Significance
Damage-associated molecular patterns (DAMPs), released from host tissues as a consequence of pathogen attack, have been proposed as endogenous activators of immune responses in both animals and plants. Oligogalacturonides (OGs), oligomers of α-1,4–linked galacturonic acid generated in vitro by the partial hydrolysis of pectin, have been shown to function as potent elicitors of immunity when they are applied exogenously to plant tissues. However, there is no direct evidence that OGs can be produced in vivo or that they function as immune elicitors. This report provides the missing evidence that OGs can be generated in planta and can function as DAMPs in the activation of plant immunity.
Keywords: plant immunity, DAMPs, polygalacturonase, PGIP, oligogalacturonides
Abstract
Oligogalacturonides (OGs) are fragments of pectin that activate plant innate immunity by functioning as damage-associated molecular patterns (DAMPs). We set out to test the hypothesis that OGs are generated in planta by partial inhibition of pathogen-encoded polygalacturonases (PGs). A gene encoding a fungal PG was fused with a gene encoding a plant polygalacturonase-inhibiting protein (PGIP) and expressed in transgenic Arabidopsis plants. We show that expression of the PGIP–PG chimera results in the in vivo production of OGs that can be detected by mass spectrometric analysis. Transgenic plants expressing the chimera under control of a pathogen-inducible promoter are more resistant to the phytopathogens Botrytis cinerea, Pectobacterium carotovorum, and Pseudomonas syringae. These data provide strong evidence for the hypothesis that OGs released in vivo act as a DAMP signal to trigger plant immunity and suggest that controlled release of these molecules upon infection may be a valuable tool to protect plants against infectious diseases. On the other hand, elevated levels of expression of the chimera cause the accumulation of salicylic acid, reduced growth, and eventually lead to plant death, consistent with the current notion that trade-off occurs between growth and defense.
In both plants and animals, recognition of invading microbes activates an innate immune response that restricts the growth of pathogens. Both plant and animal immunity is triggered by microbe-associated molecular pattern molecules (MAMPs), such as eubacterial flagellin and peptidoglycan or fungal chitin (1). Plants also recognize pathogens indirectly by the disruption of cellular homeostatic processes caused by pathogen-encoded virulence factors [referred to as effector-triggered immunity (2)], and in Drosophila melanogaster and Caenorhabditis elegans, microbial virulence factors that block host signaling pathways or protein synthesis have recently been shown to activate immune responses (3–5). Similarly, in mammalian macrophages, inhibition of protein synthesis by Legionella pneumophila effector proteins and concurrent exposure to MAMPs leads to up-regulation of immune response genes (6). In mammals, the presence of pathogens can also be sensed indirectly by the release of low molecular weight molecules, such as DNA, uric acid, or ATP from damaged host tissues during microbial infections (7, 8), or alternatively, upon tissue injury by the release of oligosaccharides, such as hyaluronan fragments from the extracellular matrix (9). These molecules are referred to as “patterns-of-pathogenesis” (1) or “damage-associated molecular patterns” (DAMPs), a term coined in 2003 (10). In mammals, the first experimental evidence for the activation of immunity by endogenous molecules during tissue injury was provided in 1994 (11; reviewed in refs. 12–14). In plants, evidence for DAMP activity in damaged tissue extracts was first published in 1974 (15). In 1981, oligosaccharides, which were solubilized from plant cell walls by acid hydrolysis and that are rich in galacturonic acid, were shown to induce the synthesis of phytoalexins, low molecular weight antimicrobial compounds (16). Two years later the eliciting oligosaccharides were identified as oligogalacturonides (OGs), oligomers of α-1,4–linked galacturonic acid released by partial hydrolysis of homogalacturonan, a major component of pectin in the plant cell wall (17). During microbial infections, OGs are expected to be released through the action of pathogen-encoded enzymes, such as polygalacturonases (PGs) (18). In vitro, the generation of elicitor-active OGs is promoted by plant-encoded PG-inhibiting proteins (PGIPs), key components of the plant defense response, which block the complete hydrolysis of homogalacturonan to galacturonic acid (19, 20). However, the hypothesis that PGIPs are responsible for the production of OGs in vivo and, in turn, that OGs act as endogenous DAMPs during infection, has never been proven directly and relies on evidence based on the exogenous application of elicitors obtained from commercial sources of pectin.
Exogenously applied OGs with a degree of polymerization (DP) between 10 and 15 activate a wide range of defense responses, including the accumulation of phytoalexins (21, 22), the expression of defense-related genes (23), and the production of reactive oxygen species (24, 25). Notably, application of exogenous OGs protects plants against subsequent fungal infection (26, 27). OGs can bind the extracellular domain of the Arabidopsis wall-associated receptor kinase 1 (WAK1) (28–30), and construction of chimeric receptors showed that activation of WAK1 by OGs triggers downstream defense responses (31), indicating that WAK1 acts as a receptor for OGs.
The signal transduction pathway linking OG perception to the activation of the immune response has been extensively studied. OGs activate the phosphorylation of the Arabidopsis mitogen-activated protein (MAP) kinases AtMPK3 and AtMPK6 (32), and trigger a robust oxidative burst, which is required for the accumulation of callose in the cell wall (25, 32). In addition, OGs activate gene expression and induced resistance against pathogen infection independently of the defense-related hormones ethylene, salicylic acid (SA), and jasmonic acid (27). In addition to their effects on defense, OGs have been proposed to be released at low levels during plant growth by endogenous PGs and to affect a variety of physiological and developmental responses (18).
It remains to be proven, however, whether endogenously generated OGs accumulate to significant concentrations and function as signaling molecules either in intact or in pathogen-infected tissues. We therefore devised an experimental strategy to test the hypothesis that the accumulation of OGs in vivo is a consequence of the interaction of microbial PGs with plant PGIPs. We reasoned that transgenic plants engineered to accumulate equimolar levels of a fungal PG and a plant PGIP would generate endogenous OGs that would activate defense-related responses. To realize this goal, we constructed a chimeric protein by fusing PG from the fungal pathogen Fusarium phyllophilum (FpPG) to PvPGIP2, a PGIP from common bean (Phaseolus vulgaris). PvPGIP2 is the only known effective inhibitor of FpPG (33). The rationale for constructing the chimeric PGIP–PG was that the stoichiometric and simultaneous accumulation of the two proteins in the same plant cannot be easily obtained, either by crossing transgenic plants separately expressing either PG or PGIP, or by using a construct that independently expresses PG and PGIP as individual genes/proteins, because their rate of transcription, translation, posttranslational modification, and turnover would likely be different. We show here that transgenic Arabidopsis plants expressing a particular PGIP-PG chimera, referred to as the “OG machine” (OGM), accumulate OGs and exhibit enhanced resistance to a variety of pathogens, thereby providing direct evidence for the function of OGs as in vivo elicitors of the plant defense response.
Results
Different forms of chimeras, in which PvPGIP2 and FpPG were fused into a single peptide with linkers comprising seven to nine repeats of a Gly4Ser1 module, were expressed in Pichia pastoris and Arabidopsis. Relatively long linkers were used to construct these chimeras to allow intramolecular enzyme–inhibitor interactions. However, in both Pichia and Arabidopsis, the chimeras were not stable and their expression caused severe growth defects, presumably because of the proteolytic release of a highly active FpPG moiety. To circumvent this problem, a chimeric protein in which PvPGIP2 and FpPG were linked by three alanine residues was engineered. Although this linker is too short to permit intramolecular enzyme–inhibitor interactions (33), it should allow intermolecular interactions between the PGIP and PG moieties (Fig. 1A). The PGIP-(Ala)3-PG fusion protein (henceforth, PGIP–PG) was expressed in P. pastoris as a single, intact polypeptide of the expected size (Fig. 1B). The protein was purified by affinity chromatography using a Sepharose resin conjugated to the Aspergillus niger PG, PGAII, which is capable of binding the PGIP portion of the fusion protein with higher affinity than FpPG (34) (Fig. 1C). The PG activity of the purified fusion protein was strongly attenuated compared with the native FpPG, suggesting that the activity of the PG enzyme is markedly inhibited by the linked PGIP as a consequence of intermolecular interactions between different PGIP–PG chimeras (Fig. 1B). Consistent with this hypothesis, PG-specific activity [expressed as reducing group (RGU) units per milligram of protein] decreased with increasing concentrations of the chimera (Fig. S1). Previous work demonstrated that FpPG and PvPGIP2 can be specifically cross-linked by formaldehyde in vitro as a heterodimeric complex (33, 35). In vitro cross-linking of the purified PGIP–PG chimera resulted in complexes ranging from homodimers (∼160 kDa) to homotetramers (∼320 kDa), supporting the conclusion that the fusion protein is able to establish intermolecular interactions (Fig. 1C).
Fig. 1.
Biochemical characterization of the PGIP–PG chimera expressed in P. pastoris. (A) Representation of two PGIP–PG chimeric proteins forming a homodimer. PvPGIP2 (in green), FpPG (in purple), and the (Ala)3 linker are indicated. (B) (Upper) PG activity of purified PGIP–PG (220 ng) and FpPG (1 ng) evaluated by agar diffusion assay; (Lower) immunoblot analysis of PGIP–PG (220 ng) and FpPG (1 ng) samples using an antibody against FpPG. The expected molecular mass of PGIP–PG (80 kDa) and FpPG (37 kDa) are indicated. (C) SDS/PAGE analysis of purified PGIP-PG eluted from a PGAII affinity column and after chemical cross-linking by formaldehyde (OGM-CL). The molecular weight marker (M, Amersham High Molecular Weight Calibration Kit) and the calculated molecular mass of PGIP-PG monomer and multimers are indicated.
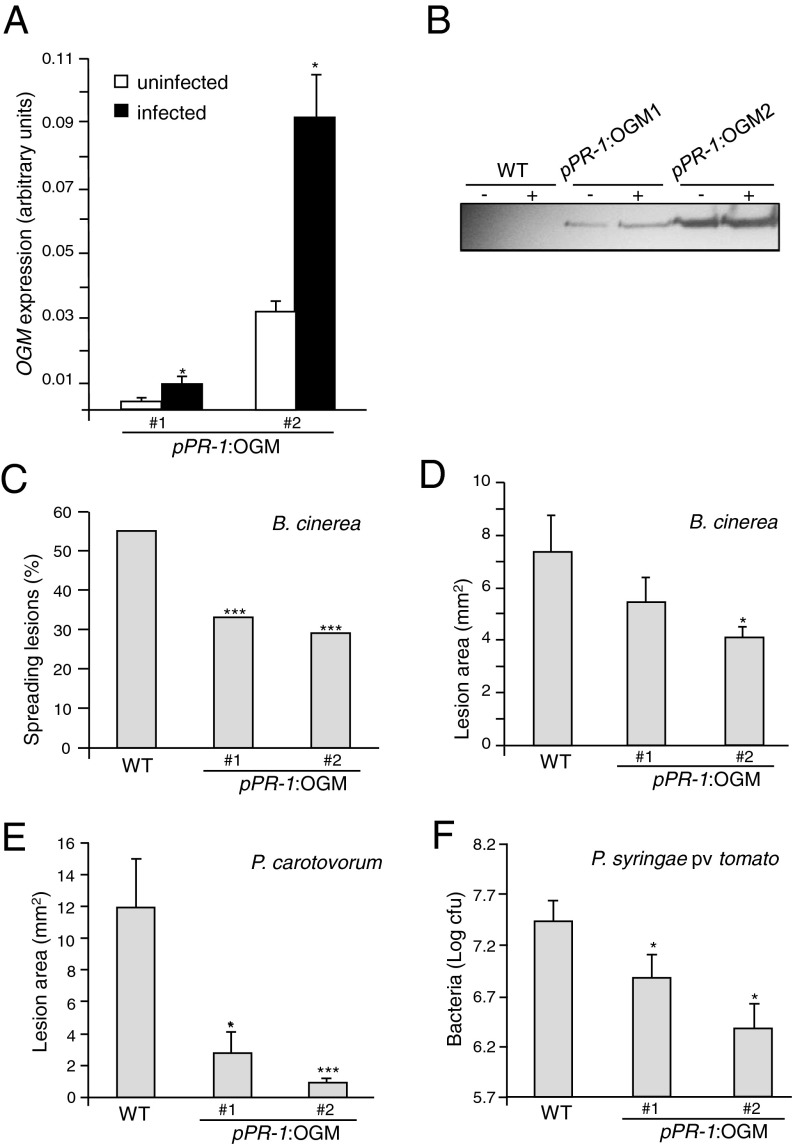
The chimeric PGIP–PG protein was expressed in Arabidopsis under the control of a β-estradiol–inducible promoter. Increasing levels of accumulation of the protein were observed in leaves of transgenic plants between 14 and 170 h after β-estradiol treatment (Fig. 2A). Increasing expression of the fusion protein was accompanied by the accumulation of increasing PG activity in crude extracts (Fig. 2B). Noninduced transgenic plants showed very low PG activity after 170 h and did not show obvious physiological effects compared with the wild-type. In contrast, at 170 h after induction with β-estradiol, leaves expressing the PGIP–PG chimera exhibited discoloration and chlorosis (Fig. 2C). β-Estradiol treatment of the transgenic plants also activated defense responses, including the accumulation of callose (Fig. 2D) and the expression of two genes previously shown to be strongly up-regulated by OGs (23), the gene At1g26380, encoding a protein with homology with reticuline oxidase, hereon indicated as RetOx, and WRKY40, encoding a transcription factor involved in the regulation of the plant immune response (36) (Fig. 2E). The same responses have been observed upon exogenous application of 100 µg/mL OGs (23, 25). β-Estradiol did not induce any defense response in wild-type or empty-vector control plants.
Fig. 2.
Expression of the PGIP–PG chimera induces defense responses. (A) PGIP–PG chimera levels determined by immunoblot upon induction with 50 µM β-estradiol. (B) PG activity evaluated by agar diffusion assay in protein extracts of Arabidopsis nontreated (−) and treated (+) with 50 µM β-estradiol. (C) A representative PGIP–PG transgenic plant without treatment or after 170 h of induction. (D) Callose staining after 140 h of induction. Both images are at the same scale. (Magnification: 10×.) (E) Defense gene induction upon treatment with 50 µM β-estradiol. The photographs shown are representative of typical results.
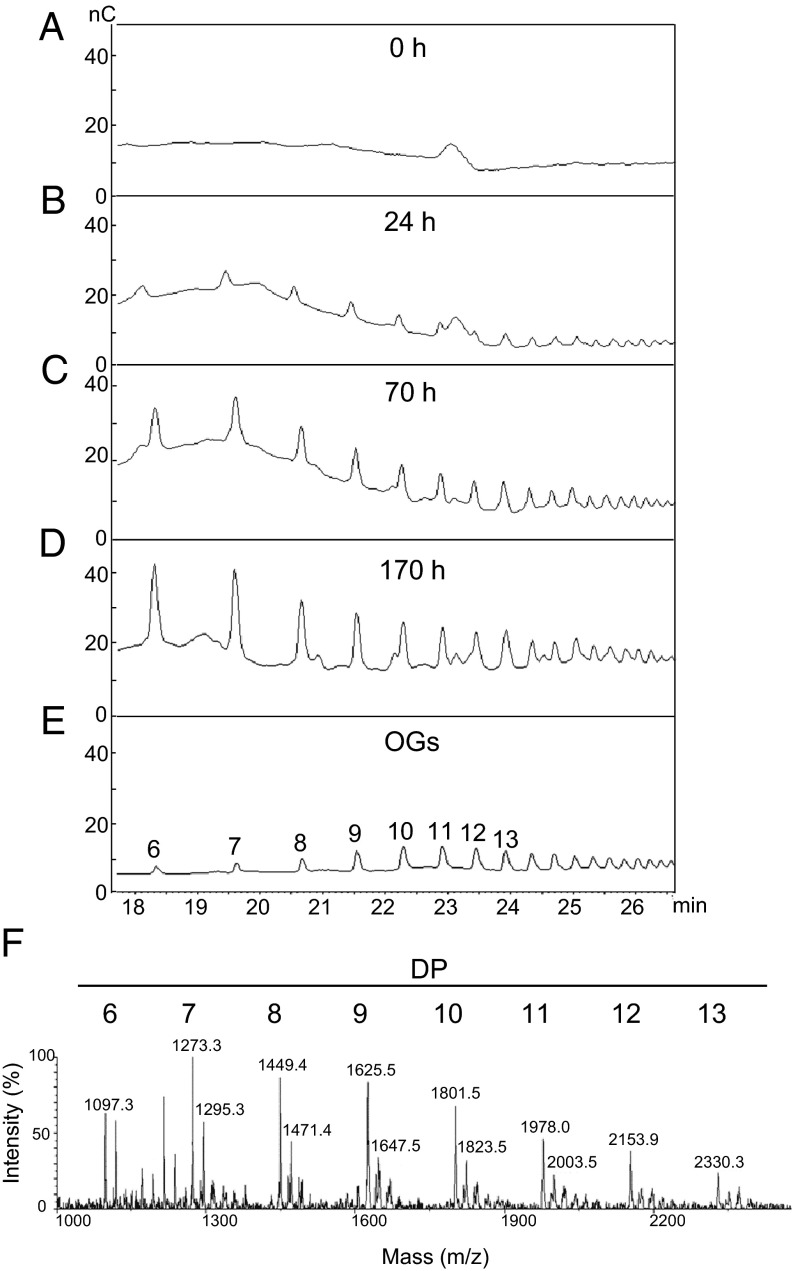
To determine whether transgenic plants expressing the PGIP–PG chimera generate OGs in vivo, pectin-enriched cell wall extracts were extracted from transgenic leaves at 0, 24, 70, and 170 h after treatment with β-estradiol. Extracts were subsequently analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), which revealed the presence of peaks with retention times comparable to those of a mixture of OGs with DP higher than 6 (Fig. 3 A–E). Mass spectrometric analysis indicated that these peaks corresponded to sodium adducts of OGs with DP 6–13 (Fig. 3F). Treatment of the fractions with FpPG strongly reduced peak areas, confirming their structure as galacturonic acid oligomers (Fig. S2). These data indicate that the PGIP–PG fusion protein, hereafter referred to as the OGM, releases active OGs when expressed in plant tissues.
Fig. 3.
Inducible release of OGs in Arabidopsis expressing the PGIP–PG chimera. (A–D) Plants expressing the inducible PGIP–PG chimera were treated for the indicated times with 50 µM β-estradiol and oligosaccharides in the cell wall pectin fraction were analyzed by HPAEC-PAD. Chromatograms indicate signal intensity (nC) at each retention time (min). (E) Representative chromatogram of purified OGs; the numbers indicate the DP. (F) MALDI-TOF analysis of the fraction in D. Numbers above peaks indicate their mass (m/z). Mass values correspond to OG sodium adducts. Numbers above mass peaks indicate the calculated DP of the corresponding oligomer.
We also generated Arabidopsis plants expressing the OGM under the control of the promoter of the Arabidopsis pathogenesis-related protein 1 (PR-1) gene, which is strongly induced by a variety of pathogens (37). Nine independent transgenic lines were obtained that did not show any obvious morphological defects either on plates or in soil. Two independent lines (pPR-1:OGM 1 and 2) were selected for further analysis and displayed, in the absence of pathogens, a detectable basal expression of the transgene, which was higher in line 2 than in line 1 (Fig. 4 A and B). After inoculation with the fungal pathogen Botrytis cinerea, a marked increase of OGM transcript levels and a less marked increase in protein levels were observed in both lines (Fig. 4 A and B). Notably, the number of successful infections by B. cinerea was significantly reduced in both transgenic lines compared with the wild-type (Fig. 4C). The average area of the lesions was also significantly smaller in the two pPR-1:OGM lines (Fig. 4D). Transgenic plants also showed a strong reduction of symptoms in response to the bacterial necrotroph Pectobacterium carotovorum (Fig. 4E), and supported 3- to 10-fold less growth than the wild-type of the bacterial hemibiotroph Pseudomonas syringae pv. tomato (Fig. 4F). Disease symptoms caused by P. syringae were markedly reduced in transgenic plants (Fig. S3). Notably, pPR-1:OGM transgenic plants displayed only symptoms typical of the infection caused by the inoculated pathogens, albeit at reduced levels compared with wild-type plants, but did not develop the chlorosis and necrosis observed in the estradiol-inducible OGM lines (Fig. 2C).
Fig. 4.
Increased resistance of plants expressing a pathogen-inducible OGM (PGIP–PG chimera). (A–D) pPR-1:OGM lines 1 and 2 were inoculated with B. cinerea. OGM transcript (A) and protein (B) levels were analyzed at 0 (−) and 48 h postinfection (hpi) (+). Bars, average ± SD, n = 3. (C) Percentage of spreading lesions (n > 60 in four combined independent experiments) and (D) average lesion area ± SE (n > 12) at 72 hpi. (E) Average lesion area ± SE, n > 12, after P. carotovorum infection. (F) P. syringae pv. tomato DC3000 growth (average ± SE, n > 6) in wild-type (WT) and transgenic plants at 72 hpi. *P < 0.05; ***P < 0.01, Fischer’s exact test (C) or Student’s t test (D–F). All experiments were repeated at least twice with similar results.
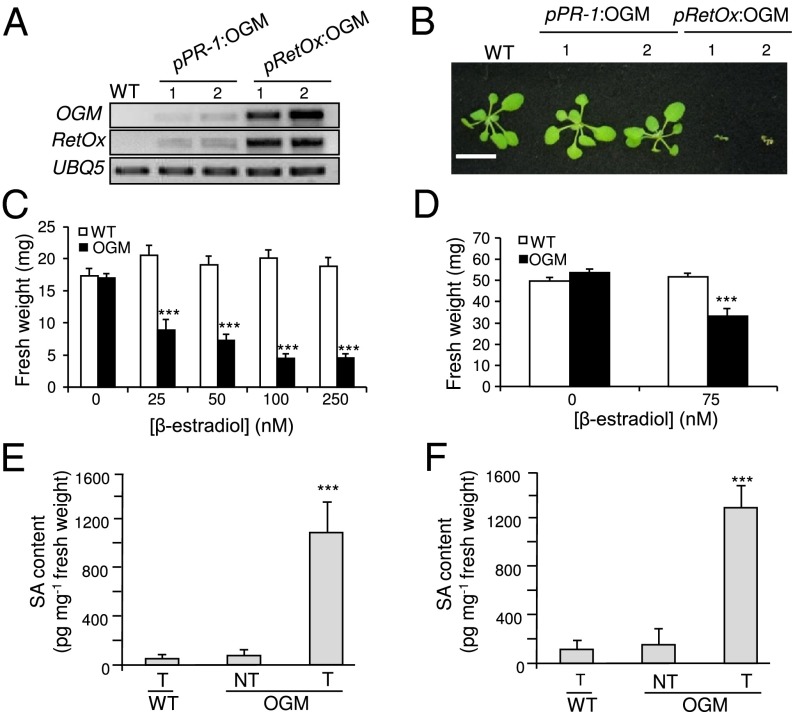
In an attempt to amplify the OG-mediated immune response via a feed-forward process, we transformed Arabidopsis with a construct for the expression of the OGM under the control of the promoter of the RetOx gene, which is strongly and rapidly induced by OGs (23). Fifteen primary pRetOx:OGM transformants were obtained. In contrast to wild-type plants, all of the T1 transgenic plants accumulated high levels of OGM and RetOx transcripts, indicating that OGs generated by the OGM were activating the RetOx promoter and that feed-forward amplification of RetOx and pRetOx:OGM transcription was occurring (Fig. 5A). However, the pRetOx:OGM transgenic plants displayed a marked dwarfism, curled leaves and reduced stem elongation, and died between 2 and 4 wk after transfer to soil (Fig. 5B).
Fig. 5.
High level of expression of OGM (PGIP–PG chimera) reduces plant growth and promotes SA accumulation. (A) Levels of OGM and RetOx transcripts in 2-wk-old wild-type (WT) plants, pPR-1:OGM 1 and 2 plants, and in two representative pRetOx:OGM T1 plants. UBQ5 was used as reference. (B) Representative photo of the same plants used in A. (Scale bar, 2 cm.) (C and D) Plants expressing the inducible OGM were germinated in the presence of β-estradiol (C) or were treated with β-estradiol 5 d after germination (D), and fresh weight was measured after 10 d (C) or 5 d (D). Bars: average (n > 10) ± SE; ***P < 0.001, statistical difference between control- and estradiol-treated seedlings (Student’s t test). (E and F) Accumulation of SA in plants expressing the inducible OGM. Four-week-old plants (E) or 10-d-old seedlings (F) were treated with β-estradiol (T) or DMSO (NT) and SA levels were determined after 24 h. Asterisks indicate statistically significant differences with β-estradiol–treated wild-type (WT) plants, according to Student’s t test (P < 0.01).
To verify that high levels of expression of the OGM impair plant growth, seeds of the transgenic line described above carrying the β-estradiol–inducible OGM construct were germinated in the presence of increasing concentrations of β-estradiol. Transgenic seedlings showed reduced biomass proportional to the level of induction (Fig. 5C), confirming that high levels of OGM expression negatively affect growth. A similar result was obtained when seedlings germinated in the absence of β-estradiol were subsequently treated with the inducer when they were 5-d-old and then harvested 5 d later (Fig. 5D). These data suggest that high levels of OGs lead to reduced growth.
The observed reduction of growth in plants expressing high levels of OGM may be a consequence of an exaggerated activation of defense responses. In particular, it is known that the phytohormone SA, which plays a key role in the response to biotic stress (38), also has significant effects on growth (39). Arabidopsis mutants that have constitutively high levels of SA, such as cpr5 (constitutive expressor of PR5) (40) and acd6-1 (accelerated cell death 6-1) (41), are dwarfs, whereas NahG (naphthalene-degrading salicylate 1-hydroxylase) transgenic plants, which accumulate lower than normal levels of SA as a consequence of NahG-mediated degradation of SA, have a higher growth rate and leaf biomass, compared with untransformed plants (42). When plants expressing the β-estradiol–inducible OGM were treated with the inducer, an almost 10-fold increase in SA levels was observed, both at the adult and at the seedling stage, within 24 h from the treatment (Fig. 5 E and F), supporting the hypothesis that the reduced biomass of these plants is caused by the OG-mediated activation of an immune response.
Discussion
By demonstrating that OGs released in planta activate immune responses and inhibit plant growth at high doses, our work provides support for the hypothesis that the extracellular matrix of plants contains signal molecules “entrapped” in a complex network of polysaccharides (43). Our strategy of constructing transgenic plants that enzymatically generate specific oligosaccharides also provides strong supporting evidence for the DAMP hypothesis in the activation of plant immunity and illustrates how hosts can distinguish pathogens from innocuous or beneficial microbes by monitoring the consequences of pathogenesis in addition to MAMPs.
Previous reports have shown either that host-derived molecules are released upon tissue damage (often referred to as DAMPs), or that these molecules, when exogenously applied, activate an immune response (reviewed in ref. 18). However, our work provides, to our knowledge, the first direct evidence of in vivo activation of an innate immune response upon release of DAMPs from the extracellular matrix of either plant or animal cells. Here, using a chimeric protein comprising a PG that degrades homogalacturonan in the plant cell wall combined with a specific PGIP, we demonstrate that the expression of the chimeric PGIP–PG protein causes the in vivo release of fragments of the plant extracellular matrix (OGs), which trigger a defense response similar to that activated when OGs are exogenously applied. The rationale for constructing the PGIP–PG chimera and expressing the chimera in transgenic plants was based on the hypothesis that PGIPs evolved not only to inactivate pathogen-encoded PGs, but also to regulate the activity of PGs so that they generate OGs of specific DP that function as elicitors of the plant defense response.
Expression of the PGIP–PG chimeric protein under the control of a pathogen-induced promoter resulted in increased resistance to infection, pointing also to a possible biotechnological strategy to protect plants against microbial diseases. A strategy that uses the OGM to generate OGs that function as DAMPs could potentially be used to engineer crops to be more resistant to microbial pathogens. Our results indicate that expression of the OGM under the control of a suitable promoter releases elicitor-active OGs in planta that leads to enhanced resistance to both fungi and bacteria. Importantly, because the OGM confers resistance against multiple pathogens with different lifestyles, it may be particularly useful for agronomic applications (44). The most widely used disease-resistance genes in traditional breeding programs, as well as for genetic engineering of crops, encode the so-called nucleotide binding site–leucine-rich repeat resistance (R) proteins, which trigger an immune response upon recognition of specific pathogen genotypes. However, resistance conferred by R genes that encode nucleotide binding site–leucine-rich repeat proteins, typically lacks durability because pathogens continually evolve genotypes that are able to evade recognition (44). Furthermore, R gene-mediated resistance is usually effective only against biotrophic pathogens but not against necrotrophic fungi and bacteria, including field and postharvest pathogens and saprophytes that cause important crop losses and mycotoxin contamination (45). Indeed, most necrotrophic pathogens attack a wide range of plants and acquisition of resistance through a single host gene is uncommon. In addition to the use of R genes to engineer enhanced disease resistance, the Arabidopsis elongation factor Tu receptor (EFR), which recognizes the bacterial MAMP EF-Tu (elongation factor thermo-unstable) (46), has been used in genetic engineering applications to confer enhanced pathogen resistance to solanaceous species (47). However, this latter strategy is likely to function only against bacterial pathogens that express EF-Tu.
In contrast to the use of immune receptors to engineer disease resistance, engineering the endogenous production of DAMPs may broaden the range of pathogens that can be targeted and may be useful in conferring a broad-spectrum resistance against many microbes. The case of OGs is paradigmatic because OG-mediated activation of defenses occurs in many plants and is effective against many microbes (18, 48–50). The majority of pathogenic fungi and bacteria need to breach the cell wall to infect or extract nutrients from plant tissues and, therefore, produce an array of pectic and other cell wall-degrading enzymes. Both dicots and monocots have evolved PGIPs that counteract the activities of microbial PGs (51). PGIPs have been used to protect plants but their utilization is limited by the restricted specificity of these inhibitors (52). The use of the OGM, however, may circumvent susceptibility to those microbial pathogens producing PGs that are not recognized and inhibited by the PGIPs occurring in their corresponding host plants.
It is noteworthy that when the OGM was induced at high levels, plant growth was significantly reduced or arrested, indicating that high concentrations of endogenous OGs interfere with normal developmental programs. Like in animals, it appears that an exaggerated release of endogenous danger signals leads to a deleterious hyperimmune response. This finding is consistent with the current notion that trade-off occurs between growth potential and the capacity for defense. Maintenance of immunity is costly and immune responses are typically counterbalanced by decreasing the allocation of resources to biomass production (53). For example, it has been previously shown that alterations of the biosynthesis of lignin precursors that cause the accumulation of SA in Arabidopsis strongly reduce plant growth (54), similar to what we observed upon the overexpression of the OGM.
Although it seems likely that the effects on plant growth accompanying overexpression of the OGM are a direct consequence of OG-elicited production of SA, OGs have also been proposed to function directly as growth regulators (18). The plant cell wall is constantly subjected to remodeling and OGs may normally be released at low doses by endogenous PGs to regulate growth processes. In the experiments reported here, we cannot determine whether OGs can also function directly as growth regulators because we only observed an effect on growth when the OGM was overexpressed, conditions that also resulted in the production of abnormally high levels of SA. Growth regulators that are involved in development are also key elements of immune response cascades and immune elicitors often inhibit auxin responses (18). Plants that constitutively express defense responses are often dwarf (55, 56).
In conclusion, the data presented in this report show that in vivo-generated OGs activate a potent immune response that confers resistance to pathogen attack, but at the same time can negatively affect plant growth and development. Importantly, the molecular mechanisms underlying the effects of OGs are still poorly understood and more work is needed to distinguish whether OGs function directly as bona fide growth regulators or whether the effects of OGs on growth and development are a secondary consequence of the activation of an immune response. The OGM may be a useful tool in this regard. For example, developmentally controlled expression of the OGM may be used to further study the roles of OGs in defense and development under physiological conditions.
Materials and Methods
Bioassays.
Callose deposition was analyzed by UV epifluorescence in leaves of 4-wk-old plants stained with Aniline Blue. For Arabidopsis growth assay, β-estradiol was supplied to the medium either before germination or after 5 d of growth and fresh biomass was collected after 10 or 5 d, respectively, for evaluation.
Infection Assays.
Pathogenicity assays with B. cinerea and P. syringae DC3000 were performed on leaves of 4-wk-old plants. Pathogenicity assay with P. carotovorum was carried out on detached leaves of 4-wk-old plants inoculated with a 5 × 107 cfu/mL bacterial suspension.
Isolation and Detection of Oligogalacturonides.
The AIS and pectin were prepared from about 100 mg of leaf tissue of wild-type and transgenic plants treated with 50 μM β-estradiol. Oligomers in the pectin fractions were analyzed by HPAEC-PAD chromatography and by mass spectrometric analysis.
Extraction of SA and Detection by LC-MS.
SA was extracted from leaves of 4-wk-old plants and seedlings (∼50–100 mg) 24 h after induction with β-estradiol. Samples were dissolved in methanol at 1:1 ratio [tissue (mg):methanol (μL)] and analyzed by liquid chromatography coupled to mass spectrometry.
Further information is provided in SI Materials and Methods. See Table S1 for primers used for the construction of the different OGM cassettes.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council Advanced Grant 233083 “FUEL-PATH” (to F.C.); the Ministero delle Politiche Agricole, Alimentari e Forestali Grant BIOMASSVAL (to F.C.); Grant FITOLISI (to G.D.L.); Ministero dell’Università e della Ricerca Scientifica Grant PRIN2009 (to G.D.L.); the Institute Pasteur–Fondazione Cenci Bolognetti; a Banting Postdoctoral fellowship (to Z.C.); and National Science Foundation Grant IOS-0929226 and National Institutes of Health Grant R37-GM48707 (to F.M.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504154112/-/DCSupplemental.
References
- 1.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Boyer L, et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35(4):536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11(4):364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11(4):375–386. doi: 10.1016/j.chom.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontana MF, et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7(2):e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8(4):279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer L. Complexity of danger: The diverse nature of damage-associated molecular patterns. J Biol Chem. 2014;289(51):35237–35245. doi: 10.1074/jbc.R114.619304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheibner KA, et al. Hyaluronan fragments act as an endogenous danger signal by engaging TLR2. J Immunol. 2006;177(2):1272–1281. doi: 10.4049/jimmunol.177.2.1272. [DOI] [PubMed] [Google Scholar]
- 10.Land W. Allograft injury mediated by reactive oxygen species: from conserved proteins of Drosophila to acute and chronic rejection of human transplants. III. Interaction of (oxidative) stress-induced heat shock proteins with Toll-like receptor-bearing cells of innate immunity and itsconsequences for the development of acute and chronic allograft rejection. Transplant Rev. 2003;17(2):67–86. [Google Scholar]
- 11.Land W, et al. The beneficial effect of human recombinant superoxide dismutase on acute and chronic rejection events in recipients of cadaveric renal transplants. Transplantation. 1994;57(2):211–217. doi: 10.1097/00007890-199401001-00010. [DOI] [PubMed] [Google Scholar]
- 12.Matzinger P. Friendly and dangerous signals: Is the tissue in control? Nat Immunol. 2007;8(1):11–13. doi: 10.1038/ni0107-11. [DOI] [PubMed] [Google Scholar]
- 13.Land WG, Messmer K. The danger theory in view of the injury hypothesis: 20 years later. Front Immunol. 2012;3:349. doi: 10.3389/fimmu.2012.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pradeu T, Cooper EL. The danger theory: 20 years later. Front Immunol. 2012;3:287. doi: 10.3389/fimmu.2012.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFarland D, Ryan CA. Proteinase inhibitor-inducing factor in plant leaves: A phylogenetic survey. Plant Physiol. 1974;54(5):706–708. doi: 10.1104/pp.54.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn MG, Darvill AG, Albersheim P. Host-pathogen interactions: XIX. The endogenous elicitor, a fragment of a plant cell wall polysaccharide that elicits phytoalexin accumulation in soybeas. Plant Physiol. 1981;68(5):1161–1169. doi: 10.1104/pp.68.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nothnagel EA, McNeil M, Albersheim P, Dell A. Host-pathogen interactions: XXII. A galacturonic acid oligosaccharide from plant cell walls elicits phytoalexins. Plant Physiol. 1983;71(4):916–926. doi: 10.1104/pp.71.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari S, et al. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci. 2013;4:49. doi: 10.3389/fpls.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cervone F, De Lorenzo G, Degrà L, Salvi G, Bergami M. Purification and characterization of a polygalacturonase-inhibiting protein from Phaseolus vulgaris L. Plant Physiol. 1987;85(3):631–637. doi: 10.1104/pp.85.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervone F, Hahn MG, De Lorenzo G, Darvill A, Albersheim P. Host-pathogen interactions: XXXIII. A plant protein converts a fungal pathogenesis factor into an elicitor of plant defense responses. Plant Physiol. 1989;90(2):542–548. doi: 10.1104/pp.90.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis KR, Darvill AG, Albersheim P, Dell A. Host-pathogen interactions: XXIX. Oligogalacturonides released from sodium polypectate by endopolygalacturonic acid lyase are elicitors of phytoalexins in soybean. Plant Physiol. 1986;80(2):568–577. doi: 10.1104/pp.80.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin DF, West CA. Characteristics of galacturonic acid oligomers as elicitors of casbene synthetase activity in castor bean seedlings. Plant Physiol. 1984;74(4):989–992. doi: 10.1104/pp.74.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denoux C, et al. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant. 2008;1(3):423–445. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellincampi D, Dipierro N, Salvi G, Cervone F, De Lorenzo G. Extracellular H(2)O(2) induced by oligogalacturonides is not involved in the inhibition of the auxin-regulated rolB gene expression in tobacco leaf explants. Plant Physiol. 2000;122(4):1379–1385. doi: 10.1104/pp.122.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galletti R, et al. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 2008;148(3):1695–1706. doi: 10.1104/pp.108.127845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aziz A, Heyraud A, Lambert B. Oligogalacturonide signal transduction, induction of defense-related responses and protection of grapevine against Botrytis cinerea. Planta. 2004;218(5):767–774. doi: 10.1007/s00425-003-1153-x. [DOI] [PubMed] [Google Scholar]
- 27.Ferrari S, et al. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007;144(1):367–379. doi: 10.1104/pp.107.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He ZH, Fujiki M, Kohorn BD. A cell wall-associated, receptor-like protein kinase. J Biol Chem. 1996;271(33):19789–19793. doi: 10.1074/jbc.271.33.19789. [DOI] [PubMed] [Google Scholar]
- 29.Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13(2):303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decreux A, Messiaen J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005;46(2):268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- 31.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA. 2010;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galletti R, Ferrari S, De Lorenzo G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011;157(2):804–814. doi: 10.1104/pp.111.174003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benedetti M, et al. Structural resolution of the complex between a fungal polygalacturonase and a plant polygalacturonase-inhibiting protein by small-angle X-ray scattering. Plant Physiol. 2011;157(2):599–607. doi: 10.1104/pp.111.181057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leckie F, et al. The specificity of polygalacturonase-inhibiting protein (PGIP): A single amino acid substitution in the solvent-exposed beta-strand/beta-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J. 1999;18(9):2352–2363. doi: 10.1093/emboj/18.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benedetti M, et al. A single amino-acid substitution allows endo-polygalacturonase of Fusarium verticillioides to acquire recognition by PGIP2 from Phaseolus vulgaris. PLoS ONE. 2013;8(11):e80610. doi: 10.1371/journal.pone.0080610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18(5):1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6(11):1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 39.Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot. 2011;62(10):3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 40.Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9(9):1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 1999;11(9):1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abreu ME, Munné-Bosch S. Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J Exp Bot. 2009;60(4):1261–1271. doi: 10.1093/jxb/ern363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darvill A, et al. Oligosaccharins involved in plant growth and host-pathogen interactions. Biochem Soc Symp. 1994;60:89–94. [PubMed] [Google Scholar]
- 44.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341(6147):746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mengiste T. Plant immunity to necrotrophs. Annu Rev Phytopathol. 2012;50:267–294. doi: 10.1146/annurev-phyto-081211-172955. [DOI] [PubMed] [Google Scholar]
- 46.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 47.Lacombe S, et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol. 2010;28(4):365–369. doi: 10.1038/nbt.1613. [DOI] [PubMed] [Google Scholar]
- 48.Ridley BL, O’Neill MA, Mohnen D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry. 2001;57(6):929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 49.Mohnen D, Hahn MG. Cell wall carbohydrates as signals in plants. Semin Cell Biol. 1993;4(2):93–102. doi: 10.1006/scel.1993.1012. [DOI] [PubMed] [Google Scholar]
- 50.De Lorenzo G, Ferrari S. Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr Opin Plant Biol. 2002;5(4):295–299. doi: 10.1016/s1369-5266(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 51.Casasoli M, et al. Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proc Natl Acad Sci USA. 2009;106(18):7666–7671. doi: 10.1073/pnas.0812625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borras-Hidalgo O, Caprari C, Hernandez-Estevez I, Lorenzo GD, Cervone F. A gene for plant protection: Expression of a bean polygalacturonase inhibitor in tobacco confers a strong resistance against Rhizoctonia solani and two oomycetes. Front Plant Sci. 2012;3:268. doi: 10.3389/fpls.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Todesco M, et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465(7298):632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA. Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA. 2011;108(51):20814–20819. doi: 10.1073/pnas.1117873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vos IA, Pieterse CMJ, Van Wees SCM. Costs and benefits of hormone-regulated plant defences. Plant Pathology. 2013;62(Supplement S1):43–55. [Google Scholar]
- 56.Anurag A, Agrawal RK. The Ecology and Evolution of Inducible Defenses. Princeton Univ Press; Princeton, NJ: 1999. pp. 45–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.