Significance
Microbes drive biogeochemical cycles across the globe, collectively playing a central role in shaping the biosphere. Despite their immense importance, the in situ activities of communities of microbes, in particular uncultivated lineages of “microbial dark matter,” remain poorly elucidated. In this study, we report that common temporal and ecological dynamics underpin disparate marine microbial communities, providing the first evidence that trans-Pacific diurnal transcriptional patterns in these communities may regulate ecological and biogeochemical processes across the ocean. In total, our findings indicate a remarkable regularity in the timing of community-wide activity in the ocean, and suggest that global patterns of a variety of biogeochemical transformations may be temporally predictable and governed by structured ecological determinants.
Keywords: microbial oceanography, transcriptional networks, bacterioplankton, systems biology, diel cycles
Abstract
Planktonic microbial communities in the ocean are typically dominated by several cosmopolitan clades of Bacteria, Archaea, and Eukarya characterized by their ribosomal RNA gene phylogenies and genomic features. Although the environments these communities inhabit range from coastal to open ocean waters, how the biological dynamics vary between such disparate habitats is not well known. To gain insight into the differential activities of microbial populations inhabiting different oceanic provinces we compared the daily metatranscriptome profiles of related microbial populations inhabiting surface waters of both a coastal California upwelling region (CC) as well as the oligotrophic North Pacific Subtropical Gyre (NPSG). Transcriptional networks revealed that the dominant photoautotrophic microbes in each environment (Ostreococcus in CC, Prochlorococcus in NPSG) were central determinants of overall community transcriptome dynamics. Furthermore, heterotrophic bacterial clades common to both ecosystems (SAR11, SAR116, SAR86, SAR406, and Roseobacter) displayed conserved, genome-wide inter- and intrataxon transcriptional patterns and diel cycles. Populations of SAR11 and SAR86 clades in particular exhibited tightly coordinated transcriptional patterns in both coastal and pelagic ecosystems, suggesting that specific biological interactions between these groups are widespread in nature. Our results identify common diurnally oscillating behaviors among diverse planktonic microbial species regardless of habitat, suggesting that highly conserved temporally phased biotic interactions are ubiquitous among planktonic microbial communities worldwide.
Understanding the relationships between microbial community dynamics and ecosystem processes remains a central goal of microbial ecology (1–4). Such linkages are beginning to be explored in ocean surface waters, where a relatively small number of broadly-defined phylogenetic groups from all three domains of life dominate planktonic microbial communities worldwide (5–7). Both culture-dependent and -independent studies are increasing our knowledge of the metabolic properties and ecological dynamics of individual clades (3, 6, 8), yet it remains largely unknown how these microorganisms interact with one other and their environment to shape whole-ecosystem processes and biogeochemical fluxes.
Although the in situ dynamics of planktonic microbial assemblages have typically been difficult to evaluate, recent development in Lagrangian robotic sampling coupled with subsequent metatranscriptomic sequencing have allowed for daily whole-community temporal dynamics to be assessed (9–12). These techniques use an Environmental Sample Processor (ESP) that autonomously collects and preserves environmental samples while floating freely with a planktonic assemblage (12). Using these approaches it is now possible to evaluate not only the activities of individual microbial populations but also the temporal dynamics that define their ecological niches. Moreover, comparison of the temporal dynamics between microbial lineages and analysis of shared intertaxon metabolic patterns permits investigation into the factors that give rise to ecosystem-level phenomena, and how these factors may vary between environments.
In this study we used a comparative approach to identify common temporal features among phylogenetically related planktonic groups that inhabit drastically disparate marine environments. Our analysis leveraged a five-day, high-resolution metatranscriptomic time-series from surface waters off the northern California coast (CC) (this report), together with a metatranscriptomic time series constructed from surface waters of the North Pacific Subtropical Gyre (NPSG) (9). Although many phylogenetically similar microbes inhabit these environments, the two ecosystems are environmentally distinct. Waters of the California Current waters experience marked seasonality, are much colder, and are relatively nutrient rich compared with the NPSG (13), which is characterized by warmer, nutrient poor surface waters with little seasonal variability (14). High temporal resolution, whole-community metatranscriptome time series from microbial populations inhabiting these environments offered a unique opportunity to compare microbial community dynamics in these different oceanic provinces.
Results and Discussion
As expected, different photoautotrophic primary producers dominated the coastal versus open ocean metatranscriptomes (Ostreococcus in CC and Prochlorococcus in NPSG) (15–17) (SI Appendix, Fig. S1 A and B and Dataset S3). In contrast, many dominant heterotrophic bacterioplankton clades were common to community transcripts in both habitats, including the SAR11, SAR86, SAR116, SAR406, and Roseobacter groups (SI Appendix, Fig. S1 A and B). We also identified populations of Flavobacteria, Marine Group II (MGII) Euryarchaeota, SAR92 and ARCTIC96-BD19 in the new coastal dataset, and a previously reported SAR324 population in the pelagic samples (Table 1 and SI Appendix, Fig. S1 A and B) (9). Mapping of reads onto reference genomes (95% identity, minimum 150 bp match length) indicated the overall phylogenetic profiles for the coastal and pelagic metatranscriptomes were largely non-overlapping (SI Appendix, Fig. S2), suggesting that although common heterotrophic clades were shared between the two habitats, their population-level genotypes were distinct. During the California coast drift, the relative abundance of reads assigned to Ostreococcus and MGII Euryarchaeota declined, whereas those assigned to Flavobacteria, SAR92, and ARCTIC96-BD19 increased. These changes were concomitant with increases in salinity and fluctuations in temperature (SI Appendix, Fig. S1C), potentially indicating movement of the ESP through distinct water masses. Similar albeit less severe variability in temperature and salinity was observed in the NPSG drift, although transcript abundance across phylogenetic groups was more stable in this dataset (SI Appendix, Fig. S1 B and D).
Table 1.
Summary of the California coast and North Pacific Subtropical Gyre metatranscriptomes
| Parameter | California coast | North Pacific Subtropical Gyre |
| Time points | 35 | 30 |
| Approx. sampling frequency | 4 h | 2 h |
| Duration of drift | 9/12–17, 2012 (5 1/2 d) | 9/7–11, 2011 (2 1/2 d) |
| No. of transcripts* | ||
| ARCTIC96-BD19 | 623 (50) | — |
| Flavobacteria | 376 (1) | — |
| GII Euryarchaeota | 689 (34) | — |
| Roseobacter | 867 (99) | 542 (165) |
| Ostreococcus | 1,602 (781) | — |
| Prochlorococcus | — | 2,052 (1,464) |
| SAR11 | 1,300 (296) | 1,223 (270) |
| SAR86 | 1,400 (31) | 795 (39) |
| SAR92 | 404 (2) | — |
| SAR116 | 969 (146) | 1,661 (146) |
| SAR324 | — | 513 (28) |
| SAR406 | 615 (0) | 490 (17) |
Total number of transcripts (those with significant 24-h periodicity).
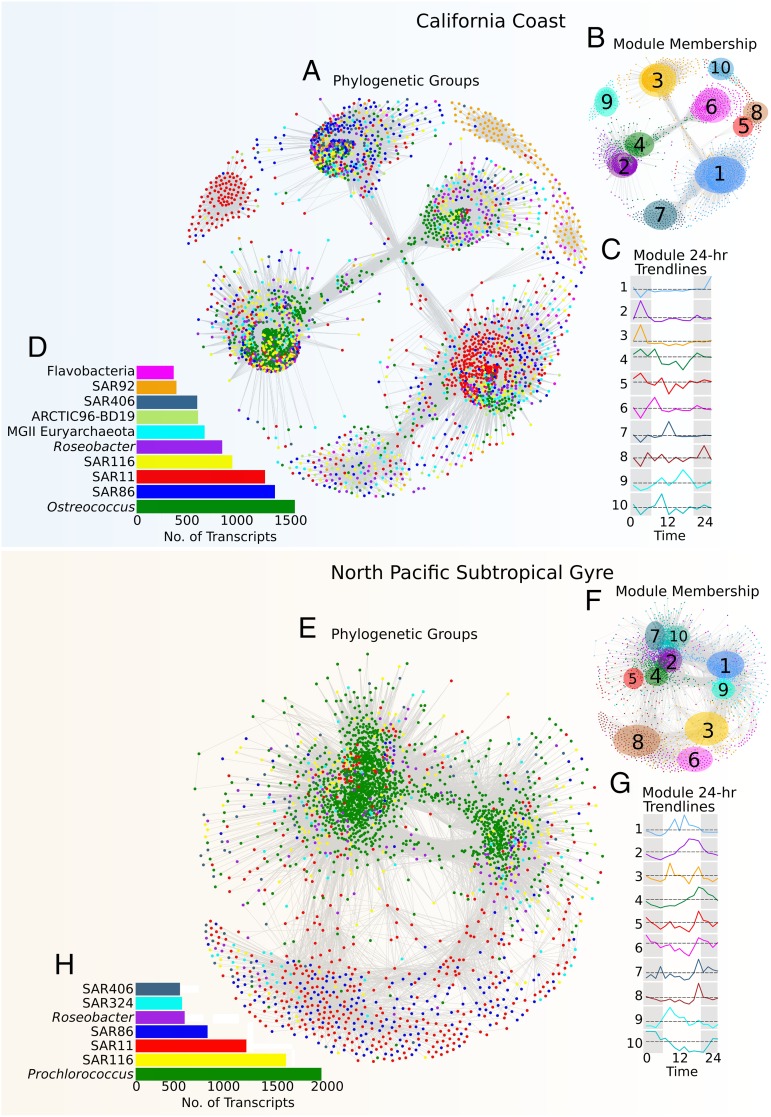
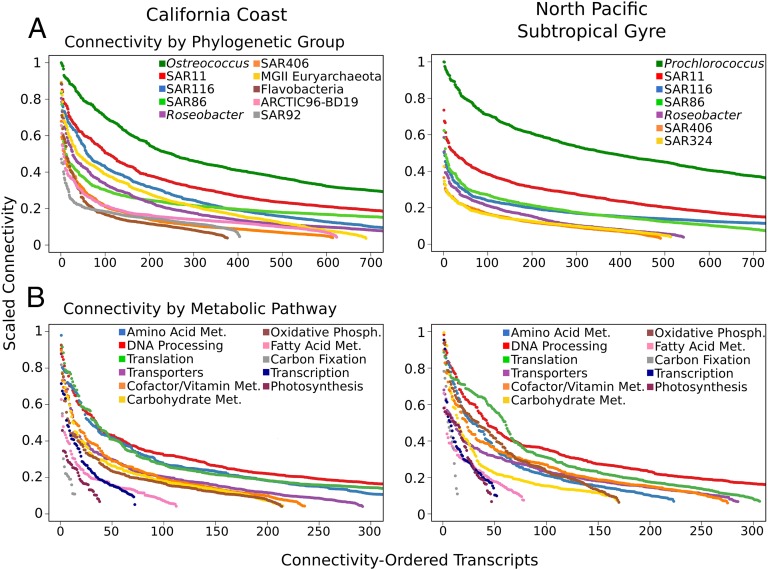
We analyzed the temporal dynamics of reads mapping to 8,844 and 7,276 orthologous protein clusters (henceforth referred to as “transcripts”) with high abundance in the coastal and pelagic metatranscriptomes, respectively, and compared the daily transcriptional patterns within and between different phylogenetic groups using a weighted transcriptomic network approach as implemented by the WGCNA package (18, 19) (Methods). Results of these analyses suggested that the majority of transcripts in both the coastal and pelagic transcriptomic networks clustered around the transcripts derived from the dominant photoautotrophic members of each community (Fig. 1 A and D). Moreover, Ostreococcus (CC) and Prochlorococcus (NPSG) transcripts were found to consistently have the highest connectivity (a.k.a. degree centrality) of all nodes in the networks (Fig. 2A and Dataset S1), likely reflecting the central role of photoautotrophic community members in determining the overall community transcriptional profiles in both environments. Analysis of sets of transcripts with similar temporal profiles (termed modules; colored groupings in Fig. 1 B and E) revealed distinct patterns of 24-h periodic expression (Fig. 1 C and F), consistent with the underlying structuring of the transcriptomic networks around the well defined diel cycles of the photoautotrophic members.
Fig. 1.
Whole-community transcriptional networks in coastal and pelagic marine microbial plankton are predominantly structured around the 24-h metabolic cycles of the photoautotrophic members. Whole-community transcriptome networks represent correlations across transcripts identified in California coast (CC) and North Pacific Subtropical Gyre (NPSG) planktonic microbial assemblages. Networks are colored according to microbial group (CC: A; NPSG: E) and module (i.e., set of coexpressed transcripts; CC: B; NPSG: F). Nodes in the network represent transcripts, whereas edges represent high pair-wise topological overlap scores between transcripts. Inset bar charts represent the total number of high abundance transcripts to which reads could be assigned in each dataset (Methods). Trendlines representing the average 24-h expression profile for the 10 largest transcriptional modules are given (CC: C; NPSG: G), with colors matching module assignments and dashed lines indicating profile averages. Both correlated and anticorrelated transcripts are present in each module, with the overall profile aligned to the predominant expression value identified. The overall number of transcripts identified in each phylogenetic group is given in the bar charts (CC: D; NPSG: H), with colors matching those shown in the main networks (A and E).
Fig. 2.
Transcripts from both the California coast and NPSG metatranscriptomes belonging to photoautotrophic community members (A) and those involved in key metabolic processes such as amino acid metabolism, DNA processing, and translation (B) have the highest connectivity (degree centrality) in transcriptional networks. The scaled connectivity (degree centrality) of transcripts is given on the y-axes, with transcripts ordered highest to lowest based on taxonomic assignment and KEGG annotation. Met: Metabolism; Phosph.: Phosphorylation.
All transcriptional modules identified within weighted networks (34 in CC, 18 in NPSG) included representative transcripts from numerous phylogenetic groups, indicating that similar, highly regular transcriptional coordination was shared between microbial populations (SI Appendix, Fig. S3). Moreover, each module contained a broad representation of functional annotations both within and between taxa, reflecting coordinated regulation across metabolic pathways in different phylogenetic groups (SI Appendix, Figs. S4 and S5). Transcripts from different taxa involved in cofactor and vitamin metabolism and DNA processing often clustered in the same modules, as did those involved in oxidative phosphorylation and protein and RNA processing (SI Appendix, Figs. S4 and S5). Analysis of the functional annotation of transcripts across phylogenetic groups revealed that amino acid metabolism, DNA processing, translation, transporters, and vitamin metabolism consistently had the highest connectivity, indicating that these are potentially among the most tightly regulated metabolic processes (Fig. 2B). This pattern could be the result of intense competition for resources in both coastal and pelagic surface waters, which would render nutrient availability and the timing of growth and DNA replication as key determinants of the temporal dynamics in these planktonic assemblages. The abundance and connectivity of transcripts appeared to be associated in the pelagic but not coastal datasets (SI Appendix, Fig. S6), possibly reflecting a propensity of central metabolic genes critical for cellular processes to be highly expressed, consistent with previous findings (20). These patterns may also be due in part to a tendency for connectivity patterns to be more easily identified in highly expressed transcripts, although a lack of connectivity-abundance association in the coastal dataset suggests this is not an overriding factor. Together with the high connectivity of transcripts belonging to photoautotrophic groups, these results suggest that transcriptomic networks of both coastal and pelagic environments are largely structured by a combination of the diel cycles of the photoautotrophic members together with resource limitation common to both habitats.
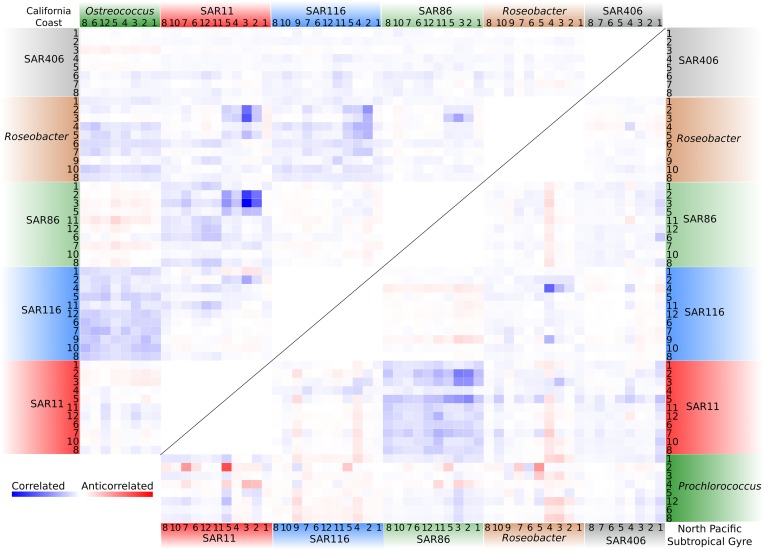
In addition to shared, intrahabitat community-wide metatranscriptome dynamics, similar transcriptional patterns were also observed between the heterotrophic bacterioplankton of coastal and pelagic environments. Using the transcriptomic networks as a foundation, we calculated the correlation of nodes belonging to the two photoautotrophic groups and five shared heterotrophic clades across different metabolic pathways (Fig. 3). We found that patterns of intertaxa co-correlation in the coastal and pelagic metatranscriptomes mirrored each other, with close coordination across similar metabolic pathways in the related heterotrophic populations of both ecosystems. This was particularly apparent for the SAR11 and SAR86 populations, which in both environments displayed tightly linked transcriptional profiles across a variety of functions including translation, transcription, and oxidative phosphorylation (Fig. 3). A similar but less apparent relationship was also found between the SAR116 and Roseobacter clades in both habitats (Fig. 3). In contrast, although SAR116 transcription patterns appear closely linked to Ostreococcus in the California coast, a similar relationship between SAR116 and Prochlorococcus was not observed in pelagic samples, suggesting that interactions between this group and different photoautotrophic primary producers may not be highly conserved.
Fig. 3.
Patterns of interspecies transcriptional coordination are conserved across coastal and pelagic planktonic microbial communities. Correlated and anticorrelated functional pathways in the photoautotrophic and shared heterotrophic groups are shown for the California coast (CC, Upper Left) and North Pacific Subtropical Gyre (NPSG, Lower Right) metatranscriptomes. Pathway correlations were calculated from scaled adjacency matrixes by averaging across the correlations of the cognate transcripts (Methods). Pathways: 1: Transporters; 2: Translation; 3: Transcription: 4: Protein/RNA processing; 5: Oxidative phosphorylation; 6: Cofactor and vitamin metabolism; 7: Citrate cycle; 8: Amino acid metabolism; 9: Chemotaxis and flagellar assembly; 10: Central carbon metabolism; 11: Glycolysis; 12: DNA and chromosome processing.
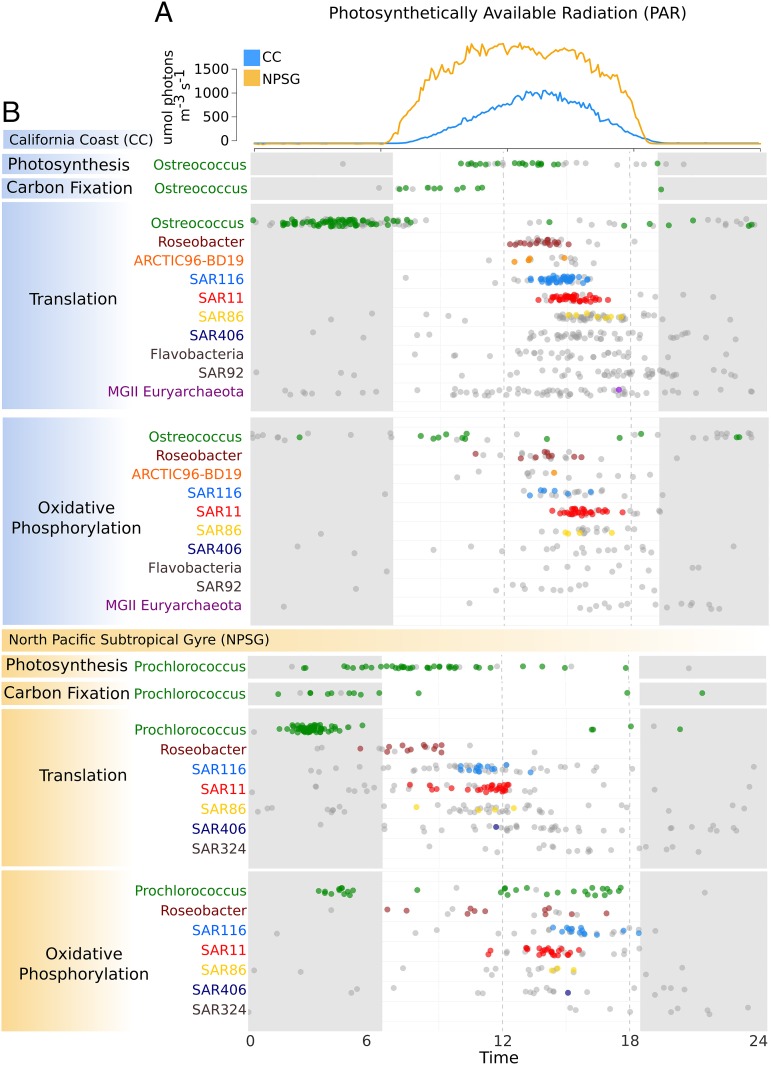
We identified numerous transcripts from heterotrophic groups that displayed significant 24-h periodicity in both coastal and pelagic metatranscriptome time series, consistent with a recent report for the NPSG dataset (9) (Table 1). The diel signal in heterotrophic groups in both environments was most prevalent in transcripts associated with translation and oxidative phosphorylation (Fig. 4B), which are useful indicators of overall metabolic activity (21). In both the coastal and pelagic habitat, the peak abundance of photoautotroph carbon fixation and photosynthesis transcripts was followed by a tight “cascade” of translation and oxidative phosphorylation transcript maxima from heterotrophic groups (Fig. 4B). This pattern of daily transcript maxima that proceeded temporally from Roseobacter to SAR116 to SAR11 and SAR86 was strikingly similar between coastal and pelagic environments, suggesting conservation of temporal associations in community-wide diel cascades among these phylogenetic groups across the Pacific Ocean basin. The pattern was less compressed and occurred earlier in the day in pelagic populations, probably due to the earlier daily light influx (Photosynthetically Available Radiation, PAR; Fig. 3A) that signals the start of photosynthesis in this habitat. Although periodic patterns in lower abundance groups were more difficult to discern, their general timing of translation and oxidative phosphorylation transcript maxima were similar to the other more abundant heterotrophic groups. Overall, the timing of these events suggested that regardless of taxon, the activities of abundant photoautotrophs in both coastal and pelagic habitats “set” the whole-community diel cycle.
Fig. 4.
Community-wide transcriptional cascades in key metabolic processes are conserved in the coastal and pelagic planktonic microbial communities sampled. Timing on the x-axis is given over a 24-h interval using data aggregated throughout the sampling periods. Photosynthetically Available Radiation profiles averaged over the duration of the ESP drifts (PAR, A) and metabolic transcriptional profiles calculated using harmonic regression in the California coast (B, in blue) and NPSG (B, in orange) planktonic groups are given. Colored dots represent the peak times of transcripts with statistically significant 24-h periodicity, whereas gray dots represent the peak times of transcripts without a significant diel component to their expression. Nighttime periods are indicated with gray shading.
We postulate that for heterotrophs this pattern of translation/oxidative phosphorylation transcript maxima is initiated and sustained by the progressive release of key nutrients from primary producers, followed by successive, temporally ordered metabolic conversions mediated by different heterotrophic community members throughout the day. For heterotrophic groups, the same transcriptional modules that contained the most translation and oxidative phosphorylation transcripts (9 and 15 in CC, 3 in NPSG) also contained a number of transcripts belonging to predicted sugar and amino acid transporters, peptidases, and genes involved in amino acid metabolism and the citric acid cycle (SI Appendix, Figs. S4 and S5 and Dataset S1), suggesting that an increase in the availability of certain compounds in marine dissolved organic matter (DOM) may spur their transcriptional upsurges. Supporting this hypothesis, distinct transcriptional responses to elevated DOM have been shown to occur in heterotrophic bacterioplankton studied in microcosm experiments (22). Given that Ostreococcus and Prochlorococcus likely produce distinct suites of dissolved organic matter (23), this would suggest that the temporal succession of microbial activity may exist regardless of the specific compounds involved.
Metabolic patterns in heterotrophic groups could also be inferred from other pathways. In particular, reads mapping to the glyoxylate shunt enzymes isocitrate lyase and malate synthase were highly abundant in both metatranscriptomes (SI Appendix, Fig. S7), consistent with findings that this pathway plays a key role in carbon provisioning in bacterioplankton (24). Overall, transcripts mapping to ribosomal proteins, oxidative phosphorylation enzymes, and diverse transporters were relatively more abundant in coastal than pelagic SAR11, SAR116, SAR86, SAR406, and Roseobacter populations (Mann–Whitney U Test, P < 0.005) (SI Appendix, Fig. S8 and Dataset S2), likely reflecting the higher nutrient availabilities in the coastal system. In contrast, transcripts mapping to proteins involved in light stress attenuation (photo-lyases), free radical detoxification (catalase, peroxiredoxins, thioredoxins), photoactive damage of coenzymes (biotin sulfoxide reductase), and heat shock in all of these same groups were more highly expressed in pelagic than coastal populations (SI Appendix, Fig. S8). These results are consistent with the higher solar radiation and temperatures that typify surface waters of the NPSG, supporting the recently recognized importance of heterotrophic free radical detoxification processes in this ecosystem (25, 26). Transcripts involved in phosphonate uptake were also enriched in pelagic versus coastal SAR11 and SAR116 populations (Dataset S2), reflecting their importance as a source of phosphate in oligotrophic environments (27, 28).
Significant progress has been made over the past decades in characterizing the phylogenetic diversity of eukaryotic, bacterial and archaeal groups inhabiting the world’s oceans (3, 29, 30), but the in situ metabolic activities and interactions of these groups that combine to create cohesive, resilient community structure and ecosystem-level processes remain largely undetermined. The extraordinary similarity of transcriptional temporal dynamics and coordination that is shared between microbial populations of disparate marine ecosystems as shown here illustrates the importance of short-term temporal interactions in defining the ecological niches of microbial plankton. Moreover, the rapid turnover of nutrients in the upper ocean (14, 31) together with the observed commonalities in whole-community diel cycles among all three domains of life in these disparate habitats implies that biogeochemical fluxes mediated by microbial community metabolism are diurnally phased across entire ocean basins. Identifying the precise mechanisms and metabolic fluxes that regulate the specific microbial interactions giving rise to whole-community metabolism is a major challenge for microbial oceanography in the future. Our findings underscore the importance of gaining deeper mechanistic perspective into the tightly coordinated ecosystem-level dynamics that orchestrate daily biogeochemical fluxes, and how they may respond in the face of rapid anthropogenic environmental change.
Materials and Methods
ESP Deployment and Sample Collection.
For our coastal metatranscriptomes, samples were collected by an Environmental Sample Processor (ESP) (12) suspended at a depth of 10 m from September 12–17, 2012 off the coast of California near Monterey (initial deployment: −122.7°W, 36.4°N). As previously described, planktonic microbial assemblages were collected in the 0.22–5 µm size range at 4-h intervals and preserved aboard the ESP (10, 12). Filter samples collected by the ESPs were stored at −80 °C within 24 h of their retrieval. RNA was extracted, cDNA was generated, and Illumina sequencing (32) was performed as described (33–35). Details regarding sequence processing and classification can be found in the Supplementary Appendix. Sequences generated in this study have been deposited in the NCBI Sequence Read Archive under accession number SRP050269. Collection and processing of the NPSG samples has been described (9).
Transcriptomic Network Analyses.
For both the coastal and pelagic metatranscriptomes reads were mapped to ortholog cluters of proteins constructed from the phylogenetic groups of interest, and a weighted transcriptomic network approach using the R packages WGCNA and igraph was used to analyze the resulting count tables. Details regarding the soft thresholds used in network construction, the calculation of adjacency matrixes, and the partitioning of transcripts into modules can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank B. Barone, D. Mende, T. Nielsen, T. Palden, A. Romano, S. Wilson, and members of the E.F.D. laboratory for helpful comments, J. Robidart for collecting seawater samples for rRNA subtraction-probe construction, and E. Ottesen for sharing R code. This work was supported by grants from the Gordon and Betty Moore Foundation (to E.F.D., 3777), National Science Foundation Grant EF0424599 (to E.F.D.), and the Simons Foundation grant Simons Collaboration on Ocean Processes and Ecology (SCOPE) (to E.F.D.). Development and application of the Environmental Sample Processor has been supported by National Science Foundation Grant OCE-0314222, National Aeronautics and Space Administration Astrobiology Grants NNG06GB34G and NNX09AB78G, the Gordon and Betty Moore Foundation (to C.A.S., 2728), and the David and Lucile Packard Foundation (to C.A.S., 901204, 901205). The cruise in the California Current was part of the MBARI CANON initiative and supported by the David and Lucile Packard Foundation (to F.P.C.; 901009). This work is a contribution of the Center for Microbial Oceanography: Research and Education (C-MORE) and the Simons Collaboration on Ocean Processes and Ecology (SCOPE).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive database (accession no. SRP050269).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502883112/-/DCSupplemental.
References
- 1.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5(10):782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 2.Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459(7244):193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- 3.Zinger L, Gobet A, Pommier T. Two decades of describing the unseen majority of aquatic microbial diversity. Mol Ecol. 2012;21(8):1878–1896. doi: 10.1111/j.1365-294X.2011.05362.x. [DOI] [PubMed] [Google Scholar]
- 4.Yokokawa T, Nagata T. Linking bacterial community structure to carbon fluxes in marine environments. J Oceanogr. 2010;66(1):1–12. [Google Scholar]
- 5.Giovanonni SJ, Rappe MM. Evolution, diversity, and molecular ecology of marine prokaryotes. In: Kirchman DL, editor. Microbial Ecology of the Oceans. 1st Ed. Wiley-Liss; New York, NY: 2000. pp. 47–85. [Google Scholar]
- 6.Giovannoni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437(7057):343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- 7.DeLong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437(7057):336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 8.DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459(7244):200–206. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- 9.Ottesen EA, et al. Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345(6193):207–212. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 10.Ottesen EA, et al. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 2011;5(12):1881–1895. doi: 10.1038/ismej.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ottesen EA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110(6):E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholin CA, et al. Remote detection of marine microbes, small invertebrates, harmful algae and biotoxins using the Environmental Sample Processor (ESP) Oceanography (Wash DC) 2009;22:158–167. [Google Scholar]
- 13.Chavez FP, Messié M. A comparison of Eastern Boundary Upwelling Ecosystems. Prog Oceanogr. 2009;83(1-4):80–96. [Google Scholar]
- 14.Karl DM, Church MJ. Microbial oceanography and the Hawaii Ocean Time-series programme. Nat Rev Microbiol. 2014;12(10):699–713. doi: 10.1038/nrmicro3333. [DOI] [PubMed] [Google Scholar]
- 15.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63(1):106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell L, Vaulot D. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (station ALOHA) Deep Sea Res Part Oceanogr Res Pap. 1993;40(10):2043–2060. [Google Scholar]
- 17.Demir-Hilton E, et al. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 2011;5(7):1095–1107. doi: 10.1038/ismej.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1):e17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart FJ, Sharma AK, Bryant JA, Eppley JM, DeLong EF. Community transcriptomics reveals universal patterns of protein sequence conservation in natural microbial communities. Genome Biol. 2011;12(3):R26. doi: 10.1186/gb-2011-12-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gifford SM, Sharma S, Moran MA. 2014. Linking activity and function to ecosystem dynamics in a coastal bacterioplankton community. Front Microbiol 5. Available at: journal.frontiersin.org/Journal/10.3389/fmicb.2014.00185/full [Accessed October 11, 2014]
- 22.Sharma AK, et al. Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of Prochlorococcus, Pelagibacter and the OM60 clade. Environ Microbiol. 2014;16(9):2815–2830. doi: 10.1111/1462-2920.12254. [DOI] [PubMed] [Google Scholar]
- 23.Becker JW, et al. Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol. 2014;5:111. doi: 10.3389/fmicb.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palovaara J, et al. Stimulation of growth by proteorhodopsin phototrophy involves regulation of central metabolic pathways in marine planktonic bacteria. Proc Natl Acad Sci USA. 2014;111(35):E3650–E3658. doi: 10.1073/pnas.1402617111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. MBio. 2012;3(2):e00036–e12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS ONE. 2011;6(2):e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karl DM, et al. Aerobic production of methane in the sea. Nat Geosci. 2008;1(7):473–478. [Google Scholar]
- 28.Metcalf WW, et al. Synthesis of methylphosphonic acid by marine microbes: A source for methane in the aerobic ocean. Science. 2012;337(6098):1104–1107. doi: 10.1126/science.1219875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276(5313):734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 30.Temperton B, Giovannoni SJ. Metagenomics: Microbial diversity through a scratched lens. Curr Opin Microbiol. 2012;15(5):605–612. doi: 10.1016/j.mib.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Karl DM. Nutrient dynamics in the deep blue sea. Trends Microbiol. 2002;10(9):410–418. doi: 10.1016/s0966-842x(02)02430-7. [DOI] [PubMed] [Google Scholar]
- 32.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5(4):433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 33.Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4(7):896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- 34.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105(10):3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5(6):999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.