Significance
African henipaviruses (HNVs) may be responsible for the misdiagnosis of encephalitis-associated outbreaks of malaria. Host-cell infection by an African HNV relies on the initial interaction between a virally encoded surface glycoprotein and a host-cell receptor. Here, we provide a structural description of how a bat-borne Ghanaian HNV hijacks human ephrinB2 to facilitate cross-species transmission. We demonstrate that, although the Ghanian HNV is sequence dissimilar (<30% sequence identity) and displays a receptor-binding scaffold that differs significantly in structure to pathogenic HNV relatives from Asia, it adopts a nearly identical primary ephrinB2 binding mode. These data provide a molecular-level explanation for previously observed spillover of African HNVs into human populations.
Keywords: emerging virus, viral attachment, glycoprotein, henipavirus, structure
Abstract
The discovery of African henipaviruses (HNVs) related to pathogenic Hendra virus (HeV) and Nipah virus (NiV) from Southeast Asia and Australia presents an open-ended health risk. Cell receptor use by emerging African HNVs at the stage of host-cell entry is a key parameter when considering the potential for spillover and infection of human populations. The attachment glycoprotein from a Ghanaian bat isolate (GhV-G) exhibits <30% sequence identity with Asiatic NiV-G/HeV-G. Here, through functional and structural analysis of GhV-G, we show how this African HNV targets the same human cell-surface receptor (ephrinB2) as the Asiatic HNVs. We first characterized this virus−receptor interaction crystallographically. Compared with extant HNV-G–ephrinB2 structures, there was significant structural variation in the six-bladed β-propeller scaffold of the GhV-G receptor-binding domain, but not the Greek key fold of the bound ephrinB2. Analysis revealed a surprisingly conserved mode of ephrinB2 interaction that reflects an ongoing evolutionary constraint among geographically distal and phylogenetically divergent HNVs to maintain the functionality of ephrinB2 recognition during virus–host entry. Interestingly, unlike NiV-G/HeV-G, we could not detect binding of GhV-G to ephrinB3. Comparative structure–function analysis further revealed several distinguishing features of HNV-G function: a secondary ephrinB2 interaction site that contributes to more efficient ephrinB2-mediated entry in NiV-G relative to GhV-G and cognate residues at the very C terminus of GhV-G (absent in Asiatic HNV-Gs) that are vital for efficient receptor-induced fusion, but not receptor binding per se. These data provide molecular-level details for evaluating the likelihood of African HNVs to spill over into human populations.
The emergence of negative-sense, single-stranded RNA viruses belonging to the genus Henipavirus, family Paramyxoviridae, epitomizes the increasing threat of zoonotic viruses to human health (1). Since the discovery of the highly pathogenic Nipah virus (NiV) and Hendra virus (HeV) in the 1990s, >20 henipaviruses (HNVs) have been detected throughout Africa, Asia, Australia, and Central America (2, 3). NiV is the prototypic member of this group and enzoonitically resides (>50% seroprevalence in some instances) in Old World fruit bat populations throughout Australasia (4). Zoonotic transfer of NiV to human populations from these natural reservoirs (5), sometimes through an animal intermediary such as pigs (6), leads to rapid-onset encephalitis with case-fatality rates >90% (1). Following zoonosis, some cases of person-to-person transmission have been observed (7, 8). Although bats are the predominant host reservoir, a putative henipa-like virus (HNLV), Mojiang virus (MojV), associated with severe pneumonia and three case fatalities, has also been isolated from rats in China (9). The extreme pathogenicity and potential for misuse has led to the designation of NiV and HeV as high-priority agents that require handling under biosafety-level-four conditions.
The detection of 19 distinct clades of HNV in Africa (2, 10, 11) correlates with the broad geographic distribution and wide-ranging migrational patterns of the fruit bat host reservoir, Eidolon helvum (11). The remarkably high seroprevalence (∼40%) of HNV cross-reactive antibodies and the localization of many E. helvum communities near African towns and cities underscores the potential risk of spillover events into human populations (11, 12). Indeed, NiV cross-neutralizing antibodies have been detected in the sera of humans living in Cameroon (12). That these antibodies were found exclusively in individuals at high risk for zoonotic transmission, such as those that slaughter bats for bushmeat consumption and sale, suggests that such spillover events can occur. Whether or not African HNVs are as pathogenic to humans as NiV or HeV remains to be determined. Although it has also been suggested that these viruses may be the causative agent of misdiagnosed encephalitis-associated malaria (2, 13, 14), it is likely that the divergent clades of African HNVs are also diverse in their pathogenic potential.
HNV entry into a host cell is a pH-independent process orchestrated by two membrane-anchored glycoproteins, HNV-G and -F (15). These viral glycoproteins interdependently facilitate cellular attachment and fusion, whereby receptor recognition by HNV-G at the cell surface triggers rearrangements in the HNV-F fusion glycoprotein (16). HNV-G is an oligomeric membrane protein, consisting of a short N-terminal cytoplasmic tail, a transmembrane region, an oligomerization-inducing stalk region, and a receptor-binding C-terminal six-bladed β-propeller. Identification of the ubiquitously expressed cell-surface signaling glycoproteins, ephrinB2 and ephrinB3, as functional receptors used during viral attachment by NiV and HeV has been key to understanding the broad tissue tropism of these viruses (16–21). Structural investigations of these ephrins in complex with NiV- and HeV-G have revealed the molecular determinants for host-cell recognition and zoonosis (22–26).
In contrast to the wealth of available NiV and HeV genome sequences, only one African HNV has been sequenced to entirety, but it has not yet been isolated (2, 27). The sequence of this putative HNV (Gh-M74a; termed here as GhV) was derived from a bat in Ghana and is genetically distinct from Asiatic HeV and NiV (2). In contrast to NiV- and HeV-G, which are genetically quite similar (80% sequence identity), the putative GhV attachment glycoprotein from this virus, GhV-G, exhibits very limited sequence identity (<30%) with its Asiatic counterparts. Despite this genetic distance, ephrinB2 has been suggested as a functional interaction partner for this virus (27, 28). The conserved use of this receptor by GhV-G and Asiatic HNVs supports a general mechanism for HNV zoonosis in human populations. The likelihood of zoonotic transmission and the pathogenicity of such zoonotic viruses may depend, at least in part, on what adaptations are necessary for efficient use of the host receptor(s).
Here, we determined the molecular basis for the interaction between GhV-G and ephrinB2 by X-ray crystallographic analysis. Despite the varied architecture of the henipaviral β-propeller scaffold between GhV-G and Asiatic HNV-Gs, we observed a highly conserved mode of ephrinB2 engagement. However, we also identify a secondary ephrinB2 interaction site that contributes to the more efficient receptor-mediated entry exhibited by NiV-G relative to GhV-G. These data verify a conserved HNV cell-attachment strategy for African and pathogenic Asiatic HNVs and establish a mechanism by which humans may be susceptible to African HNV infection.
Results and Discussion
GhV-G Is Antigenically Distant from Asiatic Clades HNV-Gs.
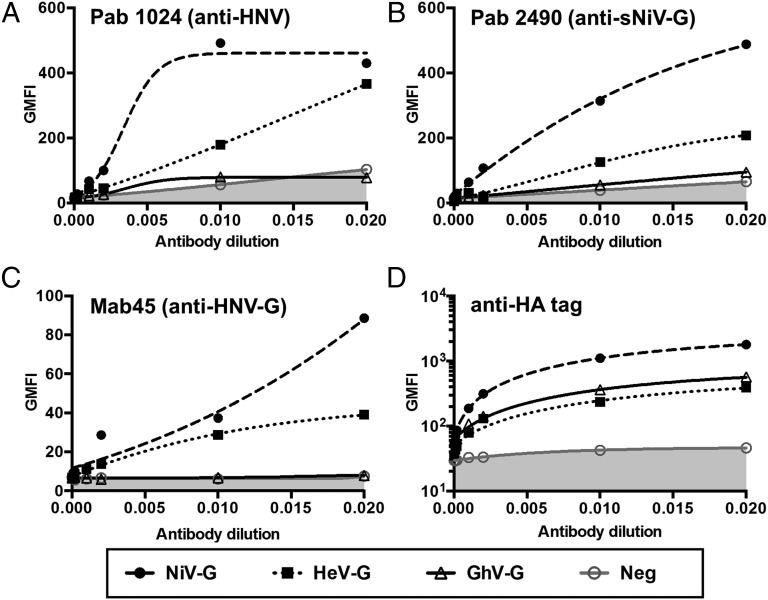
Both NiV and HeV use ephrinB2 as entry receptors. Antibodies against one can exhibit heterologous cross-reactivity against the other. Because GhV-G binds to ephrinB2 (27, 28), we asked whether GhV-G is also antigenically related to HeV- and NiV-G. We transfected C-terminally HA-tagged NiV-, HeV-, and GhV-G into 293T cells and detected relative cell-surface expression with a panel of polyclonal and monoclonal antibodies. All of these antibodies bound to NiV- and HeV-G with varying degrees of cross-reactivity (Fig. 1 A–C). However, they showed little, if any, cross-reactivity with GhV-G, even though GhV-G was expressed at least as well as HeV-G (Fig. 1D). Despite this antigenic distance and the low sequence identity between the Asiatic and African clades of HNVs, the common use of ephrinB2 suggest a conserved mode of receptor interactions (12). To gain molecular-level insights into how NiV-G/HeV-G and GhV-G are antigenically dissimilar yet use the same receptor, we determined the crystal structure of GhV-G in complex with ephrinB2.
Fig. 1.
GhV-G is antigenically distant from Asiatic clade HNV-Gs. Representative polyclonal and monoclonal antibodies made against NiV envelope proteins were tested for cross-reactivity against HeV- and GhV-G. The 293T cells were transfected with C-terminally HA-tagged NiV-, HeV-, and GhV-G expression vectors. pCAGS-transfected cells served as vector controls for nonspecific staining by primary and/or secondary antibodies. At 16 h after transfection, equivalent samples were stained with the serial dilutions of the indicated antibodies and analyzed by FACS. (A and B) Rabbit polyclonal antibodies made by genetic immunization with Nipah viral-like particles (NiVLPs; NiV-M +F +G) and soluble NiV-G (A) or soluble NiV-G alone (B). (C) Mab45 is a well-characterized rabbit monoclonal antibody that recognizes a conserved receptor-binding enhanced (RBE) epitope in NiV- and HeV-G (46). (D) Staining with rabbit polyclonal anti-HA antibodies shows relative surface expression of NiV-G > HeV-G ∼ GhV-G.
Structure of the GhV-G–EphrinB2 Complex.
The crystal structure of the receptor-binding domain of GhV-G was solved in complex with the ectodomain of human ephrinB2 by molecular replacement to 1.7-Å resolution using the structure of NiV-G bound to ephrinB2 [Protein Data Bank (PDB) ID code 2VSM (22)] as the search model (Table 1). Two nearly identical GhV-G–ephrinB2 complexes were observed in the asymmetric unit [0.3-Å root-mean-square deviation (rmsd) (29) over 552 equivalent Cα atoms], and no molecular contacts consistent with the formation of higher-order dimers or tetramers, such as those present in HeV-G (24) and many paramyxovirus HNs, were observed (30–34). The paucity of higher-order oligomers in the GhV-G–ephrinB2 crystal is consistent with the observation that the stalk region, which was absent from the crystallized GhV-G construct, is predominantly responsible for driving dimerization and tetramerization of paramyxoviral attachment glycoproteins (22, 35–40).
Table 1.
Crystallographic data and refinement statistics
| Data collection | |
| Beamline | Diamond I04 |
| Resolution range, Å | 100.0–1.70 (1.74–1.70)* |
| Space group | I222 |
| Cell dimensions | |
| a, b, c, Å | 127.2, 152.5, 163.3 |
| α, β, γ, ° | 90.0, 90.0, 120.0 |
| Wavelength, Å | 0.979 |
| Unique reflections | 172,833 (12,602) |
| Completeness, % | 99.8 (99.3) |
| Rmerge,* % | 4.2 (88.4) |
| I/σI | 18.2 (1.8) |
| Ave. redundancy | 4.9 (4.8) |
| CC1/2† | 0.999 (0.697) |
| Refinement | |
| Resolution range, Å | 100.0–1.70 (1.74–1.70) |
| Number of reflections | 162,840 (11,797) |
| Rwork,‡ % | 17.5 |
| Rfree,§ % | 19.9 |
| rmsd | |
| Bonds, Å | 0.010 |
| Angles, ° | 1.5 |
| Complexes per a.s.u. | 2 |
| Atoms per a.s.u., protein/sugar/water | 9,035/154/1,020 |
| Average B factors, Å2, protein/sugar/water | 33.6/55.9/42.4 |
| Ramachandran plot quality | |
| Most favored region, % | 97.1 |
| Allowed region, % | 2.9 |
Parentheses refer to the relevant outer resolution shell. a.s.u., asymmetric unit.
Rmerge = Σhkl Σi|I(hkl;i) − <I(hkl)>|/Σhkl ΣiI(hkl;i), where I(hkl;i) is the intensity of an individual measurement and <I(hkl)> is the average intensity of multiple observations.
CC1/2 is defined by Karplus and Diederichs in ref. 58.
Rwork = Σhkl||Fobs| − k|Fcalc||/Σhkl |Fobs|.
Rfree is calculated as for Rwork, but using only 5% of the data which were sequestered before refinement.
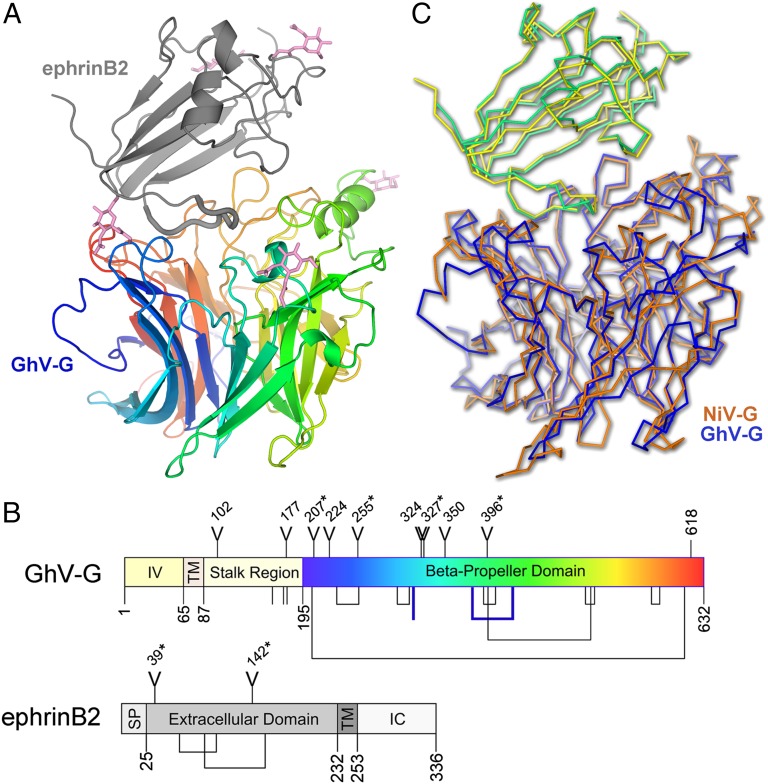
Characteristic of attachment (G), hemagglutinin (H), and hemagglutinin–neuraminidase (HN) glycoproteins from the Paramyxovirinae (41), the receptor-binding domain of GhV-G displays a classical six-bladed β-propeller topology, with four antiparallel β-strands per blade forming a toroidal arrangement around a central axis (Fig. 2A). The fold is stabilized by eight disulfide bonds, seven of which are present in the β-propeller scaffold of NiV- and HeV-G (Fig. 2B and SI Appendix, Fig. S1). The additional disulfide bond not present in NiV- or HeV-G stabilizes the third blade of the β-propeller between residues Cys-378 and -422. An unpaired cysteine (Cys-314) is also located near the solvent-accessible surface of the second blade of the β-propeller, distal from the receptor-binding site and previously predicted regions of oligomerization (24). The unpaired C314 appears stable and is partially buried under a GlcNAc moiety attached to Asn-327 (SI Appendix, Fig. S2). Mutagenesis of this “aberrant” free cysteine to alanine had no effect on cell surface expression or ephrinB2 binding (SI Appendix, Fig. S3). Curiously, a more homologous C314M mutant was intracellularly retained and not expressed at the cell surface, suggesting that the amino acid at this position contributes to proper folding of GhV-G and is not completely tolerant to substitutions.
Fig. 2.
GhV-G and recognition of ephrinB2. (A) Crystal structure of GhV-G in complex with ephrinB2. The six-bladed β-propeller of GhV-G is shown as a rainbow ramped from blue (N terminus) to red (C terminus). The Greek-key fold of ephrinB2 is shown as a gray cartoon. Asparagine-linked N-acetylglucosamine moieties are shown as pink sticks. (B) Domain schematic of GhV-G (Upper) and ephrinB2 (Lower). IV, intravirion domain; TM, transmembrane domain; SP, signal peptide; IC, intracellular domain. Y-shaped symbols designate N-linked glycosylation sites; those sites observed to be occupied in the crystal structure are marked with an asterisk. Lines and connected lines below each diagram correspond to cysteines and disulfide bonds, respectively. A cysteine and a disulfide bond present in GhV-G, but not conserved in NiV- and HeV-G, are highlighted in bold blue. Only cysteines contained in the mature protein and exposed to the oxidizing extracellular environment are annotated. Residues 618–632 in GhV-G were disordered and not built in the crystal structure. Diagrams were produced by using DOG software (Version 2.0; ref. 72). (C) Structural comparison of African and Asiatic HNV-Gs in complex with human ephrinB2. Structures of GhV-G–ephrinB2 (GhV-G, blue; ephrinB2, yellow) and NiV-G–ephrinB2 (NiV-G, orange; ephrinB2, green; PDB ID code 2VSM) are shown in a Cα trace representation superimposed on the viral receptor component of the complexes.
The receptor-binding β-propeller of GhV-G encodes seven putative N-linked glycosylation sequons; however, none of these sites appear to play a role in the GhV-G–ephrinB2 receptor surface. Electron density corresponding to asparagine-linked acetylglucosamine residues at four of seven of these sites was well ordered (N207, N255, N327, and N396; Fig. 2A and SI Appendix, Fig. S4). No clear electron density was observed at the remaining N-linked sites (N224, N324, and N350), suggestive that the glycans at these sites were either too flexible to be observed or that the corresponding residues were not glycosylated during protein folding. One of these sites, N224, lies on the first blade of the β-propeller near the putative dimerization site observed in the crystal structure of unliganded HeV-G (24) (SI Appendix, Fig. S5). An N-glycan at this site might be detrimental to receptor-mediated fusion because cognate interactions between head domains of oligomeric paramyxovirus attachment proteins are important for receptor-mediated fusion. However, site-directed mutagenesis of this residue to positively (N223R) or negatively (N224D) charged amino acids had no obvious effect on fusogenicity (SI Appendix, Fig. S6). These data suggest that, if an N-linked glycan does occupy the N224 site, it is unlikely to play a functional role in receptor-induced fusion. Furthermore, it does not appear that the N224 residue is in the critical dimer interface area, as posited for HeV-G (and NiV-G) (24).
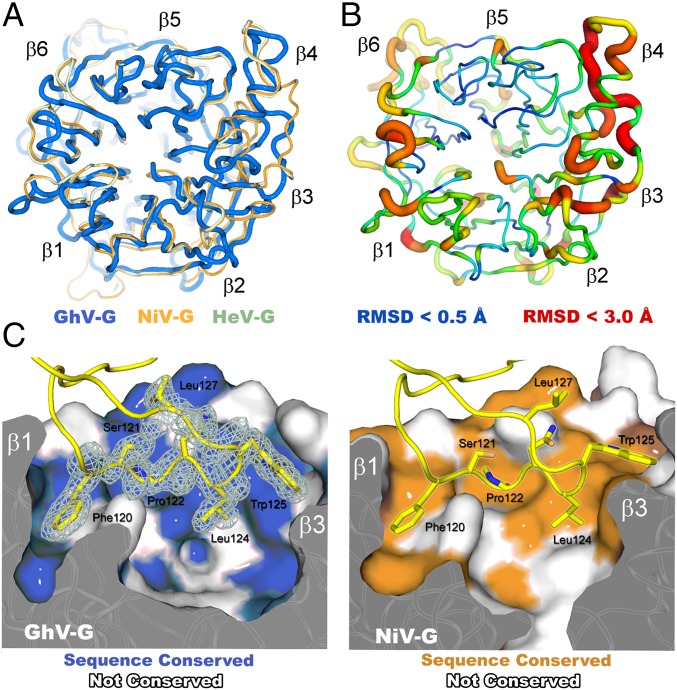
GhV-G maintains <30% sequence identity with NiV- and HeV-G. As anticipated (42), this low level of genetic conservation is reflected structurally, where GhV-G exhibits significant structural variation in the β-propeller scaffold with respect to related NiV- and HeV-G [Figs. 2C and 3; 1.7-Å rms (29) deviation over 374 equivalent Cα atoms]. Differences in structure are particularly evident at peripheral solvent accessible regions in the second, third, and fourth blades of the β-propeller (Fig. 3 A and B), and these regions map to areas with high structural diversity and propensity for N-linked glycosylation across the Paramyxovirinae (23). SI Appendix, Table S1 summarizes and compares the key features of our GhV-G–ephrin2 crystal structure with extant structures of NiV-G and HeV-G–ephrinB2 complexes.
Fig. 3.
Structural variation of viral and receptor scaffolds. (A) Overlay of HNV six-bladed β-propeller receptor-binding domains: GhV-G (blue), NiV-G (orange; PDB ID code 2VSM), and HeV-G (mint; PDB ID code 2VSK). (B) Cartoon representation of the HNV-G β-propeller with rms displacement of equivalent residues between GhV- and NiV-G mapped onto the Cα trace of GhV-G. The tube radius and color represent the RMS displacement (ramped from blue to red). Regions with the highest deviations between GhV- and NiV-G are thick and colored red. Regions with low deviation are thin and colored blue. (C) Conserved mode of ephrinB2 receptor binding between African and Asiatic HNV-Gs. Comparison of the GhV-G–ephrinB2 and NiV-G–ephrinB2 interfaces. Van der Waals surface representations of GhV-G (Left) and NiV-G (Right) in complex with ephrinB2 are shown. EphrinB2 residues belonging to the GH loop are shown as yellow sticks with maximum-likelihood-weighted 2Fo − Fc electron density shown for the GhV-G–ephrinB2 structure. Residues on the surface colored blue (Left) and orange (Right) are conserved between NiV- and GhV-G, respectively.
The GhV-G–EphrinB2 Interface.
EphrinB2 forms a classical Greek key fold and sits at the top of the β-propeller in a 1:1 interaction. This protein–protein interface occludes ∼2,400 Å2 of solvent-accessible surface [as calculated by the PISA EBI server (43)] and is stabilized by 18 hydrogen bonds. Although extensive, this interface area is ∼300–400 Å2 smaller than the ephrinB2 interface area with NiV- and HeV-G. The Greek key fold of ephrinB2 is almost identical in structure to the NiV-G bound form [Fig. 2C; 0.5-Å rmsd (29) over 136 equivalent Cα atoms]. The primary interaction site of the GhV-G–ephrinB2 complex is located at the top-center of the β-propeller, where residues from the GHephrinB2 loop (residues 116–130) form a shallow but extensive protein–protein interface (Fig. 3C). Despite significant structural differences between GhV- and NiV-G β-propellers scaffolds (Figs. 2C and 3 A and B), the conformation of the GHephrinB2 loop is unexpectedly similar to NiV-G– and GhV-G–bound forms, where Phe-120ephrinB2 and Trp-125ephrinB2 are buried deeply into hydrophobic pockets on the GhV-G surface (Fig. 3C).
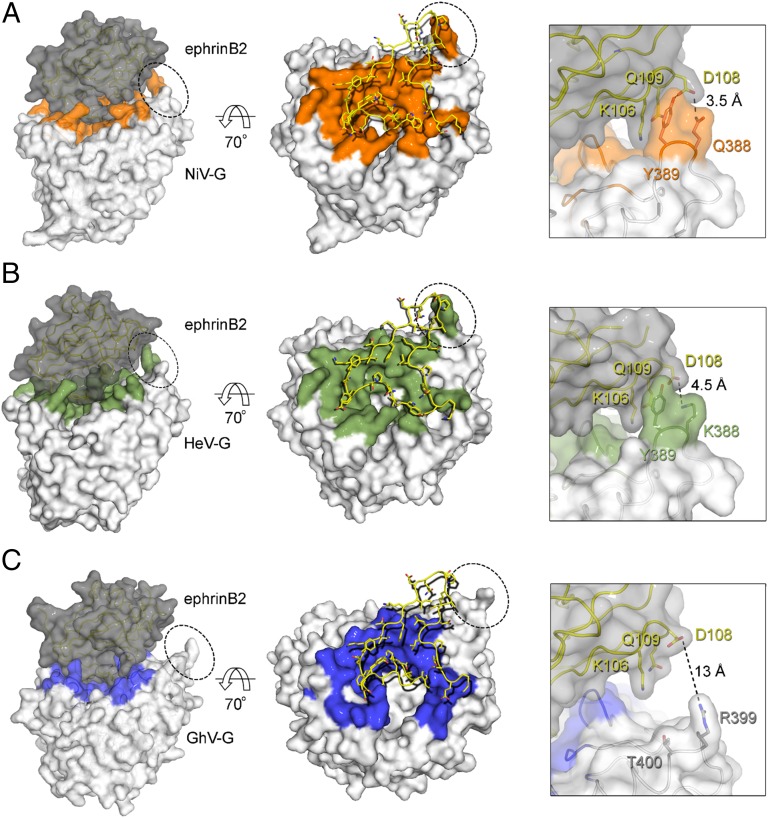
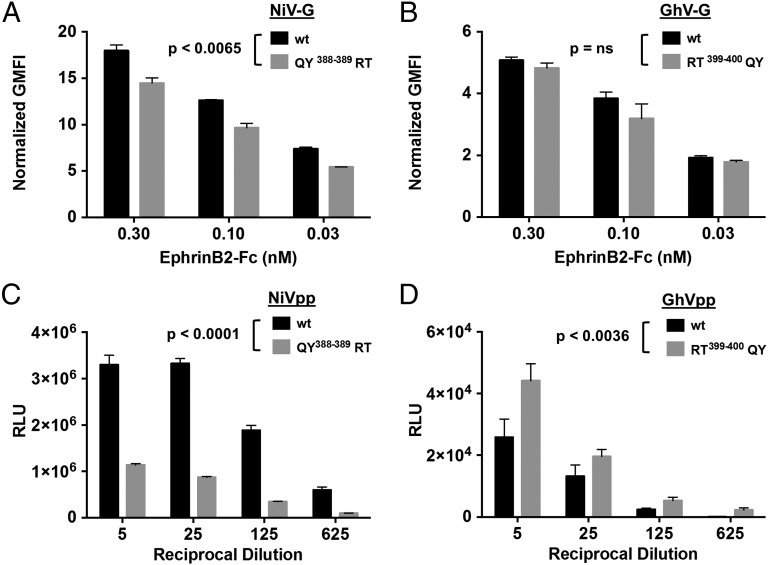
The number of protein–protein contacts in the GhV-G–ephrinB2 interface is ∼15% and 12% less than that observed in the Asiatic NiV-G–ephrinB2 (Fig. 4A) and HeV-G–ephrinB2 (Fig. 4B) structures, respectively. This reduced binding surface is chiefly due to the absence of a secondary interaction, distal from the primary central ephrin-binding site, at the tip of the fourth blade of the GhV-G β-propeller (Fig. 4C). Residues QY388–389 and KY388–389 in NiV- and HeV-G, respectively, form hydrogen bonds with ephrinB2, whereas the equivalent RT399–400 residues in GhV-G do not form these same protein–protein contacts (Fig. 4 Insets). The absence of this secondary binding site in GhV-G correlated with decreased ephrinB2 binding, with respect to NiV-G; cell-surface GhV-G reproducibly bound soluble ephrinB2–Fc at 25–30% of NiV-G levels (note the difference in the y axis scale between Fig. 5 A and B). Reciprocal exchange of the QY388–389 residues in NiV-G with the analogous RT399–400 residues in GhV-G significantly reduced and enhanced ephrinB2-mediated entry for NiV- and GhV-G, respectively (Fig. 5 C and D). Notably, the “loss-of-function” mutant in NiV-G (QY388–389RT) and its counterpart “gain-of-function” mutant in GhV-G (RT399–400QY) had less obvious phenotypes in receptor-binding (Fig. 5 A and B) vs. infectivity assays (Fig. 5 C and D). These data indicate that the hydrogen bonds at the secondary binding site impart affinity and stabilize the receptor-bound complex and that even small differences in receptor binding can translate into significant differences in the efficiency of infection. Such structural differences provide a basis for the different modes of receptor engagement by African GhV and known Asiatic HNVs, outside the highly conserved primary interaction interface. In summary, our data suggest that GhV-G does not mediate human ephrinB2-dependent entry as efficiently as NiV-G and that further adaptations in the β-propeller scaffold of GhV-G are required to optimize the secondary ephrinB2 binding site interactions that contribute to the more efficient ephrinB2-dependent entry mediated by NiV-G.
Fig. 4.
The amino acid footprint of human ephrinB2 on HNV-G. (Left) NiV-G–ephrinB2 (PDB ID code 2VSM) (A), HeV-G–ephrinB2 (PDB ID code 2VSK) (B), and GhV-G–ephrinB2 (C) complexes shown in van der Waals surface representation with the G proteins colored white and ephrinB2 gray. Residues on NiV-, HeV-, and GhV-G, which form contacts with ephrinB2, are colored orange, green, and blue, respectively. (Center) Looking down at the ephrinB2 binding site of HNV-G with residues from ephrinB2 shown as yellow sticks. (Right) Close-up view illustrating the absence of a secondary interaction interface in the GhV-G–ephrinB2 complex. The region of the close-up view is indicated by the dotted circles in Left and Center. Note that the secondary ephrinB2 interaction interface is present in both the NiV-G–ephrinB2 and HeV-G–ephrinB2 complexes, but not in GhV-G–ephrinB2.
Fig. 5.
GhV-G uses ephrinB2 less efficiently. (A and B) EphrinB2 binding to WT NiV-G (A) or GhV-G (B) and the respective QY388-389 (A) or RT399-400 (B) mutants involving the secondary ephrinB2-binding interface. Soluble EphrinB2–Fc binding to cell surface HNV-G was background subtracted and normalized to HNV-G cell-surface expression as detected by anti-HA staining. Data are shown as the mean normalized geometric mean fluorescent intensity (GMFI) ± SD of duplicate experiments. One representative experiment of three is shown. (C and D) Effects of reciprocal NiV-G (QY388-389RT) and GhV-G (RT399-400QY) mutations in the context of an infectivity assay using VSV-based pseudotyped particles, prepared as described in Materials and Methods. (C and D) NiVpp (C) and GhVpp (D) bearing WT F/G or the indicated mutant G proteins were used to infect U87 glioblastoma target cells over a range of viral inoculum (indicated by a dilution series of the viral stock preparation). Entry of NiVpp/GhVpp results in production of luciferase by the reporter virus backbone (VSV-ΔGLuc). Data are shown as mean ± SD from quadruplicate samples. One representative experiment of two is shown. QY388-389RT mutations in NiV-G (C) and RT399-400QY mutations in GhV-G (D) significantly decreased (P < 0.0001) and increased (P < 0.0036) infectivity with respect to their parental WT NiVpp and GhVpp (two-way ANOVA for multiple comparisons across the dilution series examined). In contrast, only the NiV-G QY388-389RT mutant (A) showed moderate but significantly decreased ephrinB2 binding with respect to WT NiV-G (P < 0.0065; two-way ANOVA for multiple comparisons). Expression of the various F and G proteins on pseudotyped particles and the cell surface are shown in SI Appendix, Fig. S7.
GhV-G Does Not Recognize EphrinB3 Cell Surface Receptor.
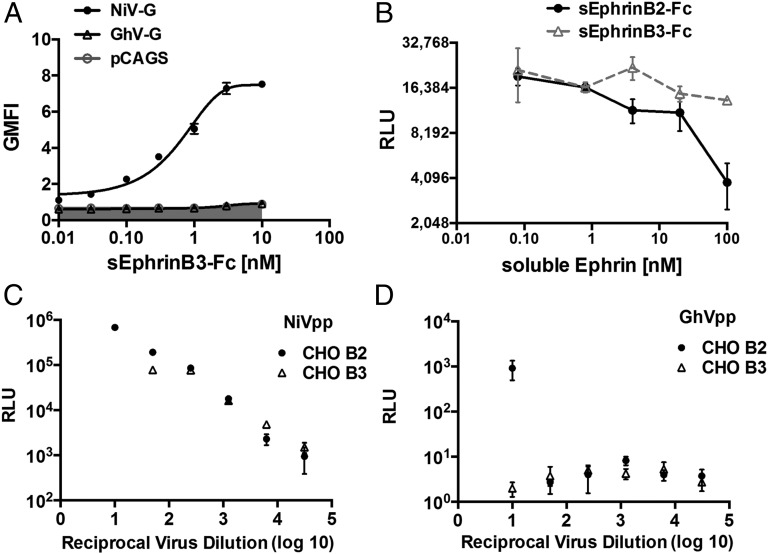
EphrinB3 is an established alternative receptor for NiV and HeV (19). Previous structural studies have revealed how the high sequence similarity between ephrinB2 and B3, primarily in the GH interaction loops, leads to similar modes of NiV-G binding (22, 26). Given the near-identical conformations of ephrinB2 binding to African and Asiatic HNVs (Fig. 3C), we sought to investigate whether GhV-G also exploits ephrinB3 as an alternate cell-surface receptor. Surprisingly, we could not detect binding of soluble ephrinB3–Fc to cell-surface GhV-G, even at concentrations up to 10 nM, whereas the EC50 for soluble ephrinB3–Fc binding to NiV-G was <1 nM (Fig. 6A). In addition, soluble ephrinB2–Fc, but not ephrinB3–Fc, inhibited GhV pseudotyped virus particle (GhVpp) entry into highly permissive U87 cells (Fig. 6B). Lastly, whereas NiVpp efficiently entered CHO cells stably expressing ephrinB2 (CHO-B2) or ephrinB3 (CHO-B3) (20), GhVpp only showed entry into CHO-B2 cells (Fig. 6 C and D).
Fig. 6.
GhV-G does not use ephrinB3 as a cell-surface receptor. (A) Soluble ephrinB3–Fc binds NiV-G but not GhV-G. NiV- or GhV-G was transfected into CHO cells. At 18 h after transfection, cells were stained with increasing amounts of recombinant soluble ephrinB3–Fc and processed for FACS analysis after secondary staining with PE-conjugated anti-human Fc antibodies. Data are shown as mean ± SD of experiments performed in duplicates. One representative experiment of two is shown. (B) sEphrinB2–Fc but not sEphrinB3–Fc, blocks GhVpp infection of U87 cells. GhVpp prepared as described in Fig. 5 was used to infect U87 cells in the presence of increasing amounts of soluble ephrinB2–Fc or ephrinB3–Fc as indicated. Data are shown as mean ± SD of quadruplicate infection experiments. (C and D) NiVpp (C) and GhVpp (D) were used to infect CHO cells stably expressing ephrinB2 (CHO-B2) or ephrinB3 (CHO-B3) over a fivefold dilution series of viral inoculum ranging from 1:10 to 1:31,250 (reciprocal virus dilution indicated on the x axis). Entry of NiVpp/GhVpp results in production of luciferase by the reporter virus backbone (VSV-ΔGLuc). Data are shown as mean ± SD from quadruplicate samples. One representative experiment of two is shown.
Although it is possible that the interaction of GhV-G with ephrinB3 is too inefficient to be detectable by our assays, we note that another HNLV recently isolated from pteropid bats (Cedar virus; CedV) also appeared to use ephrinB2 but not ephrinB3 (3). CedV-G exhibits low sequence identity with NiV- and HeV-G (∼30%), but appears even more distantly related to GhV-G (24% sequence identity). Because ephrinB3 is expressed at higher levels in the brainstem than ephrinB2, it has been proposed that ephrinB3-mediated entry may be the ultimate cause of brainstem neuronal dysfunction that is a hallmark of fatal HNV encephalitis (44). Future comparative structure–function studies on these HNVs with differential receptor tropism are critical for determining whether ephrinB3 use is a correlate of efficient ephrinB2 use and/or pathogenicity.
Determinants of Receptor-Mediated Fusion.
The initial step of receptor-mediated triggering of the HNV fusion cascade is dependent upon binding of HNV-G to host-cell ephrins (45). Highly conserved amino acid residues thought to play a key role in this triggering process have been identified at the base of the first and sixth blades of the NiV- and HeV-G β-propeller domains, proximal to the proposed region of HNV-G dimerization (24, 46). Residues in this region contribute to a well-characterized receptor-binding–enhanced (RBE) epitope recognized by Mab45. However, these residues are not conserved in GhV-G (SI Appendix, Fig. S1, shaded yellow box), and Mab45 does not cross-react with GhV-G (Fig. 1C). Thus, there may be an alternative motif that contributes to receptor-mediated triggering of the fusion cascade for this HNV.
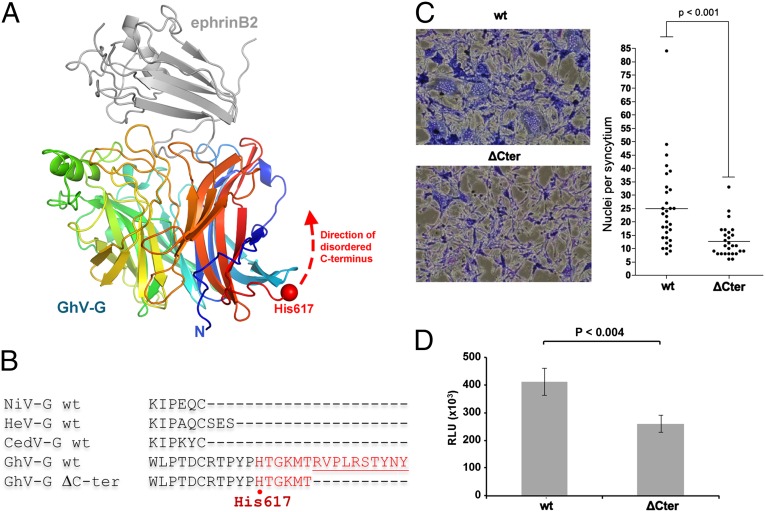
The GhV-G attachment glycoprotein also encodes a 17- to 20-amino-acid C-terminal extension, which is not present in NiV-G, HeV-G, or the more recently described CedPV-G (SI Appendix, Fig. S1). Because C-terminal extensions have an established role in autoinhibition of receptor-mediated fusion of avirulent Newcastle Disease virus HN-mediated entry (NDV-HN) (47–49), we hypothesized that the extension observed in GhV-G may likewise modulate the viral fusion cascade. However, although the C-terminal extension in the HN of the NDV Ulster strain was well ordered (49), we were unable to detect clear electron density for this region in the structure of GhV-G, an observation consistent with structure disorder prediction analysis, which reveals that this region may be natively disordered (Fig. 7 A and B and SI Appendix, Fig. S8) (50). To assess whether these residues may have a functional role in receptor-mediated fusion and virus entry, we generated a minimal C-terminal truncation mutant (GhV-GΔC) that was expressed and bound human ephrinB2 at wild-type (WT) levels (SI Appendix, Fig. S7 B and C) and determined its ability to mediate fusion and infection. When cotransfected with WT GhV-F, GhV-GΔC was significantly deficient in syncytia formation compared with WT GhV-G (Fig. 7C). This fusion deficiency was also reflected in decreased infectivity in the context of pseudotyped virus infection (Fig. 7D), confirming that the C-terminal residues on GhV-G play a role in efficient virus entry. We speculate that the C terminus of GhV-G may play a similar role as the Mab45 RBE in NiV- and HeV-G with regards to receptor-mediated allosteric triggering of fusion in the genus Henipavirus.
Fig. 7.
The C terminus of GhV-G plays a functional role in fusion and entry. (A) GhV-G residues Thr-618–Tyr632 at the C terminus are disordered and likely directed back along the first and sixth blade of the β-propeller scaffold. GhV-G and ephrinB2 are depicted as in Fig. 2A. (B) C-terminal sequence alignment of NiV-G, HeV-G, CedV, and GhV-G showing that the C terminus of GhV-G is 17–20 amino acids longer than the other HNV-Gs. (C and D) Truncation of the C-terminal 9 amino acids of GhV-G (GhV-G ΔC-ter) does not affect cell-surface expression, sEphrinB2–Fc binding, or incorporation into pseudovirions (SI Appendix, Fig. S7), but significantly affects its ability to mediate syncytia formation (C) and infectivity (D) when paired with WT GhV-F. Syncytia formation and GhVpp infections were performed as described in Materials and Methods. P values were determined by unpaired Student’s t test.
Structurally Inferred Evolutionary Relationship of GhV with Other Paramyxoviruses.
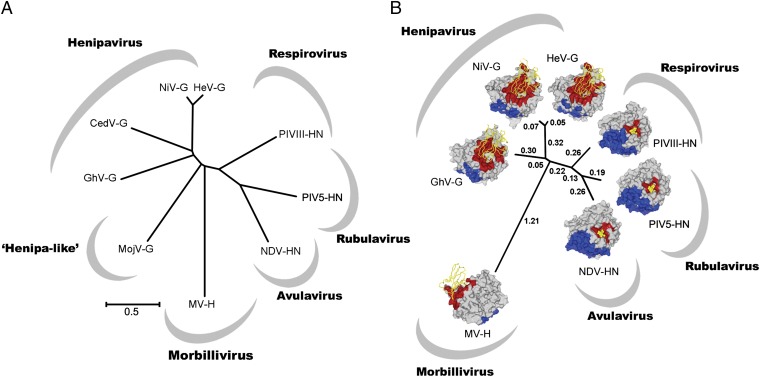
Structure-based phylogenic analysis by pairwise distance calculation is a useful tool for exploring evolutionary relationships between genetically diverse, but structurally homologous, viral proteins (51). When applied to paramyxoviral attachment glycoprotein structures, this approach has revealed multiple evolutionary events by which paramyxovirus H and G glycoproteins have derived protein-binding specificity from their glycan-binding precursors (22, 41). Given the relatively low sequence identity of GhV-G with Asiatic NiV- and HeV-G (Fig. 8A), we sought to infer the structural relationship of GhV-G with HNV-Gs and other paramyxovirus attachment glycoproteins. We performed a side-by-side structural and genetic phylogeny analysis (Fig. 8 A and B) and observed that, consistent with known functionality, paramyxoviruses separate according to receptor specificity, whereby ephrin-binding HNV-Gs segregate from sialic acid-binding HNs and protein-binding measles virus (MV). Although GhV-G maintains low sequence identity with the receptor-binding domains of NiV- and HeV-G (<30%), structural phylogenetic analysis confirms that GhV-G allies to the Asiatic HNV-G cluster of the structural phylogeny. We also note that, consistent with sequence-based analysis (Fig. 8A), GhV-G and Asiatic NiV-G/HeV-G are both almost equidistant in structure from glycan-binding paramyxoviruses. These data are suggestive of separate structural evolution of GhV-G from Asiatic HNVs, after bifurcation from a common ephrin-binding HNV ancestor.
Fig. 8.
Structural phylogeny reveals that both African GhV-G and Asiatic NiV-G/HeV-G are almost equidistant in structure from glycan-binding paramyxoviruses. (A) Unrooted sequence phylogeny of extant HNV-Gs with attachment glycoprotein homologs from viruses representing each of the Paramyxovrinae genera. Virus name (abbreviation) and GenBank accession nos. are as follows: MojV KF278639; GhV HQ660129; CedPV JQ001776; NiV NC_002728; HeV NC_001906; human parainfluenza virus 3 (PIV3) AY283063; parainfluenza virus 5 (PIV5) NC_006430; NDV AF212323; and MV AB583749. (B) Structure-based phylogeny of receptor-bound six-bladed β-propellers. A pairwise distance matrix between structures of GhV-G, NiV-G (PDB ID code 2VSM), HeV-G (PDB ID 2VSK), PIV3-HN (PDB ID code 1E8V), PIV5-HN (PDB ID code 1Z4V), NDV-HN (PDB ID code 1E8V), and MV-H (PDB ID code 3ALX) was constructed by using SHP (66) and plotted as an unrooted tree by using the program PHYLIP (67). Structure-based evolutionary distances, as defined by Stuart and coworkers (51, 66), are annotated. The structure of each β-propeller is shown as a surface representation. Residues involved in receptor binding and dimerization were identified with the PISA EBI server (43) and are colored red and blue, respectively. Residues forming homodimeric contacts in NiV- and GhV-G were predicted by structure-based alignment with the dimeric HeV-G structure. Cell-surface receptors are illustrated as yellow ribbons (henipavirus and morbillivirus) and sticks (avulavirus, respirovirus, and rubulavirus).
Conclusions
Recent surveillance studies have recognized that bats are major reservoirs for paramyxoviruses (2, 11). Although sequence information revealed the presence of novel paramyxoviruses from all major mammalian Paramyxovirinae genera, the putative identification of at least 19 novel and divergent clades of HNLVs in Africa is arguably the most surprising (2). These studies have led to a reassessment of the risk that emerging HNVs or HNLVs pose to human populations (2, 12). The early symptomology of HNV infections—fever and headache—cannot be easily differentiated from the symptomatic noise inherent in the pathogen-rich areas of tropical Africa. Thus, although the extent of the biomedical impact of these viruses upon human health has yet to be established, it is plausible that African HNV-related illnesses have been misdiagnosed as malaria-associated encephalitis (2, 13) or otherwise have been attributed to the innumerable cases of unresolved fevers of unknown origin in that part of the world. Here, we provide molecular-level details for evaluating the likelihood of spillover of African HNVs into human populations.
We demonstrate that GhV, the only African HNV for which envelope sequence information is available, akin to highly pathogenic Asiatic HNVs, hijacks species conserved ephrinB2 cell-signaling receptors during cell entry. X-ray crystallographic investigation of a GhV-G–ephrinB2 complex reveals that this African HNV and Asiatic HNVs display HNV-G receptor-binding scaffolds, which differ significantly in structure but are capable of accommodating nearly identical primary ephrinB2-binding modes. However, our studies also revealed that the GhV-G–ephrinB2 complex has a significantly lower level of buried surface at the envelope-receptor interface compared with NiV-G/HeV-G (2,400 Å2 vs. 2,700–2,800 Å2, respectively). This lower surface area, largely due to the lack of a secondary ephrinB2-binding site (Fig. 4), may account for the less efficient use of ephrinB2; GhV-G mediates lower ephrinB2-mediated infectivity than NiV-G in pseudotyped virus entry assays and forms smaller syncytia than NiV- or HeV-G (Fig. 5 and ref. 27). Whether this finding argues for the decreased likelihood of zoonotic bat-to-human transmission awaits comparative analysis of the various HNV-G interactions with bat and human ephrinB2. However, we note that bat and human ephrinB2 differ by only three conserved amino-acid substitutions in the contact residues with GhV-G and thus likely maintain very similar modes of binding (SI Appendix, Fig. S9).
In a therapeutic context, unlike the tractable and structurally conserved sialic-acid binding sites of paramyxovirus HNs (52), the apparent structural plasticity (23, 24) and low level of sequence conservation (<30% sequence identity) on the HNV-G receptor-binding β-propeller scaffold are likely to prove an obstacle in the rational design of vaccines that induce broadly neutralizing antibodies to the ever-expanding spectrum of HNVs or HNLVs. Notably, none of our polyclonal or monoclonal antibodies made against NiV cross-reacted with GhV-G (Fig. 1) or cross-neutralized GhVpp infection (12). Similarly, anti-NiV/-HeV polyclonal and monoclonal neutralizing antibodies also did not neutralize CedV, another HNLV also distantly related to NiV/HeV (≤30% sequence identity in the G protein) (3). However, because GhV- and CedV-G both mediate ephrinB2-dependent entry, and the primary ephrinB2-binding site appears conserved, despite the low overall sequence identity, immune focusing by prime-boost vaccination (53, 54) may be a strategy for eliciting broadly neutralizing antibodies that target the primary ephrinB2-interacting site on divergent HNV-Gs.
In summary, our data reveal a unified strategy for genetically and geographically diverse HNVs to enter host cells. These data not only broaden the known viral architectures that can bind ephrins, but also provide a molecular rationale for spillover of newly emerging HNVs into human populations.
Materials and Methods
Protein Expression and Purification.
The putative six-bladed β-propeller domain of the GhV-G attachment glycoprotein (residues 199–632; GenBank accession no. AFH96011.1) and receptor-binding domain of human ephrinB2 (residues 25–168; GenBank accession no. NM_004093) were cloned into the pHLsec mammalian expression vector (55). GhV-G and ephrinB2 were coexpressed in HEK293T cells in the presence of the α-mannosidase inhibitor, kifunensine, with 2 mg of DNA per liter of cell culture in a 3:1 ratio of GhV-G to ephrinB2. Cell supernatant containing secreted GhV-G–ephrinB2 were collected 6 d after transfection, clarified, and diafiltrated against a buffer containing 10 mM Tris (pH 8.0) and 150 mM NaCl. GhV-G–ephrinB2 complex was purified by immobilized metal-affinity chromatography and partially deglycosylated with endoglycosidaseF1 (75 μg⋅mg−1 protein, 16 h, 21 °C). After deglycosylation, GhV-G–ephrinB2 was purified by size-exclusion chromatography using a Superdex 200 10/30 column equilibrated against 10 mM Tris (pH 8.0) and 150 mM NaCl. The approximate yield of purified, deglycosylated GhV-G–ephrinB2 was 2.0 mg per liter of cell culture.
Crystallization and Structure Determination.
GhV-G–ephrinB2 complex was crystallized at a concentration of 13.2 mg⋅mL−1 using the sitting-drop vapor-diffusion method (56), by mixing 100 nL of protein and 100 nL of precipitant containing 12% (wt/vol) PEG 4000, 0.1 M sodium acetate (pH 5.5), 0.1 M LiSO4, and 0.1 M NaCl. Crystal drops were equilibrated against 95 µL of a precipitant-containing reservoir for 3 d at 22 °C. Crystals were cryoprotected with 25% (vol/vol) ethylene glycol plus precipitant followed by rapid cooling in liquid nitrogen. X-ray data were recorded at Beamline I04 at Diamond Light Source (Didcot, U.K.) on a Dectris Pilatus 6M-F detector. X-ray data were indexed, integrated, and scaled with XIA2 (57). The high-resolution cutoff for the data was determined by analysis of CC1/2, as defined by Karplus and Diederichs (58). The structure of the GhV-G–ephrinB2 complex was phased by molecular replacement with PHASER (59) using the crystal structure of NiV-G in complex with ephrinB2 as a search model (PDB ID code 2VSM) (22). The initial model was built with ARP/wARP (60) and then manually rebuilt with COOT (61). Structure refinement was performed with Refmac5 (62) in the CCP4 suite and included restrained refinement with TLS (62, 63) and locally defined noncrystallographic symmetry. The final refined structure was validated with Molprobity (64).
Cells and Plasmids.
U87 and HEK293T (ATCC) cells were grown in DMEM with 10% (vol/vol) FCS and 1% penicillin/streptomycin (Life Technology). Previously described CHO–ephrinB2 and CHO–ephrinB3 cells (44) were maintained in DMEM/F12 medium (Life Technology) with 10% (vol/vol) FCS, 1% penicillin/streptomycin, and 1.0 mg/mL G418. Codon-optimized sequences of GhV-G and the corrected GhV-F tagged with a C-terminal HA or Au1, respectively, were cloned in pcDNA3.1 vector and used for glycoprotein expression (as described in ref. 27). Glycoprotein surface expression was compared by transfecting 293T cells with GhV-G (or associated GhV-G mutants) expression vector using Lipofectamine 2000 (Life Technology). At 24 h postinfection (hpi), cells were detached by gentle pipetting, fixed in 2% (vol/vol) paraformaldehyde, and stained with anti–HA-PE 1/500 (MACS Molecular). Cells were then analyzed by using a FACS Calibur cytometer.
Syncytia Assay.
Syncytia assays were performed by transfecting U87 cells with GhV-F and -G expression vector as described above. At 48 hpi, cells were fixed for 10 min in methanol and stained for 45 min with Giemsa. Cells were then washed with water and allowed to dry. Stained cells were photographed, and nuclei-per-syncytium ratio was determined using ImageJ with the Cell Counter plug-in.
Pseudotyped Virus.
NiV and GhV pseudotyped particles (NiVpp and GhVpp) were produced as described (12, 19, 65). Glycoprotein expression on NiVpp and GhVpp was determined by Western blotting for F and G using anti-AU1 and -HA tag antibodies, respectively. Briefly, NiVpp and GhVpp purified by ultracentrifugation through a 20% (wt/vol) sucrose cushion were lysed in 6× Laemmli buffer [5% (vol/vol) β-mercaptoethanol final]. Samples were then boiled for 10 min, run on 12% (wt/vol) polyacrylamide gel with SDS and transferred onto a PVDF membrane. The membrane was blocked in Li-Cor buffer and stained with rabbit α-HA 1/15,000 in Li-Cor buffer and mouse α-AU1 (1/5,000) in Li-Cor buffer plus 0.1% Tween 20. The membrane was then washed in PBS plus 0.1% Tween 20 and incubated with Li-Cor goat α-mouse 800 and goat α-rabbit 700 diluted to 1/10,000 and 1/20,000, respectively. Membrane was finally washed and scanned by using a Li-Cor Odyssey Scanner. Protein amount was determined by using the Li-Cor Odyssey software.
Target cells plated in 96-well plates were infected with the indicated pseudoviruses serially diluted in DMEM for 2 h at 37 °C. For inhibition experiments, the indicated amount of soluble ephrin–Fc (Sigma) was added together with a predetermined amount of pseudotyped virus stock during this incubation period. Cells were then washed twice with PBS and incubated at 37 °C in the appropriate medium. After 36 h, cells were washed with PBS, lysed, and processed by using Renilla Luciferase Detection kit according to the manufacturer's directions (Promega). Luminescence intensity was read on a Tecan M1000 luminometer.
Construction of Structure-Based Phylogenies.
Structural alignments were run with the program SHP (66) to optimize the localized fit between equivalent Cα positions, where the probability of equivalence between Cα pairs of residues is greatest (51). By alignment of available paramyxoviral H, HN, and G structures, a pairwise evolutionary distance matrix (51) was constructed and plotted as an unrooted tree with PHYLIP (67).
Phylogenetic Analysis of G/H/HN Amino Acid Sequences of Selected Paramyxoviruses.
The sequences of nine paramyxovirus G/H/HN proteins were aligned by using the Muscle (68) program of the MEGA6 software suite (69). The evolutionary history was inferred by using the neighbor-joining method (70). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by using the Poisson correction method (71) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 516 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (69).
Data Deposition.
Coordinates and structure factors of the GhV-G–ephrinB2 complex were deposited in the PDB (PDB ID code 4UF7). Crystallographic data collection and refinement statistics are presented in Table 1.
Supplementary Material
Acknowledgments
We thank W. Lu for help with tissue culture; K. Harlos for data collection; the staff of beamline I04 at Diamond Light Source for support; and Yvonne Jones, David Stuart, and Max Crispin for helpful discussions. This work was supported by Wellcome Trust Grant 090532/Z/09/Z; Medical Research Council Grant MR/L009528/1 (to T.A.B.); and NIH Grants NIAID R01 AI069317, U54 AI065359, and R21 AI115226 (to B.L.), K08 AI093676 (to A.A.A.), and T32 AI07323 (to S.M.B.). B.L. also acknowledges the Ward Coleman estate for endowing the Ward-Coleman Chairs at the Icahn School of Medicine at Mount Sinai.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4UF7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501690112/-/DCSupplemental.
References
- 1.Eaton BT, Broder CC, Middleton D, Wang LF. Hendra and Nipah viruses: Different and dangerous. Nat Rev Microbiol. 2006;4(1):23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drexler JF, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh GA, et al. Cedar virus: A novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8(8):e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breed AC, et al. The distribution of henipaviruses in Southeast Asia and Australasia: Is Wallace’s line a barrier to Nipah virus? PLoS ONE. 2013;8(4):e61316. doi: 10.1371/journal.pone.0061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halpin K, et al. Henipavirus Ecology Research Group Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am J Trop Med Hyg. 2011;85(5):946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua KB, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354(9186):1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- 7.Gurley ES, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13(7):1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum LS, Khan R, Nahar N, Breiman RF. In-depth assessment of an outbreak of Nipah encephalitis with person-to-person transmission in Bangladesh: Implications for prevention and control strategies. Am J Trop Med Hyg. 2009;80(1):96–102. [PubMed] [Google Scholar]
- 9.Wu Z, et al. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg Infect Dis. 2014;20(6):1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iehlé C, et al. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg Infect Dis. 2007;13(1):159–161. doi: 10.3201/eid1301.060791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peel AJ, et al. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nat Commun. 2013;4:2770. doi: 10.1038/ncomms3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pernet O, et al. Evidence for henipavirus spillover into human populations in Africa. Nat Commun. 2014;5:5342. doi: 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathers CD, Ezzati M, Lopez AD. Measuring the burden of neglected tropical diseases: The global burden of disease framework. PLoS Negl Trop Dis. 2007;1(2):e114. doi: 10.1371/journal.pntd.0000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayman DT, et al. Evidence of henipavirus infection in West African fruit bats. PLoS ONE. 2008;3(7):e2739. doi: 10.1371/journal.pone.0002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B, Ataman ZA. Modes of paramyxovirus fusion: A Henipavirus perspective. Trends Microbiol. 2011;19(8):389–399. doi: 10.1016/j.tim.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pernet O, Wang YE, Lee B. Henipavirus receptor usage and tropism. Curr Top Microbiol Immunol. 2012;359:59–78. doi: 10.1007/82_2012_222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hafner C, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50(3):490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- 18.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6(6):462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 19.Negrete OA, et al. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2(2):e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negrete OA, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436(7049):401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 21.Bonaparte MI, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci USA. 2005;102(30):10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden TA, et al. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15(6):567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 23.Bowden TA, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: A template for antiviral and vaccine design. J Virol. 2008;82(23):11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: Evidence for a conserved mode of assembly. J Virol. 2010;84(12):6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden TA, Jones EY, Stuart DI. Cells under siege: Viral glycoprotein interactions at the cell surface. J Struct Biol. 2011;175(2):120–126. doi: 10.1016/j.jsb.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu K, et al. Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA. 2008;105(29):9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pernet O, Beaty S, Lee B. Functional rectification of the newly described African henipavirus fusion glycoprotein (Gh-M74a) J Virol. 2014;88(9):5171–5176. doi: 10.1128/JVI.03655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weis M, et al. Characterization of African bat henipavirus GH-M74a glycoproteins. J Gen Virol. 2014;95(Pt 3):539–548. doi: 10.1099/vir.0.060632-0. [DOI] [PubMed] [Google Scholar]
- 29.Hurchla MA, et al. The epoxyketone-based proteasome inhibitors carfilzomib and orally bioavailable oprozomib have anti-resorptive and bone-anabolic activity in addition to anti-myeloma effects. Leukemia. 2013;27(2):430–440. doi: 10.1038/leu.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch BD, et al. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog. 2013;9(8):e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan P, et al. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci USA. 2011;108(36):14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7(11):1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence MC, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335(5):1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 34.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13(5):803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Bishop KA, et al. Residues in the stalk domain of the hendra virus g glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82(22):11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison TG. Structure, function, and intracellular processing of paramyxovirus membrane proteins. Virus Res. 1988;10(2-3):113–135. doi: 10.1016/0168-1702(88)90010-x. [DOI] [PubMed] [Google Scholar]
- 37.Markwell MA, Fox CF. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J Virol. 1980;33(1):152–166. doi: 10.1128/jvi.33.1.152-166.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parks GD, Lamb RA. Folding and oligomerization properties of a soluble and secreted form of the paramyxovirus hemagglutinin-neuraminidase glycoprotein. Virology. 1990;178(2):498–508. doi: 10.1016/0042-6822(90)90347-t. [DOI] [PubMed] [Google Scholar]
- 39.Thompson SD, Laver WG, Murti KG, Portner A. Isolation of a biologically active soluble form of the hemagglutinin-neuraminidase protein of Sendai virus. J Virol. 1988;62(12):4653–4660. doi: 10.1128/jvi.62.12.4653-4660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378(2):282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowden TA, Crispin M, Jones EY, Stuart DI. Shared paramyxoviral glycoprotein architecture is adapted for diverse attachment strategies. Biochem Soc Trans. 2010;38(5):1349–1355. doi: 10.1042/BST0381349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Negrete OA, Chu D, Aguilar HC, Lee B. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J Virol. 2007;81(19):10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee B, Ataman ZA, Jin L. Evil versus ‘eph-ective’ use of ephrin-B2. Nat Struct Mol Biol. 2008;15(6):540–542. doi: 10.1038/nsmb0608-540. [DOI] [PubMed] [Google Scholar]
- 46.Aguilar HC, et al. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F) J Biol Chem. 2009;284(3):1628–1635. doi: 10.1074/jbc.M807469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorman JJ, Nestorowicz A, Mitchell SJ, Corino GL, Selleck PW. Characterization of the sites of proteolytic activation of Newcastle disease virus membrane glycoprotein precursors. J Biol Chem. 1988;263(25):12522–12531. [PubMed] [Google Scholar]
- 48.Sakaguchi T, et al. Newcastle disease virus evolution. I. Multiple lineages defined by sequence variability of the hemagglutinin-neuraminidase gene. Virology. 1989;169(2):260–272. doi: 10.1016/0042-6822(89)90151-7. [DOI] [PubMed] [Google Scholar]
- 49.Yuan P, Paterson RG, Leser GP, Lamb RA, Jardetzky TS. Structure of the ulster strain newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulence. PLoS Pathog. 2012;8(8):e1002855. doi: 10.1371/journal.ppat.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21(16):3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 51.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15(6):655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 52.Ryan C, et al. Structural analysis of a designed inhibitor complexed with the hemagglutinin-neuraminidase of Newcastle disease virus. Glycoconj J. 2006;23(1-2):135–141. doi: 10.1007/s10719-006-5446-8. [DOI] [PubMed] [Google Scholar]
- 53.Sabarth N, et al. Comparison of single, homologous prime-boost and heterologous prime-boost immunization strategies against H5N1 influenza virus in a mouse challenge model. Vaccine. 2010;28(3):650–656. doi: 10.1016/j.vaccine.2009.10.105. [DOI] [PubMed] [Google Scholar]
- 54.Eda Y, et al. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J Virol. 2006;80(11):5552–5562. doi: 10.1128/JVI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 10):1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 56.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 6):651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winter G. xia2: An expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 58.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 63.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 64.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garner OB, et al. Timing of galectin-1 exposure differentially modulates nipah virus entry and syncytium formation in endothelial cells. J Virol. 2015;89(5):2520–2529. doi: 10.1128/JVI.02435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 67.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 68.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 71.Zuckerkandl E, Pauling L. Evolutionary Divergence and Convergence in Proteins. Academic; New York: 1965. [Google Scholar]
- 72.Ren J, et al. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009;19(2):271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.