Significance
EVfold_bb predicts all-atom 3D models of large transmembrane β-barrel proteins that are notoriously hard to determine experimentally. In some cases the algorithm can identify interactions between large extracellular loops, plugs, and the transmembrane β-strands. The major advance is the combination of evolutionary couplings between amino acid residues from protein family sequence alignments with prediction of β-strands to ascertain residue pairs that are hydrogen bonded between adjacent β-strands. The method will enable biological research into the function and druggability of outer-membrane proteins.
Keywords: transmembrane β-barrels, de novo 3D structure prediction, evolutionary couplings, maximum-entropy analysis, hydrogen bonding
Abstract
Transmembrane β-barrels (TMBs) carry out major functions in substrate transport and protein biogenesis but experimental determination of their 3D structure is challenging. Encouraged by successful de novo 3D structure prediction of globular and α-helical membrane proteins from sequence alignments alone, we developed an approach to predict the 3D structure of TMBs. The approach combines the maximum-entropy evolutionary coupling method for predicting residue contacts (EVfold) with a machine-learning approach (boctopus2) for predicting β-strands in the barrel. In a blinded test for 19 TMB proteins of known structure that have a sufficient number of diverse homologous sequences available, this combined method (EVfold_bb) predicts hydrogen-bonded residue pairs between adjacent β-strands at an accuracy of ∼70%. This accuracy is sufficient for the generation of all-atom 3D models. In the transmembrane barrel region, the average 3D structure accuracy [template-modeling (TM) score] of top-ranked models is 0.54 (ranging from 0.36 to 0.85), with a higher (44%) number of residue pairs in correct strand–strand registration than in earlier methods (18%). Although the nonbarrel regions are predicted less accurately overall, the evolutionary couplings identify some highly constrained loop residues and, for FecA protein, the barrel including the structure of a plug domain can be accurately modeled (TM score = 0.68). Lower prediction accuracy tends to be associated with insufficient sequence information and we therefore expect increasing numbers of β-barrel families to become accessible to accurate 3D structure prediction as the number of available sequences increases.
Transmembrane β-barrels (TMBs) constitute 2–3% of all genes in Gram-negative bacterial genomes (1) and are also found in eukaryotes (2). There has been increasing interest in this class of proteins, as their roles have been uncovered in a wide range of biomedical fields. These roles include outer-membrane protein biogenesis (3, 4), antibiotic resistance (5), vaccine design, translocation of virulence factors, and the design of cancer therapeutics (6). In many of these examples, the 3D structure of the TMB has been crucial in elucidating the mechanisms of, for instance, substrate transport and voltage gating and in aiding therapeutic design.
Existing computational approaches can successfully identify the location of β-strands (7, 8), but 3D-modeling techniques such as tobmodel and 3d-spot (9, 10) cannot account for the nonsymmetrical, noncircular shape of the barrel pore or the barrel/plug or the transmembrane β-strand/loop interactions. Recent work has shown that 3D structures of globular (11) and α-helical membrane proteins (12, 13) can be successfully predicted from the identification of coevolved residues in multiple-sequence alignments (MSA). The idea is that spatially close residues coevolve to maintain structural and functional integrity of the protein (11). Although this approach was first suggested and tried in 1994 (14–17), only recent methods using a global statistical model identify sufficiently accurate residue–residue contacts from evolutionary covariation to successfully fold proteins de novo (11, 18–20). The key innovation was to distinguish direct from indirect correlations, using maximum-entropy or related statistical approaches under the constraints of the data.
Here, we present a hybrid method based on evolutionary couplings for contact prediction obtained from EVFold-PLM (11, 19) together with an improved β-strand prediction method based on boctopus2, a topology prediction method for transmembrane β-barrels (7). The method predicts consistent sets of backbone hydrogen-bonding restraints that can be used to fold large TMBs. Our approach relies on structural features that are common to the known 3D structures of bacterial TMBs, such as the antiparallel arrangement of transmembrane β-strands and the facts that the first strand, as far as is known, always traverses from the inner-membrane region to the extracellular side and pairs of β-strands have a right-handed twist when viewed along the direction of the strand. In addition to these features, our algorithm uses evolutionary couplings (ECs) in transmembrane β-strands to infer the optimal strand registration between pairs of adjacent β-strands and, by implication, backbone hydrogen-bonded residue pairs. The method achieves reasonably correct strand registration between adjacent β-strand pairs and all-atom 3D structures of TMBs and, where applicable, predicts interactions between the β-barrel and plug domains and between transmembrane β-strands and long extracellular loops. Moreover, in a few proteins we show that ECs can be used to detect functionally important residues.
Results
De Novo Folding of Large Transmembrane β-Barrels.
Accuracy of predicted contacts from evolutionary constraints.
EVFold-PLM (11, 19) was used to calculate ECs for all proteins in the dataset (Fig. 1). Predicted ECs involving residue pairs that are in contact in the known 3D structure (shortest interresidue atom–atom distance ≤5 Å) were considered correctly predicted. With this definition, ∼48% of the top L/2 ECs (in order of decreasing EC value; L is the number of amino acid residues in the protein), were correctly predicted (Table 1 and Fig. S1). Moreover, ∼62% of the top L/2 ECs between adjacent β-strands were correctly predicted (Fig. S1). Outside the β-barrels, accuracy is lower. Indeed, for 15 proteins in the dataset, about 60% of all incorrectly predicted ECs involved loop−loop or loop−strand contacts.
Fig. 1.
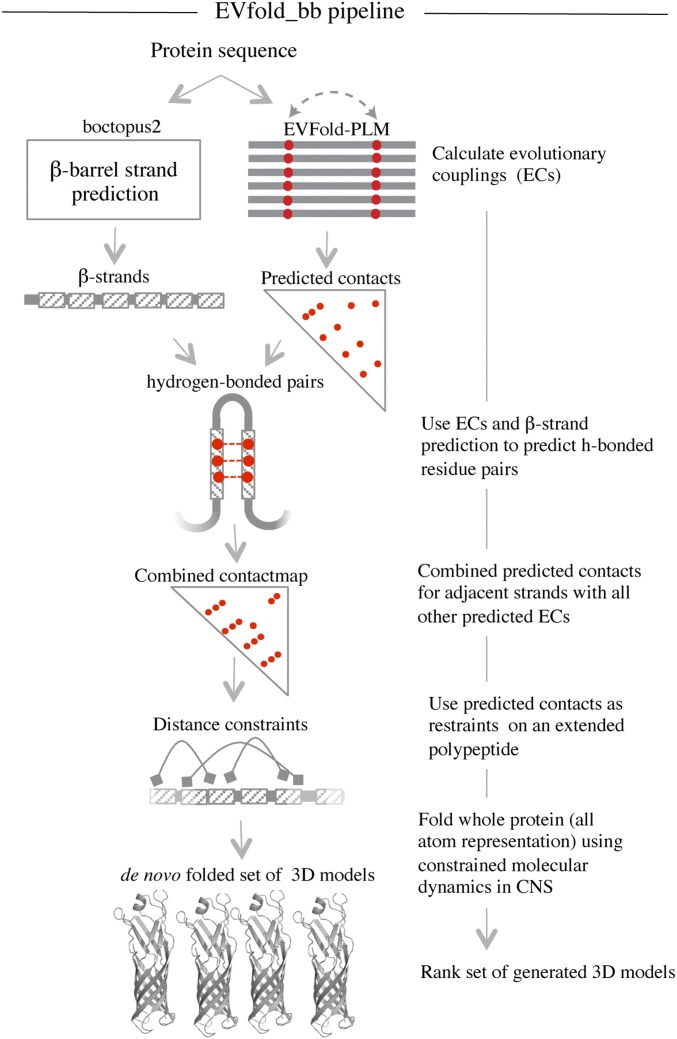
EVfold_bb pipeline to de novo fold transmembrane β-barrels. EVFold-PLM is used to generate evolutionary couplings (ECs) from a multiple-sequence alignment of the target protein. Boctopus2 is used to assign β-strands. Alternative strand registrations are compared for successive relative shifts of adjacent strands up to plus or minus three residues. The configuration with the largest sum of EC values is chosen and distance constraints are applied on N–O atoms of alternate residue pairs. In addition, other nonstrand–strand constraints are used to de novo fold the protein. Multiple models are generated and blindly ranked.
Table 1.
Prediction performance in a blinded test on known structures
| PDB ID (no. strands) | Gene name | Amino acid count | No. sequences in alignment | No. sequences per amino acid | PPV of top L/2 ECs | PPV of top L/2 ECs between adjacent strands | rmsd of the top-ranked model† | TM score of the top-ranked model† |
| 1p4t (8) | NspA | 160 | 19,458 | 121.6 | 0.66 | 0.72 | 1.59 (87) | 0.85 |
| 1kmp (22) | FecA | 535 | 48,590 | 90.8 | 0.79 | 0.82 | 4.48 (217) | 0.67 |
| 3kvn (12) | EstA | 287 | 20,273 | 70.6 | 0.7 | 0.72 | 3.12 (124) | 0.68 |
| 2j1n (16) | OmpC | 353 | 18,764 | 53.2 | 0.64 | 0.77 | 4.65 (152) | 0.58 |
| 3ohn (24) | FimD | 534 | 15,557 | 29.1 | 0.59 | 0.6 | 4.77 (150) | 0.46 |
| 4k3c (16) | BamA | 384 | 10,014 | 26.1 | 0.65 | 0.67 | 5.22 (141) | 0.48 |
| 4e1t (12) | InvA | 251 | 5,778 | 23.0 | 0.42 | 0.57 | 4.49 (93) | 0.45 |
| 1t16 (14) | FadL | 387 | 5,422 | 14.0 | 0.63 | 0.69 | 3.86 (147) | 0.66 |
| 3syb (18) | OpdP | 430 | 5,031 | 11.7 | 0.58 | 0.65 | 4.15 (162) | 0.62 |
| 2wjr (12) | NanC | 223 | 2,552 | 11.4 | 0.5 | 0.66 | 4.38 (109) | 0.50 |
| 1thq* (8) | PagP | 161 | 1,836 | 11.4 | 0.34 | 0.55 | NA | NA |
| 1qd6 (12) | OmpLA | 250 | 2,367 | 9.5 | 0.46 | 0.58 | 3.5 (112) | 0.61 |
| 4q35 (26) | LptD | 585 | 5,285 | 9.0 | 0.44 | 0.62 | 6.54 (174) | 0.36 |
| 1a0s (18) | ScrY | 423 | 3,299 | 7.8 | 0.2 | 0.55 | 6.12 (139) | 0.39 |
| 1tly (12) | Tsx | 270 | 2,039 | 7.5 | 0.27 | 0.56 | 5.06 (103) | 0.41 |
| 2jk4 (19) | VDAC | 283 | 1,914 | 6.8 | 0.22 | 0.5 | 6.18 (157) | 0.45 |
| 2erv (8) | PagL | 159 | 1,006 | 6.3 | 0.25 | 0.48 | 2.55 (79) | 0.67 |
| 4gey* (16) | OprB | 420 | 2,376 | 5.7 | 0.47 | 0.56 | NA | NA |
| 2o4v (16) | OprP | 397 | 2,182 | 5.5 | 0.3 | 0.52 | 4.98 (109) | 0.40 |
Seventeen of the 19 proteins with a known structure are folded in a blinded test such that no known structural information is used for folding. Positive predictive value (PPV) is the ratio of correct predictions (shortest distance between two residues in the known structure ≤ 5 Å) and all predictions made. Three-dimensional models are compared with the known structure, using a TM score that ranges from 0 to 1 and an rmsd value. The closer the TM score is to 1, the more similar the model is to the known structure. rmsd values closer to 0 signify high structure similarity. rmsd values of residues aligned (shown in brackets) is reported in angstroms. PDB, Protein Data Bank.
PagP and OprB are not applicable (NA) cases for folding, as boctopus2 strand assignment is incorrect.
TM score and rmsd of the predicted transmembrane β-barrel region excluding the loops.
Identification of residue pairs that are hydrogen bonded between β-strands.
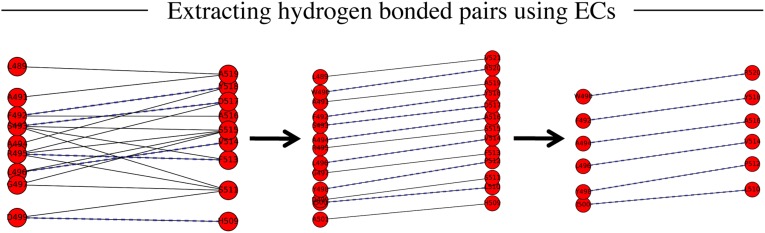
Assuming up-and-down antiparallel β-strand interactions in all barrels, a key prediction step is to determine the precise strand–strand registration between adjacent strands. Similar in spirit to an earlier attempt to optimize strand–strand registration based on residue pairing scores (21), we implemented an algorithm that uses the evolutionary couplings between adjacent strands to predict the hydrogen-bonding pattern (Fig. 2). Briefly, the algorithm predicts residue pairs that are in hydrogen-bonded contact on adjacent β-strands, using the top L ECs located on adjacent predicted β-strands. Starting with the first strand, each strand is paired with the next adjacent strand and the strands in the pair are shifted relative to each other to find the optimal registration such that the pairwise summed EC score of residues between the two strands is maximized (Fig. 2). A similar technique of shifting strands to predict the optimal strand registration was used in earlier work (21). Once the optimal strand registration has been calculated for all of the pairs of adjacent strand pairs, alternate residues are chosen in each strand pair for placing hydrogen bond constraints, consistent with known β-sheet geometry. The refinement step in the algorithm resulted in the identification of 674 of 983 (69%) β-barrel hydrogen-bonded residue pairs.
Fig. 2.
Evolutionary couplings give residue pairs that are hydrogen bonded. From the evolutionary couplings (ECs) (black) predicted between residues (red) on strands 5 and 6 of EstA (Left), a subset of hydrogen-bonded pairs (dashed lines, where N–O distance ≤ 3.4 Å) is extracted. Predicted adjacent β-strands are shifted plus or minus three residues relative to each other to generate alternate configurations of residue pairs (Center). To select the best configuration, the EC strength of all pairs is summed and the highest-scoring configuration is selected; then distance constraints are applied to N–O atoms of alternate residue pairs (Right) to reflect N–H ... O = C hydrogen bonds.
One technical difficulty arises from the fact that no high-ranking (up to top L) ECs were detected between the first and last strands for 11 of 17 proteins (Fig. S2). The fact that ECs between the first and the last strands are missed in some cases is plausibly caused by the lack of terminal sequence coverage in the alignment. We address this issue to close the barrel by adding hydrogen-bonding pair constraints between the first and last strand that best pair open h-bonding valences, derived from the left–right alteration of h bonds on antiparallel β-strands.
De novo 3D structure prediction and assessment of 3D accuracy.
Starting with the target protein in an extended polypeptide conformation, distance constraints on hydrogen-bonded residue pairs, other EC-derived distance constraints, and secondary structure constraints were used to fold the target protein (Fig. 3), using CNS software (22). For each set of constraints, 20 models were generated, each with a different random start in the distance geometry calculation in CNS. All models were ranked using a model likelihood score, which assesses the number of hydrogen-bonded constraints satisfied in the folded model as well as a measure of strand–strand twist (11). To assess the overall accuracy of the 3D structure, these models were then compared with the known 3D structure in blinded mode, using the template-modeling (TM) score (23) as well as the positional root-mean-square deviation (rmsd) (24).
Fig. 3.
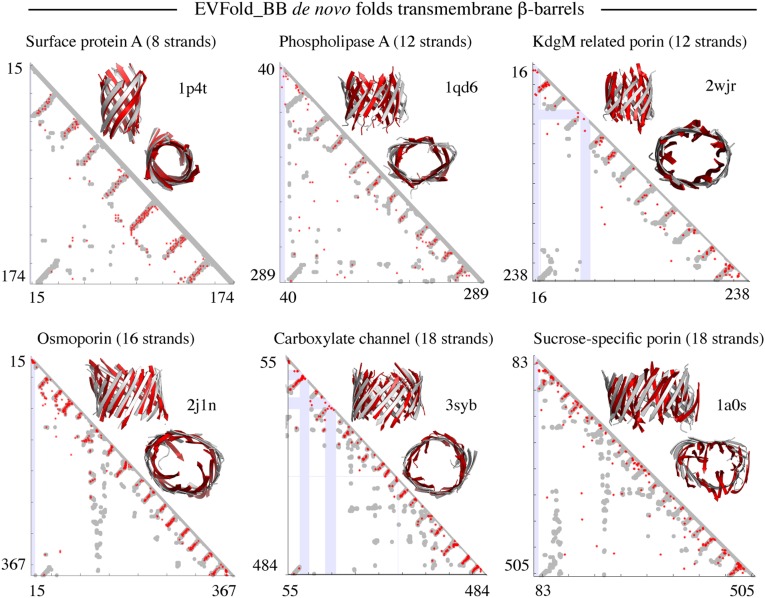
Blinded benchmark de novo 3D models of transmembrane β-barrels. Shown are predicted contact maps (red, ECs; gray, crystal contacts ≤ 5 Å; blue, gaps in crystal structure) and front and top views of folded structures (red, de novo folded; gray, crystal structure) for six proteins in the dataset.
For each protein, the quality of the 3D models of the barrel region was evaluated for the top-ranked model (TM score 0.36–0.85) and the overall best possible of all models generated (TM score 0.42–0.87). Overall, the accuracy tends to decrease with the number of sequences in the MSA normalized by length L of the protein sequence (Tables S1 and S2). Two structures are often considered to have similar folds if their TM score is greater than 0.5 (25). With this cutoff, nine proteins were predicted accurately (Table 1); and of the seven protein families with a TM score of less than 0.5, five have fewer than 10 sequences per residue. The average TM score of the top-ranked (0.54) and the overall best possible model (0.59) suggests that although there are slightly better models in the ensemble of models generated, our top-ranked model often comes reasonably close to the overall best possible model (Tables S1 and S2).
The average TM scores for the full proteins (i.e., β-barrel plus nonbarrel regions such as loops) are consistently lower than for the barrels alone (Tables S1 and S2). Our method does not do as well in the loop regions as it does in the β-barrel region. Poor performance in folding loops is probably due to lack of sufficiently well-predicted contacts in those regions (Fig. S2). Whereas the TM score and positional rmsd values give a reasonable quantitative assessment of the quality of the predicted 3D models, visual inspection of the atomic coordinates of the barrel region and full protein reveals further details about where EVfold_bb modeling is more or less accurate. For OprP, VDAC, Tsx, ScrY, OmpLA, and BamA, the EVfold_bb protocol fails to generate well-folded structures, which could be due to a suboptimal folding procedure or to fewer accurate contacts.
Blind prediction of LptD.
The 3D structure or the number of strands in the outer membrane protein LptD, which is essential for lipopolysaccharide (LPS) transport, was not known at the outset of this study (26). Boctopus2 (and boctopus) predicts 26 strands, whereas the other methods predict fewer strands (Fig. S3). We used the overlap with predicted ECs (excluding the first and the last strands) to discriminate different topology predictions and found that the boctopus2 topology completely agrees with the ECs (Fig. S3). Comparison of our predictions with the two subsequently published LptD crystal structures shows mixed results. On the positive side, the number and the location of the predicted β-strands correctly match with those in the crystal structures (27, 28). In addition, the disulfide bond between C724 and C725 was correctly predicted to lie on the inner loop between strands 24 and 25 (Fig. S3). However, 3D structure comparison of the top-ranked predicted 3D model with the crystal structure shows that β-strands in the crystal structure are more tilted than in the model and that the predicted model is more circular and not kidney shaped. This results in a low TM score (0.36) and high rmsd (6.54 Å over 174 of 267 residues). Taken together, our de novo 3D modeling of the LptD barrel pore was quite poor. The known structures of LptD contain its binding partner LtpE that is located inside the barrel pore and affects its native shape (27, 28). We were not able to account for structural changes in the barrel pore region that might occur due to LptD–LptE interaction, as the interprotein sequence coverage was too low to identify the interacting residues.
Comparison with an alternate method.
The main conceptual and practical advantage of EVfold_bb over previous methods, such as tobmodel (9), is the prediction of residue–residue interactions from the calculation of evolutionarily constrained residue pairs from sequence information and the generation of nonidealized all-atom coordinate models, both for the β-barrel and for other regions, such as loops and plug domains. For example, the plug domain of FecA and the long L6 loop in BamA interact with the barrel and have important functional roles and are therefore of considerable interest. In contrast, tobmodel is a machine-learning–based method for predicting the topological arrangement of transmembrane β-strands and the generation of idealized Cα 3D models of the barrel region (9). Although these models may be approximately accurate, they cannot represent any deviation from ideal barrel geometry and do not reveal information about functional couplings, either within the barrel or between loops and plug domains.
For the β-barrel region, one can compare the accuracy of EVfold_bb models with those of tobmodel models at a shared level of information, i.e., focused on the accuracy of strand–strand register between adjacent β-strands, which both models use as crucial input to the generation of models. For each residue in strand S1, its corresponding in-register partner is defined as the closed (Cα–Cα distance) residue in strand S2, where S1 and S2 are adjacent strands. The average TM scores of the models generated by the two methods are similar, but there are notable differences in the number of residue pairs found in correct registration (Table S3) between adjacent β-strands, as quantified by comparing the number of in-register residue pairs found in the model with that of the known structure. The average number of residue pairs in correct registration in EVfold_bb models (44%) and tobmodel models (18%) is low (Table S3). However, 65% and 41% of residue pairs are within plus or minus one residue of correct registration in EVfold_bb and tobmodel models, respectively, which shows the advantage of the use of evolutionary couplings in EVfold_bb for modeling 3D structures of transmembrane β-barrels.
Interaction of the Barrel Domain with Plugs and Extracellular Loops.
In addition to folding TMBs, barrel/plug domain interactions can be predicted for some proteins with good accuracy, especially when a sufficient number of sequences are available, but on average the overall accuracy of these predictions remains inadequate. The technical challenge is related to the fact that TMBs have long and flexible extracellular loops and that these loops make only a few contacts with the barrel domain as they protrude away from the membrane center. However, in crystal structures some of these loops are in the barrel pore and their interaction with transmembrane β-strands appears to aid in substrate transfer (29, 30) and gating (3).
Prediction of interactions with a plug domain.
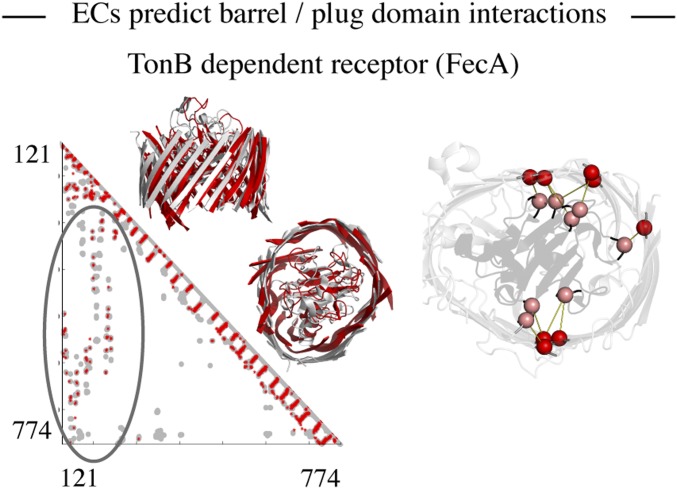
FecA is a 22-stranded active outer membrane transporter with a large plug domain (∼126 residues) that is implicated in signal transduction and is involved in its gating mechanism (31). Residue pairs in 9 of 10 top couplings between the barrel and the plug domain ECs make contact (d ≤ 5 Å) in the known structure (Fig. 4). In addition, the positive predictive value (PPV) over the top 50 plug/barrel ECs is ∼0.7, suggesting that most of the interdomain interactions are captured by ECs. The TM score (23) for the top-ranked FecA model is excellent for the barrel alone (0.67) and for the barrel plus the plug domain (0.68). Consistent with our prediction, it is known that interaction of plug domain residues R150 and R196 with E541 and E587 located on β-strands is of functional importance in fixing the plug within the barrel (32). Computational evidence for strong evolutionary constraints comes from the fact that all four residues occur in multiple high-ranking coevolutionary pairs. This illustrates the potential utility of evolutionary couplings not only for constraining interactions in the computational folding process, but also in revealing highly constrained functionally important interactions.
Fig. 4.
ECs predict interactions between loops/plugs and the barrel domain. Interactions between the barrel (245–774) and the plug (121–244) domain in FecA are highlighted in the predicted contact map (red, ECs; gray, crystal contacts ≤ 5 Å). Top 10 interdomain contacts between the barrel (red) and the plug domain (pink) are shown on the crystal structure and have a PPV of 0.9.
Interactions with extracellular loops.
In BamA, the flexible extracellular loop L6 (residues 639−691) contains the conserved VRGF/Y motif and undergoes large conformational changes during its activation and inactivation (33). We found that 11 of the 20 top-ranking ECs between β-strands and extracellular loops in top L/4 predictions lie on L6 and the residues in the RGF motif are ranked high in the EC list (Fig. S4). Moreover, residue F662 is ranked high (fourth) on the list of residues with multiple coevolutionary partners. Residue R660 on L6 stabilizes the transformation from a closed to an open state (3). Residues G754, P719, and E698 that are predicted to coevolve with R660 are located near the cytosolic end of strands 12−14 (Fig. S4), which could be sites for L6 interaction after a conformation change.
In the substrate-specific channel (OprB) and the carboxylate channel (OpdP), 10 of 14 and 6 of 8 top-ranked ECs between β-strands and extracellular loops in the top L/4 predictions are within 5 Å in the known crystal structure, respectively (Fig. S4). Four of these 10 residues in OprB (141, 142, 150, and 155) lie on the extracellular loop L3 that forms a constriction zone in the barrel (30). In the eukaryotic voltage-dependent anion channel (VDAC), the conformation of the N-terminal helix (residues 1−26) within the barrel pore and its impact on gating are not clear (34). We found 5 ECs between the helix and pore-forming β-strands within top 3L/4 EC predictions (Fig. S4). All these residue pairs are more than 5 Å away in the crystal structure. However, when superimposed on the structure, they suggest an alternative conformation of the helix possibly important for the gating mechanism.
Relation Between Coevolution and Functionally Relevant Sites.
Residues involved in multiple evolutionarily constrained interactions plausibly are in functionally important sites (13). For example, in LptD, the disulfide bond-forming residues between the two domains, C173 and C725, appear within the top 20 interdomain ECs (Fig. S3). The presence of a disulfide bond between the two domains is essential for LtpD/E interaction with LptA (35). In addition, three conserved (36) proline residues (P231, P246, and P261) are predicted to lie on three adjacent β-strands (Fig. S3). Additionally, P246 is the top-ranked residue in terms of interaction partners (10) within the top L/2 predictions. On comparison with recent crystal structures (27, 28), our predictions correctly capture the location of the “exit portal” necessary for lateral diffusion of LPS, which is observed between strands 1 and 2 and is formed due to interaction between the three proline residues (Fig. S3). In Tsx, a nucleoside transporter, residues F27 and G28 in the nucleoside-binding site Nuc1 are involved in six highly ranked couplings (rank 8). Interestingly, F27L and G28R mutants are defective in nucleoside transport (37). In OprB, residue Q396 that is known to stabilize extracellular loops L2 and L3 is the highest-ranked residue with eight interaction partners in the top L predictions (30). In the long-chain fatty acid transporter FadL, residue S370, which is required for optimal long-chain fatty acid transport, occurs in seven high-ranking EC pairs (ranked sixth) (Fig. S4).
Potential Application to Protein Families Without a Known 3D Structure.
To estimate the number of transmembrane β-barrel families without a known 3D structure, we analyzed 15,483 sequences predicted by Freeman and Wimley (38) to be TMBs and mapped them to domain families in the PFAM database when possible (39). A sensitive search for remote homologs, using HHSearch (version 2013) (40), reduces the number of β-barrel domain families without a known 3D structure to 172. Of these, 63 (∼37%) have enough sequences to predict contacts. However, only three [YP_861842.1 (region 694–832), YP_001305047.1 (region 122–317), and NP_754081.1 (region 25–248)] predicted contact maps have the characteristic antiparallel β-strand pattern (by visual inspection). Thus, our analysis indicates that current methods (38) for identifying novel transmembrane β-barrels probably result in many false positives. The 109 putative transmembrane β-barrel families for which insufficient numbers of homologous sequences were obtained using current sequence databases can be analyzed in the future when more sequences become available.
Discussion
Evolutionary couplings between residues together with the prediction of β-strands and their topological up–down arrangement can be used to predict optimal strand registration and identify residue pairs that are hydrogen bonded between adjacent β-strands in TMBs. These β-sheet pairing constraints together with other highly ranked ECs are sufficient to fold large TMBs de novo. Given sufficient sequence coverage, EVfold_bb can be used to determine the location and interaction between the barrel and plug domains. The main advantage of using EVfold_bb is the generation of evolutionarily derived residue–residue contacts used to compute full-atom, realistic 3D models of the transmembrane barrel region of TMBs. The resolution of models generated by EVfold_bb in many cases is sufficient for determining the spatial location of known functionally interesting regions and inferring physico-chemical properties of the barrel domain, but modeling long loops is still an open issue. For a few proteins, we show that ECs can identify previously unknown functionally important residues (Fig. S4). Other studies estimate that more than 5L sequences are necessary for accurate contact prediction (40). By this estimate, currently EVfold_bb can be applied to only 37% of the PFAM β-barrel domain families without a known 3D structure. With more genomes being sequenced, we anticipate that more TMB sequences will be available soon and contact prediction methods based on coevolutionary analysis can be applied to more TMB protein families. There has also recently been some progress toward decreasing the need for the number of sequences by combining deep learning and direct information (41). Although the current analysis focused on proteins with known TMB domain structures, a fully automated method for de novo prediction of TMB structures from sequence would require implementation of a method to reliably detect TMB domains in proteins. Also, EC-based strategies could be developed to identify β-barrel domains and identify the precise location of the β-strands in putative TMBs. In addition, we propose that our approach of extracting sets of hydrogen-bonded residue pairs from predicted ECs could be generalized and extended to other β-sheet–containing proteins (21).
Methods
Benchmark Dataset.
The benchmark dataset contained 19 proteins, representing all known TMB families with a sufficient number of sequences in their MSA (Table 1). EVfold_bb was used to de novo model 17 of 19 proteins. Two families (PagP and OprB) were not folded because of failure in topology prediction by boctopus2 (Tables S4 and S5).
Topology Prediction Using Boctopus2.
Boctopus2 was developed using an almost identical strategy to that used when developing boctopus (7). The main difference is that all residues in the dataset were labeled as outer loop (o), inner loop (i), β-strand pore facing (p), and β-strand lipid facing (l), whereas in boctopus the “p” and “l” residues were grouped together. The position-specific scoring matrix (PSSM) obtained using three iterations of hhblits (version 2.0.13) (42) against the “nr” database (nr20_12Aug11) is used as the input to four separate support vector machines (SVMs) that were trained to predict the per-residue location. Together with secondary structure prediction using PSIPRED (43), a per-residue profile is generated and used as input to a hidden Markov model to predict the overall topology. Boctopus2 is trained in a 10-fold cross-validated manner, where all proteins belonging to the same family were put together in the training or the test set. In contrast to boctopus all transition probabilities could be set to 1, which means that the hidden Markov model (HMM) architecture is not trained (Fig. S5). Within the barrel domain, boctopus2 predicts the correct β-strand arrangement for 32 of 36 proteins in the benchmark dataset (Tables S4 and S5). Additionally, topologies for five proteins not in the initial boctopus2 dataset (3syb, 4k3c, 4e1t, 3ohn, and 2jk4) are predicted using boctopus2 (Tables S4 and S5).
Prediction of Evolutionary Couplings from Multiple-Sequence Alignments.
MSAs for all proteins are generated using three iterations of jackhmmer (version 3.1) (44) against the UniProt database. For all proteins, an E-value of 10−2 was used to ensure the maximum number of sequences. For LptD and FecA multidomain interaction predictions, MSAs were generated at an E-value threshold of 10−10 and 10−20, respectively, to obtain stringent alignments and ensure sequence coverage in both domains. A global statistical inference method based on pseudolikelihood maximization (19) as implemented in EVFold (evfold.org/) (11) is used to extract direct interactions from all of the observed correlations in a MSA. A ranked list of ECs is obtained by taking the average-product corrected norm of the matrix of couplings that adjusts for the phylogenetic bias (19).
De Novo Folding Using CNS and Applied Constraints.
Distance constraints are applied to one side-chain heavy atom per residue pair, O−N, N−O, and Cα−Cα atoms for residue pairs that are predicted to be hydrogen bonded (Table S6). For other ECs between nonstrand–strand residues, distance constraints are applied to side-chain heavy atoms only (Table S6). Intrastrand distance constraints are applied to O−O, N−N, Cα−Cα, Cβ−Cβ, O−N, and Cα−O atoms to maintain the structure of predicted transmembrane β-strands (Table S7). In addition, dihedral angles in predicted transmembrane β-strands are constrained with default values (ϕ = 135.0 and ψ = −139.0) for an antiparallel β-sheet. Default values for secondary structure constraints for nontransmembrane β-strand regions are obtained from Marks et al. (11). Distance constraints were also applied to other top-ranking nonstrand−strand and loop−strand residue pairs predicted to be evolutionary coupled (Table S6). To fold TMBs, we start with applying constraints only on residues predicted to be hydrogen bonded on adjacent strands. Other EC constraints are included in steps of 10 up to L, where L is the number of amino acid residues in the protein. Location of predicted β-strands is used to filter out constraints that are considered unviable as described by Marks et al. (11). For each set of constraints only 20 models are generated (Tables S1 and S2). CNS uses a distance geometry protocol followed by simulated annealing to satisfy the input constraints (22). All folding predictions start with a fully extended polypeptide chain and a square-well potential function is used to penalize constraint violations. After annealing, a short two-stage energy minimization step is used to relax generated structures and add hydrogen bonds. To facilitate folding of the ∼120-residues-long FecA nonbarrel plug domain, models are generated starting with at least 60 constraints involving the plug domain.
Blinded Model Ranking and Comparison.
Generated models are blindly ranked based on the number of constraints satisfied in the predicted barrel pore region of the folded model as well as a measure of strand–strand torsion angles along predicted helices and between predicted β-strands as described previously by Marks et al. (11). Both values are normalized before summation. A constraint on a residue pair that is hydrogen bonded is considered satisfied if the distance between the N−O atoms is in the range 2.9 ± 0.3 Å. For FecA models with the plug domain, the number of constraints satisfied in the plug domain was also considered. In addition, in a blinded test, the predicted models are compared with the known structure based on TM score and positional rmsd values (23, 24). TM score and rmsd values are measures to assess the similarity of two-protein 3D structures. A TM score of 1 means a perfect match whereas any value above 0.5 suggests similarity at protein fold level (23) (Table 1 and Tables S1–S3).
Supplementary Data.
All supplementary data and an extended methods section are available at cbio.mskcc.org/foldingproteins/transmembrane/betabarrels/.
Note Added in Proof.
During writing of this paper, two papers (27, 28) describing the crystal structure of LptD were published. No information from those publications was used for the LptD prediction in this study and the Protein Data Bank coordinates (4Q35) and (4N4R) were not available at the time of submission of this work to biorvix (biorxiv.org/content/early/2014/06/25/006577).
Supplementary Material
Acknowledgments
We thank Thomas A. Hopf for sharing some unpublished code and discussions. D.S.M., C.S., and S.H. are supported by National Institutes of Health Award R01 GM106303. A.E. is supported by grants from the Swedish Research Council (VR-NT 2012-5046 and VR-M 2010-3555), the Foundation for Strategic Research, and Vinnova through the Vinnova-JSP Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419956112/-/DCSupplemental.
References
- 1.Freeman TC, Jr, Wimley WC. A highly accurate statistical approach for the prediction of transmembrane beta-barrels. Bioinformatics. 2010;26(16):1965–1974. doi: 10.1093/bioinformatics/btq308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai K, Fujita N, Gromiha MM, Horton P. Eukaryote-wide sequence analysis of mitochondrial β-barrel outer membrane proteins. BMC Genomics. 2011;12:79. doi: 10.1186/1471-2164-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noinaj N, et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501(7467):385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schleiff E, Becker T. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 2011;12(1):48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 5.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6(12):893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 6.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 7.Hayat S, Elofsson A. BOCTOPUS: Improved topology prediction of transmembrane β barrel proteins. Bioinformatics. 2012;28(4):516–522. doi: 10.1093/bioinformatics/btr710. [DOI] [PubMed] [Google Scholar]
- 8.Singh NK, Goodman A, Walter P, Helms V, Hayat S. TMBHMM: A frequency profile based HMM for predicting the topology of transmembrane beta barrel proteins and the exposure status of transmembrane residues. Biochim Biophys Acta. 2011;1814(5):664–670. doi: 10.1016/j.bbapap.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Hayat S, Elofsson A. Ranking models of transmembrane β-barrel proteins using Z-coordinate predictions. Bioinformatics. 2012;28(12):i90–i96. doi: 10.1093/bioinformatics/bts233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naveed H, Xu Y, Jackups R, Jr, Liang J. Predicting three-dimensional structures of transmembrane domains of β-barrel membrane proteins. J Am Chem Soc. 2012;134(3):1775–1781. doi: 10.1021/ja209895m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS ONE. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nugent T, Jones DT. Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis. Proc Natl Acad Sci USA. 2012;109(24):E1540–E1547. doi: 10.1073/pnas.1120036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149(7):1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Göbel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18(4):309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 15.Shindyalov IN, Kolchanov NA, Sander C. Can three-dimensional contacts in protein structures be predicted by analysis of correlated mutations? Protein Eng. 1994;7(3):349–358. doi: 10.1093/protein/7.3.349. [DOI] [PubMed] [Google Scholar]
- 16.Neher E. How frequent are correlated changes in families of protein sequences? Proc Natl Acad Sci USA. 1994;91(1):98–102. doi: 10.1073/pnas.91.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor WR, Hatrick K. Compensating changes in protein multiple sequence alignments. Protein Eng. 1994;7(3):341–348. doi: 10.1093/protein/7.3.341. [DOI] [PubMed] [Google Scholar]
- 18.Morcos F, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA. 2011;108(49):E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87(1):012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 20.Lapedes L, Giraud B, Jarzynski C. 2012. Using sequence alignments to predict protein structure and stability with high accuracy. arXiv:1207.2484.
- 21.Lifson S, Sander C. 1979. On the mutual recognition of strands in β-sheets. Molecular Mechanisms of Biological Recognition, ed Balaban M (Elsevier/North-Holland Biomedical, Amsterdam), pp 145–155.
- 22.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57(4):702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr A. 1976;32(5):922–923. [Google Scholar]
- 25.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26(7):889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci USA. 2010;107(12):5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511(7507):108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 28.Dong H, et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature. 2014;511(7507):52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 29.Ye J, van den Berg B. Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J. 2004;23(16):3187–3195. doi: 10.1038/sj.emboj.7600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg B. Structural basis for outer membrane sugar uptake in pseudomonads. J Biol Chem. 2012;287(49):41044–41052. doi: 10.1074/jbc.M112.408518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noinaj N, et al. Structural basis for iron piracy by pathogenic Neisseria. Nature. 2012;483(7387):53–58. doi: 10.1038/nature10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauter A, Braun V. Defined inactive FecA derivatives mutated in functional domains of the outer membrane transport and signaling protein of Escherichia coli K-12. J Bacteriol. 2004;186(16):5303–5310. doi: 10.1128/JB.186.16.5303-5310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigel NW, Ricci DP, Silhavy TJ. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(13):5151–5156. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zachariae U, et al. β-Barrel mobility underlies closure of the voltage-dependent anion channel. Structure. 2012;20(9):1540–1549. doi: 10.1016/j.str.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chng SS, et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337(6102):1665–1668. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armon A, Graur D, Ben-Tal N. ConSurf: An algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307(1):447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 37.Fsihi H, Kottwitz B, Bremer E. Single amino acid substitutions affecting the substrate specificity of the Escherichia coli K-12 nucleoside-specific Tsx channel. J Biol Chem. 1993;268(23):17495–17503. [PubMed] [Google Scholar]
- 38.Freeman TC, Jr, Wimley WC. TMBB-DB: A transmembrane β-barrel proteome database. Bioinformatics. 2012;28(19):2425–2430. doi: 10.1093/bioinformatics/bts478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci USA. 2013;110(39):15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skwark MJ, Raimondi D, Michel M, Elofsson A. Improved contact predictions using the recognition of protein like contact patterns. PLoS Comput Biol. 2014;10(11):e1003889. doi: 10.1371/journal.pcbi.1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remmert M, Biegert A, Hauser A, Söding J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 43.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 44.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.