Significance
Triple-negative breast cancers (TNBCs) are aggressive with poor clinical outcomes. Understanding the pathways that control their aggressive growth may reveal new targets for therapeutic intervention. TNBCs are highly glycolytic, providing fuel for growth promoting biosynthetic pathways. We establish that the c-Myc transcription factor drives this metabolic phenotype. Classically, the c-Myc proto-oncogene drives glycolysis by activating target genes encoding glycolytic enzymes and glucose transporters; however, we show here that c-Myc represses the expression of thioredoxin-interacting protein (TXNIP), which is a potent blocker of glucose utilization. Thus, c-Myc’s repression of TXNIP provides an additional route to c-Myc–driven glucose metabolism. Highlighting the clinical significance of our finding, a Mychigh/TXNIPlow gene signature correlates with poor overall survival in TNBC but not in other subclasses of breast cancer.
Keywords: Myc, MondoA, thioredoxin-interacting protein, glycolysis, triple-negative breast cancer
Abstract
Triple-negative breast cancers (TNBCs) are aggressive and lack targeted therapies. Understanding how nutrients are used in TNBCs may provide new targets for therapeutic intervention. We demonstrate that the transcription factor c-Myc drives glucose metabolism in TNBC cells but does so by a previously unappreciated mechanism that involves direct repression of thioredoxin-interacting protein (TXNIP). TXNIP is a potent negative regulator of glucose uptake, aerobic glycolysis, and glycolytic gene expression; thus its repression by c-Myc provides an alternate route to c-Myc–driven glucose metabolism. c-Myc reduces TXNIP gene expression by binding to an E-box–containing region in the TXNIP promoter, possibly competing with the related transcription factor MondoA. TXNIP suppression increases glucose uptake and drives a dependence on glycolysis. Ectopic TXNIP expression decreases glucose uptake, reduces cell proliferation, and increases apoptosis. Supporting the biological significance of the reciprocal relationship between c-Myc and TXNIP, a Mychigh/TXNIPlow gene signature correlates with decreased overall survival and decreased metastasis-free survival in breast cancer. The correlation between the Mychigh/TXNIPlow gene signature and poor clinical outcome is evident only in TNBC, not in other breast cancer subclasses. Mutation of TP53, which is a defining molecular feature of TNBC, enhances the correlation between the Mychigh/TXNIPlow gene signature and death from breast cancer. Because Myc drives nutrient utilization and TXNIP restricts glucose availability, we propose that the Mychigh/TXNIPlow gene signature coordinates nutrient utilization with nutrient availability. Further, our data suggest that loss of the p53 tumor suppressor cooperates with Mychigh/TXNIPlow-driven metabolic dysregulation to drive the aggressive clinical behavior of TNBC.
Breast cancer is a heterogeneous disease with multiple subtypes. Triple-negative breast cancers (TNBC) lack expression of the estrogen receptor (ER), the progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Compared with the other breast cancer subtypes, TNBCs tend to occur at a younger age and have a higher rate of reoccurrence and worse outcomes (1). Because ER, PR, and HER2 are lacking, no targeted therapies are currently available for TNBC, and treatment options are restricted to surgery, radiation therapy, and chemotherapy. Approximately 15–20% of all breast cancers are triple negative, and about 75% of TNBCs also fall into the basal subclass of breast cancer, which is defined by a gene-expression signature. The aggressiveness of TNBC and the lack of targeted therapies highlight the need to understand the pathways required for TNBC growth and survival.
Cancer cells up-regulate glucose and glutamine metabolism to fuel their bioenergetic and biosynthetic demands (2). TNBC is no exception to this generality, having elevated glucose uptake and a glycolytic gene-expression signature (3). Furthermore, TNBCs are more sensitive to glutamine depletion and have higher glutamine consumption than other breast cancer subtypes, suggesting that glutaminolysis is also up-regulated (4). The transcriptional drivers of increased glucose metabolism in TNBC are not well characterized, although dysregulation of the transcription factor c-Myc (hereafter, Myc) is thought to play a critical role. For example, Myc overexpression and Myc-dependent gene signatures are features of TNBCs (3, 5, 6), and Myc is a known driver of glycolytic gene expression, glucose uptake, and aerobic glycolysis (7).
Myc is a member of the basic region helix–loop–helix leucine zipper (bHLHZip) family of transcription factors and requires interaction with another bHLHZip protein, Max, for its transcriptional and transforming activity (8). Myc:Max complexes stimulate aerobic glycolysis by driving the expression of glycolytic target genes and glucose transporters (7), providing carbon backbones for anabolic biosynthesis (2, 9). Many of these biosynthetic pathways, as well as ribosomal biogenesis, are under Myc transcriptional control (7, 10). We previously identified another bHLHZip transcription complex, the MondoA:Mlx complex, which controls the availability of glucose and coordinates the utilization of glucose and glutamine (11).
MondoA:Mlx complexes are under tight regulatory control, and they accumulate in the nucleus on target gene promoters only in the presence of high glycolytic flux. MondoA:Mlx complexes are important, and perhaps the principal, regulators of glucose-induced gene expression (12). One glucose-induced and direct transcriptional target of MondoA:Mlx complexes is thioredoxin-interacting protein (TXNIP) (12). Among its many functions, TXNIP is a potent negative regulator of glucose uptake and aerobic glycolysis (13–15). Thus, the glucose- and MondoA-dependent induction of TXNIP triggers a negative feedback circuit that normalizes glycolytic flux.
In addition to blocking glucose uptake and restricting aerobic glycolysis, TXNIP has other antiproliferative activities (11), suggesting that TXNIP may be a tumor suppressor. Multiple reports support this contention (11, 16–19). Here we investigate how Myc controls nutrient utilization in TNBC and report that Myc represses TXNIP expression, stimulating TNBC glucose metabolism to support the growth and survival of this aggressive breast cancer subtype.
Results
Myc Represses TXNIP in TNBC.
To investigate Myc’s role in TNBC metabolism, we reduced its levels in the triple-negative (TN) cell line MDA-MB-157 using an siRNA pool (Fig. 1A). Consistent with Myc’s well-established role in glycolysis, glucose uptake was reduced in knockdown cells relative to controls (Fig. 1B). Myc knockdown in this same cell line did not reduce glutamine uptake (Fig. 1C), suggesting that Myc is not a primary driver of glutamine uptake in TNBC. Consistent with this conclusion, a recent study found no correlation between Myc levels or Myc-activity score and the sensitivity of more than 40 breast cancer cell lines, including TNBC lines, to glutamine deprivation (4). Similarly, we found no correlation between Myc-activity score and the sensitivity of 15 TNBC cell lines to the glutaminase inhibitor CB-839 (Fig. S1A) (20).
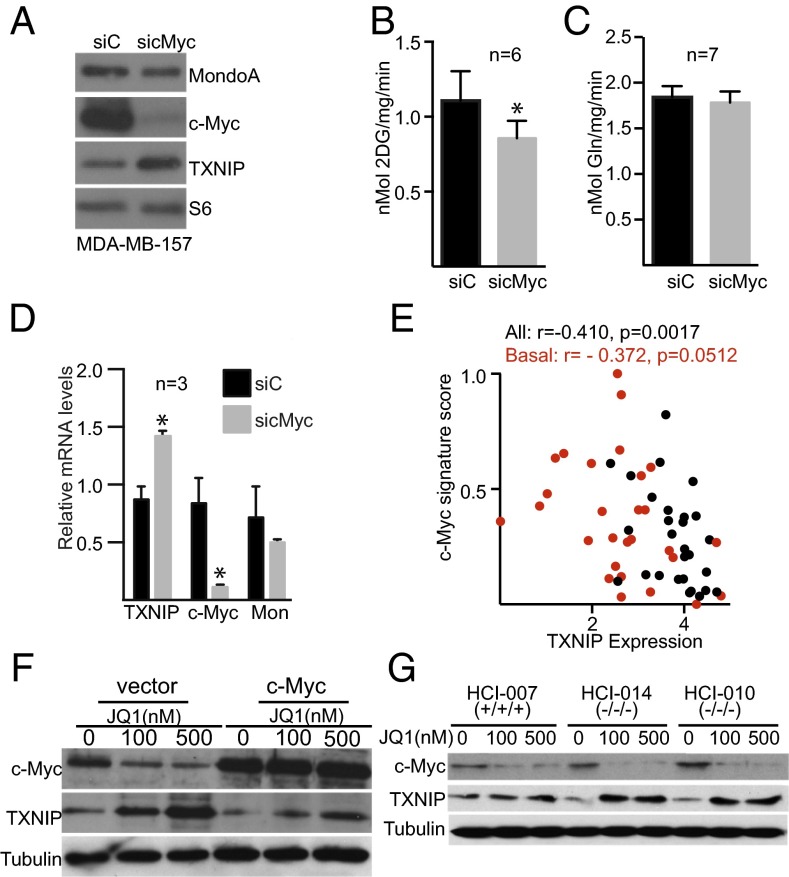
Fig. 1.
Myc regulates glycolysis and represses TXNIP in TNBC. (A–D) c-Myc levels were reduced in MDA-MB-157 cells by siRNA transfection. siC, control siRNA transfection. (A) Levels of the indicated proteins were determined by Western blotting 72 h after transfection. (B and C) The rates of glucose (B) and glutamine (C) uptake were determined in control or c-Myc–knockdown cells. (D) Levels of the indicated mRNAs were determined from control or c-Myc–knockdown cells by qPCR. (E) Myc activity as determined by gene-pathway signature is plotted against TXNIP mRNA level in 56 breast cancer cell lines, and the Pearson’s correlation coefficient was determined. Basal breast cancers are shown as red circles.(F) MDA-MB-157 cells with or without Myc overexpression were treated with the indicated amount of JQ1 for 24 h, and levels of the indicated proteins were determined by Western blotting. (G) Breast tumor explant cells HCI-010, HCI-014, and HCI-007 were treated for 24 h with the indicated dose of JQ1, and the levels of the indicated proteins were determined by Western blotting. *P < 0.05 as measured using unpaired t test; n, number of independent biological replicates. Values are reported as mean ± SEM.
Classically, Myc regulates glycolysis by direct regulation of genes encoding glycolytic enzymes. However, given the similarities between Myc and MondoA (11), we wondered whether Myc might also drive glycolysis by repressing TXNIP. Several pieces of evidence support this hypothesis. First, TXNIP protein levels increased following Myc knockdown (Fig. 1A). TXNIP mRNA levels also increased after Myc knockdown, suggesting that Myc regulates TXNIP transcription rather than TXNIP translation or protein stability (Fig. 1D). Second, we identified an inverse relationship between Myc activity as determined by gene-signature score and TXNIP mRNA levels in a panel of 56 breast cancer cell lines (Fig. 1E and Table S1). Third, Myc knockdown in the TN cell line MDA-MB-231 also increased TXNIP mRNA levels and the mRNA levels of the TXNIP paralog, ARRDC4 (Fig. S1B). Fourth, low levels of the bromodomain inhibitor JQ1 reduced Myc levels and increased TXNIP levels in MDA-MB-157 and MDA-MB-231 cells (Fig. 1F and Fig. S1 C and D). Importantly, TXNIP induction by JQ1 was dose dependent and was suppressed almost completely by ectopic Myc expression (Fig. 1F), suggesting that JQ1 induces TXNIP primarily via Myc down-regulation and not by an off-target effect. Taken together, these results indicate that Myc can repress TXNIP expression, suggesting another route to Myc-driven aerobic glycolysis in TNBC.
To investigate the relationship between Myc and TXNIP further, we reduced Myc levels in primary tumor cell cultures isolated from patient-derived breast cancer xenografts and cultured minimally on plastic (21). Because these primary cells are not readily transfectable, we reduced Myc levels with JQ1 in three different cell populations. TXNIP levels increased in each cell population, with more pronounced induction in the two-cell populations isolated from TNBCs (HCI-010 and HCI-014) than in cells isolated from a triple-positive breast tumor (HCI-007) (Fig. 1G). JQ1 also induced TXNIP mRNA more robustly in TNBC cell lines than in non-TNBC cell lines (Fig. S1E). However, TXNIP induction by JQ1 did not correlate with Myc-activity score (Table S1), suggesting that the differences in Myc activity in TNBC and triple-positive cell lines do not dictate all JQ1 responsiveness. Consistent with Myc’s regulation of glucose but not glutamine uptake in TNBC (Fig. 1C), treatment of HCI-014 cells with 100 nM JQ1 reduced the glycolytic intermediates glucose 6-phosphate, 3-phosphoglycerate, and 2-phosphoglycerate (Fig. S1 F–H) but did not reduce levels of glutamate (Fig. S1I).
TXNIP Induction Requires MondoA.
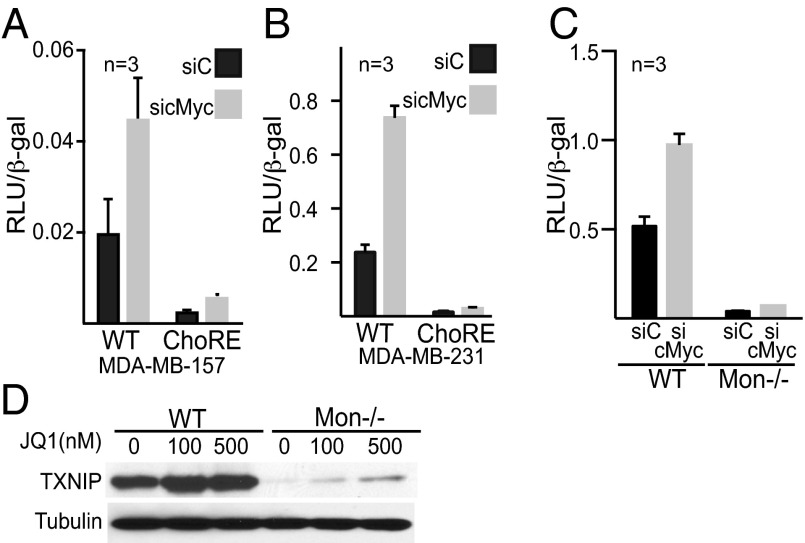
We next determined whether up-regulation of TXNIP following reduction of Myc requires MondoA. In either TN MDA-MB-157 or MDA-MB-231 cells, siRNA-mediated knockdown of Myc or JQ1 treatment increased the activity of a wild-type TXNIP luciferase reporter twofold (Fig. 2 A and B and Fig. S2 A and B). This transcriptional up-regulation was abolished by mutations in the double E-box carbohydrate response element (ChoRE), which is the known MondoA:Mlx binding site in the TXNIP promoter (Fig. 2 A and B and Fig. S2 A and B). Further, induction of TXNIP luciferase reporter activity, endogenous TXNIP message, and TXNIP protein in response to Myc knockdown or JQ1 treatment was abolished almost completely in MondoA-null murine embryonic fibroblasts (MEFs) (Fig. 2 C and D and Fig. S2 C and D). Together, these results demonstrate that TXNIP up-regulation following Myc suppression is dependent on MondoA.
Fig. 2.
TXNIP up-regulation following Myc suppression requires MondoA. (A–C) c-Myc levels were decreased by transfection of a c-Myc–specific siRNA pool. Representative experiments performed in triplicate are shown. (A and B) MDA-MB-157 (A) or MDA-MB-231 (B) cells were transfected with either a wild-type TXNIP luciferase reporter construct or a TXNIP promoter construct carrying mutations in the ChoRE MondoA:Mlx binding site. (C) Wild-type MEFs or MondoA-null (Mon−/−) MEFs were transfected with a wild-type TXNIP luciferase reporter construct. (D) Wild-type or MondoA-null MEFs were treated for 24 h with the indicated concentrations of JQ1, and the levels of the indicated proteins were determined by Western blotting. n, number of independent biological replicates. Values are reported as mean ± SEM.
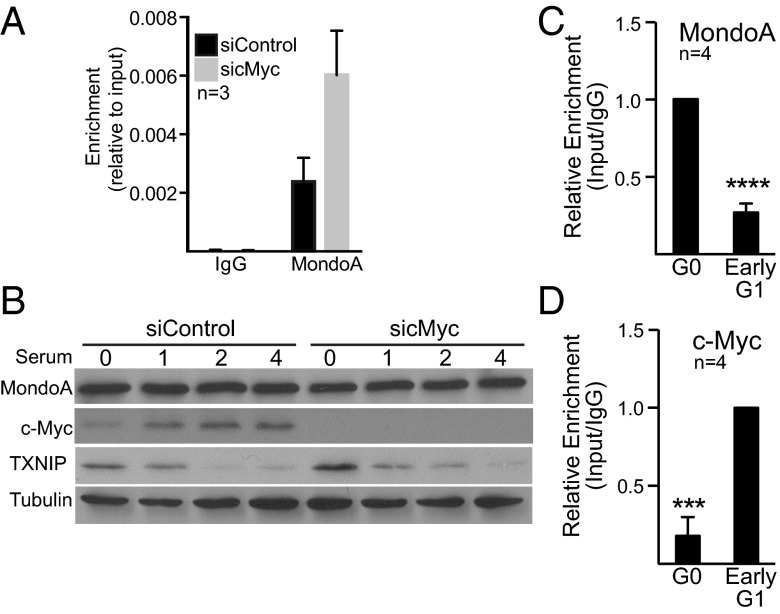
Three pieces of genomic data suggest that Myc represses TXNIP expression directly. First, ENCODE ChIP-sequencing (ChIP-seq) datasets (University of California, Santa Cruz Genome browser), including one from the breast cancer cell line MCF7, demonstrate that Myc can occupy a region just upstream of the TXNIP transcriptional start site. Second, Max also can occupy this region of the TXNIP promoter, although these studies were not conducted in MCF7s. Third, Myc:Max complexes recruit Miz-1 to repress transcription, and Myc and Miz-1 can co-occupy the TXNIP promoter in T cells and MEFs (22). Because MondoA:Mlx and Myc:Max affect TXNIP gene expression in opposite directions, we determined whether they show reciprocal binding to the TXNIP promoter. When we knocked down Myc in MDA-MB-157 cells, conditions that induce TXNIP expression, we observed increased occupancy of MondoA at the TXNIP promoter (Fig. 3A). These data suggest that, following a reduction of Myc, increased binding to the TXNIP promoter by MondoA:Mlx complexes increased TXNIP expression.
Fig. 3.
Myc represses TXNIP expression directly. (A) c-Myc levels were decreased in MDA-MB-157 cells using a c-Myc–specific siRNA pool. The amount of MondoA bound to the TXNIP promoter was determined by ChIP 72 h after transfection. (B) MDA-MB-157 cells were serum starved for 72 h with Myc levels being reduced for 48 h with a c-Myc–specific siRNA pool before serum treatment. The different cell populations were then serum stimulated for the indicated number of hours. Levels of the indicated proteins were determined by Western blotting. (C and D) The amount of MondoA (C) or Myc (D) bound to the TXNIP promoter in quiescent (G0) or early G1 (4 h after serum release) in MDA-MB-157 cells was determined by ChIP. ***P < 0.001; ****P < 0.0001 as determined using paired t tests. n, number of independent biological replicates. Values are reported as mean ± SEM.
Myc Represses TXNIP Directly.
We next determined Myc and MondoA occupancy at the TXNIP promoter under a physiological cell transition in which both Myc and TXNIP are dynamically regulated (8, 23), namely the G0-to-G1 transition. As expected c-Myc and TXNIP were up-regulated and down-regulated, respectively, by serum stimulation of quiescent MDA-MB-157 cells (Fig. 3B and Fig. S2E), but MondoA levels did not change during the G0-to-G1 transition (Fig. 3B). TXNIP down-regulation during early G1 was reduced by Myc knockdown or JQ1 treatment, suggesting that Myc is required to repress TXNIP in early G1 (Fig. 3B and Fig. S2F). Relative to the serum-starved controls, MondoA levels decreased and Myc levels increased on the TXNIP promoter after serum treatment (Fig. 3 C and D). Thus, Myc occupancy of the TXNIP promoter correlates with low TXNIP expression, whereas MondoA occupancy correlates with high TXNIP expression. We conclude that Myc binds the TXNIP promoter, suppressing TXNIP expression by a direct mechanism.
Myc and TXNIP Control TNBC Glucose Metabolism.
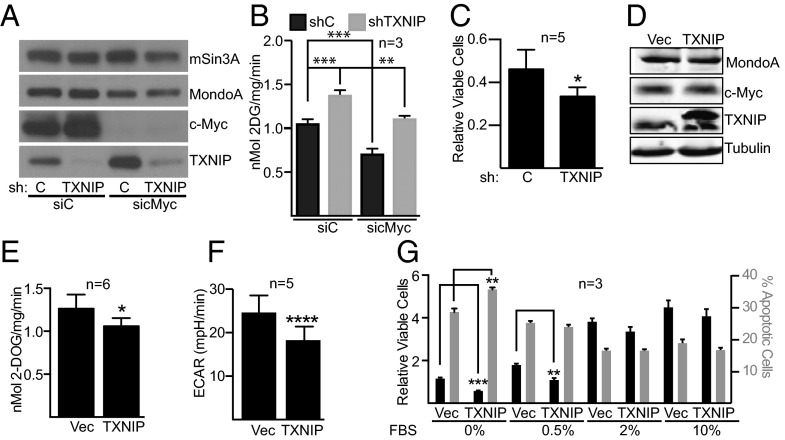
To understand the interplay between Myc and TXNIP in controlling the growth and survival of TNBC cells, we manipulated their levels and measured different metabolic and growth parameters. First, we reduced TXNIP levels in MDA-MB-157 cells using a lentiviral-delivered TXNIP-specific shRNA. We then transfected a Myc-specific siRNA pool to reduce Myc levels in TXNIP-knockdown or control cells. As above (Fig. 1A), Myc knockdown increased TXNIP expression (Fig. 4A). Glucose uptake increased in cells with TXNIP knockdown, and glucose uptake returned to control levels in TXNIP-knockdown cells that also had Myc knockdown (Fig. 4B). Conversely, glucose uptake was suppressed in Myc-knockdown cells, and glucose uptake returned to control levels in Myc-knockdown cells that also had TXNIP knockdown. TXNIP-knockdown cells were more sensitive to the glucose analog and glycolytic inhibitor 2-deoxyglucose, suggesting that low TXNIP levels drive a dependence on glycolysis (Fig. 4C). Despite the increase in glucose uptake, TXNIP-knockdown cells did not have an elevated extracellular acidification rate (ECAR) or changes in the oxygen consumption rate (OCR) (Fig. S2 G and H). This finding suggests that TXNIP loss propels glucose-derived carbons into biosynthetic pathways rather than driving the generation of lactate.
Fig. 4.
TXNIP is a suppressor of TNBC glucose metabolism. We reduced TXNIP levels in MDA-MB-157 cells using a lentivirus expressing a TXNIP-specific shRNA. We used transfection of a c-Myc–specific siRNA pool to reduce c-Myc levels in each cell population. (A) Levels of the indicated proteins were determined by Western blotting. (B) Rates of glucose uptake in the different cell populations were determined. (C) We determined the number of viable cells in control or TXNIP-knockdown cells following 3 d of growth in glucose-free medium that contained 20 mM 2-deoxyglucose. The number of viable cells is expressed relative to the number of cells seeded on day 1. (D) Control (Vec) or TXNIP-inducible MDA-MB-157 cells were treated with 5 ng/µL doxycycline for 48 h, and the levels of the indicated proteins were determined by Western blotting. (E–G) We determined the rates of glucose uptake (E) and ECAR (F) and the relative number of viable cells or percentage apoptotic cells (G) in control (Vec) or TXNIP-induced cells. In G, the different cell populations were grown in the indicated amounts of serum for 3 d. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. In B, statistical significance was determined using ordinary one-way ANOVA. In C, E, F, and G, statistical significance was determined using t tests. n, number of independent biological replicates. Values are reported as mean ± SEM in B and G and as mean ± SD in C, E, and F.
To determine how TXNIP overexpression affects TNBC cell growth, we generated an MDA-MB-157 cell population that carries a doxycycline-inducible V5-tagged TXNIP allele (Fig. 4D). TXNIP induction reduced glucose uptake, as expected, and reduced ECAR (Fig. 4 E and F). TXNIP overexpression did not alter glutamine consumption or the OCR (Fig. S2 I and J). Furthermore, TXNIP induction reduced the number of viable cells by blocking cell proliferation and increasing apoptosis, particularly in cells grown in low serum to reduce signaling through progrowth pathways (Fig. 4G). Together, these experiments suggest that TXNIP loss promotes proliferation/survival and that TXNIP overexpression restricts proliferation/survival of MDA-MB-157 cells.
Low TXNIP and High Myc Expression Levels Predict Poor Outcome in TNBC.
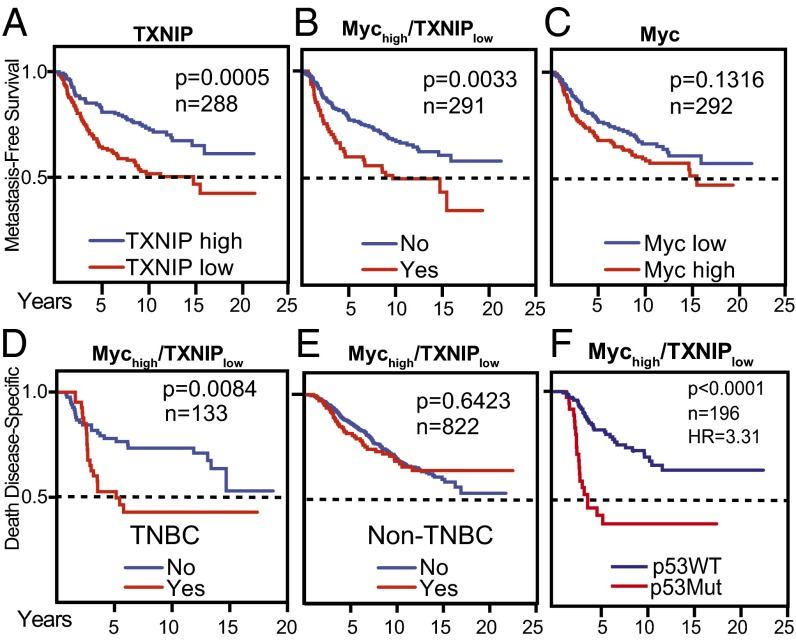
To evaluate the clinical significance of the reciprocal relationship between Myc and TXNIP, we correlated their expression levels with patient outcome. We examined their expression in tumors collected from 295 patients, all under the age of 53 y, with stage I or stage II breast cancer (24). We confirmed previous findings that low TXNIP levels correlated with decreased overall survival (Fig. S3A) (16, 25) and also identified a correlation between low TXNIP and decreased metastasis-free survival (Fig. 5A). Furthermore, low TXNIP expression correlated with decreased overall survival and decreased metastasis-free survival in patients whose tumors also had high Myc expression (Fig. 5B and Fig. S3A). This finding does not simply reflect a strong tumorigenic drive of high Myc expression, because Myc levels alone had no correlative power in terms of metastasis-free survival (Fig. 5C). No correlative relationship was evident in tumors that had low TXNIP and low Myc expression (Fig. S3 A and B). We confirmed each of these findings in a large compendium dataset with more than 1,200 samples that was generated by normalizing and combining expression levels from five unique datasets (26) and in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset (Fig. S3 B and C). We conclude that low TXNIP expression correlates with poor prognosis; however, the negative effect of low TXNIP expression is most evident when Myc levels are high also.
Fig. 5.
Low TXNIP expression and a Mychigh/TXNIPlow gene-expression signature correlate with poor patient outcome. Kaplan–Meier plots indicate the clinical outcomes for the gene-expression patterns given at the top of each panel. For TXNIP and Myc, high expression indicates expression above the mean, calculated across all samples. Conversely, low expression indicates expression below the mean. Mychigh/TXNIPlow indicates Myc expression above the mean in combination with TXNIP expression below the mean. n indicates the number of patient samples evaluated in each analysis. (A–C) Data available from the Netherlands Cancer Institute were analyzed (24). (D–F) Clinical outcomes for the Mychigh/TXNIPlow signature were evaluated in the METABRIC dataset (41). (D and E) The signature was evaluated in TNBCs (D) and non-TNBCs (E). (F) The Mychigh/TXNIPlow signature was correlated with clinical outcome based on p53 mutation status. In F, the 95% confidence interval of the hazard ratio is 1.91–5.70. P values were calculated using the Mantel–Cox log-rank test.
We next correlated the Mychigh/TXNIPlow gene signature with outcomes in different clinical subtypes of breast cancer. We subdivided the METABRIC data into TNBC (133 tumors) and non-TNBC (822 tumors). The Mychigh/TXNIPlow gene signature correlated with death from TNBC but had no correlative power in non-TNBCs (Fig. 5 D and E). Subdividing the METABRIC data into the intrinsic breast cancer subtypes revealed that the Mychigh/TXNIPlow gene signature correlated with death from basal breast cancer but did not correlate with clinical outcome in luminal A, luminal B, or HER2+ breast cancers (Fig. S3D). To understand better why the Mychigh/TXNIPlow gene signature correlated with poor clinical outcome only in TNBC, we stratified the clinical data by p53 status, which is mutated in ∼60–90% of TNBCs (27). We made three observations: First, the correlation between the Mychigh/TXNIPlow gene signature and poor clinical outcome was observed only in tumors with mutant p53 (Fig. S3E); second, p53 mutation enhanced the correlation between the Mychigh/TXNIPlow gene signature and poor clinical outcome (Fig. 5F); third, the interaction between mutant p53 and the Mychigh/TXNIPlow gene signature depended primarily on low TXNIP expression and not on high Myc expression (Fig. S3 F and G).
Discussion
Classically, Myc increases aerobic glycolysis by regulating the expression of glycolytic target genes and glucose transporters (7). However, because knockdown or deletion of TXNIP is sufficient to reprogram metabolism toward aerobic glycolysis (13, 14), our discovery that Myc can repress TXNIP directly provides an additional route to Myc-driven aerobic glycolysis. We propose that the overall level of Myc-driven aerobic glycolysis is a combination of its direct activation of glycolytic target genes and its repression of TXNIP. Supporting this conclusion, the increase in glucose uptake stemming from TXNIP knockdown requires Myc, and, conversely, the decrease in glucose uptake stemming from Myc knockdown requires TXNIP (Fig. 4B). Further, Myc or TXNIP knockdown alters glucose uptake to a similar degree, suggesting that Myc’s repression of TXNIP contributes significantly to Myc-dependent aerobic glycolysis. Our focus is on TXNIP’s role as a repressor of glucose uptake and aerobic glycolysis; however, its down-regulation by Myc presumably also restricts its other growth-suppressive activities (11). Thus, Myc-dependent repression of TXNIP may limit a number of antiproliferative functions. In this study we show that Myc represses TXNIP in TNBC cells; however, c-Myc increases MondoA expression in a number of lymphocyte cell lines, and N-Myc increases MondoA-dependent TXNIP expression in Tet21N neuroblastoma cells (28). Thus, whether Myc family members repress or activate TXNIP expression appears to be paralog or cell-type specific, perhaps reflecting differences in how nutrients are used or sensed in different cell lineages.
Our study provides a mechanistic framework to understand better earlier work that implicated Myc in controlling glucose metabolism in TNBC (3). Myc suppression increases TXNIP expression in two TNBC cell lines and in cell cultures derived from two independent patient TN tumors. Further, Myc activity is inversely correlated with TXNIP expression in a large collection of breast cancer cell lines. Thus, we conclude that Myc’s repression of TXNIP and the resultant increased glucose metabolism is a general feature of TNBC. In contrast, previous studies showed that TNBCs also rely on glutamine for growth; however, we found no correlation between Myc activity and sensitivity to inhibition of glutaminolysis. Thus, Myc may not be the primary driver of glutamine dependence in TNBC. The Gray laboratory (4) suggested that the uptake of cystine by the xCT antiporter, which requires glutamine, may underlie the glutamine dependence of TNBC, providing one potential Myc-independent mechanism of glutamine addiction.
Myc occupies a region just upstream of the TXNIP transcriptional start site; thus, we conclude that Myc complexes reduce TXNIP expression directly. There are two double E-Box elements spaced about 100 bp apart in the Myc-occupied region of the TXNIP promoter that may function as direct Myc:Max binding sites (29). MondoA:Mlx complexes function primarily through the start site-proximal double E-box ChoRE (30). Myc can repress transcription by a variety of mechanisms (31), so it is possible that Myc:Max complexes recruit repression machinery to the distal E-box pair to repress TXNIP expression. However, we demonstrate that MondoA binding to the TXNIP promoter is inversely correlated with Myc binding and that the binding of MondoA, whose levels are not under Myc control, to the TXNIP promoter increases in Myc-knockdown cells. These data raise the possibility that Myc:Max complexes displace strongly activating MondoA:Mlx complexes (32, 33) from the TXNIP promoter, resulting in repression of TXNIP expression. Alternatively, because suppression of Myc levels rewires intracellular metabolism, Myc knockdown may increase the nuclear activity of MondoA:Mlx complexes and their occupancy of the TXNIP promoter by an indirect mechanism. Our study sets the stage to distinguish between these regulatory mechanisms.
TXNIP expression above the mean (TXNIPhigh) correlates with better breast cancer outcome in two reports, one of which examined data from four independent studies (16, 25). We confirmed this result in two additional independent datasets and in a compendium dataset comprising expression and outcome data from five independent studies. We also discovered that high TXNIP expression correlates with increased metastasis-free survival in younger women with early-stage cancers (24). Thus, decreased TXNIP expression appears to be a relatively early event in tumorigenesis that is predictive of worse prognosis later in disease progression. Finding that TXNIP is a putative metastasis suppressor in breast cancer supports a number of in vitro experiments showing that it can block metastatic phenotypes (19, 34, 35).
Myc overexpression is a feature of TNBC (5, 6, 36), and a previous study showed that the highest tumor expression of a Myc pathway gene signature correlated with worse clinical outcome and resistance to chemotherapy (37). However, we found that high Myc levels correlated with lower overall survival only in one dataset. In contrast, a Mychigh/TXNIPlow gene signature correlated with reduced overall survival in all datasets examined and with decreased metastasis-free survival in the one dataset with available clinical data. These results validate the clinical significance of the reciprocal relationship between Myc and TXNIP described here and suggest that the deleterious effects of low TXNIP expression are exacerbated by high Myc expression. We propose that tumors with low TXNIP expression have elevated glucose uptake and aerobic glycolysis, which help fuel anabolic biosynthesis driven by high Myc expression. The Mychigh/TXNIPlow gene signature correlated with worse outcome in TNBCs but not in non-TNBCs, suggesting that the Mychigh/TXNIPlow gene signature contributes to the aggressive behavior of this breast tumor subclass. Our data suggest that the Mychigh/TXNIPlow gene signature is restricted to or is enriched in TNBC, because p53 is mutated in the majority of these cancers (27). How p53 loss cooperates with the Mychigh/TXNIPlow gene signature to promote the growth and survival of this aggressive tumor type merits further investigation.
Materials and Methods
Cell Culture and Conditions.
Cells were maintained at 37 °C in 5% CO2 in medium containing penicillin/streptomycin, glutamine, and 10% (vol/vol) standard FBS (Invitrogen) unless otherwise indicated. Wild-type and MondoA-KO MEFs were generated as described previously (38). MCF7, MDA-MB-361, MDA-MB-157, and MEF cells were grown in DMEM. MDA-MB-231 cells were grown in RPMI. The primary breast tumor cells (HCI-007, HCI-010, and HCI-014) were isolated from human tumor explants and grown in DMEM/F12 as described previously (21). For serum starvation, cells were incubated in DMEM containing 0.1% BSA and penicillin/streptomycin for 72 h. Serum starvation medium was replaced with growth medium for the times indicated.
Plasmids and Viruses.
The TXNIP-luciferase reporter constructs were described previously (38). Lentivirus producing TXNIP shRNA were as described (39). A TXNIP-V5–tagged cDNA was cloned into the pLVX-Tet-One-Puro vector (Clontech), and the resulting virus was used to generate MDA-MB-157 cells with doxycycline-inducible expression of TXNIP-V5.
Western Blotting.
Cells were collected and lysed in ice-cold RIPA buffer containing protease and phosphatase inhibitors. We used the Bio-Rad Protein Assay to determine protein concentration (Bio-Rad Laboratories). Forty micrograms of protein lysate were analyzed. Primary antibodies were used at dilutions of 1:500 for anti-MondoA (38), 1:1,000 for anti-VDUP1 (TXNIP) (Medical and Biological Laboratories), 1:500 for anti-Myc (Santa Cruz Biotechnology or Abcam), or 1:10,000 for anti-tubulin (Sigma). Secondary antibodies (Amersham Biosciences) were used at a 1:5,000 dilution. Western Lightning Chemiluminescence Plus (PerkinElmer) was used for detection.
Quantitative PCR.
For expression analysis, total RNA was extracted from cells using RNAeasy Mini Kit (Qiagen), and cDNA was generated from 1 µg RNA using GoScript Reverse Transcription System (Promega). Quantitative PCR (qPCR) was performed with SYBR Green PCR Mix, the CFX connect Real Time System, and CFX Manager 3.1 software (Bio-Rad) to quantify RNA levels. In our analysis, mRNA levels were determined using a standard curve. Values listed for specific mRNAs are expressed relative to corresponding β-actin levels. Measurements represent the average and SD of three biological replicates for RNA analysis. Primer sequences are available on request.
Cell-Based Assays.
Luciferase reporter assays were performed as described previously (40). The number of viable and apoptotic cells in the cell-viability assays were determined using the Guava ViaCount Assay (Millipore). Uptake of tritiated deoxy-d-glucose, 2-[1,2-3H(N)] (PerkinElmer) or glutamine, l-[2,3,4-3H] (American Radiolabeled Chemicals) was performed essentially as described using 1 µCi of either compound in a 1-mL labeling reaction (38). OCR and ECAR were determined on a Seahorse XF24 by the University of Utah’s Metabolic Phenotyping Core. ON-TARGETplus SMARTpool MYC (l-003282-00-0005) and ON-TARGETplus Nontargeting pool (d-001810-10-5) were from Dharmacon. Five microliters of a 20-µM siRNA stock were mixed with 5 µL of Lipofectamine RNAiMax (Life Technologies) and 200 µL of Opti-MEM (Life Technologies). The final concentration in the transfection was 50 nM siRNA. To determine steady-state metabolites, extracts were prepared from 2–6 × 106 cells with methanol containing internal standard solution of d4-succinate (Human Metabolome Technologies) and analyzed by the University of Utah Metabolomics Core. GC-MS analysis was performed with a Waters GCT Premier mass spectrometer (GERSTEL) fitted with an Agilent 6890 gas chromatograph and a GERSTEL MPS2 autosampler. Data were collected using MassLynx 4.1 software (Waters). Metabolites were identified and their peak area was recorded using QuanLynx. Metabolite identity was established using a combination of an in-house metabolite library developed using pure purchased standards and the commercially available National Institute of Standards and Technology library. All samples were normalized to the internal standard d4-succinate.
ChIP.
One 15-cm plate of MDA-MB-157 cells was used for each condition. Cross-linked and sheared chromatin was prepared as described (38) and was incubated overnight at 4 °C with 2 µg anti-MondoA (Proteintech) or anti-Myc (NeoMarkers) antibodies. Immunocomplexes were captured with 10 µL Dynabeads M-280 sheep anti-rabbit/mouse (Invitrogen) for 1 h at 4 °C. After washing, reversal of crosslinks, and DNA purification, transcription factor binding was determined by normalizing to input or input + IgG controls and was expressed relative to the highest enriched condition. Primers used for qPCR were reported previously (23).
Informatics.
The METABRIC data consist of clinical data and data derived from transcriptional profiling (41). MYC and TXNIP gene-expression values were extracted from normalized data available from the European Genome-Phenome Archive. Construction of the breast cancer compendium has been described (26). All survival data were extracted from original publications. The Kaplan–Meier estimate was used for plotting survival curves, and P values were calculated using the Mantel–Cox log-rank test. MYC activity for individual cell lines was determined using a well-described oncogenic pathway signature applied in a binary regression analysis (42). Gene-expression data for the MYC signature were obtained from Gene Expression Omnibus GSE3151 (43). We obtained exon array gene-expression data on breast cancer cell lines from the Gray cell line panel deposited at ArrayExpress E-MTAB-181 (44). Both datasets were normalized using the SCAN algorithm, and a distant weighted discriminant method was applied (45).
Supplementary Material
Acknowledgments
We thank Stephen Lessnick and Elizabeth Leibold for comments on the manuscript. D.E.A. was supported by National Institutes of Health Grants 5R01GM055668-15 and 5R01DK084425-03, by developmental funds from the Huntsman Cancer Foundation, and by Cancer Center Support Grant P30 CA42014. L.S. was supported China Scholarship Council Grant CSC-2011659013.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501555112/-/DCSupplemental.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Palaskas N, et al. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Res. 2011;71(15):5164–5174. doi: 10.1158/0008-5472.CAN-10-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmerman LA, et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24(4):450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alles MC, et al. Meta-analysis and gene set enrichment relative to er status reveal elevated activity of MYC and E2F in the “basal” breast cancer subgroup. PLoS ONE. 2009;4(3):e4710. doi: 10.1371/journal.pone.0004710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandriani S, et al. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS ONE. 2009;4(8):e6693. doi: 10.1371/journal.pone.0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 9.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28(27):2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Shea JM, Ayer DE. 2013. Coordination of nutrient availability and utilization by MAX- and MLX-centered transcription networks. Cold Spring Harb Perspect Med 3(9):241–256.
- 12.Stoltzman CA, et al. Glucose sensing by MondoA:Mlx complexes: A role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci USA. 2008;105(19):6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaadige MR, Looper RE, Kamalanaadhan S, Ayer DE. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci USA. 2009;106(35):14878–14883. doi: 10.1073/pnas.0901221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hui ST, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA. 2008;105(10):3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu N, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49(6):1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JL, et al. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLoS Genet. 2010;6(9):e1001093. doi: 10.1371/journal.pgen.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikarashi M, et al. Vitamin D3 up-regulated protein 1 (VDUP1) expression in gastrointestinal cancer and its relation to stage of disease. Anticancer Res. 2002;22(6C):4045–4048. [PubMed] [Google Scholar]
- 18.Sheth SS, et al. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene. 2006;25(25):3528–3536. doi: 10.1038/sj.onc.1209394. [DOI] [PubMed] [Google Scholar]
- 19.Shin D, et al. VDUP1 mediates nuclear export of HIF1alpha via CRM1-dependent pathway. Biochim Biophys Acta. 2008;1783(5):838–848. doi: 10.1016/j.bbamcr.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Gross MI, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 21.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511(7510):483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elgort MG, O’Shea JM, Jiang Y, Ayer DE. Transcriptional and translational downregulation of Thioredoxin Interacting Protein is required for metabolic reprogramming during G1. Genes Cancer. 2010;1(9):893–907. doi: 10.1177/1947601910389604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 25.Cadenas C, et al. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010;12(3):R44. doi: 10.1186/bcr2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunha S, et al. The RON receptor tyrosine kinase promotes metastasis by triggering MBD4-dependent DNA methylation reprogramming. Cell Reports. 2014;6(1):141–154. doi: 10.1016/j.celrep.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner N, et al. Targeting triple negative breast cancer: Is p53 the answer? Cancer Treat Rev. 2013;39(5):541–550. doi: 10.1016/j.ctrv.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Carroll PA, et al. Deregulated Myc Requires MondoA/Mlx for Metabolic Reprogramming and Tumorigenesis. Cancer Cell. 2015;27(2):271–285. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu FX, Luo Y. Tandem ChoRE and CCAAT motifs and associated factors regulate Txnip expression in response to glucose or adenosine-containing molecules. PLoS ONE. 2009;4(12):e8397. doi: 10.1371/journal.pone.0008397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoltzman CA, Kaadige MR, Peterson CW, Ayer DE. MondoA senses non-glucose sugars: Regulation of thioredoxin-interacting protein (TXNIP) and the hexose transport curb. J Biol Chem. 2011;286(44):38027–38034. doi: 10.1074/jbc.M111.275503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herkert B, Eilers M. Transcriptional repression: The dark side of myc. Genes Cancer. 2010;1(6):580–586. doi: 10.1177/1947601910379012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20(23):8845–8854. doi: 10.1128/mcb.20.23.8845-8854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole MD, Nikiforov MA. Transcriptional activation by the Myc oncoprotein. Curr Top Microbiol Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg SF, et al. Melanoma metastasis suppression by chromosome 6: Evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63(2):432–440. [PubMed] [Google Scholar]
- 35.Masaki S, Masutani H, Yoshihara E, Yodoi J. Deficiency of thioredoxin binding protein-2 (TBP-2) enhances TGF-β signaling and promotes epithelial to mesenchymal transition. PLoS ONE. 2012;7(6):e39900. doi: 10.1371/journal.pone.0039900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatza ML, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA. 2010;107(15):6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horiuchi D, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209(4):679–696. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson CW, Stoltzman CA, Sighinolfi MP, Han KS, Ayer DE. Glucose controls nuclear accumulation, promoter binding, and transcriptional activity of the MondoA-Mlx heterodimer. Mol Cell Biol. 2010;30(12):2887–2895. doi: 10.1128/MCB.01613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chutkow WA, Patwari P, Yoshioka J, Lee RT. Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. J Biol Chem. 2008;283(4):2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- 40.Billin AN, Eilers AL, Queva C, Ayer DE. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J Biol Chem. 1999;274(51):36344–36350. doi: 10.1074/jbc.274.51.36344. [DOI] [PubMed] [Google Scholar]
- 41.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 43.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piccolo SR, Withers MR, Francis OE, Bild AH, Johnson WE. Multiplatform single-sample estimates of transcriptional activation. Proc Natl Acad Sci USA. 2013;110(44):17778–17783. doi: 10.1073/pnas.1305823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.