Significance
The anaphase-promoting complex/cyclosome (APC) is a multisubunit RING E3 ubiquitin (Ub) ligase that regulates mitosis, meiosis, and numerous facets of neurobiology by targeting key regulatory proteins for Ub-mediated degradation. Despite great importance, it remains unclear how APC, or most of the other 600 RING E3s in humans, targets Ub to lysines in disordered substrates. Here, we report the structural and molecular basis for substrate ubiquitination by APC and its partner E2, UBCH10. UBCH10 is recruited to APC, activated for ubiquitination, and positioned for substrate targeting through multisite interactions with the APC cullin–RING core. We propose that many RING E3–E2 assemblies work similarly, with multisite interactions establishing specificity, harnessing ubiquitination machineries to accelerate searching for target lysines, and facilitating regulation.
Keywords: ubiquitin, anaphase-promoting complex, cullin, E3, electron microscopy
Abstract
For many E3 ligases, a mobile RING (Really Interesting New Gene) domain stimulates ubiquitin (Ub) transfer from a thioester-linked E2∼Ub intermediate to a lysine on a remotely bound disordered substrate. One such E3 is the gigantic, multisubunit 1.2-MDa anaphase-promoting complex/cyclosome (APC), which controls cell division by ubiquitinating cell cycle regulators to drive their timely degradation. Intrinsically disordered substrates are typically recruited via their KEN-box, D-box, and/or other motifs binding to APC and a coactivator such as CDH1. On the opposite side of the APC, the dynamic catalytic core contains the cullin-like subunit APC2 and its RING partner APC11, which collaborates with the E2 UBCH10 (UBE2C) to ubiquitinate substrates. However, how dynamic RING–E2∼Ub catalytic modules such as APC11–UBCH10∼Ub collide with distally tethered disordered substrates remains poorly understood. We report structural mechanisms of UBCH10 recruitment to APCCDH1 and substrate ubiquitination. Unexpectedly, in addition to binding APC11’s RING, UBCH10 is corecruited via interactions with APC2, which we visualized in a trapped complex representing an APCCDH1–UBCH10∼Ub–substrate intermediate by cryo-electron microscopy, and in isolation by X-ray crystallography. To our knowledge, this is the first structural view of APC, or any cullin–RING E3, with E2 and substrate juxtaposed, and it reveals how tripartite cullin–RING–E2 interactions establish APC’s specificity for UBCH10 and harness a flexible catalytic module to drive ubiquitination of lysines within an accessible zone. We propose that multisite interactions reduce the degrees of freedom available to dynamic RING E3–E2∼Ub catalytic modules, condense the search radius for target lysines, increase the chance of active-site collision with conformationally fluctuating substrates, and enable regulation.
Modification by ubiquitin (Ub) and Ub-like proteins (Ubls) is a central mechanism controlling eukaryotic protein properties such as half-life, localization, conformation, and interactions. Regulation is conferred by E1–E2–E3 trienzyme cascades, where E3 ligase enzymes ultimately select proteins for Ub modification. This most commonly involves E3s in the “RING” (Really Interesting New Gene) family. E3 RING domains typically recruit and activate specific E2∼Ub intermediates in which Ub’s C terminus is thioester-bonded to the E2 catalytic cysteine. In general, RING E3s stimulate Ub transfer from an E2’s active site to a lysine on a distally recruited substrate, or on a substrate-linked Ub during polyubiquitination.
The massive number of RING E3s (∼600 in humans) and E2s (∼40 in humans) underscores the significance of this pathway (1). A particularly fascinating and mysterious RING E3 is the gigantic (1.2-MDa), multisubunit anaphase-promoting complex/cyclosome (APC). APC catalyzes Ub-dependent proteolysis of numerous key cell cycle regulators to control eukaryotic cell division (2, 3). APC and an associated coactivator (CDC20 or CDH1) together recruit KEN- and/or D-box motifs in substrates such as Cyclin B, Securin, and yeast Hsl1 (4–7). Polyubiquitination is achieved by APC collaborating sequentially with distinct E2s, typically UBCH10 and UBE2S in humans (Fig. 1A) (8–13). APC11 RING’s canonical E2-binding surface activates UBCH10 to transfer Ub to APCCDH1-bound substrates (14). For some substrates, the linked Ub is then captured by a discrete APC11 RING surface for polyubiquitination by UBE2S (14, 15). Other substrates can be degraded even in the absence of UBE2S, via multiple mono-Ubs or short Ub chains generated by APC and UBCH10 alone (16).
Fig. 1.
Cryo-EM reconstruction of APCCDH1–UBCH10 poised for substrate ubiquitination. (A) APC ubiquitination mechanism. APC and the E2 UBCH10 ligate Ub directly to substrates. APC and the E2 UBE2S grow a poly-Ub chain on a premodified substrate. (B) Scheme for preparing complex representing APCCDH1–UBCH10–Ub–substrate intermediate. APC–CDH1 was mixed with three-way cross-linked UBCH10–Ub–substrate peptide (derived from Hsl1) complex for copurification. (C) Cryo-EM reconstruction of the APCCDH1–UBCH10–Ub–substrate complex, showing nearly complete secondary structure model of APC subunits as described for APCCDH1–substrate complex (29), and highlighting unassigned domain binding UBCH10 backside. (D) Close-up of cryo-EM density highlighting that the substrate (Hsl1 peptide) acceptor site approaches active site of E2 UBCH10 in a manner parallel to crystallographically defined Ubl transfer intermediates. EM density corresponds to the substrate peptide (orange) cross-linked to the catalytic Cys (green sphere) of UBCH10 (cyan). CDH1–substrate were modeled by fitting S. cerevisiae CDH1 complexed with KEN- and D-box sequences into the EM map using Chimera (33, 36), with the remainder of the peptide approximated using Coot (58). (E) Portion of structure of a non-RING E3–E2∼SUMO–substrate intermediate, highlighting the substrate RanGAP1 (orange) and its acceptor relative to E2 UBC9 (cyan), and E2 backside binding by the E3 RanBP2 (navy) (37). (F) Portion of structure representing a RING E3–E2∼NEDD8–substrate–Co E3 intermediate, highlighting the substrate CUL1 (orange) and its acceptor relative to active site of E2 UBC12 (cyan) and the RING domain from the E3 RBX1 (navy) (20).
Despite its fundamental importance, how the APC11 RING domain and E2 UBCH10 direct Ub-mediated proteolysis of APC substrates remains incompletely understood. Previous studies on other E3–E2 complexes provide some insights into features of the APC–UBCH10 catalytic assembly. For example, structures of RING E3–E2∼Ub intermediates have revealed an archetypal catalytic architecture where the RING, E2, and Ub all interact (17–19). The only two RING E3–E2–substrate complexes that have been structurally defined—RBX1–UBC12 targeting the Ubl NEDD8 to CUL1, and PRC1–UBCH5 targeting Ub to nucleosomal Histone H2A—are exceptionally specific and ligate only one Ub/Ubl molecule to a single substrate (20, 21). In both cases, the RING–E2∼Ub/Ubl catalytic modules are targeted to their specific substrate lysines in part by auxiliary buttressing subunits (a co-E3 in the case of neddylation; DNA in the case of H2A) and steric features of the folded substrates. However, these characteristics are not shared by APC or many other RING E3s during ubiquitination of numerous lysines on various disordered substrates.
Some clues may come from parallels between APC and cullin–RING ligases (CRLs), which likewise display broad substrate targeting. Canonical CRLs are nucleated by a cullin (CUL1–5) and RING protein (RBX1 or 2) (22, 23), which are homologous to the APC catalytic core subunits APC2 and APC11, respectively. Some interactions between APC2 and APC11 mirror those in canonical CRLs. Strong sequence homology indicates that the APC2–APC11 interaction involves APC2’s central α/β-domain embracing APC11’s N-terminal strand and tethering APC11’s RING like cullins bind RBX1 (24–27). CRLs are activated by modification of the cullin’s C-terminal “winged-helix B” (WHB) domain with the Ubl NEDD8, which relieves inhibitory interactions and enables the RING to swing into various positions (28). Although APC2 lacks a neddylation site, previous cryo-electron microscopy (cryo-EM) data suggested conceptually similar APC activation by CDH1 indirectly setting the APC2–APC11 catalytic core in motion (29). However, it remains unknown how a dynamically tethered cullin-bound RING–E2∼Ub complex—from canonical CRLs or APC—is harnessed to target distally bound disordered substrates. Although it was postulated that APC’s large central cavity would accommodate ubiquitination (30), details for how this might occur are elusive.
A major challenge is to visualize elements within E3–E2–substrate assemblies that enable specific E2s to ubiquitinate target lysines. Here, we address this problem using hybrid structural approaches involving cryo-EM, NMR, and X-ray crystallography, in tandem with enzymology and protein degradation assays. We report a cryo-EM reconstruction representing a human APCCDH1–UBCH10–substrate ubiquitination intermediate at 8-Å resolution that shows UBCH10’s active site juxtaposed with the target. This active conformation is achieved by APC2’s WHB and APC11’s RING corecruiting UBCH10, to present the active site to a CDH1-bound substrate. Overall, our data offer the first glimpse (to our knowledge) of how APC achieves Ub ligation and provide a framework for understanding how multisite E2 binding drives RING E3-mediated ubiquitination.
Results and Discussion
Visualizing an APC–UBCH10 Complex Poised for Substrate Modification.
To visualize a ubiquitination complex, we trapped an assembly mimicking an APCCDH1–UBCH10∼Ub–substrate intermediate by cross-linking. Briefly, we adapted our method for trapping a HECT E3 in action (31) to simultaneously three-way cross-link UBCH10’s active-site cysteine, the C terminus of a donor Ub, and a substrate. A KEN- and D-box–containing peptide derived from the well-characterized APCCDH1 substrate yeast Hsl1 was synthesized with azidohomoalanine in place of a preferred target Lys (Fig. S1). This was linked by click chemistry to a homobifunctional maleimide cross-linker. One arm was cross-linked to UBCH10 with a single catalytic Cys114, and the other to a Cys replacement for Gly75 of Ub. The three-way cross-linked UBCH10–Ub–substrate complex was assembled with APC generated by coexpressing all 14 subunits in insect cells (32) and CDH1 for structural studies (Fig. 1B).
We used cryo-EM to determine a three-dimensional reconstruction of the APCCDH1–UBCH10–Ub–substrate complex to 8-Å resolution (Fig. 1C). Comparison with prior EM maps of apo APC and an APCCDH1–Hsl1 complex lacking an E2 (29) unambiguously showed extra density in APC’s central cavity corresponding to the substrate and E2 (Fig. S2).
The overall trajectory of the peptide sequence was readily modeled—from the KEN-box, continuing to the cross-link with UBCH10’s active site, and then to the D-box—based on prior structures of coactivator complexes with KEN- and D-boxes (Fig. 1D) (33, 34). Furthermore, automated fitting of UBCH10’s isolated catalytic domain (UBCH10cat) using Chimera (35, 36) placed the active site adjacent to the acceptor from the peptide substrate (Fig. 1 C and D). APC11’s RING domain was readily observed, aligned to activate UBCH10 based on homologous RING–E2∼Ub structures (14, 17–19).
Substrate and E2 Juxtaposed via Multisite Interactions with APCCDH1.
Notably, the substrate is poised for the acceptor to approach UBCH10’s active site in a manner parallel to other Ub/Ubl transfer intermediates (20, 37, 38) (Fig. 1 D–F). This geometry is likely established by multiple constraints, including by APCCDH1 binding the substrate at two sites, CDH1 engaging the KEN-box, and CDH1 and APC10 together corecruiting the D-box (6, 7), and by APCCDH1 placement of UBCH10.
To understand mechanisms establishing the catalytic architecture, we generated a near-complete secondary structure model for the majority of the complex by docking prior structures and homology models into our map. Some subunits were modeled by placing helices based on clearer density in a cryo-EM map of APCCDH1–Hsl1 (29), although the low resolution precluded determining directions of many secondary structures or assigning sequences to them (Fig. 1C). Although the globular domain from the donor Ub would readily fit into the complex (17–19), it is not visible in the EM map, potentially for a variety of reasons (Fig. S2I). First, the cross-linker may hinder E2∼Ub interactions, despite its successful use in trapping the active HECT E3∼Ub conformation (31). Second, the position of the donor Ub may fluctuate, as was observed for another RING–E2∼Ub intermediate (39). A third intriguing possibility is raised by docking a donor Ub. The UBCH10 surfaces flanking the modeled donor Ub are predominantly acidic, and could potentially interact with a basic “initiation motif” found in some APC–UBCH10 substrates but lacking in Hsl1 (40). Future studies will be required to determine whether initiation motifs serve as priming elements that stabilize the closed UBCH10∼Ub conformation.
Unexpectedly, the EM map showed that UBCH10’s catalytic domain interacts with multiple regions from APCCDH1, not only the APC11 RING domain (Fig. 1D). Another small domain was observed to bind the so-called “backside” surface of UBCH10. This backside surface is remote from UBCH10’s catalytic Cys114, and distinct from E2 regions that contact the RING domain or thioester-linked Ub. Interestingly, the corresponding region on other E2s often binds supplemental E2-interacting, non-RING domains from E3s or other binding partners (37, 41–44) (Fig. S3). These additional E2 backside–E3 contacts are thought to secure E2–E3 interactions, establish specificity, and enable regulation. For UBCH10–APC interactions, we determined that the cullin, APC2, binds the E2 backside in a distinctive cullin–RING activation mechanism, as described below.
APC Cullin and RING Synergize for APC–UBCH10 Ubiquitination.
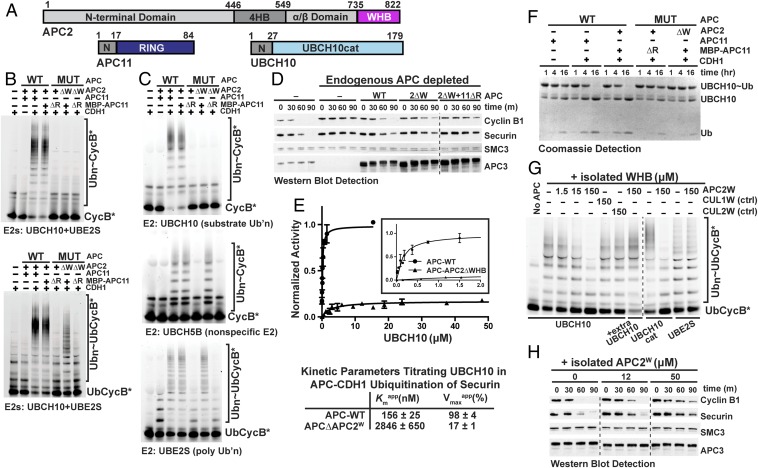
The cryo-EM map showed UBCH10’s backside-binding domain proximal to APC2’s α/β-domain, which as in other cullins both connects to the C-terminal WHB domain and also binds the RING subunit, here APC11 (Fig. 2A) (27). Although APC2’s WHB has not been identified in prior EM maps, the corresponding domain in canonical CRLs is neddylated and regulates ubiquitination (22, 23). Thus, APC2’s WHB domain was an excellent candidate for interacting with UBCH10.
Fig. 2.
Recruitment of UBCH10 to APCCDH1 via multisite binding to the APC2–APC11 cullin–RING-like catalytic core. (A) Primary domain structures of APC2 [N-terminal domain; 4-helix bundle (4HB); APC11 interacting domain (α/β domain); and winged-helix B (WHB)], APC11 [N-terminal APC2-binding domain (N); RING (navy)] and UBCH10 [N-terminal extension (N) and catalytic core domain conserved among E2s (UBCH10cat; cyan)]. (B) Roles of APC2 WHB (W) and APC11 RING (R) domains in ubiquitination by both E2s UBCH10 and UBE2S together, tested with wild type or indicated deletion mutant versions of recombinant APCCDH1. Fluorescence scans detect substrates Cyclin B N-terminal domain or a Ub-fused version. (C) Assays as in B, except testing effects of deleting APC2’s WHB and APC11’s RING on APCCDH1 activity with individual E2s (UBCH10, UBCH5B, and UBE2S). (D) Roles of APC2 WHB (W) and APC11 RING (R) domains in APC-dependent Ub-mediated proteolysis in X. laevis egg extracts. After immunodepleting endogenous APC, the wild type and mutant versions of recombinant APC were added to extracts and activated with nondegradable ∆90 CyclinB1. Degradation time course for added full-length Cyclin B1 and Securin was monitored by Western blot, as were controls for levels of APC (APC3) and of protein not ubiquitinated by APC (SMC). (E) Curve fits and kinetic parameters from titrating UBCH10 to test the role of APC2 WHB domain on catalytic efficiency of Securin ubiquitination by APCCDH1. SEM, n ≥ 3. (F) Role of APC2 WHB (W) in APC11 RING (R)-dependent intrinsic activation of an UBCH10∼Ub intermediate. The indicated APC complexes and mutants were tested for activating hydrolysis of an oxyster-linked UBCH10∼Ub complex to generate free UbcH10 and Ub, detected over time with Coomassie-stained SDS/PAGE gels. (G) Assays as in C testing whether titrating the isolated, pure WHB domain from APC2 (APC2W) inhibits ubiquitination by APCCDH1 and UBCH10, the UBCH10 catalytic domain (UBCH10cat), or UBE2S. Specificity controls for APC2 examined structurally homologous but distinct, non-APC WHB domains from CUL1 and CUL2. (H) Assays as in D but without immunodepletion testing whether titrating the indicated amounts of isolated APC2W domain inhibits endogenous APC-dependent Cyclin B1 and Securin turnover in X. laevis egg extracts.
Numerous experiments examining wild-type and mutant versions of recombinant human APC indicated that APC2’s WHB domain is essential and specific for APC–UBCH10-dependent substrate modification and degradation. A deletion mutant version of APC was generated that lacks APC2’s WHB domain, and assayed alongside controls lacking APC11’s RING domain or with both domains deleted (Fig. S4A). Experiments performed with different E2s distinguish direct ligation to substrate and formation of short Ub chains (UBCH10) (Fig. 1A), Ub chain elongation or branching (UBE2S) (Fig. 1A), or activity with a nonspecific E2 (UBCH5) that supports ubiquitination by numerous E3s including APC (2, 9). Experiments with different substrates distinguish direct modification by UBCH10 or UBCH5 (Cyclin B or Securin), or UBE2S-catalyzed polyubiquitination in the absence of a Ub-priming E2 (Ub-fused Cyclin B or Securin) (9–13).
First, we found that APC2’s WHB domain is specifically required for ubiquitination by UBCH10 and is dispensable for polyubiquitination by APC’s other partner E2, UBE2S. In reactions containing both UBCH10 (to prime substrates with Ub) and UBE2S (to build a poly-Ub chain on a substrate-linked Ub), APCCDH1 or APCCDC20 lacking APC2’s WHB domain failed to ubiquitinate Cyclin B or Securin (Fig. 2B and Fig. S4 B and C). However, Ub-fusion substrates were substantially modified, suggesting that the defect might arise during direct substrate modification by UBCH10. Second, assays with individual E2s demonstrated that deleting APC2’s WHB domain specifically impaired activity with UBCH10, but did not affect UBE2S or UBCH5 (Fig. 2C). This revealed selectivity for UBCH10 and proper APC assembly with the APC2 mutant. Importantly, control reactions with mutants lacking APC11’s RING domain showed no ubiquitination with any of the E2s (Fig. 2 B and C and Fig. S4 B and C) (14). Third, these defects were mirrored in APC substrate degradation in Xenopus laevis egg extracts. After immunodepleting endogenous APC from the extracts, degradation of Cyclin B1 and Securin was restored by wild-type recombinant human APC. However, with APC harboring the mutant lacking APC2’s WHB domain, substrate degradation was relatively slowed, with only minor further defects manifested upon also deleting the RING domain (Fig. 2D).
The critical role of APC2 in recruiting UBCH10 for RING-dependent activation is supported by several lines of evidence. First, measuring kinetic parameters for UBCH10 showed that an intact APC2 WHB domain contributes ∼100-fold to the catalytic efficiency of Securin ubiquitination. Deleting APC2’s WHB caused an 18-fold increase in the Kmapp for UBCH10, and an approximately sixfold decrease in Vmaxapp (Fig. 2E and Fig. S5A). This demonstrates a major role of APC2 in recruiting and positioning UBCH10∼Ub for catalysis. Second, the key role of APC2 is manifested in assays without substrate that monitor intrinsic ability of a RING domain to activate reactivity of an E2∼Ub intermediate (Fig. S5B) (17). Briefly, an oxyester-bonded UBCH10∼Ub complex was purified with Ub linked to a Ser replacement for UBCH10’s catalytic Cys, mixed with various versions of APC or the APC2–APC11 subcomplex, and catalysis was monitored based on hydrolysis of the E2∼Ub oxyester linkage (Fig. 2F and Fig. S5 B–E). Importantly, control reactions demonstrated that hydrolysis depends on a known catalytic element: the reaction is impaired in the absence of the coactivator CDH1, or by CDH1 mutations known to hinder APC activation (45) (Fig. 2F and Fig. S5C).
Activation of UBCH10∼Ub depends on both the APC2 WHB and APC11 RING domains being present in a single complex. A stoichiometric amount of purified APC2–APC11 stimulated hydrolysis of oxyester-linked UBCH10∼Ub on the timescale observed for another RING E3–E2 complex (17) (Fig. S5D). This depended on intact APC2 WHB and APC11 RING domains, and was not recapitulated by adding the isolated domains alone or together, even in fivefold molar excess (Fig. S5D). Moreover, these features are recapitulated by APCCDH1 activation of UBCH10∼Ub, albeit over a longer time course due to our only achieving a 1:20 ratio of APCCDH1:UBCH10∼Ub (Fig. 2F and Fig. S5 C and E). Importantly, this assay also enabled our validating that the three-way cross-linked UBCH10–Ub–substrate complex properly mimics UBCH10 binding to APCCDH1, as it inhibits this APC-catalyzed reaction (Fig. S5E).
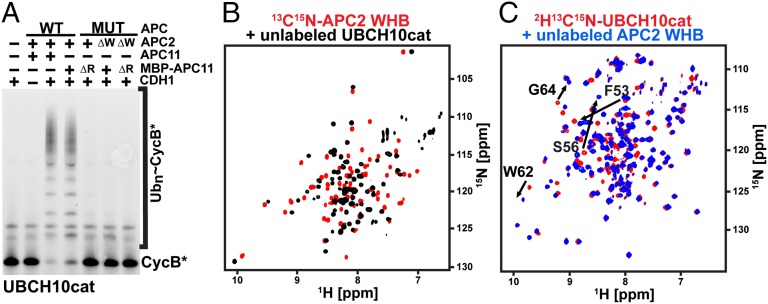
Direct Interactions Between APC2 WHB and the UBCH10 Catalytic E2 Domain.
We mapped the binding site of APC2’s WHB domain to UBCH10’s conserved E2 catalytic domain. First, as with full-length UBCH10, APCCDH1-dependent ubiquitination with UBCH10cat also depends on an intact APC2 WHB domain (Fig. 3A). Second, NMR experiments demonstrated direct interactions between the isolated UBCH10cat and APC2 WHB domain (APC2W) domains. Adding unlabeled APC2W perturbed chemical shifts in the 15N-1H transverse relaxation optimized spectroscopy (TROSY) spectrum of 15N-labeled UBCH10cat, and vice versa (Fig. 3 B and C). Third, adding the isolated APC2W in trans inhibited UBCH10-dependent substrate ubiquitination by wild-type APCCDH1 with a Kiapp of 19 μM (Fig. 2G and Fig. S4 D–F). This inhibition was specific to APC2’s WHB domain and was not observed with homologous domains from other cullin proteins (CUL1W or CUL2W). Similarly, adding isolated APC2W domain to Xenopus laevis egg extracts inhibited substrate degradation catalyzed by endogenous APC (Fig. 2H).
Fig. 3.
APC2W interacts with the catalytic core domain of UBCH10 (UBCH10cat). (A) Fluorescence scan of assay testing effects of deleting APC2’s WHB domain (ΔW) and/or the APC11’s RING domain (ΔR) on fluorescent Cyclin B (CycB*) ubiquitination by APCCDH1 with isolated catalytic domain from UBCH10 (UBCH10cat). (B) TROSY spectra for 0.5 mM 13C,15N-APC2W alone (red) or mixed with 1 mM UBCH10cat (black) showing specific chemical shift perturbations. (C) TROSY spectra for 0.5 mM 13C,15N-UBCH10cat alone (red) or mixed with 2 mM APC2W (black) showing specific chemical shift perturbations.
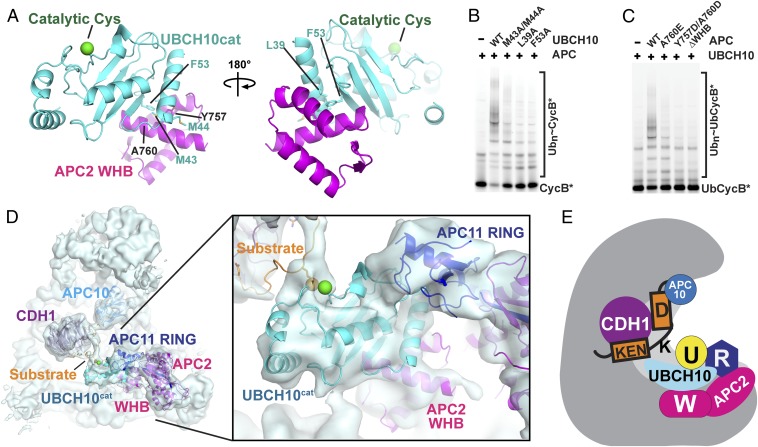
Crystal Structure of APC2W–UBCH10cat.
To visualize the interactions in atomic detail, we determined the crystal structure of a human APC2W–UBCH10cat complex at 1.8-Å resolution (Fig. 4A, Fig. S6A, and Table S1). The interaction involves an extensive hydrophobic surface emanating from the first and second helices of APC2’s winged-helix fold binding a complementary surface from UBCH10cat involving α-helix-1, the β1- and β2-strands, and the β1/β2-loop. The interface is consistent with NMR chemical shift perturbation data and is largely conserved (Fig. S7). Importantly, mutating UBCH10’s APC2-binding residues and vice versa decreases APCCDH1-catalyzed substrate ubiquitination (Fig. 4 B and C and Figs. S4A and S6 B and C).
Fig. 4.
Structural basis for distinctive cullin–RING mechanism of substrate ubiquitination by APCCDH1 and UBCH10. (A) Crystal structure of complex between APC2 WHB (magenta)–UBCH10cat (cyan) domains. APC2 WHB binds UBCH10’s backside, remote from the catalytic cysteine (green sphere). Interacting residues mutated in B are shown as sticks. (B) Effects of UBCH10 point mutations in residues contacting APC2 on APCCDH1–UBCH10-dependent ubiquitination of CycB*. (C) Effects of APC2 point mutations in residues contacting UBCH10 on APCCDH1–UBCH10-dependent ubiquitination of UbCycB*. (D) Cryo-EM reconstruction of the APCCDH1–UBCH10–Ub–substrate complex, showing cullin and RING mechanism for juxtaposing UBCH10 active site with substrate. APC2W–UBCH10cat structure docked with APC11 RING domain was fit into the EM map using Chimera (36). (E) Model for ubiquitination by APCCDH1 and UBCH10. UBCH10∼Ub is recruited via multisite interactions with the APC2–APC11 catalytic core. Substrate is corecruited at a distance, via KEN- and D-box binding to CDH1 and the APC core. Multisite E2 and substrate interactions establish specificity, reduce degrees of freedom for flexibly tethered reactants to promote catalysis, and enable regulation.
Visualizing APC’s Distinctive Cullin–RING Mechanism.
Fitting the APC2W–UBCH10cat structure into the EM map of the APCCDH1–UBCH10–Ub–substrate complex using Chimera (36) confirmed the placement of the E2 catalytic domain, and allowed visualizing the key cullin–RING elements coordinating UBCH10’s catalytic domain and substrate for ubiquitination (Fig. 4D). Support for the positioning of the APC2–APC11–UBCH10 catalytic core comes from proximity of the docked APC2 WHB and APC11 RING domains to their connection points in the modeled APC2–APC11 interaction α/β-domain. Taken together, the structural data showed how APC’s catalytic module is restrained by multiple contacts, with APC2’s WHB and APC11’s RING resembling two sides of a clamp coengaging UBCH10.
Implications for Regulation of APC: Modulation of the APC2–APC11 Cullin–RING Core for Different Functions.
To gain insights into regulation of APC’s distinctive cullin–RING ligase activity, we compared EM reconstructions and structures of various APC and E2 complexes, which shed light on how ubiquitination is achieved, and on how activity is attenuated during interphase (Fig. S2). APC activity requires association with a coactivator subunit. The coactivators CDC20 and CDH1 not only bind substrates, but they also activate catalysis (4, 5, 45, 46). Prior EM studies showed that coactivator binding triggers substantial reorientation of the APC2–APC11 catalytic core and several flanking subunits (APC1, APC4, APC5) (29, 30). Our data are consistent with prior suggestions that CDH1 binding activates catalysis by transmitting conformational changes to the catalytic core. Indeed, we found that CDH1 activates intrinsic catalysis for human APC, as adding CDH1 massively stimulated hydrolysis of the UBCH10∼Ub oxyester-linked complex (Fig. 2F). Accordingly, the APCCDH1–UBCH10–Ub–substrate architecture differs from apo-APC and globally resembles that of the APCCDH1–Hsl1 complex, with similar locations for APC2’s N-terminal domain, APC1, APC4, and APC5 (Figs. S2 and S8A).
One striking difference is that the catalytic core is visible in the APCCDH1–UBCH10–Ub–substrate complex. This contrasts with the structure lacking the E2, or its linkage to substrate and Ub; APC11’s RING domain, the APC11–APC2 interaction α/β-domain, and APC2’s WHB domain were not visible in the prior refined map of APCCDH1–Hsl1 (29). The prior data were interpreted to mean that the positions of the activated APC11 RING and its associated UBCH10∼Ub intermediate fluctuate and that various substrates and/or lysines could be ubiquitinated through essentially infinite, varying catalytic geometries (29). However, our structural data suggest the alternative possibility that multisite interactions with UBCH10 harness the flexible catalytic assembly to direct Ub ligation to the substrate.
Our data also extend insights into how substrate ubiquitination is blocked by the interphase inhibitor, EMI1–SKP1. Prior studies showed how EMI1–SKP1 blocks D-box substrate binding to APCCDH1, as well as Ub chain elongation by UBE2S (47–49). Superimposing the EM maps, along with our identification of the cullin–RING interacting, WHB, and RING domains of APC2 and APC11 in the cryo-EM reconstruction, shows EMI1–SKP1 binding this region (Fig. S2). This would block access to UBCH10. Indeed, EMI1–SKP1 inhibits APCCDH1-dependent hydrolysis of the oxyester-linked UBCH10∼Ub complex (Fig. S5E). These data likely also explain how EMI1’s meiotic paralog, EMI2, inhibits APC activity with UBCH10 (50).
Implications for Regulation of UBCH10.
UBCH10’s unique N-terminal extension has been reported to restrict activity through various mechanisms, including attenuating UBCH10 charging by E1, confining activity to APC substrates, and mediating APC-dependent UBCH10 autoubiquitination in the absence of substrates (51–54). Consistent with the proposed competition between APC substrates and UBCH10’s N terminus for access to its active site (51), it was not possible for us to resolve this region in the map of the APCCDH1–UBCH10–Ub–substrate complex. Although the N terminus of UBCH10’s catalytic domain is localized near the junction of APC1, APC2, and APC11 (Fig. S8B), future studies will be required to determine whether UBCH10’s N terminus also modulates catalysis by contacting this region or by other means.
APC Mechanism, Target Lysine Prioritization, and Implications for RING E3-Catalyzed Substrate Ubiquitination.
Understanding how a large cohort of RING E3–E2 complexes target numerous lysines on their many disordered substrates has been hampered by lack of structural data. A complication of existing structural models for APC, CRLs, and many other E3s has been an enormous gap separating the substrate-binding site and the active site on the RING-bound E2. Two different mechanisms have been proposed for how the RING–E2∼Ub active site and target lysines could become juxtaposed. At one extreme, the E3 would have a rigid architecture that holds the RING–E2 assembly in a fixed orientation, and flexible substrates would reach across the gap to present their target lysines to the active site on the opposite side of the complex (22, 23, 27). The other extreme model suggests that both the RING–E2∼Ub intermediate and the substrate are flexibly tethered to the E3 (22, 23, 29). In this scenario, ubiquitination requires the moving active site and dynamic substrate to somehow collide.
Insights into how the catalytic RING–E2∼Ub intermediate could be directed to substrates may be gleaned by our visualization of an APCCDH1–UBCH10–Ub–substrate complex (Figs. 1 and 4), which to our knowledge shows for the first time the APC in a conformation compatible with substrate modification. The structural data, taken together with recent data for related CRLs (20), supports a hybrid mechanism for substrate targeting by dynamic E3s with broad substrate profiles. We postulate that mobility of the RING domain enables regulation but that harnessing the RING into different relative positions defines various E3 catalytic activities, differential interactions with partner proteins, and inhibition.
It seems that the bipartite binding to UBCH10 both establishes specificity for the E2 that directly ligates Ub to substrates and also reduces the degrees of freedom available to the catalytic assembly. Substrates also often bind APC at multiple sites, such as via KEN- and D-boxes as in our structure, or through other APC/coactivator-binding motifs that are being identified at a rapid pace (14, 55). Multisite binding by the E2, either alone or in combination with the substrate, could harness the flexible assemblies to reduce the entropy of the search space and increase the probability of catalytic encounter (Fig. 4E).
Considering that apo- and EMI-bound forms of APC are inactive, comparing EM data for the APCCDH1–UBCH10–Ub–substrate complex suggested the APC11 RING and APC2 WHB domains may be less accessible to UBCH10 in apo-APC (29) and the catalytic core rigidified and blocked by EMI1 in interphase until its degradation (Fig. S2) (48). In the absence of EMI1, CDH1-binding sets the APC2–APC11 assembly in motion (29), although our data show that APC2’s WHB domain and the APC11 together clasping UBCH10 is essential to activate ubiquitination (Fig. 4D). Following Ub transfer to the substrate, UBCH10 must be released for another mono-Ub transfer, because interactions with APC2’s WHB domain prevent UBCH10 charging by E1 (Fig. S9). Although APC11’s RING is closely associated with the APC11–APC2 interaction domain as in other APC structures (Fig. 4D and Fig. S2), density connecting the WHB to the remainder of APC2 is lacking. We speculate that mobility of the WHB serves to open and close the clamp on UBCH10. This could be analogous to motions in canonical CRLs, where the helix connecting the WHB to the rest of the cullin can rotate nearly 180° to move the WHB into various positions for different CRL functions (20, 27, 28).
At this point, we do not know whether the catalytic module is flexibly restrained or in a fixed architecture for substrate ubiquitination. Nonetheless, several observations are consistent with restriction of the orientations available to the active site. First, during our attempts to design an optimal Hsl1 construct for structural studies, we characterized APCCDH1–UBCH10-dependent multimonoubiquitination using a lysineless Ub (UbK0). Notably, the number of Ubs transferred to the different substrates correlates with the number of lysines that could access the active site in the structurally observed APCCDH1–UBCH10 position (Fig. S8). Second, it is compelling that the CDH1-bound substrate approaches UBCH10’s active site from the catalytic orientation that was crystallographically defined for other E2∼Ubl–substrate intermediates (20, 37). In principle the cross-link between UBCH10 and substrate would not physically dictate their angle of encounter if the APC-bound UBCH10 were free to rotate into numerous positions (Fig. 1 D–F). Third, although its connection to the remainder of APC2 seems flexible, the location of APC2’s WHB domain appears at least somewhat constrained through contacts to APC1 on one side and the APC4/APC5/APC15 region on the other (Fig. S8). Understanding potential roles of these connections will require future high-resolution structural studies identifying the key residues involved.
Multisite E2 binding is emerging as a general feature directing RING E3s to substrates. Although auxiliary subunits served this role in the two other structurally characterized E3–E2–substrate complexes (20, 21), there are indications that many RING E3s could function much like APC. To date, these have only been structurally characterized in isolation, although many E3s do display non-RING domains that like APC2’s WHB bind the backsides of their specific partner E2s (37, 41–44) (Fig. S3). Likewise, many E2s display N- or C-terminal extensions beyond their catalytic core domains that serve as additional E3 binding elements. These include highly charged tails from UBE2S and CDC34, which bind the catalytic cores of APC and CRLs, respectively, to mediate polyubiquitination (11–15, 56). Although these E2 tails have been implicated in mediating dynamic yet high-affinity E2–E3 interactions, and enabling E1 charging of E2s while bound to their E3s, it is plausible that E2–tail–E3 interactions also serve to stabilize distinct cullin–RING conformations for Ub chain elongation. Indeed, like APC, CRLs are also under conformational control. Numerous elements position the mobile RING for NEDD8 ligation to a canonical cullin, and after neddylation the catalytic core displays conformational flexibility (23). We speculate that future studies will reveal exciting mechanisms—potentially involving roles for the neddylated cullin WHB domain akin to APC2’s WHB—for how the dynamic CRL catalytic core is directed to specific substrate lysines. It seems likely that multisite E2–E3 interactions will prove to be generally important in enabling regulation, establishing specificity, and directing catalytic conformations of highly dynamic RING E3s.
Materials and Methods
Details can be found in SI Materials and Methods. Proteins described are human, except Hsl1 from Saccharomyces cerevisiae. Recombinant APC, CDH1, the APC2–APC11 subcomplex, and E1 were expressed in insect cells, and all other proteins in Escherichia coli as described previously (14, 57). Methods for generating the three-way cross-linked UbcH10–Ub–substrate complex were adapted from ref. 31, and for assembling the complex with APC for cryo-EM, from ref. 14. Ubiquitination reactions, NMR, cryo-EM, and substrate degradation assays in X. laevis egg extracts were largely performed as described previously (14, 57). Assays monitoring hydrolysis of the oxyester-linked UBCH10∼Ub complex were performed by purifying UBCH10∼Ub and then mixing with different versions of the APC2–APC11 subcomplex or APC. Persistence/hydrolysis of the oxyester-bonded UBCH10∼Ub was monitored as a function of time by SDS/PAGE stained with Coomassie blue. For X-ray crystallography, UBCH10cat and APC2W were purified individually, mixed at a 1:1 ratio, and crystallized by the hanging-drop vapor diffusion method. Crystallography data were collected at Advanced Photon Source (APS) Northeastern Collaborative Access Team (NECAT) ID-24-C.
Supplementary Material
Acknowledgments
We thank G. Petzold, P. Neumann, I. Davidson, S. Bozeman, D. W. Miller, D. J. Miller, J. Bollinger, R. Kriwacki, K. Kavdia, J. Peng, and R. Pappu for assistance and helpful discussions. For funding, we thank the Jane Coffin Childs Foundation (N.G.B.); Japan Society for the Promotion of Science (M.Y.); American Cancer Society Research Scholar Grant RSG CDD-120969 (to N.F.); Boehringer Ingelheim, the Laura Bassi Centre for Optimized Structural Studies, European Union Seventh Framework Programme Grant 227764 MitoSys, and the Austrian Research Fund (to J.-M.P.); Deutsche Forschungsgemeinschaft Sonderforschungsbereich 860 (to H.S.); and American Lebanese Syrian Associated Charities, NIH Grants R37GM065930 and P30CA021765, and Howard Hughes Medical Institute (to B.A.S.). B.A.S. is an investigator of the Howard Hughes Medical Institute. NECAT and APS were supported by NIH Grant P41 GM103403 and Department of Energy Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography (coordinates and structure factors), NMR (chemical shifts), and electron microscropy (map) data have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4YII.pdb), Biological Magnetic Resonance Data Bank, www.bmrb.wisc.edu (BMRB codes 26526, 26527, 26528, and 26529), and EMDataBank, emdatabank.org (EMD ID code 2929), respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504161112/-/DCSupplemental.
References
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.King RW, et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81(2):279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 3.Sudakin V, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6(2):185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15(18):2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18(5):543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 6.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470(7333):274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buschhorn BA, et al. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18(1):6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aristarkhov A, et al. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci USA. 1996;93(9):4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6(4):455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130(1):127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Garnett MJ, et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11(11):1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williamson A, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106(43):18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu T, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci USA. 2010;107(4):1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown NG, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol Cell. 2014;56(2):246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol Cell. 2014;56(2):232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Dimova NV, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14(2):168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489(7414):115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19(9):876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruneda JN, et al. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47(6):933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott DC, et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157(7):1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514(7524):591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20(6):714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duda DM, et al. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2011;21(2):257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, et al. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279(5354):1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 25.Tang Z, et al. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell. 2001;12(12):3839–3851. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zachariae W, et al. Mass spectrometric analysis of the anaphase-promoting complex from yeast: Identification of a subunit related to cullins. Science. 1998;279(5354):1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 27.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 28.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell. 2008;134(6):995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513(7518):388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20(6):867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19(11):1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He J, et al. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol Cell. 2013;50(5):649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian W, et al. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci USA. 2012;109(45):18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Hwang WC, Basavappa R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. J Biol Chem. 2002;277(24):21913–21921. doi: 10.1074/jbc.M109398200. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13(10):915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 39.Soss SE, Klevit RE, Chazin WJ. Activation of UbcH5c∼Ub is the result of a shift in interdomain motions of the conjugate bound to U-box E3 ligase E4B. Biochemistry. 2013;52(17):2991–2999. doi: 10.1021/bi3015949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson A, et al. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol Cell. 2011;42(6):744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21(6):873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Das R, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34(6):674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci USA. 2011;108(14):5590–5595. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzger MB, et al. A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol Cell. 2013;50(4):516–527. doi: 10.1016/j.molcel.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol Cell. 2008;32(4):576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 46.Van Voorhis VA, Morgan DO. Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency. Curr Biol. 2014;24(13):1556–1562. doi: 10.1016/j.cub.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JJ, et al. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20(17):2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frye JJ, et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol. 2013;20(7):827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat Cell Biol. 2013;15(7):797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shoji S, et al. The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation. FEBS Open Bio. 2014;4:689–703. doi: 10.1016/j.fob.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432(7017):588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 52.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124(1):89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 53.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31(4):544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 2008;15(3):280–287. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- 55.Di Fiore B, et al. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell. 2015;32(3):358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139(5):957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi M, et al. Structure of an APC3-APC16 complex: Insights into assembly of the anaphase-promoting complex/cyclosome. J Mol Biol. 2015;427(8):1748–1764. doi: 10.1016/j.jmb.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.