
RACE-Ready cDNA was produced according to the protocol in the Clontech SMARTer RACE 5′/3′ Kit. Forward primers used to generate RACE-Ready cDNA are listed in the Table S1. RACE-Ready cDNA was quantified using low-cycle RT-PCR as described above.
Northern Blots.
Fifteen micrograms of total RNA from JSL1 cells treated for indicated times with PMA were run on a 1% agarose formaldehyde gel using the NorthernMax kit (Life Technologies). The gel was transferred to Hybond-N+ by capillary transfer, then hybridized to the indicated 5′ 32P end-labeled oligonucleotides probes in UltraHyb-Oligo buffer at 42 °C overnight. Blots were washed 2× 30 min in 2× SSC+0.1%SDS and exposed to a PhosphorImager for quantification. The intron (I) probe is 5′-CCGTCTCTCTTGGTAAAAGTTGGCAATGTGG. The Exon 14 (E14) probe is 5′-CAACTACACGGTTATGTTCACAATGAGGACAG.
RASL-Seq.
RASL-Seq was performed as previously described using a set of probes that interrogate ∼5,000 specific splicing events (33). In brief, total RNA was harvested from biologic triplicate samples of wild-type and CELF-depleted JSL1 cells grown under normal conditions, or stimulated with PMA or from DN and DP cells isolated from three donors. These RNA samples were individually hybridized to the probe set and selected by oligo-dT. Juxtaposed probes annealed to selected RNAs were then ligated and amplified and barcoded by PCR for subsequent multiplexed sequencing on a HiSeq2000. On average, 2 million reads were obtained for each RNA sample. Splicing events were filtered for a minimum of 10 reads averaged across all biologic replicates and conditions and then isoform ratios were calculated by comparing number of reads representing the longest isoform to the number of total reads for that splicing event (PSI = percent spliced in of variable exon). The change in PSI (ΔPSI) was then calculated as the difference between the average PSI across the three biologic replicates of RNA from PMA-stimulated cells versus the three replicates of unstimulated cells. PMA-induced splicing events that are dependent on CELF2 were identified as splicing events for which the absolute value of ΔPSI in wild-type cells was ≥10, with P < 0.05 (unpaired Student’s t test) and the ΔPSI in CELF2 depleted cells was <0.4(ΔPSI wild-type). Two different sequences within the CELF2 mRNA were targeted for knock-down (see CELF2 Depletion and cDNA Expression, above) and cells containing each shRNA were processed separately, such that significant CELF2-dependent events are only those altered by both hairpin sequences. For thymocyte data, P values were calculated using a paired Student’s t test.
Supplementary Material
Acknowledgments
We thank Paul Halberg and the University of Pennsylvania biohazard cell sorting facility for the thymic sorts; the Cooperative Human Tissue Network for providing the human thymus samples; Amy DeMicco and Craig Bassig (University of Pennsylvania) for assistance with quantitative PCR; and Nick Conrad (University of Texas Southwestern) for advice for the Click-IT assay. This work was supported by Grants GM103383 (to K.W.L.) and GM052872 (to X.-D.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423695112/-/DCSupplemental.
References
- 1.Smale ST. Transcriptional regulation in the innate immune system. Curr Opin Immunol. 2012;24(1):51–57. doi: 10.1016/j.coi.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14(6):361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Jangi M, Sharp PA. Building robust transcriptomes with master splicing factors. Cell. 2014;159(3):487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh RK, et al. Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol Cell. 2014;55(4):592–603. doi: 10.1016/j.molcel.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj B, et al. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol Cell. 2014;56(1):90–103. doi: 10.1016/j.molcel.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ip JY, et al. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13(4):563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- 10.Martinez NM, et al. Alternative splicing networks regulated by signaling in human T cells. RNA. 2012;18(5):1029–1040. doi: 10.1261/rna.032243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24(7):653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47(3):422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warzecha CC, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29(19):3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21(13):1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105(51):20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallory MJ, et al. Signal- and development-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Mol Cell Biol. 2011;31(11):2184–2195. doi: 10.1128/MCB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane LP, Shapiro VS, Stokoe D, Weiss A. Induction of NF-kappaB by the Akt/PKB kinase. Curr Biol. 1999;9(11):601–604. doi: 10.1016/s0960-9822(99)80265-6. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro VS, Mollenauer MN, Greene WC, Weiss A. c-rel regulation of IL-2 gene expression may be mediated through activation of AP-1. J Exp Med. 1996;184(5):1663–1669. doi: 10.1084/jem.184.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yablonski D, Kane LP, Qian D, Weiss A. A Nck-Pak1 signaling module is required for T-cell receptor-mediated activation of NFAT, but not of JNK. EMBO J. 1998;17(19):5647–5657. doi: 10.1093/emboj/17.19.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waelchli R, et al. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorg Med Chem Lett. 2006;16(1):108–112. doi: 10.1016/j.bmcl.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 21.Pierce JW, et al. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272(34):21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 22.Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271(44):27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 23.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 24.Kunz J, Hall MN. Cyclosporin A, FK506 and rapamycin: More than just immunosuppression. Trends Biochem Sci. 1993;18(9):334–338. doi: 10.1016/0968-0004(93)90069-y. [DOI] [PubMed] [Google Scholar]
- 25.Favata MF, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273(29):18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 26.Shin M, Yan C, Boyd D. An inhibitor of c-jun aminoterminal kinase (SP600125) represses c-Jun activation, DNA-binding and PMA-inducible 92-kDa type IV collagenase expression. Biochim Biophys Acta. 2002;1589(3):311–316. doi: 10.1016/s0167-4889(02)00195-7. [DOI] [PubMed] [Google Scholar]
- 27.Grosveld GC. Gamma-secretase inhibitors: Notch so bad. Nat Med. 2009;15(1):20–21. doi: 10.1038/nm0109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: Implications for physiology and disease. Biochim Biophys Acta. 2013;1829(6-7):624–633. doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21(6):708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 31.Yap K, Makeyev EV. Regulation of gene expression in mammalian nervous system through alternative pre-mRNA splicing coupled with RNA quality control mechanisms. Mol Cell Neurosci. 2013;56:420–428. doi: 10.1016/j.mcn.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Pandya-Jones A, et al. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA. 2013;19(6):811–827. doi: 10.1261/rna.039081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 2012 doi: 10.1002/0471142727.mb0413s98. Chapter 4:Unit 4.13.1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez NM, Lynch KW. Control of alternative splicing in immune responses: Many regulators, many predictions, much still to learn. Immunol Rev. 2013;253(1):216–236. doi: 10.1111/imr.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dembowski JA, Grabowski PJ. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet. 2009;5(8):e1000595. doi: 10.1371/journal.pgen.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saltzman AL, et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28(13):4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossbach O, et al. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29(6):1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Änkö ML, et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13(3):R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Dea E, Hoffmann A. NF-κB signaling. Wiley Interdiscip Rev Syst Biol Med. 2009;1(1):107–115. doi: 10.1002/wsbm.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voll RE, et al. NF-kappa B activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity. 2000;13(5):677–689. doi: 10.1016/s1074-7613(00)00067-4. [DOI] [PubMed] [Google Scholar]
- 41.Aifantis I, Gounari F, Scorrano L, Borowski C, von Boehmer H. Constitutive pre-TCR signaling promotes differentiation through Ca2+ mobilization and activation of NF-kappaB and NFAT. Nat Immunol. 2001;2(5):403–409. doi: 10.1038/87704. [DOI] [PubMed] [Google Scholar]
- 42.Cheadle C, et al. Control of gene expression during T cell activation: Alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blencowe B, Brenner S, Hughes T, Morris Q. Post-transcriptional gene regulation: RNA-protein interactions, RNA processing, mRNA stability and localization. Pac Symp Biocomput. 2009;2009:545–548. [PubMed] [Google Scholar]
- 44.Pasquinelli AE. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 45.Dooley J, Linterman MA, Liston A. MicroRNA regulation of T-cell development. Immunol Rev. 2013;253(1):53–64. doi: 10.1111/imr.12049. [DOI] [PubMed] [Google Scholar]
- 46.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gudikote JP, Wilkinson MF. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 2002;21(1-2):125–134. doi: 10.1093/emboj/21.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reznik B, Lykke-Andersen J. Regulated and quality-control mRNA turnover pathways in eukaryotes. Biochem Soc Trans. 2010;38(6):1506–1510. doi: 10.1042/BST0381506. [DOI] [PubMed] [Google Scholar]
- 49.Ladd AN. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol Cell Neurosci. 2013;56:456–464. doi: 10.1016/j.mcn.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21(4):1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenardo MJ. Molecular regulation of T lymphocyte homeostasis in the healthy and diseased immune system. Immunol Res. 2003;27(2-3):387–398. doi: 10.1385/IR:27:2-3:387. [DOI] [PubMed] [Google Scholar]
- 52.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huelga SC, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Reports. 2012;1(2):167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28(6):637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang HY, et al. SRPK2: A differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140(4):737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122(3):191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35(1):1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adamson P, Etienne S, Couraud PO, Calder V, Greenwood J. Lymphocyte migration through brain endothelial cell monolayers involves signaling through endothelial ICAM-1 via a rho-dependent pathway. J Immunol. 1999;162(5):2964–2973. [PubMed] [Google Scholar]
- 59.Gattazzo C, et al. Cortactin, another player in the Lyn signaling pathway, is over-expressed and alternatively spliced in leukemic cells from patients with B-cell chronic lymphocytic leukemia. Haematologica. 2014;99(6):1069–1077. doi: 10.3324/haematol.2013.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirkbride KC, Sung BH, Sinha S, Weaver AM. Cortactin: A multifunctional regulator of cellular invasiveness. Cell Adhes Migr. 2011;5(2):187–198. doi: 10.4161/cam.5.2.14773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L, et al. OPA1 mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability. J Am Heart Assoc. 2012;1(5):e003012. doi: 10.1161/JAHA.112.003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicolas M, Hassan BA. Amyloid precursor protein and neural development. Development. 2014;141(13):2543–2548. doi: 10.1242/dev.108712. [DOI] [PubMed] [Google Scholar]
- 63.Deng H, Gao K, Jankovic J. The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol. 2014;10(6):337–348. doi: 10.1038/nrneurol.2014.78. [DOI] [PubMed] [Google Scholar]
- 64.Lynch KW, Weiss A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol Cell Biol. 2000;20(1):70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bresson SM, Conrad NK. The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 2013;9(10):e1003893. doi: 10.1371/journal.pgen.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothrock C, Cannon B, Hahm B, Lynch KW. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell. 2003;12(5):1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- 67.Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell. 2010;40(1):126–137. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.