Significance
The cyclic dinucleotides c-di-GMP and c-di-AMP are responsible for controlling broad changes in cell phenotypes during the life cycles of many bacterial species. To date, riboswitches that sense c-di-GMP and c-di-AMP have been discovered. The genomic locations of riboswitches reveal numerous genes that are controlled by these signaling compounds and thereby reveal the biological processes that are regulated. Herein, we report that a subset of conserved noncoding RNA domains previously annotated as c-di-GMP-I riboswitches in fact bind the newly discovered second messenger c-AMP-GMP. These riboswitches control many genes involved in the utilization of extracellular iron(III) oxide as an electron sink by various Geobacter species. Our findings reveal additional roles for this recently discovered signaling molecule.
Keywords: riboswitch, Geobacter, GEMM, c-di-GMP, second messenger
Abstract
Major changes in bacterial physiology including biofilm and spore formation involve signaling by the cyclic dinucleotides c-di-GMP and c-di-AMP. Recently, another second messenger dinucleotide, c-AMP-GMP, was found to control chemotaxis and colonization by Vibrio cholerae. We have identified a superregulon of genes controlled by c-AMP-GMP in numerous Deltaproteobacteria, including Geobacter species that use extracellular insoluble metal oxides as terminal electron acceptors. This exoelectrogenic process has been studied for its possible utility in energy production and bioremediation. Many genes involved in adhesion, pilin formation, and others that are important for exoelectrogenesis are controlled by members of a variant riboswitch class that selectively bind c-AMP-GMP. These RNAs constitute, to our knowledge, the first known specific receptors for c-AMP-GMP and reveal that this molecule is used by many bacteria to control specialized physiological processes.
Three major types of cyclic dinucleotide second messengers have been discovered in biological systems. The first to be found was c-di-GMP (Fig. 1A, Top), which serves as a regulator of diverse physiological changes in most bacterial lineages (1). The second, c-di-AMP (2), is an important signaling compound during bacterial sporulation and germination (3) and has also been found to regulate osmotic shock responses in Gram-positive bacteria (4, 5). More recently, a third type of cyclic dinucleotide called c-AMP-GMP (Fig. 1A, Bottom) was found to be required for Vibrio cholera virulence (6).
Fig. 1.
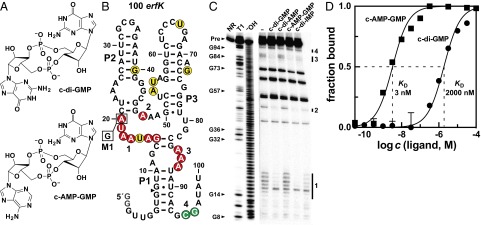
Selective recognition of c-AMP-GMP by a natural aptamer. (A) Chemical structures of c-di-GMP and c-AMP-GMP. (B) Sequence and secondary structure of the 100 erfK RNA from G. metallireducens. P1, P2, and P3 identify base-paired substructures. M1 designates a mutant construct wherein position 20 is changed to G. Regions of constant, decreasing, and increasing RNA cleavage upon addition of ligand are indicated by yellow, red, and green circles, respectively. The arrowhead indicates the start of the RNA structure stability data derived from C. (C) PAGE analysis of in-line probing assays of 5′ 32P-labeled 100 erfK in the presence (10 µM) of various cyclic dinucleotides. NR, T1, and ‒OH designate lanes loaded with precursor RNA (Pre), RNA partially digested with RNase T1 (resulting in cleavage after G residues), and RNA partially digested with alkali (resulting in cleavage after every residue). Several RNase T1 cleavage product bands are labeled. Regions undergoing substantial change in spontaneous cleavage rates are labeled 1–4. The inosine-based analog of c-di-GMP is labeled c-di–IMP. (D) Plot of the fraction of riboswitch bound to ligand versus the log of the molar concentration (c) of the ligand. Data are derived from Fig. S2, and each point is the average of the normalized fraction of modulation at sites 1 and 2. Error bars indicate the SD of the average. Included are theoretical curves expected for one-to-one interaction between ligand and RNA for the KD values given.
Although bacterial c-AMP-GMP is formed via two 3′,5′-phosphodiester linkages, metazoans produce an analog with one 2′,5′-phosphodiester linkage and one 3′,5′-phosphodiester linkage (7–9). This natural structural variant has been identified as important for triggering innate immune responses in metazoans (7–11). Far greater diversity of cyclic dinucleotides is possible by varying either the nucleotide composition or the manner in which dinucleotides are joined. Therefore, many additional types of cyclic dinucleotides might await discovery in nature where they might regulate other critical cellular processes.
Our approach to uncovering the chemical structures and biological functions of cyclic dinucleotide signaling compounds is to discover their RNA receptors. Three different riboswitch receptor classes have already been discovered that respond to c-di-GMP (12, 13) and c-di-AMP (5). Riboswitches are structured noncoding RNA (ncRNA) domains that selectively bind a small molecule or ion and thereby trigger a change in the expression of associated genes (14, 15). The majority of riboswitch ligands are composed of or derived from nucleotides, which supports the hypothesis that RNA-based metabolites and their riboswitch partners might descend from primordial RNA world organisms (16). If true, then cyclic dinucleotides and their receptors might be modern reflections of ancient signaling pathways. Regardless of their origins, the discovery of each new regulatory RNA motif that selectively recognizes a second messenger also reveals a superregulon for the signaling compound and the biology that it controls (5, 12, 13).
Given that the diversity of natural cyclic dinucleotides might be greater than is currently known, and given that some riboswitch classes have undergone subtle changes to adapt to different ligands (17–20), we re-examined known cyclic dinucleotide riboswitch classes and their associated genes for evidence of unidentified ligands or riboswitch classes. Of special interest were those RNAs that both carry mutations in the core of the ligand-binding aptamer and have gene associations considered unusual for the parent riboswitch class. Through these efforts, we identified a subset of riboswitches within the c-di-GMP-I riboswitch class (12) that we hypothesized might bind a different ligand. In the current study, we demonstrate that these riboswitches recognize c-AMP-GMP and control a set of genes that are important for the utilization of iron(III) oxide in exoelectrogenesis.
Results and Discussion
Variant c-di-GMP-I Riboswitch Aptamers Recognize c-AMP-GMP.
Approximately 6,800 sequences conform to the c-di-GMP-I riboswitch class consensus (12), which was formerly called the Genes for the Environment, for Membranes and for Motility (GEMM) motif (21). Predominantly found in species of Bacillales, Clostridia, Deltaproteobacteria, and Gammaproteobacteria (Fig. S1), the vast majority have aptamer sequences and gene associations that are typical for c-di-GMP binding and control. For example, c-di-GMP riboswitches commonly control genes involved in pilin formation, chemotaxis, signaling, and the synthesis and degradation of c-di-GMP (12, 13). However, numerous variant aptamer sequences that have unusual gene associations were identified in Deltaproteobacteria. The expression of some of these gene types, such as those related to peptidoglycan biosynthesis or remodeling, are known to be controlled by c-di-AMP riboswitches in Gram-positive bacteria (5). Therefore, we hypothesized that the variant RNAs might have a different ligand specificity compared with the more common c-di-GMP-responsive riboswitches.
Much of the ligand specificity of c-di-GMP-I riboswitches is derived from two conserved nucleotides within the aptamer (22–25), hereafter called G20 and C92. All of the aptamers with the unusual gene associations noted above carry a G20A mutation, which also suggests a potential change in ligand specificity. To determine the ligand specificity of the variant aptamers, an aptamer construct called 100 erfK (Fig. 1B) and derived from Geobacter metallireducens was subjected to in-line probing (26) in the presence of various cyclic dinucleotides. In-line probing exploits the propensity of phosphodiester linkages to spontaneously cleave with speeds that depend on local RNA structure (27). Both c-di-GMP and c-AMP-GMP at 10 μM considerably decrease the amounts of RNA products generated by spontaneous cleavage in regions comprising the ligand-binding core of the aptamer (Fig. 1C), which reveals that both second messengers modulate the structure of this RNA construct. However, the 100 erfK aptamer is capable of strongly discriminating against c-di-GMP (KD ∼2,000 nM) while favoring c-AMP-GMP (KD ∼3.2 nM) by nearly three orders of magnitude (Fig. 1D and Fig. S2).
Several variant riboswitch representatives from other Deltaproteobacteria species were tested to determine if they preferentially recognize c-AMP-GMP. Indeed, all such RNAs examined that carry the G20A variation are strongly selective for c-AMP-GMP, whereas a representative that retains the G20 sequence is selective for c-di-GMP (Fig. 2A). These data strongly suggest that the 100 erfK construct and the other variant aptamers from Deltaproteobacteria represent a distinct riboswitch class that is triggered by c-AMP-GMP, rather than by c-di-GMP. This conclusion is also supported by the findings of another recent study (28).
Fig. 2.
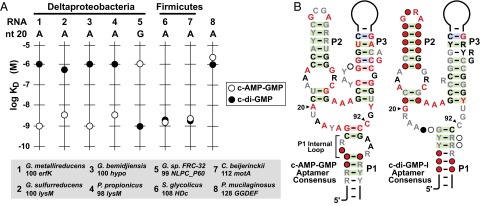
Numerous aptamers with a G20A variation bind c-AMP-GMP and discriminate against c-di-GMP. (A) KD values for various natural riboswitch aptamer variants. (B) Consensus sequences and secondary structures of RNAs predicted to selectively bind c-AMP-GMP (Left) and the c-di-GMP-I riboswitch aptamer. Nucleotides depicted in red, black, and gray, respectively, are present in at least 97, 90, and 75% of representatives. The c-AMP-GMP consensus comprises 107 sequences, whereas the c-di-GMP-I consensus comprises 99 sequences from Deltaproteobacteria. Circles designate the presence of a nucleotide at the indicated position with frequencies of 97, 90, 75, and 50% as depicted by red, black, gray, and open circles, respectively. Base pairs shaded in green exhibit phylogenetic evidence of covariation. The nucleotides corresponding to positions 20 and 92 are indicated by arrowheads. Additional annotations are as described for Fig. S1. Validated and predicted c-AMP-GMP riboswitches typically carry an internal loop in P1.
Mutational analysis and other experiments reveal that the nucleotide at position 20 is not the only determinant of ligand selectivity for c-AMP-GMP over c-di-GMP. Reversion of the adenosine at position 20 of 100 erfK to guanosine yields a construct (M1, Fig. 1B) that recognizes both c-di-GMP and c-AMP-GMP with nanomolar affinity (Fig. S3 and Table S1). This result is similar to that for c-di-GMP-I riboswitches, where incorporation of an adenosine in place of guanosine at this same position also leads to nonselective binding (29). Thus, although the G20A alteration is necessary to improve binding of c-AMP-GMP, changes elsewhere in the aptamer likely assist in discrimination against c-di-GMP.
Riboswitches That Sense c-AMP-GMP Are Found in Deltaproteobacteria.
Interestingly, some WT RNA aptamer representatives from Firmicutes that carry the G20A variation do not discriminate well between c-di-GMP and c-AMP-GMP (Fig. 2A). It is possible that G20A riboswitches residing in other bacterial lineages are naturally bispecific and capable of gene regulation in response to either second messenger. Alternatively, if c-AMP-GMP is not being produced by most species outside of Proteobacteria, there would likely be no selective pressure on the riboswitch to discriminate against c-AMP-GMP. Several observations support the latter possibility. For example, we find that the G20A variation is enriched in Deltaproteobacteria, where more than 50% of all GEMM motif RNAs carry an adenosine at position 20 (Fig. S4). These variants typically are associated with genes not known to be controlled by c-di-GMP (Dataset S1). In contrast, only 15% of GEMM motif RNAs from other lineages carry the G20A mutation, and the majority of these RNAs remain associated with genes that are commonly controlled by c-di-GMP. Furthermore, homologs of the dinucleotide cyclase DncV, which has been demonstrated to synthesize c-AMP-GMP in V. cholerae, have to date been identified only in Proteobacteria (6). However, given how little is known about c-AMP-GMP and the enzyme(s) that synthesize it, the possibilities that c-AMP-GMP is present in other bacterial lineages or that other populations of c-AMP-GMP riboswitches exist cannot be ruled out.
To search for other nucleotide changes needed to increase selectivity for c-AMP-GMP, we created a consensus sequence and structural model for c-AMP-GMP aptamers by comparing ∼100 representatives from Deltaproteobacteria (Fig. 2B). Several additional differences are evident in c-AMP-GMP aptamers relative to those for c-di-GMP-I (Fig. 2B and Fig. S4), and additional mutational analyses suggest that a combination of these changes is necessary to create highly selective riboswitches for c-AMP-GMP (Fig. S5). For example, reversion of position 20 to a G and deletion of a portion of the internal loop in P1 yields a construct (M8, Fig. S5) that is more selective for c-di-GMP than for c-AMP-GMP (SI Text).
Control of Gene Expression by c-AMP-GMP Riboswitches.
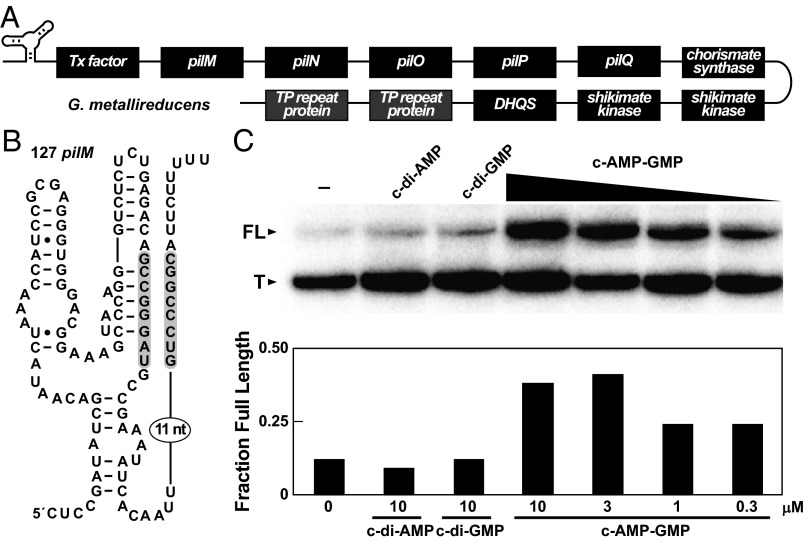
Most c-AMP-GMP riboswitches are predicted to carry an intrinsic terminator stem (30) that partially overlaps the P1 and P3 stems of the aptamer domain. Thus, c-AMP-GMP binding is expected to preclude formation of the terminator stem and increase expression by promoting transcription of the adjoining ORF. One c-AMP-GMP riboswitch with this arrangement (Fig. 3A) is found upstream of the pilMNOPQ operon in G. metallireducens (Fig. 3B). Addition of c-AMP-GMP to single-round in vitro transcription assays triggers a large increase in transcript elongation (Fig. 3C), which is consistent with regulation of transcription termination. In contrast, no increase was observed with the addition of c-di-GMP or c-di-AMP. Similar findings were observed for a second riboswitch from G. metallireducens found upstream of a gene encoding a cytochrome c protein (Fig. S6). These results demonstrate that these riboswitches regulate gene expression in response to c-AMP-GMP and confirm the binding data indicating strong discrimination against other cyclic dinucleotide second messengers.
Fig. 3.
Regulation of transcription by a c-AMP-GMP riboswitch. (A) The genes downstream of the pilM riboswitch in G. metallireducens. Genes in black boxes are putatively involved in exoelectrogenesis. Gene sizes and gaps are not drawn to scale. (B) Predicted sequence and secondary structure of 127 pilM from G. metallireducens in the presence of c-AMP-GMP. The predicted expression platform consists of an intrinsic terminator stem highlighted in gray followed by a run of U residues. (C) Single-round in vitro transcription assays using a DNA template corresponding to the riboswitch depicted in B in the absence (‒) or presence of various cyclic dinucleotides at the concentrations indicated. (Top) PAGE separation of transcripts where FL and T designate full-length and terminated products, respectively. (Bottom) Plot of the fraction of full-length RNA transcripts produced as determined from the PAGE image.
Genes Involved in Exoelectrogenesis Are Controlled by c-AMP-GMP.
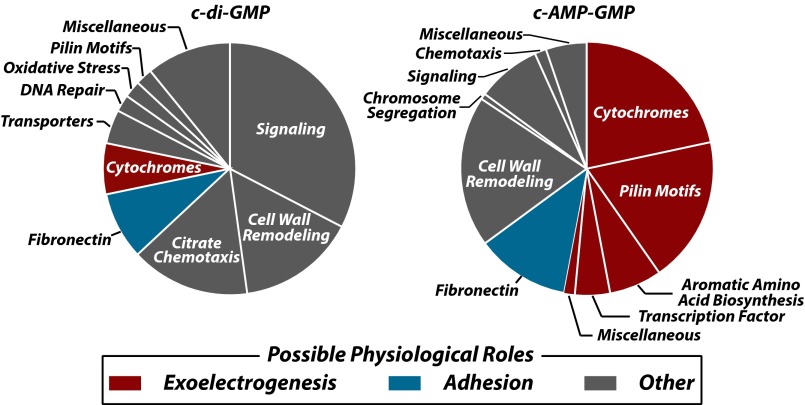
Previous evidence indicates that c-AMP-GMP is important for virulence in V. cholerae and that overexpression of dncV leads to diminished bacterial motility (6). By examining the genes controlled by c-AMP-GMP riboswitches, the roles for this signaling molecule in Deltaproteobacteria can be predicted. Genes encoding numerous cytochrome c proteins, including OmcS and OmcT (31, 32), as well as proteins involved in the formation of pilin motifs and aromatic amino acid biosynthesis (Fig. S7), reside immediately downstream of c-AMP-GMP riboswitches (Fig. 4). These genes are almost never associated with predicted c-di-GMP-responsive riboswitches and therefore likely constitute a distinct superregulon controlled by c-AMP-GMP.
Fig. 4.
The superregulons for c-di-GMP and c-AMP-GMP in Deltaproteobacteria. Genes controlled by riboswitches with a guanosine at position 20 are assigned to c-di-GMP control (46 genes), whereas those with an adenosine are assigned to the c-AMP-GMP control (134 genes). Hypothetical genes are not listed. See Table S2 for the assignment of genes to various categories.
Many of these gene products belong to protein families that are responsible for the extracellular reduction of insoluble metal complexes such as iron(III) oxide (31, 33–35). Indeed, G. sulfurreducens strains selected for improved rates of iron(III) oxide reduction carry mutations within a riboswitch that increase expression of the adjoining cytochrome c gene and improve electric current generation (36). Based on the sequence and binding characteristics of the WT aptamer (Fig. S8), we conclude that this RNA is part of a c-AMP-GMP riboswitch. Numerous additional links between c-AMP-GMP riboswitches and exoelectrogenesis-related genes are evident (SI Text). Other genes associated with c-AMP-GMP riboswitches also are likely to be important for exoelectrogenesis, and therefore we speculate that c-AMP-GMP functions as a second messenger to trigger or enhance exoelectrogenesis in many Geobacter species.
The identification of a c-AMP-GMP-responsive regulon that controls exoelectrogenesis in Geobacter spp. suggests similarities between colonization of humans by V. cholerae (6) and utilization of iron oxide by Geobacter spp. in soil. In both instances, expression of various pilin motifs is affected, and there are indications that c-AMP-GMP might indirectly control the initial stages of adherence in both organisms. In V. cholerae, mannose-sensitive hemagglutinin (MSHA) pilus expression is repressed by accumulation of c-AMP-GMP (6), whereas, in Geobacter, expression of the pilMNOPQ operon is induced by it (Fig. 3C). Both the MSHA pilin and pili in G. sulfurreducens have been shown to be important for agglutination (37, 38). Perhaps in these organisms the initial stages of adherence to an insoluble metal oxide or a eukaryotic cell might rely on the same general signaling mechanisms.
Opportunities for Additional Discoveries Regarding Cyclic Dinucleotide Signaling in Bacteria.
Our findings suggest that a major role of c-AMP-GMP in Deltaproteobacteria is to regulate the onset of exoelectrogenesis, similar to the roles that c-di-GMP plays in biofilm formation and that c-di-AMP plays in sporulation. However, some species likely use c-AMP-GMP to regulate other specialized physiological processes. Intriguingly, some RNAs examined in our study that were more diverged from the original GEMM consensus or that carried mutations at position 92 failed to bind any known cyclic dinucleotide (SI Text). Some of these rare riboswitch variants also have unusual gene associations (Dataset S1), suggesting that more receptors for undiscovered second messengers might exist.
Over the past 6 y, three previously unidentified signaling molecules and their riboswitch receptors have been identified, making it likely, if not inevitable, that more such RNA-derived signaling molecules and riboswitch partners remain to be discovered. Obvious candidates for signaling molecules include the other possible combinations of circularized dinucleotides. Additional RNA-derived or nucleotide-like second messengers include the long-known compounds cAMP, cGMP, ppGpp, and a more recent validated signaling molecule called ZTP (39, 40). Although there are no known riboswitch classes for cAMP, cGMP, and ppGpp, a widespread bacterial riboswitch has recently been discovered that senses ZTP (40).
Conclusions.
We have identified a class of riboswitches that recognizes the second messenger c-AMP-GMP in Deltaproteobacteria. These RNAs have a similar secondary structure to the previously discovered c-di-GMP-I riboswitch class, but carry some key differences. Most notably, a nucleotide that normally directly contacts the c-di-GMP ligand is altered in RNAs that bind c-AMP-GMP. These variant riboswitches are also found upstream of a set of genes that are largely distinct from genes commonly controlled by c-di-GMP-I riboswitches. These genes are important for exoelectrogenesis in various Geobacter spp. and lead us to speculate that one role of c-AMP-GMP is to regulate this important bacterial process.
Our findings also suggest that additional cyclic dinucleotides and riboswitch receptors remain to be discovered (SI Text). The presence of signaling molecules composed of RNA, as well as widespread RNA-based receptors for some of these compounds, suggests that such signaling mechanisms may have been very common during the latter stages of the RNA world (16). Over the past 12 years, we and others have identified more than 30 different riboswitch classes, and more than two-thirds of them recognize compounds composed of or derived from RNA. Given the widespread distribution of some of these signaling compounds, it is possible that certain riboswitch-ligand partners are modern descendants of ancient signaling networks.
Regardless of the origin of the known riboswitch classes, we speculate that many additional riboswitch and signaling compound pairs remain to be discovered, albeit they might be rarer than those already known and they also might have emerged through evolutionary processes long after most RNA world systems were replaced by protein systems. As the more common riboswitch classes continue to be discovered, the pace of discovery likely will begin to slacken. However, we are confident that additional, structured ncRNAs will continue to be identified, illustrating the many and varied roles that RNA is capable of in our modern world and was capable of when life first evolved.
Materials and Methods
Chemicals and Reagents.
Cyclic dinucleotides were purchased from Biolog Life Science Institute. [γ-32P]ATP was obtained from Perkin-Elmer and used within 3 wk of receipt. Bulk chemicals were purchased from J. T. Baker unless otherwise noted. Enzymes were purchased from New England Biolabs unless otherwise noted. All solutions were prepared using deionized water (dH2O) and either autoclaved or filter-sterilized (using 0.22-μm filters, Millipore) before use.
Bioinformatic Analysis of GEMM Motif RNAs.
An updated list of RNAs conforming to the GEMM RNA motif consensus was acquired using a previously published alignment (21) as an input and Infernal 1.1 (41). The search was performed using the bacterial subset of RefSeq version 56 and metagenomic sequences from acid mine drainage, soil and whale fall, human gut, mouse gut, gutless sea worms, sludge, Global Ocean Survey scaffolds, other marine sequences, and termite hindgut (42–47). The new list of representatives contained ∼6,800 hits, which is a more than 10-fold increase from the original GEMM list. Incomplete sequences or sequences that did not reasonably fit the consensus model were removed, and the alignments of the remaining sequences were manually adjusted. Sequences within the resulting alignment were then grouped according to bacterial lineage. Because most candidates appeared within Bacillales, Clostridia, Deltaproteobacteria, and Gammaproteobacteria, these four groups were further studied as follows. Using R2R (48), the riboswitch candidates of each phylum were divided into three groups based on nucleotide identity at the 20th position: G (most common), A, or Y (U or C). Dataset S1 was then assembled listing the sequence and genomic location of each RNA, along with the genes it presumably controls.
Genes hypothesized to be controlled by c-AMP-GMP and c-di-GMP-I riboswitches in Deltaproteobacteria were then manually assigned to one of a number of different gene categories as presented in Table S2. Notably, in several instances, we observed genes typically controlled by c-AMP-GMP riboswitches upstream of the riboswitch, rather than downstream (Dataset S1).
In-Line Probing.
Generation of radiolabeled RNA transcripts and subsequent in-line probing was performed as described previously (5, 26, 27). Briefly, synthetic DNA templates containing the desired aptamer and T7 RNA polymerase promoter sequence were assembled via primer extension of overlapping forward and reverse primers using SS II reverse transcriptase. A full list of primer sequences can be found in Table S3. The resulting template was then incubated with T7 RNA polymerase and a transcription buffer mixture (80 mM Hepes, pH 7.5 at 25 °C, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT) to generate RNA. Following PAGE purification of the desired RNA, the 5′ end of the RNA was dephosphorylated (Roche Quick Dephosphorylase) and subsequently radiolabeled using γ-[32P]ATP and T4 polynucleotide kinase. The radiolabeled RNA was purified by PAGE and stored at −20 °C until further use. For in-line probing assays, trace amounts of radiolabeled RNA were incubated with various concentrations of ligand in in-line probing buffer (20 mM MgCl2, 100 mM KCl, 50 mM Tris⋅HCl, pH 8.3 at 25 °C) in the dark for 2 d at 23 °C. Partial digestions with RNase T1 or with hydroxide were conducted as described previously (5, 26, 27), and products of spontaneous RNA cleavage were separated by PAGE and quantified by using a phosphorimager.
In Vitro Transcription Assays.
Single-round in vitro transcription assays were conducted largely as described previously (5). DNA templates were assembled by PCR and contained the lysC promoter from Bacillus subtilis to initiate transcription (49) followed by the riboswitch and approximately 60 nucleotides following the intrinsic terminator stem to aid in resolution of the two products. Approximately 2 pmol of DNA template was added to a transcription initiation buffer on ice [40 mM Tris⋅HCl (pH 7.5 at 25 °C), 150 mM KCl, 1 mM MgCl2, 10 µg mL−1 BSA, 1% glycerol, 0.04 U µL−1 E. coli RNA polymerase holoenzyme (Epicenter), 1.5 µM GTP, 2.5 µM CTP, and 2.5 µM ATP]. Approximately 5 µCi α-32P-GTP was added to the transcription mix, and the reaction was allowed to proceed at 37 °C for 10 min. The reaction mixture was placed on ice, and 8-µL aliquots were added to individual tubes. One microliter of ligand and 1 µL of elongation mix (1× initiation buffer, 150 µM ATP, GTP, and CTP, 50 µM UTP, and 1 mg/mL heparin) were then added, and the reactions were incubated at 37 °C for an additional 20 min. The reaction products were analyzed by PAGE, and the extent of elongation was determined as described previously (18).
Supplementary Material
Acknowledgments
We thank Zasha Weinberg and Ken Blount for helpful discussions and Nick Carriero and Rob Bjornson for assisting in our use of the Yale Life Sciences High Performance Computing Center (NIH Grant RR19895-02). This work was supported by National Institute of Health Grants GM022778 and DE022340. G.E.P. was supported by a George J. Schulz Fellowship (Yale University); C.E.L. was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG LU1889/1-1); and S.S. was supported by an NIH Cellular and Molecular Biology Training Grant (T32-GM007223). Research in the R.R.B. laboratory is also supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419264112/-/DCSupplemental.
References
- 1.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witte G, Hartung S, Büttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30(2):167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12(6):594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110(22):9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9(12):834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51(2):226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Reports. 2013;3(5):1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321(5887):411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329(5993):845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serganov A, Patel DJ. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4(2):1–15. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11(1):29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 18.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci USA. 2007;104(41):16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regulski EE, et al. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol Microbiol. 2008;68(4):918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JE, Jr, Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492(7427):133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35(14):4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KD, et al. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16(12):1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulshina N, Baird NJ, Ferré-D’Amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16(12):1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith KD, Strobel SA. Interactions of the c-di-GMP riboswitch with its second messenger ligand. Biochem Soc Trans. 2011;39(2):647–651. doi: 10.1042/BST0390647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanahan CA, Gaffney BL, Jones RA, Strobel SA. Differential analogue binding by two classes of c-di-GMP riboswitches. J Am Chem Soc. 2011;133(39):15578–15592. doi: 10.1021/ja204650q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 27.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5(10):1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellenberger CA. (2015) GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci USA 112:5383–5388. [DOI] [PMC free article] [PubMed]
- 29.Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J Am Chem Soc. 2013;135(13):4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3(4):495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 31.Leang C, Qian X, Mester T, Lovley DR. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl Environ Microbiol. 2010;76(12):4080–4084. doi: 10.1128/AEM.00023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voordeckers JW, Kim BC, Izallalen M, Lovley DR. Role of Geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl Environ Microbiol. 2010;76(7):2371–2375. doi: 10.1128/AEM.02250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435(7045):1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 34.Leang C, Coppi MV, Lovley DR. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol. 2003;185(7):2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas M, et al. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. MBio. 2013;4(2):e00105–e00113. doi: 10.1128/mBio.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tremblay PL, et al. A c-type cytochrome and a transcriptional regulator responsible for enhanced extracellular electron transfer in Geobacter sulfurreducens revealed by adaptive evolution. Environ Microbiol. 2011;13(1):13–23. doi: 10.1111/j.1462-2920.2010.02302.x. [DOI] [PubMed] [Google Scholar]
- 37.Jonson G, Lebens M, Holmgren J. Cloning and sequencing of Vibrio cholerae mannose-sensitive haemagglutinin pilin gene: Localization of mshA within a cluster of type 4 pilin genes. Mol Microbiol. 1994;13(1):109–118. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 38.Reguera G, Pollina RB, Nicoll JS, Lovley DR. Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J Bacteriol. 2007;189(5):2125–2127. doi: 10.1128/JB.01284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bochner BR, Ames BN. ZTP (5-amino 4-imidazole carboxamide riboside 5′-triphosphate): A proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell. 1982;29(3):929–937. doi: 10.1016/0092-8674(82)90455-x. [DOI] [PubMed] [Google Scholar]
- 40.Kim PB, Nelson JW, Breaker RR. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell. 2015;57(2):317–328. doi: 10.1016/j.molcel.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinformatics. 2009;25(10):1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36(Database issue):D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gevers D, et al. The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biol. 2012;10(8):e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markowitz VM, et al. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36(Database issue):D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer F, et al. The metagenomics RAST server: A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386–393. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: The CAMERA resource. Nucleic Acids Res. 2011;39(Database issue):D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinberg Z, Breaker RR. R2R: Software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17(21):2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.