Significance
Protein unfolding by ATP-powered enzymes is an essential aspect of many biological reactions, including degradation by AAA+ proteases such as ClpXP and ClpAP, but there have been no robust methods to assay the steady-state kinetics of these reactions. Here, we show that the kinetics of enzymatic unfolding can be monitored by the rate at which a mixture of donor-labeled and acceptor-labeled homodimers is converted into heterodimers and use this assay to characterize the activities of the AAA+ ClpX and ClpA unfoldases with and without ClpP, their proteolytic partner. Our results show differences in intrinsic unfolding activity and in ClpP stimulation and indicate that this assay should be broadly applicable for studying the mechanisms of related AAA+ enzymes and remodeling chaperones.
Keywords: protein unfolding, molecular machine, AAA+ protease, chaperone
Abstract
ATP-dependent molecular machines of the AAA+ superfamily unfold or remodel proteins in all cells. For example, AAA+ ClpX and ClpA hexamers collaborate with the self-compartmentalized ClpP peptidase to unfold and degrade specific proteins in bacteria and some eukaryotic organelles. Although degradation assays are straightforward, robust methods to assay the kinetics of enzyme-catalyzed protein unfolding in the absence of proteolysis have been lacking. Here, we describe a FRET-based assay in which enzymatic unfolding converts a mixture of donor-labeled and acceptor-labeled homodimers into heterodimers. In this assay, ClpX is a more efficient protein-unfolding machine than ClpA both kinetically and in terms of ATP consumed. However, ClpP enhances the mechanical activities of ClpA substantially, and ClpAP degrades the dimeric substrate faster than ClpXP. When ClpXP or ClpAP engage the dimeric subunit, one subunit is actively unfolded and degraded, whereas the other subunit is passively unfolded by loss of its partner and released. This assay should be broadly applicable for studying the mechanisms of AAA+ proteases and remodeling chaperones.
Protein unfolding by cellular enzymes is necessary for degradation by AAA+ proteases and for the remodeling of macromolecular complexes (1–3). In all kingdoms of life, AAA+ proteases are involved in specific regulatory tasks, including the degradation of cell-cycle components and transcription factors and housekeeping degradation of proteins that are misfolded, damaged, or the products of premature translation termination (3, 4). Bacteria contain multiple AAA+ proteases (ClpXP, ClpAP, ClpCP, HslUV, Lon, FtsH, and Mpa⋅20S); archaea have two cytoplasmic AAA+ proteases (PAN⋅20S and Cdc48⋅20S); and eukaryotes contain one cytosolic or nuclear AAA+ protease (the 26S proteasome) and homologs of AAA+ bacterial proteases in organelles. These proteolytic machines use cycles of ATP binding and hydrolysis by a hexameric AAA+ ATPase to power unfolding of a protein substrate and subsequent translocation of the denatured polypeptide into the proteolytic chamber of a partner peptidase. In some cases, a single peptidase can collaborate with several different AAA+ partners. For example the ClpP peptidase can function with the AAA+ ClpX, ClpA, or ClpC enzymes in the ClpXP, ClpAP, or ClpCP proteases, whereas the archaeal 20S peptidase can function with either the AAA+ PAN or Cdc48 enzymes (3, 5).
Certain AAA+ enzymes also display chaperone or remodeling functions in the absence of their peptidase partners (3). For instance, ClpX can disassemble tetramers of the MuA transposase bound to translocated DNA, allowing replication to proceed, and ClpA catalyzes the disassembly of RepA dimers into monomers that bind with high affinity to the origin of replication of plasmid P1 (6, 7). A ClpX ortholog in the mitochondria of Saccharomyces cerevisiae functions without ClpP, suggesting a strictly nonproteolytic function. One mechanism of remodeling is ATP-dependent unfolding of one or more subunits of a target protein. In the case of misfolded proteins, unfolding and translocation through the axial pore of an isolated AAA+ hexamer could release the denatured polypeptide into solution and allow additional chances to refold properly. Unfolding of one or more subunits of an oligomer could either free subunits to carry out other functions blocked in the multimer, as in the RepA example, or lead to disassembly of a dead-end complex, as in the case of MuA tetramers bound to translocated DNA.
Protein degradation is readily assayed by the disappearance of intact substrate on SDS gels, by the appearance of acid-soluble peptides, or by loss of native fluorescence for proteins such as GFP. More limited methods, largely involving stable naturally fluorescent proteins or single-molecule optical trapping, are available to probe unfolding by AAA+ enzymes in the absence of proteolysis but are generally poorly suited for determining steady-state kinetic parameters and the energetic efficiency of enzymatic unfolding. In this paper, we develop and test a simple and robust method to measure rates of enzymatic unfolding of a fusion protein consisting of Arc repressor and the coiled-coil region of Gcn4. Both of these domains assume a stable native fold only as dimers, and unfolding is rate limiting for subunit exchange. Thus, enzymatic unfolding of a mixture of two homodimers, one labeled with donor fluorophores and one labeled with acceptor fluorophores, results in the formation of heterodimers and Förster resonance energy transfer (FRET). Our substrate can be modified to increase or decrease stability, to require unfolding of additional domains of interest, or to be targeted to any AAA+ remodeling or degradation machine with a well-characterized recognition signal, and thus should be generally useful for interrogating the kinetics of protein unfolding and processing by these enzymes. Here, we use this new assay to address and answer several questions about the mechanism of the AAA+ ClpX and ClpA enzymes, including which of these enzymes is a more powerful and energy efficient unfoldase, how ClpP affects the intrinsic unfolding activities of these enzymes, and whether ClpXP and ClpAP degrade one or both subunits of the dimeric substrate in one cycle.
Results
Assay Design.
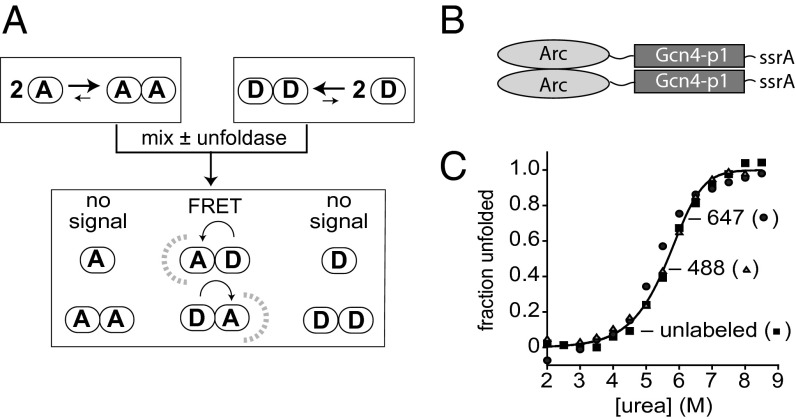
The diagram in Fig. 1A depicts an assay that allows the spontaneous or enzymatic unfolding/dissociation of a dimeric protein to be monitored by the rate of subunit exchange via FRET. This assay was previously used to characterize spontaneous unfolding/dissociation of the bacteriophage P22 Arc-repressor dimer, which is only natively folded as a dimer because both subunits contribute to a single hydrophobic core (8–10). When an Arc dimer labeled with a donor fluorophore is mixed with a dimer labeled with an acceptor fluorophore, spontaneous dimer unfolding/dissociation is rate limiting for monomer refolding/association (10). However, Arc dimers unfold/dissociate with a half-life of a few seconds (10, 11), a rate too fast to be useful for enzymatic-unfolding assays. To stabilize Arc, we fused it to the p1 peptide of Gcn4, which forms a parallel coiled coil (12). At the C terminus of the fusion protein, we placed an epitope for purification and an ssrA degron to target the protein to the ClpX and ClpA unfoldases (Fig. 1B). We refer to this substrate, which contains a single cysteine at position 23 of Arc (R23C) as Arc-Gcn4-ssrA. Separate batches of Arc-Gcn4-ssrA were individually labeled with maleimide derivatives of the Alexa-488 (donor) or Alexa-647 (acceptor) fluorescent dyes. To assess the thermodynamic stability of the resulting proteins, we monitored urea denaturation by changes in circular dichroism (Fig. 1C). Both the labeled and unlabeled proteins showed cooperative unfolding with midpoints between 5 and 6 M urea. A global fit of the data gave a ∆Gu value of ∼16 kcal/mol in the absence of denaturant, corresponding to an equilibrium constant for dimer dissociation of ∼10−12 M. These values are at least 1,000-fold lower than the values for Arc dimers (10−8 M; ref. 8) or Gcn4p1 dimers (8⋅10−9 M; ref. 13), establishing that fusion increased the equilibrium stability of each component dimer. Kinetic stability was also increased. When samples of acceptor-labeled and donor-labeled Arc-Gcn4-ssrA were mixed, ∼20% subunit exchange occurred in 30 min (Fig. 2A, no enzyme) and ∼95% mixing required overnight incubation.
Fig. 1.
Subunit-exchange assay and Arc-Gcn4-ssrA dimers. (A) Following mixing of dimers labeled with donor or acceptor fluorophores, spontaneous or enzyme-induced unfolding/dissociation allows subunit mixing and FRET. Assuming equal stabilities, donor, mixed, and acceptor dimers should be present in a 1:2:1 ratio at equilibrium. (B) Arc-Gcn4-ssrA contains the R23C Arc protein, a 10-residue linker, the Gcn4p1 peptide, and an st11-ssrA tag for purification and targeting to the ClpX and ClpA unfoldases. (C) Unlabeled Arc-Gcn4-ssrA dimers (squares; 2.5 µM), Alexa-488 labeled Arc-Gcn4-ssrA dimers (triangles; 2.5 µM), or Alexa-647 labeled Arc-Gcn4-ssrA dimers (circles; 2.5 µM) were incubated with different concentrations of urea for 20 h at room temperature and the fraction unfolded was calculated from the circular-dichroism ellipticity at 222 nm, assuming flat native and unfolded baselines. The solid line is a global fit to a N2 ⇔ 2U model with a ∆Gu of 15.9 ± 0.5 kcal/mol in the absence of denaturant (corresponding to Kdimer ∼ 10−12 M) and an m value of 1.55 ± 0.15 kcal/mol⋅M.
Fig. 2.
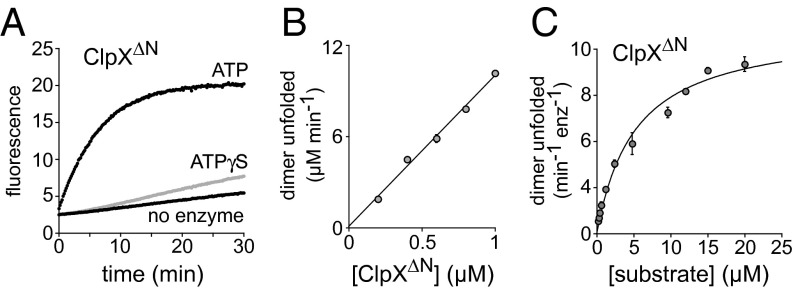
ClpX catalyzes unfolding of Arc-Gcn4-ssrA dimers. (A) Unfolding of Arc-Gcn4-ssrA by ClpX∆N was monitored by emission of acceptor fluorescence (arbitrary units). ATP supported robust unfolding relative to no enzyme, which represents spontaneous unfolding and mixing of donor- and acceptor-labeled Arc-Gcn4-ssrA dimers. ATPγS supported faster unfolding than the no-enzyme control and is hydrolyzed at ∼5% of the ATP rate (24). When present, the ClpX∆N concentration was 0.2 µM (hexamer equivalents), nucleotide concentrations were 5 mM, and donor- and acceptor-labeled Arc-Gcn4-ssrA were each present at 7.5 µM dimer equivalents. Curves are averages of two or three replicates. (B) Initial rates of Arc-Gcn4-ssrA dimer unfolding (10 µM donor labeled; 10 µM acceptor labeled) were determined using different ClpX∆N concentrations in the presence of ATP (5 mM). Data points represent averages (n = 2) ± SD. The line is a linear fit: y = 0.125 + 10x (R = 0.998). (C) Initial rates of substrate unfolding by ClpX∆N (0.2 µM) were determined for increasing concentrations of an equimolar mixture of donor-labeled and acceptor-labeled Arc-Gcn4-ssrA in the presence of ATP (5 mM). The line is a fit to the Michaelis–Menten equation (R = 0.992) with a Vmax of 11.3 ± 0.45 dimers unfolded min−1⋅enz−1 and a KM of 5.5 ± 0.7 µM. Data points are averages (n ≥ 3) ± SD.
ClpX-Catalyzed Unfolding.
Escherichia coli ClpX lacking the N-terminal domain (ClpX∆N) was used for enzymatic unfolding experiments, as it is straightforward to purify, recognizes and unfolds ssrA-tagged proteins similarly to the wild-type enzyme, and has been used in prior solution and single-molecule experiments (14–23). Following mixing of donor- and acceptor-labeled Arc-Gcn4-ssrA, the initial rate of heterodimer formation was slowest without enzyme, faster with ClpX∆N and ATPγS, and fastest with ClpX∆N and ATP (Fig. 2A). ATPγS supports binding of ssrA-tagged substrates to ClpX and ClpX∆N and is hydrolyzed at 5–10% the rate of ATP (16, 24). Thus, ClpX∆N unfolding of the fusion homodimers depends on the rate of ATP hydrolysis, as expected for a reaction requiring mechanical work.
Next, we monitored the initial rate of heterodimer formation using a constant concentration of donor-labeled and acceptor-labeled Arc-Gcn4-ssrA and increasing concentrations of ClpX∆N in the presence of ATP (Fig. 2B). The linearity of this plot shows that the enzymatic reaction is well behaved and that active hexamers are stable over the concentration range explored. Moreover, the linearity of the rate with ClpXΔN concentration shows that enzymatic unfolding and not the subsequent subunit-association step is rate limiting for formation of the substrate heterodimer. To determine steady-state kinetic parameters, we assayed initial rates of ATP-dependent heterodimer formation using a fixed concentration of ClpXΔN and increasing concentrations of Arc-Gcn4-ssrA (Fig. 2C). Fitting these data to the Michaelis–Menten equation gave a Vmax of ∼11 substrate dimers unfolded min−1⋅enz−1, and a KM of ∼5 µM.
ClpA-Catalyzed Unfolding.
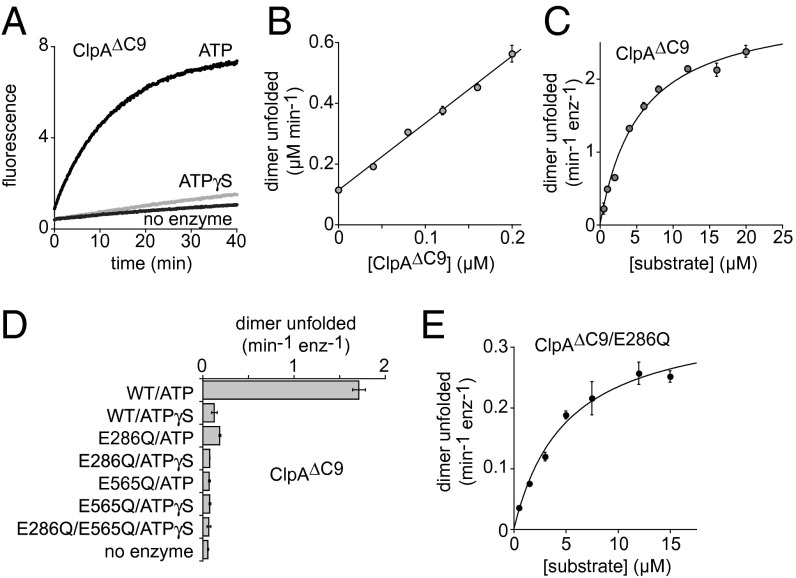
Enzymatic unfolding of Arc-Gcn4-ssrA dimers by ClpA was performed using an E. coli ClpA∆C9 variant, which is fully active but lacks a degron sequence that causes autodegradation by wild-type ClpAP (25). The rate of ClpA∆C9 unfolding of Arc-Gcn4-ssrA supported by ATP was substantially faster than the rate in the presence of ATPγS or the no-enzyme control (Fig. 3A) and varied linearly with enzyme concentration (Fig. 3B). Fitting initial ATP-supported ClpA∆C9 unfolding rates at different substrate concentrations to the Michaelis–Menten equation gave a Vmax of ∼3 dimers unfolded min−1⋅enz−1 and a KM of ∼5 µM (Fig. 3C). Thus, at substrate saturation, ClpA∆C9 unfolds Arc-Gcn4-ssrA ∼3.5 times more slowly than ClpX∆N, a somewhat surprising result given that E. coli ClpAP unfolds and degrades many substrates faster than E. coli ClpXP (26–28).
Fig. 3.
ClpA catalyzes unfolding of Arc-Gcn4-ssrA with both AAA+ rings playing important roles. (A) Unfolding of Arc-Gcn4-ssrA dimers (2.5 µM donor labeled; 2.5 µM acceptor labeled) alone and catalyzed by ClpA∆C9 (0.2 µM hexamer) in the presence of ATP or ATPγS (5 mM). Curves are averages of two experiments. Fluorescence units are arbitrary. (B) Rates of unfolding of Arc-Gcn4-ssrA dimers (10 µM donor labeled; 10 µM acceptor labeled) varied linearly with ClpA∆C9 concentration in the presence of ATP (5 mM). The line is a linear fit; y = 0.11 + 2.2x (R = 0.997). Values are averages (n ≥ 2) ± SD. (C) Increasing concentrations of equimolar donor- and acceptor-labeled Arc-Gcn4-ssrA were added to ClpA∆C9 (0.08 µM) in the presence of ATP (5 mM) and initial unfolding rates were determined. Fitting to the Michaelis–Menten equation gave a Vmax of 3.0 ± 0.09 dimers unfolded min−1⋅enz−1 and a KM of 5.5 ± 0.45 µM. Values are averages (n = 3) ± SD. (D) Rates of unfolding of Arc-Gcn4-ssrA dimers (2.5 µM donor labeled; 2.5 µM acceptor labeled) by ClpA∆C9 or variants (0.2 µM) in the presence of ATP or ATPγS (5 mM). Values are averages (n = 2) ± SD. (E) Michaelis–Menten analysis of unfolding by ClpA∆C9/E286Q (0.2 µM) and ATP (5 mM) gave a Vmax of 0.35 ± 0.02 dimers unfolded min−1⋅enz−1 and a KM of 4.9 ± 0.65 µM. Values are averages (n = 3) ± SD.
Each ClpA subunit contains two AAA+ modules, termed D1 and D2, which form distinct rings in the intact ClpA hexamer (29). To determine the contributions of each ClpA∆C9 ring to unfolding of the Arc-Gcn4-ssrA dimer, we used Walker-B mutations that inactivate ATP hydrolysis in the D1 ring (E286Q), the D2 ring (E565Q), or both rings (28, 30). Of these variants, only the E286Q enzyme unfolded the Arc-Gcn4-ssrA dimer faster than control reactions containing ATPγS or no enzyme (Fig. 3D). For ClpA∆C9/E286Q, Michaelis–Menten analysis gave an unfolding Vmax of 0.35 dimers min−1⋅enz−1 and a KM of 5 µM (Fig. 3E). The approximately eightfold lower Vmax for ClpA∆C9/E286Q unfolding compared with ClpA∆C9 unfolding shows that the D1 ring makes important contributions to unfolding, but ATP hydrolysis in this ring is not required for enzymatic unfolding. Thus, ATP hydrolysis in both rings of the ClpA∆C9 hexamer is required for robust unfolding, with hydrolysis in the D2 ring being more important.
ATPγS supports substrate binding but is hydrolyzed at less than 0.5% of the ATP rate by ClpA (31, 32). Nevertheless, ATPγS supported Arc-Gcn4-ssrA unfolding by ClpA∆C9 at a rate approximately twofold faster than expected based on the hydrolysis rate and the rate of the no-enzyme control reaction (Fig. 3D). This small enhancement in unfolding may be a consequence of substrate binding that distorts the native substrate modestly.
Degradation Releases One Subunit of the Dimeric Substrate.
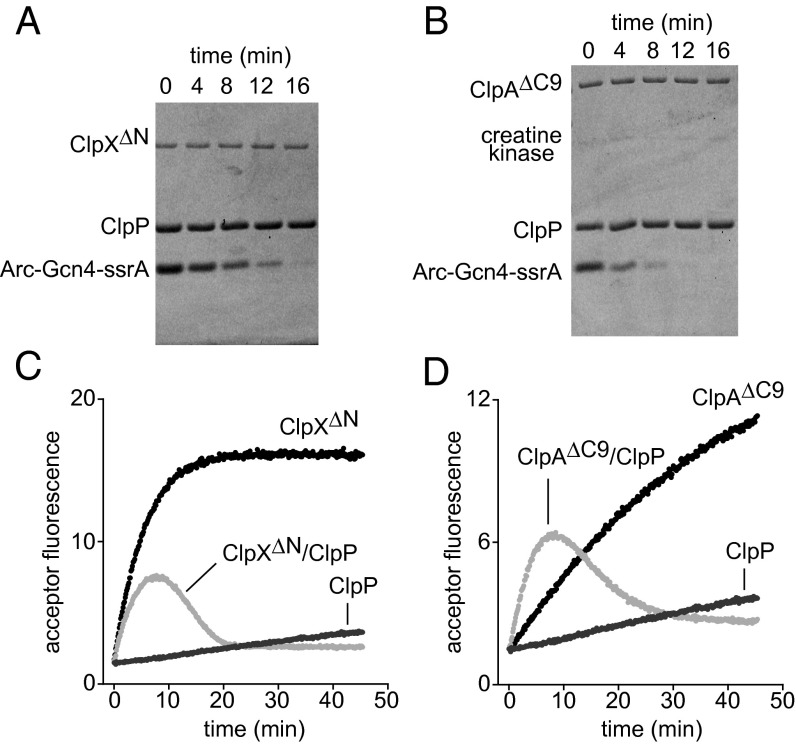
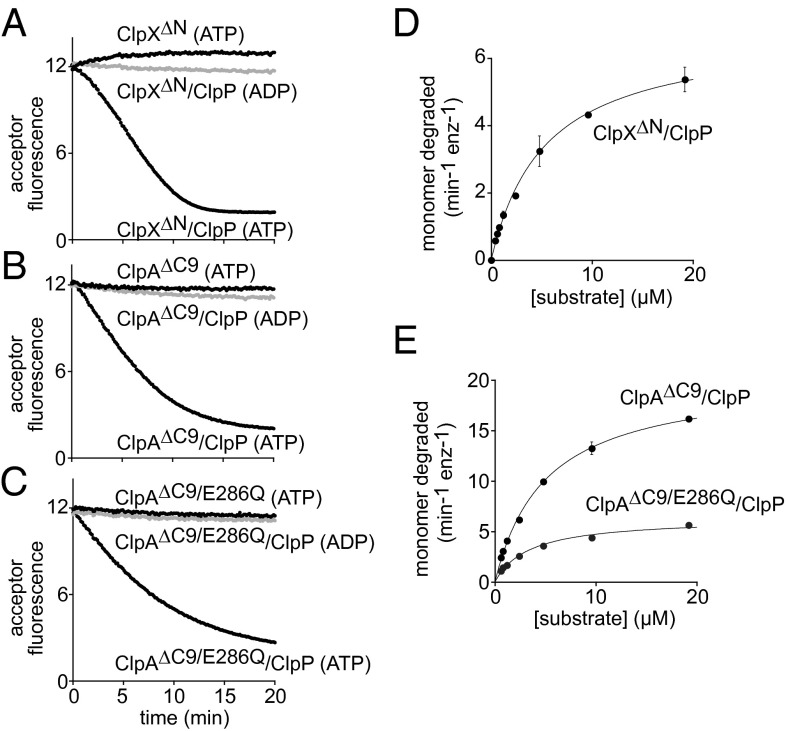
When proteolysis of Arc-Gcn4-ssrA by ClpX∆N and ClpP (Fig. 4A) or by ClpA∆C9 and ClpP (Fig. 4B) was assayed by SDS/PAGE, the reactions proceeded to completion without detectable intermediate species. Engagement of the Arc-Gcn4-ssrA dimer by these AAA+ proteases could result in two outcomes. In an independent-engagement model, only one subunit of the dimer is engaged, denatured, translocated, and degraded, and the other subunit is unfolded by loss of its partner and released into solution, where it could redimerize. In a dual-engagement model, the AAA+ protease simultaneously engages, unfolds, and degrades both subunits of the dimer. If the independent-engagement model is correct, then ClpXP or ClpAP degradation of a mixture of donor-labeled and acceptor-labeled Arc-Gcn4-ssrA should initially result in heterodimer formation and an increase in FRET, followed by a FRET decrease as heterodimers are degraded. By contrast, the dual-engagement model predicts that degradation of a mixture of donor-labeled and acceptor labeled Arc-Gcn4-ssrA should not result in increased FRET as both subunits of the dimer would be destroyed and thus preclude formation of mixed dimers. We found that incubation of a mixture of donor-labeled and acceptor labeled Arc-Gcn4-ssrA with ClpP and either ClpX∆N or ClpA∆C9 initially caused an increase in FRET, which then decreased (Fig. 4 C and D), providing strong support for the independent-engagement model. For ClpX∆N plus ClpP, the rate of monomer release was slightly slower than observed with ClpX∆N alone (Fig. 4C). For ClpA∆C9 plus ClpP, by contrast, the rate of monomer release was substantially faster than observed with ClpA alone (Fig. 4D), indicating that ClpP enhances the unfolding activity of ClpA∆C9. Substrate unfolding in the presence of ClpP alone occurred at a low rate expected for the spontaneous reaction (Fig. 4 C and D).
Fig. 4.
Degradation transiently releases one subunit of the Arc-Gcn4-ssrA dimer. (A) SDS/PAGE assay of degradation of Arc-Gcn4-ssrA dimers (9.6 µM) by ClpX∆N (0.2 µM) and ClpP (0.8 µM tetradecamer). (B) SDS/PAGE assay of degradation of Arc-Gcn4-ssrA dimers (9.6 µM) by ClpA∆C9 (0.2 µM) and ClpP (0.8 µM) (creatine kinase is part of the ATP-regeneration system). (C) Equimolar concentrations of donor- and acceptor-labeled Arc-Gcn4-ssrA dimers (5 µM each) were mixed with ClpX∆N (0.2 µM) and ClpP (0.8 µM), ClpX∆N (0.2 µM) alone, or ClpP (0.8 µM) alone, and the unfolding and release of subunits was monitored by increased FRET. (D) Same conditions as in C except that ClpA∆C9 (0.08 µM) was used in place of ClpX∆N. All reactions in this figure contained 5 mM ATP. In C and D, curves are an average of two replicates.
Degradation Rates.
To determine rates of degradation of the Arc-Gcn4-ssrA substrate, we preincubated donor-labeled and acceptor-labeled protein for 16 h at 30 °C to allow spontaneous formation of a mixture of homodimers and heterodimers, added ClpP plus ClpX∆N, ClpA∆C9, ClpA∆C9/E286Q, or ClpA∆C9/E565Q and assayed degradation by loss of FRET. Degradation by ClpP and ClpA∆C9/E565Q was barely detectable and was not quantified. For the other enzymes, proteolysis required the presence of ClpP, the AAA+ unfoldase, and ATP (Fig. 5 A–C). In each case, there was a very slow linear decrease in FRET when ClpP, the unfoldase, and ADP were present, probably as a consequence of photobleaching. For ClpX∆N, the FRET signal increased slightly when ATP was present but ClpP was absent (Fig. 5A), suggesting that mixing of donor-labeled and acceptor-labeled substrate dimers was incomplete before the addition of ClpX.
Fig. 5.
Rates of degradation of Arc-Gcn4-ssrA. For these experiments, equimolar concentrations of donor-labeled and acceptor-labeled Arc-Gcn4-ssrA dimers were mixed and allowed to equilibrate for 16 h at 30 °C. Degradation was assayed by loss of FRET. (A) Kinetic trajectories observed after mixing Arc-Gcn4-ssrA (5 µM total dimer) with ClpX∆N (0.2 µM) plus or minus ClpP (0.8 µM) and ATP or ADP (5 mM). Curves are an average of two replicates. (B) Same experiment as A but using ClpA∆C9 (0.08 µM) plus or minus ClpP (0.4 µM). (C) Same experiment as A but using ClpA∆C9/E286Q (0.2 µM) plus or minus ClpP (0.8 µM). (D) Michaelis–Menten analysis of Arc-Gcn4-ssrA degradation kinetics by ClpX∆N (0.2 µM) and ClpP (0.8 µM) in the presence of ATP (5 mM). Values are averages (n = 3) ± SD Vmax = 6.8 ± 0.22 monomers min−1⋅enz−1, KM = 2.7 ± 0.21 µM. (E) Michaelis–Menten analysis of Arc-Gcn4-ssrA degradation kinetics in the presence of 5 mM ATP by ClpA∆C9 (0.08 µM) and ClpP (0.4 µM) or ClpA∆C9/E286Q (0.2 µM) and ClpP (0.8 µM). Values are averages (n = 3) ± SD. For ClpA∆C9/ClpP degradation, Vmax was 20.3 ± 0.37 monomers min−1⋅enz−1 and KM was 2.5 ± 0.12 µM. For ClpA∆C9/E286Q/ClpP degradation, Vmax was 6.3 ± 0.21 monomers min−1⋅enz−1 and KM was 1.7 ± 0.16 µM.
Because one monomer of the Arc-Gcn4-ssrA dimer is degraded but both monomers are unfolded, we calculated degradation rates in units of monomer concentration and unfolding rates in units of dimer concentration to allow direct comparison. Based on Michaelis–Menten fitting, Vmax for ClpP degradation of Arc-Gcn4-ssrA was ∼7 monomers min−1⋅enz−1 for ClpX∆N (Fig. 5D), compared with a maximal unfolding rate of ∼11 dimers min−1⋅enz−1. Thus, ClpP modestly depresses the activity of ClpX∆N. For ClpA∆C9, the maximal degradation rate was ∼20 monomers enz−1⋅min−1 (Fig. 5E), compared with an unfolding rate of ∼3 dimers unfolded min−1⋅enz−1. Hence, ClpP enhances ClpA∆C9 activity ∼7-fold. For ClpA∆C9/E286Q, Vmax was ∼6 monomers enz−1⋅min−1 for degradation (Fig. 5E) and 0.35 dimers min−1⋅enz−1 for unfolding, an activity increase of ∼17-fold. Thus, ClpP enhancement of ClpA∆C9 activity occurs largely through effects on the peptidase-proximal D2 ring.
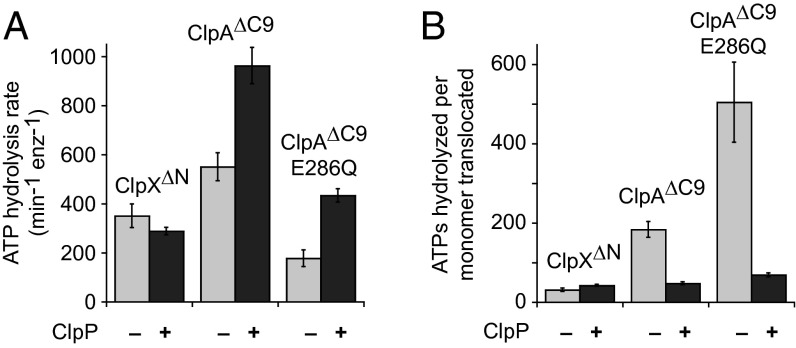
In principle, ClpP might have different effects on the activities of the ClpA and ClpX variants by differential modulation of their ATP-hydrolysis activities. To test this possibility, we measured ATPase rates in the presence of saturating Arc-Gcn4-ssrA with or without ClpP (Fig. 6A) and then calculated the number of ATP molecules hydrolyzed per monomer of Arc-Gcn4-ssrA unfolded and translocated through the axial pore during unfolding and degradation (Fig. 6B). The ATP costs for degradation were similar for ClpX∆N and ClpA∆C9 (∼45 ATPs) and were about 50% greater for ClpA∆C9/E286Q. During unfolding in the absence of ClpP, ClpX∆N was most efficient (∼30 ATPs), whereas ClpA∆C9 and ClpA∆C9/E286Q hydrolyzed 6-fold and 17-fold more ATP per monomer translocated, respectively. Thus, ClpX∆N is a faster and slightly more energetically efficient enzyme in the absence than presence of ClpP, whereas ClpA∆C9 variants are faster and substantially more energetically efficient enzymes in the presence than absence of ClpP.
Fig. 6.
Energetic efficiency of unfolding and degradation. (A) Rates of ATP hydrolysis by ClpX∆N (0.2 µM), ClpA∆C9 (0.08 µM), or ClpA∆C9/E286Q (0.2 µM) were measured in the presence of saturating concentrations of Arc-Gcn4-ssrA dimer (≥15 µM) plus or minus ClpP (0.4 or 0.8 µM). Values are averages (n = 3) ± SD. (B) The energetic efficiency of Arc-Gcn4-ssrA unfolding (minus ClpP; ATPV/unfVmax) or degradation (plus ClpP; ATPV/degVmax) for ClpX∆N, ClpA∆C9, and ClpA∆C9/E286Q, where ATPV is the rate of ATP hydrolysis in the presence of saturating substrate with or without ClpP; unfVmax is the Vmax for unfolding obtained from the Michaelis–Menten fits in Figs. 2C and 3 C and E; and degVmax is the Vmax for degradation obtained from the Michaelis–Menten fits in Fig. 5 D and E.
Discussion
Previous studies measured ClpA or ClpX unfolding of GFP-ssrA by changes in native fluorescence in the presence of a GroEL variant that traps denatured protein and prevents refolding (17, 33, 34). However, native GFP is quite stable and resists unfolding by some AAA+ enzymes, including wild-type FtsH and HslUV and mutant variants of ClpXP and ClpAP (18, 30, 35–37). Moreover, working at high substrate concentrations is difficult because of limitations in the solubility of GroELtrap and because the kinetics of trapping can affect the apparent kinetics of unfolding. Phage-λ–repressor variants labeled with donor and acceptor dyes, which are close in the native structure but farther apart in the unfolded protein, have also been used to measure ClpXP, ClpAP, or ClpA unfolding in single-turnover experiments (38). Another approach has been to measure ClpX unfolding of Kaede, a tetrameric fluorescent protein that can be clipped by exposure to light and then fails to refold following denaturation (39, 40). However, photoconversion to the clipped form of Kaede is variable and often incomplete, and unfolding of clipped Kaede by ClpX is very slow. To our knowledge, none of these methods have been used to determine the steady-state kinetic parameters for unfolding by any AAA+ enzyme. Unfolding and translocation of model domains by ClpX, ClpXP, and ClpAP variants has also been assayed by single-molecule optical trapping (20–23, 28), but there is no simple way to assess energetic efficiency or rates of substrate binding and/or engagement in these experiments. The subunit-exchange assay described here is robust, simple, and obviates the issues that limit the usefulness of other methods. Our assay would not be useful if the rate of enzymatic unfolding was faster than the rate of monomer reassociation, but the latter reaction is >90% complete within 5 s at monomer concentrations above 10 nM, and thus this situation is unlikely to be common except when using very low substrate concentrations. Another potential limitation of this assay is that the rate of enzymatic unfolding must be faster than the rate of spontaneous unfolding, although the Arc-Gcn4-ssrA substrate could be easily modified to increase or decrease protein stability by introducing, for example, mutations known to stabilize the Arc dimer. In addition, the substrate could be fused to additional protein domains to interrogate their unfolding, or targeting sequences for other AAA+ remodeling or degradation machines could be added to either protein terminus. For example, a substrate could be targeted for N-end rule or ubiquitin-dependent unfolding/degradation by addition of appropriate sequences or domains.
GroELtrap studies using single concentrations of ClpX, ClpXP, and GFP-ssrA concluded that ClpXΔN/ClpP was an approximately threefold better unfoldase than ClpX∆N, a result that assumes that enzyme–substrate complexes are formed and engaged equally well in both cases (17). Interestingly, however, in single-molecule optical-trapping experiments, the average dwell time before GFP unfolding by ClpX∆N was shorter than before unfolding by a complex of ClpX∆N and ClpP (21). We find that ClpX engages, unfolds, and translocates the Arc-Gcn4-ssrA substrate slightly faster and more efficiently in terms of ATP cost than ClpXP. Although the mechanical activities of ClpX and ClpXP may vary depending upon the specific protein substrate, it is clear that ClpX alone can function as an efficient protein-unfolding and remodeling machine.
We find that ClpAP unfolds saturating concentrations of Arc-Gcn4-ssrA approximately sevenfold faster than ClpA in the absence of ClpP. Prior studies using single enzyme concentrations and different substrates also show that ClpAP is a better unfoldase than ClpA, although rates were not quantified (33, 38). Moreover, previous studies show that ClpAP translocates peptide substrates more rapidly than ClpA, but by a factor of less than twofold (32, 41). The stimulatory effect of ClpP on ClpA unfolding activity is partially caused by a twofold increase in the rate of ATP hydrolysis. Other factors must also contribute, however, as the ATP cost of ClpAP unfolding and degradation is approximately fourfold lower than ClpA unfolding. Thus, ATP hydrolysis in the ClpAP complex must be coupled more efficiently to substrate engagement, unfolding, or translocation.
For ClpAP degradation, Weber-Ban and coworkers elegantly demonstrated that the importance of ATP hydrolysis in the D1 and D2 rings of ClpA depends on the substrate assayed (30). Specifically, they found that ATP hydrolysis in just the D1 AAA+ ring was sufficient to degrade a protein of low stability very slowly, that proteins of moderate stability could be degraded at reasonable rates when only the D2 AAA+ ring was active, and that ATP hydrolysis in both the D1 and D2 rings was necessary for efficient degradation of the most stable substrates. For Arc-Gcn4-ssrA, we found that the rate of degradation by the ClpAP variant was reduced approximately threefold when only the D2 ring was hydrolytically active, but was essentially eliminated when just the D1 ring was active. Thus, Arc-Gcn4-ssrA behaves like a moderate stability substrate. In the absence of ClpP, elimination of hydrolysis in the D2 ring effectively eliminated Arc-Gcn4-ssrA unfolding activity, whereas elimination of hydrolysis in the D1 ring reduced activity approximately ninefold. Hence, ATP hydrolysis in both rings of ClpA becomes more important for mechanical activity when ClpP is absent, a result consistent with the finding that ClpP stimulates ATP hydrolysis by the D2 ring but not the D1 ring of ClpA (30).
Previous studies demonstrated that ClpXP degrades only the tagged subunit of Arc or RepA heterodimers assembled with one degron-tagged subunit and one untagged subunit (42, 43). By contrast, ClpAP degrades the tagged and untagged subunits of a RepA heterodimer (43), and ClpX can translocate multiple covalently linked polypeptides through its axial pore and into ClpP simultaneously (42, 44). Thus, it seemed possible that both ssrA-tagged subunits of the Arc-Gcn4-ssrA dimer might be engaged and degraded together by ClpXP or ClpAP. However, our results show that degradation of the Arc-Gcn4-ssrA dimer by the ClpXP or ClpAP variants generally results in the active unfolding, translocation, and degradation of only one subunit, with passive unfolding and release of the other subunit. Moreover, both ssrA tags should be relatively close in the 3D structure of the dimer, as Gcn4p1 forms a parallel coiled coil. We conclude that engagement of the Arc-Gcn4-ssrA dimer by ClpX or ClpA almost always involves one polypeptide at a time, even when a second tagged polypeptide is nearby. For multimeric substrates, passive unfolding and release of some subunits could allow ClpXP or ClpAP to function as activating chaperones, especially if the multimeric substrate was bound more strongly than the released monomer because of avidity effects.
Materials and Methods
Cloning, Expression, and Purification.
A gene encoding ArcRC23, a GSGSGGSGGS linker, the 33-residue Gcn4p1 peptide, the st11 sequence (H6KNQHD), and the ssrA tag (AANDENYALAA) was constructed by standard PCR techniques and cloned into a pET21b vector (Novagen). DNA encoding an N-terminal His7-SUMO protein fused to E. coli ClpA missing its nine C-terminal amino acids (ClpA∆C9) was constructed and cloned into pET21a. pET23b plasmid variants encoding untagged ClpA∆C9/E286Q and ClpA∆C9/E565Q were gifts from Adrian Olivares (Massachusetts Institute of Technology, Cambridge, MA).
E. coli X90(DE3) cells transformed with the ArcRC23-Gcn4p1-st11-ssrA plasmid were grown to OD600 0.6–0.8 at 37 °C, induced with 1 mM IPTG, and shaken at room temperature for 4 h. Cells were harvested and resuspended in buffer A [50 mM Tris (pH 8.0), 300 mM NaCl, 20 mM imidazole, 1 mM DTT, 0.5 mM EDTA, complete EDTA-free protease inhibitor tablets (Roche)] and 1.5 µL of benzonase (250 units/µL, Sigma) per liter of culture was added before lysis. Cells were lysed by sonication, and insoluble material was removed by high-speed centrifugation. A total of 120 µL of 10% (wt/vol) polyethyleneimine was added to the supernatant (12 mL) and the resulting mixture was centrifuged again to remove precipitated nucleic acids. The supernatant was mixed with Ni++-NTA resin (Thermo), incubated for 1 h at 4 °C, washed with 100 mL buffer A, and eluted with 250 mM imidazole. The eluate was concentrated and chromatographed on a Superdex-75 size-exclusion column (GE Healthcare) preequilibrated in buffer B [50 mM Tris (pH 7.5), 250 mM NaCl, 10% (vol/vol) glycerol, 2 mM DTT, and 1 mM EDTA]. Fractions containing purified ArcRC23-Gcn4p1-st11-ssrA were identified by absorbance and SDS/PAGE, pooled, concentrated, and frozen at –80 °C.
E. coli BLR(DE3) cells transformed with the His7-SUMO-ClpA∆C9 expression plasmid were grown to OD600 0.6–0.8 at 37 °C, induced with 0.5 mM IPTG, and shaken at room temperature for 5 h. Cells were resuspended in buffer C [25 mM Hepes (pH 7.5), 500 mM NaCl, 10 mM imidazole (pH 7.5), and 10% (vol/vol) glycerol] and lysed by sonication. Lysates were clarified by centrifugation, and His7-SUMO-ClpA∆C9 was purified by Ni++-NTA affinity and MonoQ anion-exchange chromatography (GE Healthcare). The His7-SUMO protein was separated from ClpA∆C9 by cleavage with 1 µg Ulp1 protease per 100 µg protein for 1 h at room temperature, followed by the addition of 300 mM arginine (pH 7.0). ClpA∆C9 was further purified by size-exclusion chromatography (Superdex 200, GE Healthcare) in buffer D [50 mM Tris (pH 7.5), 200 mM NaCl, 10% (vol/vol) glycerol, 1 mM DTT, and 1 mM EDTA]. E. coli His6-ClpX∆N, E. coli ClpP-His6, and untagged E. coli ClpA∆C9 variants containing EQ mutations in the Walker-B motifs of the D1 or D2 rings were purified as described (28, 40).
Labeling of Arc-Gcn4-ssrA.
Purified Arc-Gcn4-ssrA was incubated with 10 mM DTT for 1 h at room temperature and then desalted into labeling buffer [50 mM Tris (pH 7.5), 200 mM NaCl, and 0.5 mM EDTA] using a G25 column (GE Healthcare). Alexa-488-C5-maleimide or Alexa-647-C2-maleimide (Life Technologies) was added to the desalted sample (3:1 molar ratio of fluorophore to protein in monomeric equivalents), and the reaction was incubated in the dark for 2 h at room temperature. Reactions were desalted using a PD-10 column (GE Healthcare) and then loaded onto a Superdex-75 column preequilibrated in PD buffer [25 mM Hepes (pH 7.5), 150 mM KCl, 25 mM NaCl, 10% (vol/vol) glycerol, 0.2 mM EDTA, 0.5 mM DTT]. Fractions were pooled, concentrated, and the protein concentration was determined by a Bradford assay (Bio-Rad) using unlabeled Arc-Gcn4-ssrA as the standard. Aliquots were frozen at –80 °C until use.
Equilibrium Stability Measurements.
Unlabeled or dye-labeled Arc-Gcn4-ssrA (2.5 µM dimer) was incubated in PD buffer plus different concentrations of urea at room temperature for 20 h. Circular-dichroism spectra from 200 to 300 nm were taken using an AVIV Model 420 spectrometer in a 1-mm cuvette with 3-s averaging and the ellipticity at 222 nm was used to calculate the fraction of unfolded protein.
Biochemical Assays.
Spontaneous or enzyme-mediated unfolding of donor- and acceptor-labeled Arc-Gcn4-ssrA was assayed in PD buffer plus 20 mM MgCl2 at 30 °C using a Spectramax M5 plate reader (Molecular Devices) to monitor changes in FRET (excitation 494 nm; emission 668 nm). Equimolar concentrations of donor- and acceptor-labeled Arc-Gcn4-ssrA, enzymes, and an ATP-regeneration system (16 mM creatine phosphate, 10 µg/mL creatine kinase) were incubated at 30 °C for several minutes, and reactions were started by addition of 5 mM ATP. Initial rates were calculated by dividing the initial linear slope by the total amplitude of the signal change and multiplying this value by the total substrate concentration in dimer equivalents. In cases where unfolding did not proceed to completion, ClpX was added to complete the reaction and obtain the final amplitude. Spontaneous rates of unfolding were measured in the presence of all reaction components except the AAA+ unfoldase.
Before degradation assays, donor-labeled Arc-Gcn4-ssrA dimers (7.5 µM) and acceptor-labeled Arc-Gcn4-ssrA dimers (7.5 µM) were incubated together for 16 h at 30 °C to allow equilibration and formation of mixed dimers. ClpXP or ClpAP variants were then added together with ATP (5 mM) and a regeneration system at 30 °C and loss of FRET was monitored and used to calculate the degradation rate. Rates of hydrolysis of 5 mM ATP by AAA+ proteases or unfoldases were assayed at 30 °C using an NADH-coupled assay monitored by absorbance at 340 nm (45).
Acknowledgments
We thank A. Amor, A. Olivares, K. Schmitz, B. Stinson, members of the R.T.S. and T.A.B. laboratories, and the reviewers for helpful discussions and suggestions. This work was supported by NIH Grant AI-16892. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: Common structure—diverse function. Genes Cells. 2001;6(7):575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 2.Prakash S, Matouschek A. Protein unfolding in the cell. Trends Biochem Sci. 2004;29(11):593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 4.Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol. 2009;19(2):209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Barthelme D, Sauer RT. Identification of the Cdc48•20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337(6096):843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickner S, et al. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91(25):12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton BM, Baker TA. Remodeling protein complexes: Insights from the AAA+ unfoldase ClpX and Mu transposase. Protein Sci. 2005;14(8):1945–1954. doi: 10.1110/ps.051417505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28(18):7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- 9.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by β-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367(6465):754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson T, Waldburger CD, Sauer RT. Nonlinear free energy relationships in Arc repressor unfolding imply the existence of unstable, native-like folding intermediates. Biochemistry. 1996;35(15):4795–4802. doi: 10.1021/bi953056s. [DOI] [PubMed] [Google Scholar]
- 11.Milla ME, Sauer RT. P22 Arc repressor: Folding kinetics of a single-domain, dimeric protein. Biochemistry. 1994;33(5):1125–1133. doi: 10.1021/bi00171a011. [DOI] [PubMed] [Google Scholar]
- 12.O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 13.Zitzewitz JA, Bilsel O, Luo J, Jones BE, Matthews CR. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry. 1995;34(39):12812–12819. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 14.Singh SK, et al. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem. 2001;276(31):29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- 15.Wojtyra UA, Thibault G, Tuite A, Houry WA. The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J Biol Chem. 2003;278(49):48981–48990. doi: 10.1074/jbc.M307825200. [DOI] [PubMed] [Google Scholar]
- 16.Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437(7062):1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 17.Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27(1):41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat Struct Mol Biol. 2008;15(2):139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- 19.Shin Y, et al. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc Natl Acad Sci USA. 2009;106(46):19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145(2):257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145(3):459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen M, et al. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155(3):636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158(3):647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burton RE, Baker TA, Sauer RT. Energy-dependent degradation: Linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Sci. 2003;12(5):893–902. doi: 10.1110/ps.0237603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maglica Z, Striebel F, Weber-Ban E. An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J Mol Biol. 2008;384(2):503–511. doi: 10.1016/j.jmb.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Flynn JM, et al. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;98(19):10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koodathingal P, et al. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284(28):18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivares AO, Nager AR, Iosefson O, Sauer RT, Baker TA. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat Struct Mol Biol. 2014;21(10):871–875. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273(20):12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 30.Kress W, Mutschler H, Weber-Ban E. Both ATPase domains of ClpA are critical for processing of stable protein structures. J Biol Chem. 2009;284(45):31441–31452. doi: 10.1074/jbc.M109.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pak M, Wickner S. Mechanism of protein remodeling by ClpA chaperone. Proc Natl Acad Sci USA. 1997;94(10):4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JM, Lucius AL. ATPγS competes with ATP for binding at Domain 1 but not Domain 2 during ClpA catalyzed polypeptide translocation. Biophys Chem. 2014;185:58–69. doi: 10.1016/j.bpc.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401(6748):90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 34.Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5(4):639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 35.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11(3):659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 36.Kwon AR, Trame CB, McKay DB. Kinetics of protein substrate degradation by HslUV. J Struct Biol. 2004;146(1-2):141–147. doi: 10.1016/j.jsb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Sundar S, Baker TA, Sauer RT. The I domain of the AAA+ HslUV protease coordinates substrate binding, ATP hydrolysis, and protein degradation. Protein Sci. 2012;21(2):188–198. doi: 10.1002/pro.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolygo K, et al. Studying chaperone-proteases using a real-time approach based on FRET. J Struct Biol. 2009;168(2):267–277. doi: 10.1016/j.jsb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139(4):744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stinson BM, et al. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell. 2013;153(3):628–639. doi: 10.1016/j.cell.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JM, Lin J, Li T, Lucius AL. E. coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP. J Mol Biol. 2013;425(15):2795–2812. doi: 10.1016/j.jmb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20(12):3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S, Hoskins JR, Wickner S. Binding and degradation of heterodimeric substrates by ClpAP and ClpXP. J Biol Chem. 2005;280(7):5449–5455. doi: 10.1074/jbc.M412411200. [DOI] [PubMed] [Google Scholar]
- 44.Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell. 2004;16(3):343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Nørby JG. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]