Abstract
Neuropeptides represent one of the largest classes of signaling molecules used by nervous systems to regulate a wide range of physiological processes. Over the past several years, mass spectrometry (MS)-based strategies have revolutionized the discovery of neuropeptides in numerous model organisms, especially in decapod crustaceans. Here, we focus our discussion on recent advances in the use of MS-based techniques to map neuropeptides in spatial domain and monitoring their dynamic changes in temporal domain. These MS-enabled investigations provide valuable information about the distribution, secretion and potential function of neuropeptides with high molecular specificity and sensitivity. In situ MS imaging and in vivo microdialysis are highlighted as key technologies for probing spatio-temporal dynamics of neuropeptides in the crustacean nervous system. This review summarizes the latest advancement in MS-based methodologies for neuropeptide analysis including typical workflow and sample preparation strategies as well as major neuropeptide families discovered in decapod crustaceans.
Keywords: neuropeptide, mass spectrometry, mass spectrometric imaging, crustacean, microdialysis, peptidomics
1. Overview
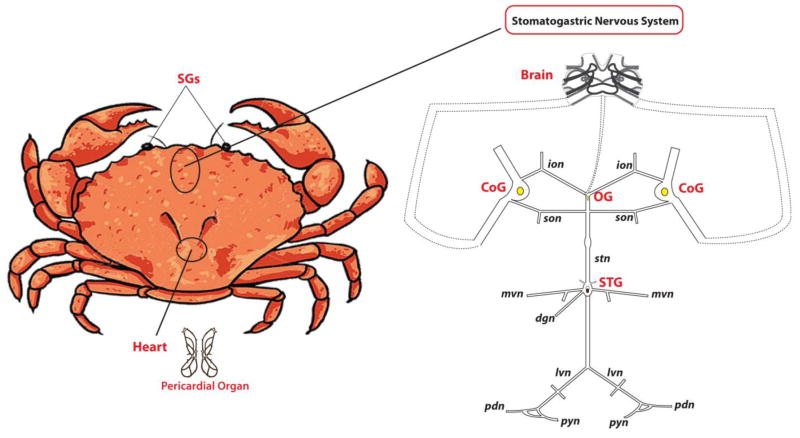
Neuropeptides represent one of the most diverse classes of signaling molecules employed by the nervous system, which can mediate numerous essential physiological processes, such as feeding, pain sensing and reproduction [1]. Neuropeptides are short chains of amino acids, which are encoded within genomes as larger precursor proteins, and undergo extensive processing and modification steps to yield bioactive forms that have regulatory roles. Extensive research has been carried out to study their structures, distributions and functions. Decapod crustacean, which has relatively simple nervous system (Figure 1) [2] and accessible electrophysiology at the single-cell and neural circuit level, serves as an attractive model preparation for neuromodulation and neuropeptide study. Our knowledge about neuropeptides in crustacean has been significantly expanded over the past decade thanks to the development and application of MS-based techniques enabling accelerated neuropeptide discovery [3, 4].
Figure 1.
Schematic drawing of the stomatogastric nervous system (STNS) of Jonah crab, Cancer borealis. The SGs (sinus glands: located in the eyestalks of the crab) and the POs (pericardial organs: located in the chamber surrounding the heart) release hormones into hemolymph. The brain is connected to the STNS via a tiny nerve called the inferior ventricular nerve (ivn), and the STNS consists of the stomatogastric ganglion (STG), oesophageal ganglion (OG), paired commissural ganglia (CoGs), which connected by motor nerves.
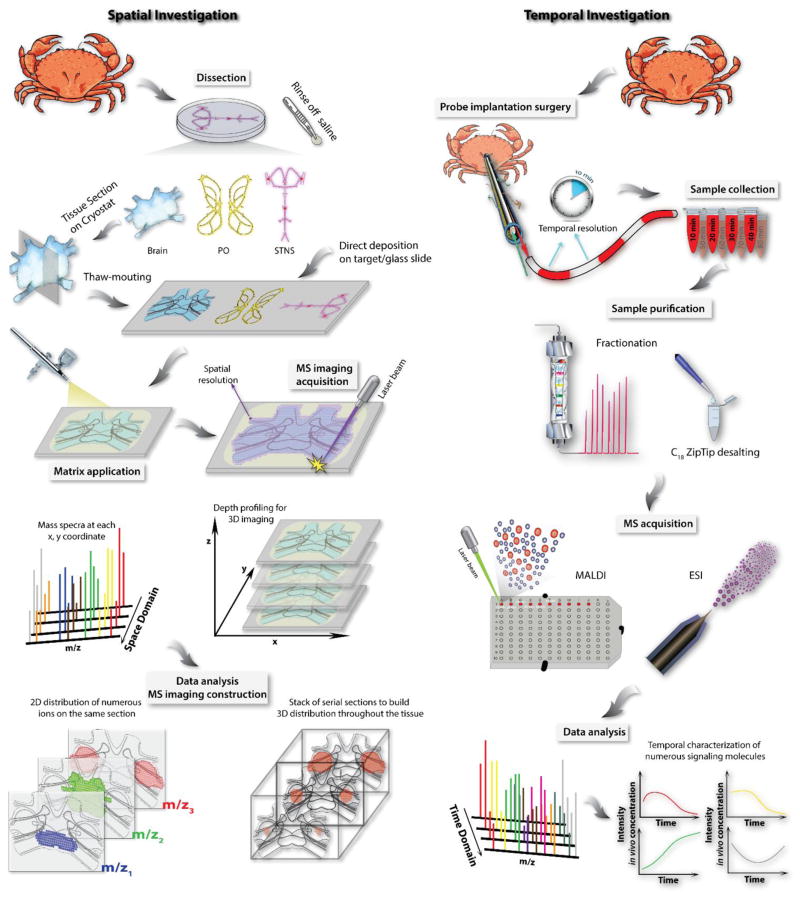
We developed a multifaceted MS-based platform for neuropeptide characterization, from sample preparation to data analysis (Figure 2) [5]. Utilizing this platform, neuropeptide content in various tissue organs [6–11] and body fluid-hemolymph [12–14] in crustaceans has been extensively studied. Despite their relatively simple nervous system, crustacean model organisms employ diverse neuropeptides, including over two dozen neuropeptide families and many isoforms from several peptide families for each species [3]. Complementary to tandem MS-based neuropeptide discovery and identification process, mass spectrometric imaging (MSI) technique has been utilized to map the spatial distribution of neuropeptide families and specific isoforms directly from tissue in an anatomical context [15, 16]. The spatial localization of specific neuropeptides and determination of their co-localization patterns will provide critical information to help elucidate the underlying mechanism of cell-cell communication via signaling neuropeptide molecules.
Figure 2.
Experimental workflow for both spatial and temporal investigation of crustacean neuropeptidomics.
In parallel to mapping the spatial distribution of these chemical messengers in tissue, monitoring the chemical dynamics in body fluids of live animals could be highly rewarding for functional investigation because it allows correlation between neurochemical content fluctuations and biological behaviors. Several techniques have been used for in vivo neurotransmitter measurement, among which, microdialysis sampling coupled to MS analysis is a popular choice for such measurements. This method has been well established for monitoring a wide range of analytes, from smaller molecular weight neurotransmitters, metabolites to larger molecule neuropeptides, and even proteins [17]. Figure 2 summarizes a typical experimental workflow for crustacean neuropeptide analysis at both spatial and temporal domains.
1.1 Brief introduction to the major crustacean neuropeptide families
Here we briefly introduce the chemical features and known biological functions of the major crustacean neuropeptide families. For more in-depth discussion, readers are suggested to refer to reviews about specific crustacean and invertebrate neuropeptides [3, 18, 19].
Allatostatin
The term allatostatin (AST) was defined based on the inhibitory effect on the insect corpora allata from a peptide [20]. In crustacean, this superfamily of neuropeptides is divided into three sub groups according to their structural features. A-type AST (AST-A) peptides share a C-terminal motif of Y/FXFGLamide (X represents a variable amino acid). Their diverse inhibitory functions are tissue specific within the nervous system [21–25]. The second family of AST (B-type AST) neuropeptides are characterized with a C-terminal sequence of W(X)6Wamide. The first native AST-B in crustacean (CbAST-B1) was found in the PO of C. borealis and C. productus [26]. This novel B-type AST was described to reduce the pyloric network frequency in crabs in a dose-dependent manner. Members of the AST-C subfamily share a C-terminal motif PISCF and a pyroglutamine blocked N-terminus. It is also noted that the disulfide bridge between the Cys residues at position 7 and 14 (in a typical 15-mer AST-C) is critical for receptor binding [27]. AST-C neuropeptides exhibit inhibitory effects on the pyloric motor output and constrain the cardiac neuromuscular system to reduce the heart rate [28].
Crustacean cardioactive peptide
Being originally isolated via heart bioassays in shore crab, C. maenas, the crustacean cardioactive peptide (CCAP) was named for its excitatory modulation on the heart [29]. This cyclic amidated neuropeptide with the sequence of PFCNAFTGCamide has a disulfide bridge between Cys3 and Cys9. In addition to its well-known cardioexcitatory properties [25, 29, 30], CCAP has also been reported to have excitatory effects on other invertebrate muscles and has been established as neuromodulator of STG and OG [3].
FMRFamides
FMRFamide-like peptides (FLPs) are the largest family of crustacean neuropeptides. They share a C-terminal motif RFamide with variable preceding sequences, such as FLRFamide, RPRFamide, RLRFamide, YLRFamide and FVRFamide. The pleiotropic physiological functions of FLPs have been extensively studied among numerous species of crustacean. Their diverse neuromodulatory roles include but not limited to muscular modulation [31], digestive system modulation [32], cardiac excitatory modulator [33] and pyloric circuit modulation [34] etc.
Orcokinins
The first orcokinin was isolated from the ventral nerve cord (VNC) of the crayfish, Orconectes limosus [35]. To the best of our knowledge, all full length decapod orcokinins are 13 amino acids long with an N-terminal motif of N/DFDEIDR. Orcokinins have been reported to enhance the frequency and amplitude of spontaneous hindgut contractions in crustacean, but the effect differs among different species [3]. Orcokinin is also demonstrated to be capable of modulating the motor output when applied exogenously to the STG of crabs and spiny lobster [36, 37].
Pyrokinin
In crustacean, pyrokinin neuropeptides possess the C-terminal motif FXPRLamide (where X is a variable amino acid). In the isolated STNS of C. borealis, two novel pyrokinins (CabPKs) discovered by MS approach exhibited effects on the pyloric and gastric mill motor circuits when bath applied to the STG. While little effect was observed on the pyloric motor pattern, profound excitatory influences on many gastric mill neurons were reported, establish its role as a modulator of the gastric mill rhythm [38].
SIFamides
To date, there are merely two full length (12-mer) SIFamide neuropeptides identified in crustacean. Both isoforms exhibit the chemical structure of XYRKPPFNGSIFamide, where X is the only variant amino acid (Gly in the former isoform and Val in the latter). In Arthropods, SIFamides have been linked to olfaction, sexual behavior and gut endocrine functions [39]. Modulation of male aggressive behavior was documented in M. rosenbergii by injection of Gly1-SIFamide into male prawn at different morphological development stages [39].
Tachykinin-related peptides (TRP)
Given the differences from vertebrate tachykinins with the C-terminal motif of FX1GX2Ramide, they are termed as tachykinin-related peptides (TRPs) in crustacean. Being the invertebrate homologs to substance P in mammal, TRPs are reported to be involved in various physiological processes [3]. TRPs have been demonstrated to colocalize with GABA in the visual system amerine neurons of crayfish suggesting its possible functions on photoreceptor sensitivity modulation [40]. In several feeding studies in crabs, TRP was released in midgut and exhibited significant elevation of expression in the brain [41].
2. Mass spectrometric imaging- the spatial probe of crustacean neuropeptides
2.1 Introduction to mass spectrometric imaging
Immunohistochemical (IHC) staining has long been a powerful tool for visualizing the distribution of neuropeptides in situ [42–46]. This antibody-based technique provides detailed spatial information of peptides with sub-cellular resolution when utilizing microscopy. In comparison to in situ hybridization, IHC staining does not require precise chemical information of the neuropeptides of interest for the recognition interaction. However, the production of specific antibodies against each peptide family could be quite costly and time-consuming [16]. It is also difficult to produce and develop highly specific antibodies that can distinguish among multiple peptide isoforms from the same family due to cross-reaction of similar neuropeptides to the antibodies. Since the pioneering work by Caprioli and co-workers [47], MSI has emerged as an attractive technology for mapping the localization of various biomolecules including metabolites, lipids, neuropeptides and proteins etc [15, 16, 48–52]. In a typical MSI study, tissue section is placed in a pre-defined 2-dimensional coordinate (x, y) system (Figure 2 left panel). To investigate the chemical content over the entire tissue slice, the sample stage carrying the tissue section is rastered against a laser beam across each position to generate a mass spectrum with its on-tissue dimensional information co-registered in the data acquisition software. This approach enables comprehensive mapping of the distributions of thousands of biomolecules simultaneously with high specificity and high throughput, which overcomes the aforementioned imperfections of IHC staining. When reconstructing MSI images from all consecutive tissue sections together, a global view of 3-dimensional localization pattern can be obtained. Confident identification of analytes could be achieved as well in MSI via in situ MS/MS fragmentation [41, 53, 54]. Thanks to the recent developments of methodology and instrumentation, it is possible to hybridize both positive and negative mode acquisitions into one MS imaging acquisition to obtain more in-depth spatial characterization with shorter experimental time [55]. A typical spatial resolution achievable in a MALDI MSI experiment is around 25 μm, which is not comparable to IHC approaches and lacks the ability to probe sub-cellular structures. Several aspects, including the diameter of a focused laser beam and the size of matrix crystals, are the key limitations. Secondary ion MS (SIMS) imaging equipped with a NanoSIMS ion beam has been developed to focus under 50 nm in diameter, which greatly improves the achievable spatial resolution of MSI [48]. The MS/MS identification process has been incorporated into MS imaging runs utilizing multiplexed setup with either targeted mass list or data dependent acquisition for more convenient molecular identification [56, 57]. Thereupon, not only the known neuropeptides could be unambiguously differentiated with their colocalization patterns, novel neuropeptides could also be revealed via on tissue de novo sequencing. Moreover, semi-quantitative information of the observed neuropeptides could be obtained with appropriate experimental conditions and post-acquisition data normalization [58–62]. Overall, MS imaging is an advantageous tool to study the spatial distribution of biomolecules in tissue with high sensitivity, specificity and throughput.
2.2 Sample preparation for MS imaging
To accurately reflect the spatial distributional characteristics of a dynamic range of neuropeptides at various abundances, it is vital to carefully handle every step of the sample preparation process.
Tissue harvest and preservation
To maintain the cell morphology, tissue dissection is usually performed in chilled artificial physiological saline. However, high concentration salt contents from extracellular matrix and saline not only interfere with the signals from neuropeptides, but also reduce the detection sensitivity of MALDI MS by suppressing the ionization and masking the signals which are especially adverse for low abundance neuropeptides. Rapid rinse of tissue in purified water immediately after isolating tissue from the animal could efficiently remove the physiological saline [10, 16, 63]. It has also been reported that changing physiological saline to dilute DHB aqueous solution during dissection could help reducing the inherent salt context [64].
Endogenous degradation caused by protease activities often poses negative effect on peptide profiles even at 3 minutes post-mortem [65, 66]. Crucial PTMs of the analytes in the brain tissue have been reported to significantly change within minutes post-mortem [67, 68]. In order to minimize and prevent the degradation of valuable biologically active molecules by proteases, tissue preservation is of great importance during and after tissue harvest. One of the most commonly adapted methods is to snap-freeze the tissue in dry ice, liquid nitrogen or cold bath using different organic solvents such as isopentane, ethanol and isopropanol to stop the enzymatic proteolysis [4]. The structural features of tissue is also well conserved at the same time (see more in tissue fixation). Generally, longer freezing time in cold-bath is more effective for preventing the tissue from cracking and keeping the tissue integrity [16]. Heat denaturation by Stabilizor T1 (Denator, Gothenburg, Sweden) has also been reported to efficiently eliminate post mortem degradation in various types of tissues [69, 70] including crustacean neuroendocrine organs [71]. Formaldehyde-fixed paraffin-embedding (FFPE) however, as a popular way to preserve tissue in biological and medical research, would cause severe crosslinking between peptides and proteins [72], which decreases its applicability in MS-based studies. Attempts have been made to successfully image FFPE tissue with a paraffin removal step prior to matrix application [72–76]. Aspects such as oxidation and tissue degradation must be taken into consideration when processing data from this type of samples. Storage at −80°C is imperative if the tissue is not used immediately after dissection [77].
Tissue sectioning and fixation
For the purpose of sectioning specimens into thin slices to be analyzed in MSI experiments, the tissue is usually embedded into scaffold materials at the time of snap-freezing to facilitate subsequent cutting. Among various supporting materials that have been reported, most of them are polymer based, such as optimal cutting temperature (OCT) compound, 2% (wt/vol) carboxymethylcellulose (CMC), Tissue-tek, and agar etc [78]. Although subsequent histological study could be conveniently carried out using these substrates [48], the strong background signals in MS from the polymer have limited their universal application to diverse biomolecules at a wide range of molecular weights [78, 79]. To eliminate the interference from embedding materials at the neuropeptide rich m/z region (typically 500 to 3000 Da) in crustacean tissue, gelatin and water are recommended [41, 80]. The thickness of the tissue is typically 10 to 20 μm [81], which is similar to the diameter of a mammalian cell, to allow for better analysis of the cellular content [78]. However, thinner sections (< 5μm) have been demonstrated to yield better results on an alternative LDI platform such as nanostructure initiator mass spectrometry (NIMS) [82–84]. To fix the tissue slice for MS imaging experiment, thaw-mounting the −20 °C cold section onto a room temperature MALDI sample plate or glass slide is the most common method [78]. Double sided tape is another popular adhesive to affix the tissue especially for samples with low water content [85]. When electrical conductive target is needed for instrumentation such as MALDI TOF MS, stainless steel target, gold-coated glass slide and indium-tin oxide coated glass slide are all excellent candidates [4]. Using transparent glass slides for tissue fixation also provide the opportunity for subsequent histological study [48]. Other than brain and thoracic ganglion (TG), most of the neural tissues in crustacean animals are too small to be sectioned, such as OG and STG. PO tissue is composed of fine interwoven nerve tubes that are difficult to section. To study the neuropeptide distribution in these organs, tissues are directly placed on the MALDI plate or glass slide followed by drying process in a desiccator at −20°C to fix the tissue [8, 53, 86].
Tissue washing
For the investigation of neuropeptides in crustacean, tissue washing with organic solvents is typically avoided to prevent neuropeptide delocalization. Many crustacean neuropeptides are relatively small and hydrophilic, making them readily extractable when using organic solvents such as methanol and ethanol. However, high abundance lipids ranging from 500 to 900 Da and from 1300 to 1600 Da in crustacean tissues often hinder the effective detection of neuropeptides within the same mass range [87]. Chloroform and xylene have been reported to effectively eliminate lipid background as washing solvent to treat various tissues [87, 88]. When the target neuropeptides are hydrophobic or have higher molecular weights such as the crustacean hyperglycemic hormone (CHH) peptides, it is more critical to carefully optimize tissue washing procedures to remove salts and small neuropeptides while preserve the spatial localization of the analytes of interest.
Matrix application
Since MALDI source is the most widely adopted MS imaging platform in crustacean neuropeptide study, the application of matrix onto the tissue prior to MS acquisition is one of the most important elements in sample preparation [89, 90]. The most common matrices in neuropeptide analysis by MALDI MS are DHB and CHCA, which are both benzene ring containing weak organic acids with the ability to absorb and transfer laser energy in the ultraviolet-wavelength range for the purpose of analyte desorption and ionization. Binary matrix mixture has been demonstrated to bring the advantages of both DHB and CHCA together and produce finer crystallization and enhanced signal intensity [80, 91]. To apply matrix, one could either spot the matrix solution onto discrete positions on the tissue or continuously spray the matrix solution over the entire tissue area. In the case of manual matrix deposition, spraying with airbrush produces much smaller crystals resulting in better lateral resolution than droplet spotting, although manual spotting would be more suitable for profiling prior to MS imaging acquisition. Robotic spotting utilizing focused acoustic dispenser has been developed to generate picoliter level droplets which greatly improve the lateral precision to around 3 μm with better reproducibility [48]. Meanwhile, the crystal size and reproducibility could both be well controlled with automated spraying devices, such as TM sprayer (pneumatic sprayer, by HTX Imaging, NC, US) and ImagePrep (vibrational sprayer, by Bruker Daltonics, Bremen, Germany). It is important to note that the matrix solution must be optimized with both concentration and solvent composition for these spraying devices to achieve best results.
Low matrix concentration typically leads to slow crystallization and consequently results in more severe lateral diffusion as the tissue has been left to be wet for extended period of time. Furthermore, without sufficient matrix coating coverage, desorption and ionization efficiency will dramatically decrease. If too much matrix is deposited on the tissue surface, fast evaporation would reduce the amount of neuropeptides to be extracted into the matrix crystal layer, which lowers the signal intensity in MS as well. With similar principles, the optimal solvent composition should be adjusted to the sweet spot of best extraction efficiency, smallest crystal size and minimum lateral delocalization.
2.3 Application of MS imaging to characterize the spatial distribution of neuropeptides in crustacean
A variety of decapod crustacean species have been extensively studied using multifaceted mass spectrometric platforms. MSI provides useful knowledge to correlate the spatial distributions of neuropeptides in the nervous system to their specific biological functions as neuromodulators. The first MS imaging study of crustacean neuropeptides was reported by DeKeyser et al [53] in Cancer borealis on a MALDI TOF/TOF MS platform. The MSI investigation in brain sections and intact PO has successfully revealed the spatial distribution of 38 neuropeptides from 10 different families via mass matching. The identities of 16 of them were validated through on-tissue MS/MS CID fragmentation. It is worth mentioning that the DHB matrix was applied differently on the tissue for MS imaging (airbrush manual spray) and the tissue for tandem MS acquisition (discrete manual spotting). In most of the distributional density maps generated by MSI, neuropeptides from the same family were similarly localized, in both brain and PO. However, exceptions did exist for neuropeptides belonging to the RFamide and orcokinin families. Different localization of neuropeptides in the nervous system suggested their potentially distinct functionalities despite their similar chemical structures. This differentiated spatial information about neuropeptide isoforms from the same family highlights the superior chemical specificity of MS imaging over IHC staining. In the brain, most of RFamides showed high intensity in the caudal region while orcokinins were found in the rostral region. SIFamide and TRP 1a displayed similar distribution patterns with those of RFamides. Although A-type and B-type AST neuropeptides were absent in the brain, they were detected in the PO with distinct spatial distributional patterns. Comparing to conventional cellular mapping of immunoreactivity in the trunks of PO, MSI study enabled the visualization of neuropeptides of this organ to the extended areas such as anterior and posterior bar regions as shown in Figure 3. Later studies by Chen et al [54] showed similar distributional patterns of RFamides and orcokinins being in contrast to each other in the brain of C. borealis.
Figure 3.
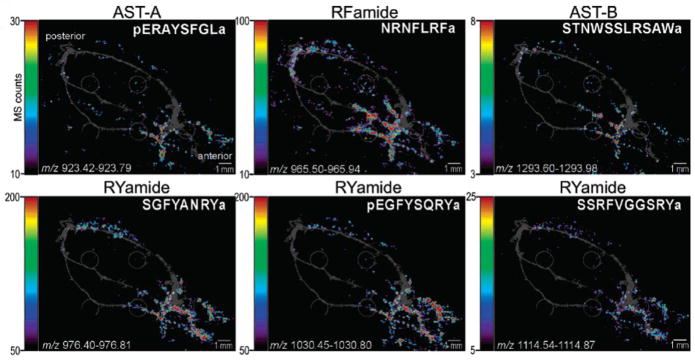
Differential localization of neuropeptide families in the pericardial organs (PO). The MSI images, colored according to associated color-intensity scale, are shown as an overlay on top of an optical image of the PO. Members of different neuropeptide families are differentially located. The most apparent contrast is observed between the RYamide family and the RFamide family. Adapted with permission from Ref. [54].
In a study by Hui et al. [92] where a novel TRP CalsTRP was reported in the blue crab Callinectes sapidus, MSI study by MALDI TOF/TOF showed its consistent localization in the brain with previously identified CabTRP 1a in both fed and unfed animals. Through comparative feeding study, the relative amounts of CalsTRP and CabTRP were suggested to be elevated in both the brain and the CoGs when evaluating the MSI distributional density maps. This semi-quantitative result was reproducible across different biological replicates and was consistent with isotopic-labeling relative quantitation performed on a CE-MALDI MS platform. With aforementioned valuable spatial information from MSI, the colocalization of CabTRP 1a and CalsTRP in the brain suggested that both TRPs were encoded in the same preprotachykinin and functionalized in the gastric mill and pyloric rhythm.
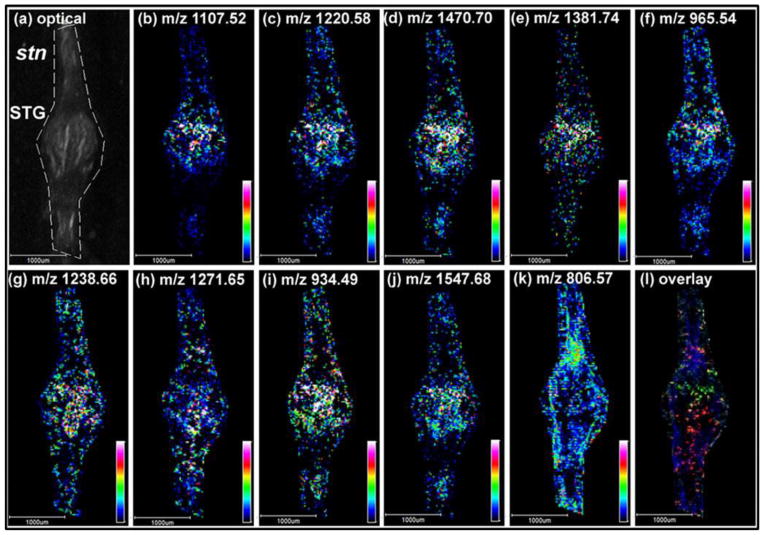
In addition to brain and PO, the entire STNS also contains multiple neuropeptides and neurotransmitters. The STG is the center of the STNS with only 26 neurons that are relatively large and well characterized with their cellular identities and electrophysiological properties. This invertebrate neuronal network provides an excellent platform to investigate general principles of neuromodulation and the functional roles of neuropeptides in circuit dynamics. Recently, Ye et al [86] were able to unambiguously map the spatial distribution of 55 neuropeptides from 10 families in the 4 major ganglia in the STNS of C. sapidus with high sensitivity, specificity and sub-neuron size spatial resolution of 25μm on an autoflex III MALDI TOF/TOF instrument. Representative MS images are presented in Figure 4. Overall, distinct expression patterns of neuropeptides of each ganglion and associated connecting nerves were revealed. For example, significantly higher abundance of two RYamides were detected in the CoGs compared to the STG, four orcokinins existed exclusively in the STG, B-type ASTs were widely distributed in the STG and stn with minimal signals in the CoGs. To further increase the confidence of identification of these highly complex neuropeptides with similar masses, a high resolving power MALDI Fourier transform ion cyclotron resonance (FT-ICR) instrument was also employed. Two ions with mass difference of 0.09Da were successfully distinguished with significantly different spatial expression patterns. Taking advantage of multifaceted platforms, a more complete coverage of the neuropeptidomes was achieved by combining instrumentation with complementary capabilities. For instance, a B-type AST GSNWSNLRGAWamide was detected by FT-ICR while absent in TOF/TOF detection, CabTRP 1a and CalsTRP on the other hand, were only seen with the TOF/TOF instrument.
Figure 4.
Representative MSI results of the neuropeptide distributions in Callinectes sapidus STG by MALDI TOF/TOF. (a) An optical image of the STG subjected to subsequent MSI acquisiton. Nine neuropeptides from five different families were shown above. B-type ASTs: (b) AGWSSMRGAWa (m/z 1107.52), (c) SGDWSSLRGAWa (m/z 1220.58), and (d) VPNDWAHFRGSWa (m/z 1470.70). SIFamide: (e) GYRKPPFNGSIFa (m/z 1381.74). RFamides: (f) NRNFLRFa (m/z 965.54), (g) SQPSKNYLRFa (m/z 1238.66), and (h) pQDLDHVFLRFa (m/z 1271.64). CabTRP 1a: APSGFLGMRa (m/z 934.49). Orcokinin: NFDEIDRSSFGFN (m/z 1547.68). For comparison, the distribution of a lipid PC (38:6) (m/z 806.57) is shown in (k). (l) is an overlaid image of (c), (h), and (k), displayed in green, red, and blue, respectively. Adapted with permission from Ref. [86].
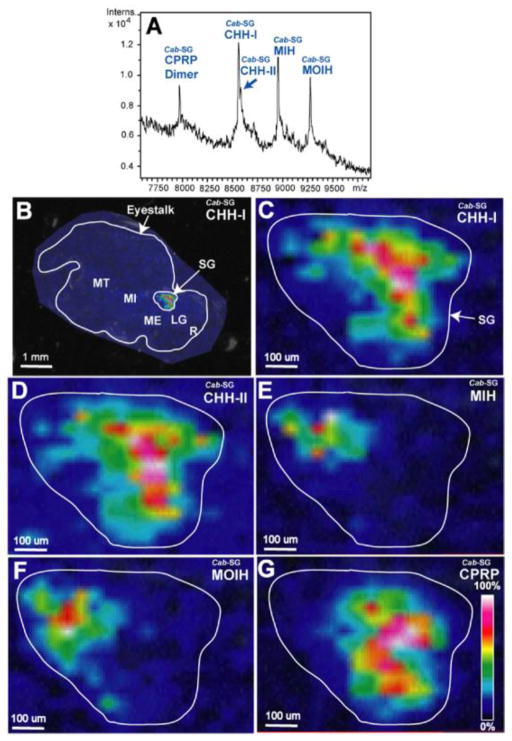
Larger neuropeptides from the CHH superfamily with molecular weights higher than 8 kDa were spatially characterized using MS imaging in the sinus gland (SG) of both C. borealis and C. sapidus for the first time by Jia et al. [93]. As shown in Figure 5, the distribution patterns of the members from two different CHH subfamilies were reported to be in stark contrast, indicating their distinct signaling pathways in the physiological process. The MALDI TOF/TOF platform is excellent for the study of large neuropeptides and neurohormones due to its ability to detect biomolecules with wide mass range. In this study, both smaller CPRPs and large CHHs were simultaneously mapped in the SG. However, due to the structural complexity of these 72-mer neuropeptides, such as various PTMs and post mortem degradation, the isolation window for plotting the spatial distribution was set to be ±10 Da to include variable isoforms of the same neuropeptide. To overcome the low specificity of MS imaging for large neuropeptides and the poor on-tissue fragmentation of singly charged large molecules, multiply charged ions have been generated for MS imaging investigation using either novel matrix [94] or electrospray based ionization platform [95, 96]. Because minimum sample tampering in MS imaging preparation tends to preserve more accurate PTM features, the production of multiply charged precursor ions in situ would be highly desirable for MS imaging and characterization of these large peptide hormones [97].
Figure 5.
Spatial distribution mapping of the CHH-family neuropeptides in C. borealis sinus gland. (A) MALDI mass spectrum of direct tissue analysis of C. borealis sinus gland. (B) MALDI-MSI image of Cab-SG-CHH-I on C. borealis eyestalk. Zoom-in MALDI images of Cab-SG-CHH-I (C), Cab-SG-CHH-II (D), Cab-SG-MIH (E), Cab-SG-MOIH (F), and Cab-SG-CPRP (G) on the sinus gland of C. borealis eyestalk. Adapted with permission from Ref. [93].
Neuropeptides of other species in decapod crustacean have also been spatially characterized using MS imaging, such as lobster Homarus americanus and giant tiger prawn Penaeus monodon. Chen et al [63] used both MALDI TOF/TOF and MALDI LTQ Orbitrap to map the distributions of neuropeptides in the H. americanus neuropeptidomic study. Single isotope in an isotopic envelope was selected for high specificity mapping of the neuropeptides with an isolation window of ±0.0025 Da from the data acquired on an Orbitrap platform. In addition to the high mass accuracy, confident and interference-free validation was granted by the ability to conduct selected reaction monitoring (SRM) MS/MS experiment in the LTQ ion trap. The abundant spatial distribution of CabTRP 1a and Val-SIFamide in the olfactory lobe (OL) and accessory lobe (AL) neuropils clearly suggested their neuroregulatory functions in the olfactory system which significantly affects the chemosensory reception in crustaceans. Moreover, the high concentration of orcokinin NFDEIDRSGFGFN in the antenna II neuropil (AnN) and lateral antennular neuropil (LAN) provided evidence of its inhibitory role in the tactile sensory. This work is an excellent example to demonstrate the great potential of MSI to reveal the physiological functions of neuropeptides by correlating their spatial distribution information. Another work by Chansela et al [98] used the ultraflex II MALDI TOF/TOF to investigate the neuropeptide distribution in P. monodon. These researchers examined paraffin-embedded tissue sections (PETs) of various neuroendocrine organs including eyestalk, brain lamina ganglionaris and TG. Among the 29 neuropeptides detected in nanoLC-ESI-QTOF, eight neuropeptides were visualized by MS imaging. PETs provide an opportunity to combine IHC staining with MSI results using minute size crustacean tissue to achieve enhanced chemical information.
For a heterogeneous tissue like a crab brain, traditional MSI analyses only provide a small 2D fraction of the complexity of various neuropils and neuronal clusters, leading to incomplete description of neuropeptide distribution in the whole brain volume. However, when 2D ion density maps of one neuropeptide are stacked in consecutive orders, comprehensive 3D spatial distribution throughout the entire tissue could provide a complete picture of neuropeptide localization in a complex neuronal structure such as brain (Figure 2). Detailed reviews about 3D MS imaging are available elsewhere [49, 99, 100]. As an example, Chen et al [41] demonstrated the reconstruction of more than 20 neuropeptides from 8 families in the 3D volume of the C. borealis brain by creating consecutive series of seven layers of brain sections with equal intervals. Moreover, a comparison between the localization of neuropeptides and phospholipids in the brain was made to highlight the distinct distributional patterns of these diverse classes of signaling molecules throughout the nervous system.
Although MALDI MSI has been the predominant technique to map the spatial distribution of biomolecules, limitations associated with the use of matrix such as background interference, possible diffusion during matrix application, and the reduced spatial resolution caused by matrix crystal size, have proven to be challenging to overcome. Efforts have been made by Sturm et al [84] to take advantage of the relatively new matrix-free NIMS imaging platform to study the neuropeptides and lipids in the brain of C. borealis. Since NIMS has been demonstrated to be capable of detecting biomolecules below 1800 Da including metabolites, lipids and drugs, most of the neuropeptides in crustacean were believed to be detectable as well since they fall in this mass range. However, only lipids from the brain were readily detected using NIMS while tissue from the same preparation studied with MALDI MS enabled the detection of neuropeptides with decent signal intensities.
In Table 1, we summarize the neuropeptides whose spatial distributions have been mapped in various crustacean species. We also categorize the types of MS instruments with which the imaging experiments were performed. It is important to point out that only the ones that were explicitly mentioned and/or plotted in the literature were included in the table. For instance, Chen et al [41] constructed 3D MS images of 20 neuropeptides in the study of C. borealis brain, but only the seven neuropeptides whose identities were revealed in the paper are listed here. In the column labeled as “in situ identification”, either on-tissue MS/MS fragmentation (labeled as MS2) or matching by accurate mass (AM) was specified as the method for identification and/or validation.
Table 1.
Neuropeptides spatially characterized via MS imaging in crustacean nervous system
| Neuropeptide Family | Sequence | Tissue and Species | MS instrumentation | In situ Identification | Reference |
|---|---|---|---|---|---|
| FMRFamides | NRNFLRFa |
C. borealis PO and brain C. sapidus STG |
MALDI-TOF/TOF | a MS2 ([41, 53, 86]) | [41, 53, 54, 86] |
| GNRNFLRFa | C. borealis PO and brain | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| SDRNFLRFa | C. borealis PO | MALDI-TOF/TOF | b AM ([53]) | [53] | |
| SQPSKNYLRFa | C. sapidus STG | MALDI-TOF/TOF | AM ([86]) | [86] | |
| GAHKNYLRFa | C. borealis PO and brain | MALDI-TOF/TOF | AM ([53, 54]) | [53, 54] | |
| GYSKNYLRFa | C. borealis PO and brain | MALDI-TOF/TOF | AM ([53]) | [53] | |
| AYNRSFLRFa | C. borealis PO and brain | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| SENRNFLRFa | C. borealis PO and brain | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| SMPSLRLRFa | C. borealis brain | MALDI-TOF/TOF | AM ([53]) | [41, 53] | |
| APQRNFLRFa | C. borealis brain | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| pQDLDHVFLRFa | C. sapidus STG | MALDI-TOF/TOF | AM ([86]) | [86] | |
| DVRTPALRLRFa | C. borealis brain | MALDI-TOF/TOF | AM ([53] [54]) | [41, 53, 54] | |
| GDRNFLRFa | P. monodon brain and TG | MALDI-TOF/TOF | AM ([98]) | [98] | |
|
| |||||
| SIFamide | GYRKPPFNGSIFa |
C. borealis brain C. sapidus STG P. monodon brain and TG |
MALDI-TOF/TOF | MS2 ([53, 98]) AM ([86]) |
[41, 53, 86, 98] |
| VYRKPPFNGSIFa | H. americanus brain | MALDI-LTQ-Orbitrap MALDI-TOF/TOF |
AM ([63]) | [63] | |
|
| |||||
| AST-A | pERAYSFGLa | C. borealis PO | MALDI-TOF/TOF | AM [53]) | [53] |
| PRDYAFGLa | C. borealis PO | MALDI-TOF/TOF | AM [53]) | [53] | |
| ANEDEDAASLFAFGLa | P. monodon brain and TG | MALDI-TOF/TOF | MS2 [98]) | [98] | |
|
| |||||
| AST-B | NWNKFQGSWa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] |
| GNWNKFQGSWa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| AGWSSMRGAWa | C. sapidus STG | MALDI-TOF/TOF | AM ([86]) | [86] | |
| NNWSKFQGSWa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| STNWSSLRSAWa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| NNNWSKFQGSWa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| SGDWSSLRGAWa | C. sapidus STG | MALDI-TOF/TOF | AM ([86]) | [86] | |
| VPNDWAHFRGSWa |
C. borealis PO C. sapidus STG |
MALDI-TOF/TOF | MS2 ([53]) AM ([86]) |
[53] [86] | |
|
| |||||
| CCAP | PFCNAFTGCa | C. borealis PO | MALDI-TOF/TOF | MS2 [53]) | [53] |
|
| |||||
| Orcokinins | NFDEIDRSGFG | C. borealis brain | MALDI-TOF/TOF | AM ([53]) | [53] |
| NFDEIDRSGFDG | P. monodon brain and TG | MALDI-TOF/TOF | AM ([98]) | [98] | |
| NFDEIDRSGFGFA | C. borealis brain | MALDI-TOF/TOF | MS2 ([41, 53, 54]) | [41, 53, 54] | |
| NFDEIDRSGFGFV |
C. borealis brain P. monodon brain and TG |
MALDI-TOF/TOF | AM ([53], [98]) | [53, 98] | |
| NFDEIDRSSFGFV | C. borealis brain | MALDI-TOF/TOF | AM ([53]) | [53] | |
| NFDEIDRSSFGFN |
C. borealis brain C. sapidus STG |
MALDI-TOF/TOF | AM ([53, 86]) | [53, 86] | |
| NFDEIDRSGFGFN | H. americanus brain | MALDI-LTQ-Orbitrap MALDI-TOF/TOF |
AM ([63]) | [63] | |
| NFDEIDRTGFGFH | C. borealis brain | MALDI-TOF/TOF | AM ([54]) | [41, 54] | |
|
| |||||
| Orcomyotropin-related | FDAFTTGFGHS | C. borealis brain | MALDI-TOF/TOF | AM ([53]) | [53] |
|
| |||||
| RYamides | FYSQRYa | C. borealis PO | MALDI-TOF/TOF | AM ([53]) | [53] |
| SGFYANRYa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
| SSRFVGGSRYa | C. borealis PO | MALDI-TOF/TOF | AM ([53]) | [53] | |
| pEGFYSQRYa | C. borealis PO | MALDI-TOF/TOF | MS2 ([53]) | [53] | |
|
| |||||
| TRPs | APSGFLGMRa |
C. borealis brain H. americanus brain C. sapidus STG |
MALDI-LTQ-Orbitrap MALDI-TOF/TOF |
AM ([41, 53, 54, 63, 86]) | [41, 53, 54, 63, 86] |
| YRSGFLGMRa | C. sapidus brain | MALDI-TOF/TOF | AM ([92]) | [92] | |
| PSGFLGMRamide | P. monodon brain and TG | MALDI-TOF/TOF | AM ([98]) | [98] | |
|
| |||||
| YRamide | HIGSLYRa | C. borealis PO | MALDI-TOF/TOF | AM ([53]) | [53] |
|
| |||||
| CHH superfamily | Cab-SG-CHH-I | C. borealis SG | MALDI-TOF/TOF | AM ([93]) | [93] |
| Cab-SG-CHH-II | C. borealis SG | MALDI-TOF/TOF | AM ([93]) | [93] | |
| Cab-SG-MIH | C. borealis SG | MALDI-TOF/TOF | AM ([93]) | [93] | |
| Cab-SG-MOIH | C. borealis SG | MALDI-TOF/TOF | AM ([93]) | [93] | |
| HEEYQAHVQTV | P. monodon brain, SG and TG | MALDI-TOF/TOF | MS2 ([98]) | [98] | |
|
| |||||
| PDH | NSELINSLLGIPK | P. monodon retina, brain and TG | MALDI-TOF/TOF | MS2 ([98]) | [98] |
|
| |||||
| Others | HI/LASLYKPR | H. americanus brain | MALDI-LTQ-Orbitrap | AM ([63]) | [63] |
MS2 indicates that the neuropeptide was identified and validated by on-tissue MS/MS fragmentation;
AM indicates that the neuropeptide was identified by accurate mass matching to existing database.
2.4 Challenges and outlook of MSI technique in the study of neuropeptides in crustacean
Hundreds of neuropeptides have been characterized in different species of crustacean providing great opportunities for a better understanding of potential correlation between location and functionalities of neuropeptides [4, 19]. Nonetheless, challenges still exist when it comes to the detection of the small neuropeptides and neurotransmitters with m/z overlapping the matrix and lipid rich regions. With the prosperous development in nanoscience, we expect to see exciting new materials to replace conventional matrices with prospective properties such as minimum background interference, excellent desorption/ionization efficiency, low to no limit to the lateral resolution etc [101–103]. Most of the known neuropeptides are de novo sequenced via LC-MS platform. On tissue de novo sequencing associated with MS imaging has only limited success for high abundance neuropeptides. To further expand the power of in situ neuropeptide discovery via de novo sequencing, such as high throughput data dependent MS/MS imaging acquisition, we need more sensitive MS instruments with better ion transmission efficiency and new matrices that enable robust production of multiply charged ions for better fragmentation efficiency. In addition, the ambient ionization enabled by the atmospheric pressure (AP)-MALDI platform [104, 105] can provide an alternative desorption/ionization mode that has the potential for detection of neuropeptides with labile modifications, such as tyrosine sulfation in sulfakinin neuropeptides [106].
3. Microdialysis- the temporal probe of crustacean neuropeptides
3.1 Introduction
Complementary to tissue imaging studies in which we can learn about the spatial locations of molecules of interest, in vivo sampling techniques, such as microdialysis and push-pull perfusion, offer the unique advantages of monitoring signaling molecules involved in biological events with temporal resolution (Figure 2).
Microdialysis has been widely employed in the field of neuroscience, due to its ability to collect neurochemicals during a physiological behavior or stimulus of interest in a time-resolved fashion with minimal disturbance to the animals, providing useful insights into the action of molecules in vivo. In this sampling technique, a microdialysis probe is implanted into the tissues of interest allowing continuously sampling from the extracellular space. Since its first introduction by Bito et al. [107], studies employing the use of microdialysis have been expanded into a wide variety of tissues and organs in the body including liver, heart, skin, blood, placenta, stomach and ear, as summarized elsewhere [108, 109]. The types of analytes that microdialysis probe could sample have also been explored and proven to be rather diverse, from low molecular weight substances such as electrolytes [110], amines and amino acids [111], to higher molecular substances such as neuropeptides [13, 14].
Like any analytical sampling technique, microdialysis has numerous advantages resulting in widespread applications, but it also suffers from several limitations. First, the small diameter of the microdialysis probe allows minimal damage to the tissue and disturbance to the animal. Consequently, long-term sampling can be achieved while the animal is awake and freely moving. Second, the microdialysis probe membrane, which has a pre-defined molecular weight cutoff can act as a physical barrier between the perfusion solution and tissue, this in turn not only protects tissue from the turbulent flow of the perfusion solution, but also excludes high-molecular-weight substances which would yield a cleaner sample. Finally, a more precise quantitative comparison of analytes could be conducted from microdialysis samples because one animal serves as its own control, minimizing individual variation.
Despite the advantages of microdialysis over other techniques, there are some disadvantages worth noting. First, microdialysis is most powerful for providing timely-resolved information. However, the temporal resolution is also limited, mostly by the sensitivity of the detection method. Second, the relatively low recovery rate of analyte using microdialysis places greater demand for sensitivity of the detection method utilized, a problem that is more severe for larger molecules like neuropeptides or proteins.
Being considered as a prime method to sample from biofluid, microdialysis has been successfully applied to several crustacean species despite the obvious obstacle of the hard shells of the animals. Cebada et al. [111] used microdialysis method to study fluctuations of non-essential amino acids, GABA, and histamine in hemolymph of crayfish and quantified these molecules on hourly basis. Two studies from our lab [13, 14] reported on the use of MS-based techniques to study neuropeptide content in the hemolymph from Jonah crab, Cancer borealis, collected via microdialysis sampling method.
3.2 Principle of microdialysis process
Among the various types of microdialysis probes that are commercially available, a common key component is the dialysis membrane at the probe tip (Figure 6). The dialysis membrane is semipermeable, which is limited by the molecular weight cutoff of the membrane. When sampling from extracellular space, it is based on gradient driven diffusion across the dialysis membrane. As perfusion solution is pushed through inlet, normally at a rate of 0.1–3.0 μl/min [112], molecules fall below the molecular weight cutoff of the membrane would diffuse across the membrane and get collected in the dialysate for analysis.
Figure 6.
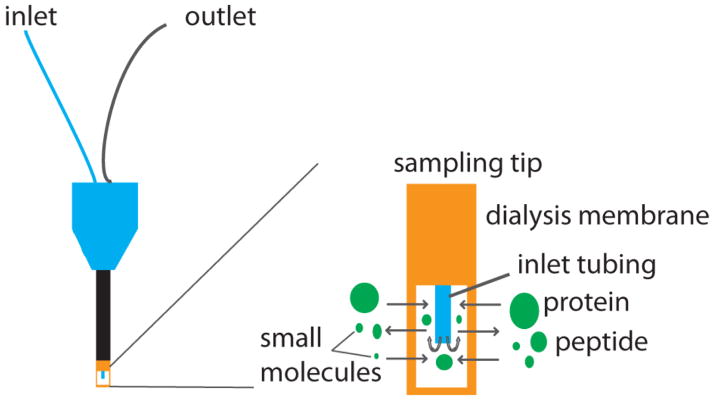
An illustration of microdialysis probe with an expanded view of the membrane tip. Analytes with a mass below the molecular weight cutoff of the membrane diffuse into the probe and get collected at the outlet.
Interpretation of microdialysis data could prove to be challenging. When sampling with microdialysis, the concentration of analyte of interest in the dialysate (Cout) will be smaller than that in extracellular space (Cext). The relative recovery (R) is defined as the ratio of Cout/Cext, and could be affected by a number of factors, such as flow rate, pore size of membrane (molecular weight cutoff), etc. Extensive efforts have been made to increase R according to the analyte of interest. For instance, by using ultraslow flow rate, which gives enough time for interaction of analytes and the membrane, R has been significantly increased for amines and drugs [113, 114]. Knowing the relative recovery is essential for estimation of external concentration of substance of interest from that in dialysate. It is easier to determine the in vitro R in dialysis of known concentration of samples and measure Cout. However, it has long been recognized that in vitro R is not an accurate representation of in vivo R due to the fact that the extracellular environment is much more complicated and highly dynamic within the animal. Several methods to measure in vivo R have been developed including manipulating the flow rate and adding an internal standard. No net flux (NNF), the concept was first introduced by Lonroth et al. in 1987 [115], soon was applied to quantitative microdialysis by several researchers [116–118] and has become one of the most popular methods. This method, also known as zero flow rate method, determines extracellular concentration by correlating perfusate flow rate with transfer coefficient. Moreover, the dialysate concentration at zero flow rate is expected to equal the extracellular concentration. Another common method is adding an internal standard, normally isotopically labeled, and then measuring the loss of the isotopically labeled internal standard from the perfusate [119, 120]. Due to the technical difficulties, most studies to date perform relative quantification of dynamic changes of signaling molecules. The absolute quantitation will require more sophisticated assays.
3.3 Microdialysis in crustacean
Neuropeptides in circulatory fluid, hemolymph, secreted from major neuroendocrine organs in decapod crustaceans, act as circulating hormones and regulate numerous physiological processes. However, the characterization of neuropeptide content in crustacean hemolymph has always been challenging. These low-level neuropeptides are difficult to detect due to high salt interference and suppression from the high abundance protein degradation products in crude hemolymph extracts. Chen et al. developed a simple and effective hemolymph preparation method suitable for MALDI MS analysis of neuropeptides, and successfully identified 10 secreted neuropeptides from several families, such as RFamide, AST, orcokinin, TRP and CCAP in Jonah crab Cancer borealis [12].
An alternative way to study hemolymph is the use of microdialysis. As described by Behrens [13] and Schmerberg [14], the microdialysis probe was implanted into the pericardial sinus of live crabs. Crustacean, unlike mammals, has an open circulatory system and the pericardial organ is a major hormonal release site. Thus, microdialysis is an ideal method to sample the circulating fluid in order to study neuropeptide release. For microdialysis sampling from crustaceans, the perfusion solution is crab saline to minimize the effect of sampling on the ionic strength of crustacean hemolymph. Members from 10 neuropeptide families including ASTs, TRP, orcokinin, RFamide, RYamide were identified from Jonah crab Cancer borealis hemolymph as reported in the first application of in vivo microdialysis method for MS analysis of neuropeptides from a freely moving crab [13]. Coupling microdialysis with MS has several advantages compared to direct hemolymph extraction method. Not only more neuropeptides have been identified with microdialysis sampling due to removal of larger protein species and producing cleaner sample matrix, but also the in vivo sampling method allows monitoring dynamic changes of neuropeptides in a timely-resolved fashion thus correlating chemistry with animal behaviors.
Comparing to small molecules, neuropeptides have a lower R value due to their relatively larger size, which would hinder passing through the dialysis membrane. Efforts have been made to improve R for larger molecules, such as sampling with microdialysis probe with larger pore size [121] yielding better recovery for proteins. An alternative way to improve R for microdialysis sampling of neuropeptides is affinity-enhanced microdialysis (AE-MD), where better recovery rate could be achieved by adding affinity agent into the perfusate. Numerous applications have employed this strategy for a wide range of analytes [122–124]. A recent study from our lab utilized RFamide antibody linked magnetic nanoparticles to increase recovery of RFamide-related peptides when sampling from Jonah crab, Cancer borealis by microdialysis [14]. In vitro R value has been increased up to 41-fold with neuropeptide standards tested, and more than 10 neuropeptides were detected in a 30 min collection sample as compared to previous 4-hour collection time without the use of AE-MD. Due to the improvement of recovery rate and subsequent increase of neuropeptide identifications, more reliable quantitation of neuropeptides during a dynamic process, such as feeding, has also been achieved.
3.4 Temporal resolution
Microdialysis has the advantage of providing temporal information over other sampling techniques. The shortest time duration over which a dynamic change event could be observed is how temporal resolution is measured. Such temporal resolution is largely dependent on detection sensitivity, where enough analytes need to be collected at certain flow rate per fraction to reach the detection limit. To monitor dynamic changes of chemicals during a specific behavior, it is critical to capture the content in a high temporal resolution fashion because concentration change of analyte could happen rather rapidly. Several highly sensitive methods are available for resolution as good as a couple of seconds for fraction collection, and for online microdialysate analysis.
With common HPLC methods, the temporal resolution for neuropeptide detection could reach 10 to 30 min [112]. As results shown from several studies, capillary electrophoresis, which could handle much smaller sample volume compared to HPLC by using microcolumn, coupled with laser-induced fluorescence detection could resolve analytes of interest in the order of seconds via online coupling with microdialysis sampling [125–127]. However, the high temporal resolution is realized using anesthetized animal at high dialysis flow rates, which leads to lower recovery rate. Further improvement of in vivo microdialysis application has been achieved by using segment flow for freely moving animals at low flow rates via microfluidic segmented outflow [128, 129], with temporal resolution of seconds. Chip-based electrophoresis provided the capability of monitoring amino acids at high temporal resolution as fast as two seconds [130, 131]. Enzymatic assay to monitor glutamate dynamic changes [132] produced a temporal resolution of 7s in vivo and 200 ms in vitro. Coupling segmented flow with electrochemical [133] and mass spectrometry [134] have been conducted.
3.5 Mass spectrometric detection of neuropeptides from microdialysis
Circulating peptide hormones are present at extremely low concentrations in extracellular space [135], ranging from pM to nM. Moreover, low recovery rate of sampling neuropeptides from the hemolymph makes the subsequent detection more difficult. Detection of microdialysis samples usually requires high sensitivity, and the capability of handling μL volumes. Traditional immunoassays, fluorometric or radioactive or enzyme, were extensively used to measure neuropeptide release collected via microdialysis [136, 137]. However, these methods lack specificity due to cross reaction, making them not suitable for complex mixture analysis and novel peptide discovery. Moreover, the number of assayable substances is also limited in each sample. The volume requirements for these assays need to be taken into consideration in regard to temporal resolution. Numerous assays have been developed for the characterization of different neurotransmitters [138].
Since first described by Emmett et al. [139], the coupling of MS detection with microdialysis sampling has become a common strategy for identification and quantitation of analytes of interest. One of the most common separation methods coupled with MS analysis is LC. The power of LC-MS has been showcased by several laboratories in the identification and quantitation of different neurotransmitters in microdialysates [140–143].
Trace level concentration and highly dynamic changes of neuropeptides in vivo make it challenging to detect these low abundance signaling molecules in the microdialysates. Capillary LC-MSn (CLC-MSn) platform has been developed by several groups towards the detection of neuropeptides. Over the past several years, the field has seen significant progress. Using such platform, neuropeptide secretion has been explored in different organisms, from crustaceans [13] to mammalian [144, 145] to primates [146]. The on-going improvement in LC-MS methods has boosted the capability of microdialysis to provide unique insight into the role of neuropeptides in behavior. The study by DiFeliceantonio et al. [147] described the first detection of opioid peptide, enkephalin, during feeding, which exhibited elevated levels in the dorsal striatum of rats. This finding about opioid peptide acting as a signal to eat was further confirmed by microinjection of the opioid peptide into the same brain region evoking feeding behavior. Quantitative measurement of neuropeptides during feeding behavior in Jonah crab, Cancer borealis by Schmerberg and Li [14] also revealed several candidates as signaling molecules involved in food intake in crustaceans.
3.6 Challenges and perspectives of probing temporal dynamics via microdialysis sampling technique
To date, application of microdialysis is more restricted to mammalian nervous systems. There are only a few publications about microdialysis in crustaceans. Technical difficulties associated with microdialysis in crustaceans are the main obstacles for its widespread use. The hard shell of the animal makes the surgery and implantation of the microdialysis probe more complicated, in addition to maintaining the microdialysis probe in working condition underwater. Other factors such as low concentration of circulating hormones in crustaceans and relatively poor recovery rate of the dialysis membrane make the detection of neuropeptides in microdialysate very challenging. Despite significant challenges, nanoflow LC coupled to tandem MS enabled detection of neuropeptides in the microdialysate collected from Jonah crab, Cancer borealis [13, 14]. AE-MD showed great potential for further improving temporal resolution by increased recovery rate. Future work will focus on further improving peptidome coverage and quantitation as well as combining multi-pronged approach to enable correlation of neurochemical and circuit dynamics with behavior.
4. Conclusions and Future Perspectives
Decapod crustaceans have made significant contributions to the study of neuropeptides and neuromodulation, both in analytical method development and in our molecular understanding of how neuropeptides can modulate circuit dynamics. MS-based techniques are optimal for studying neuropeptides, due to their high specificity when compared with other techniques. MSI is an emerging technique to map the distribution of neuropeptides in a given tissue with greater specificity than antibody-based imaging techniques and the ability to characterize the distribution of thousands of analytes in a single experiment. Complementary to tissue expression and spatial distribution of neuropeptides, in vivo microdialysis sampling on living crustacean coupled to MS detection enables correlation of neuropeptide temporal dynamics with activity.
Although recent advances in MS have significantly accelerated the pace of neuropeptide discovery and functional identification, further technological advances are under development and the full suite of crustacean neuropeptides has not yet been described completely. Advances in MS technology will greatly improve the capabilities of this technique to provide tissue localization, identity, and quantity information. In the area of MSI, methods for tissue coating with matrix are being improved to create smaller, more uniform crystals, to decrease analyte diffusion, provide better spatial resolution, increase sensitivity, and reduce “hot spots” of signal on the tissue. Improvements to laser technology will also improve the spatial resolution of this technique. For identification, faster scan rates, higher accuracy, and better resolution will improve the quality of identifications and allow less abundant compounds to be identified. Furthermore, neuropeptides can be monitored continuously with the aid of the minimally invasive sampling technique of microdialysis coupled to LC-MS/MS approach. Combined with quantitative technique, this method will narrow down the list of hundreds of neuropeptides to a few “interesting” neuropeptides in an unbiased and systematic manner. The ongoing improvement of instrument sensitivity and strategies to reduce sampling intervals and enable on-line coupling will further improve temporal resolution for monitoring of neurochemical dynamics in behaving animal. Further improvements to the analysis methods with increased spatial and temporal information that enable the study of neuropeptide functions will greatly increase our understanding of this important class of signaling molecules, both in crustaceans and in other organisms, including mammals.
Highlights.
The nervous system in crustacean is an excellent model for neuropeptide study.
Detailed workflows of MS imaging and microdialysis in crustacean are discussed.
Advances of neuropeptide analysis at the spatial and temporal domains are noted.
MS-based techniques provide comprehensive information of crustacean neuropeptides.
Acknowledgments
Preparation of this manuscript is supported in part by National Science Foundation (CHE-1413596) and National Institutes of Health through grant 1R01DK071801. L.L. acknowledges an H. I. Romnes Faculty Fellowship.
Abbreviations
- AST
allatostatin
- CCAP
crustacean cardioactive peptide
- CE
capillary electrophoresis
- CHCA
α-cyano-4-hydroxycinnamic acid
- CID
collisional induced dissociation
- CoG
commissural ganglia
- dgn
dorsal gastric nerve
- DHB
2,5-dihydroxy benzoic acid
- ESI
electrospray ionization
- FLPs
FMRFamide-like peptides
- GABA
γ-aminobutyric acid
- HPLC
high performance liquid chromatography
- ion
inferior esophageal nerve
- ivn
inferior ventricular nerve
- LC
liquid chromatography
- LDI
laser desorption ionization
- LTQ
linear trap quadrupole
- IHC
Immunohistochemical
- lvn
lateral ventricular nerve
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- MSI
mass spectrometric imaging
- MS/MS
tandem mass spectrometry
- mvn
medial ventricular nerve
- NIMS
nanostructure initiator mass spectrometry
- OG
oesophageal ganglion
- pdn
pyloric dilator nerve
- PO
pericardial organs
- PTM
post-translational modification
- pyn
pyloric nerve
- QTOF
quadrupole time of flight
- SG
sinus gland
- son
superior esophageal nerve
- STG
stomatogastric ganglion
- stn
stomatogastric nerve
- STNS
stomatogastric nervous system
- TOF
time of flight
- TRP
Tachykinin-related peptides
- VNC
ventral nerve cord
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burbach JP. What Are Neuropeptides? Methods in Molecular Biology. 2011;789:1–36. doi: 10.1007/978-1-61779-310-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Marder E, Bucher D. Understanding Circuit Dynamics Using the Stomatogastric Nervous System of Lobsters and Crabs. Annual Review of Physiology. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- 3.Christie AE, Stemmler EA, Dickinson PS. Crustacean Neuropeptides. Cellular and molecular life sciences : CMLS. 2010;67:4135–69. doi: 10.1007/s00018-010-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Q, OuYang C, Liang Z, Li L. Mass Spectrometric Characterization of the Crustacean Neuropeptidome. EuPA Open Proteomics. 2014;3:152–70. [Google Scholar]
- 5.Schmerberg CM, Li L. Function-driven Discovery of Neuropeptides with Mass Spectrometry-based Tools. Protein and Peptide Letters. 2013;20:681–94. doi: 10.2174/0929866511320060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia C, Lietz CB, Ye H, Hui L, Yu Q, Yoo S, Li L. A Multi-scale Strategy for Discovery of Novel Endogenous Neuropeptides in the Crustacean Nervous System. Journal of Proteomics. 2013;91:1–12. doi: 10.1016/j.jprot.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui L, D’Andrea BT, Jia C, Liang Z, Christie AE, Li L. Mass Spectrometric Characterization of the Neuropeptidome of the Ghost Crab Ocypode Ceratophthalma (Brachyura, Ocypodidae) General and Comparative Endocrinology. 2013;184:22–34. doi: 10.1016/j.ygcen.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hui L, Xiang F, Zhang Y, Li L. Mass Spectrometric Elucidation of the Neuropeptidome of a Crustacean Neuroendocrine Organ. Peptides. 2012;36:230–9. doi: 10.1016/j.peptides.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma M, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GL, Henry RP, Smith CM, Towle DW, Christie AE, Li L. Characterization of the Carcinus Maenas Neuropeptidome by Mass Spectrometry and Functional Genomics. General and Comparative Endocrinology. 2009;161:320–34. doi: 10.1016/j.ygcen.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma M, Wang J, Chen R, Li L. Expanding the Crustacean Neuropeptidome Using a Multifaceted Mass Spectrometric Approach. Journal of Proteome Research. 2009;8:2426–37. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Q, Kutz KK, Schmidt JJ, Hsu YW, Messinger DI, Cain SD, de la Iglesia HO, Christie AE, Li L. Hormone Complement of the Cancer productus Sinus Gland and Pericardial Organ: An Anatomical and Mass Spectrometric Investigation. The Journal of Comparative Neurology. 2005;493:607–26. doi: 10.1002/cne.20773. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, Ma M, Hui L, Zhang J, Li L. Measurement of Neuropeptides in Crustacean Hemolymph via MALDI Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2009;20:708–18. doi: 10.1016/j.jasms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behrens HL, Chen R, Li L. Combining Microdialysis, NanoLC-MS, and MALDI-TOF/TOF to Detect Neuropeptides Secreted in the Crab, Cancer borealis. Analytical Chemistry. 2008;80:6949–58. doi: 10.1021/ac800798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmerberg CM, Li L. Mass Spectrometric Detection of Neuropeptides Using Affinity-Enhanced Microdialysis with Antibody-Coated Magnetic Nanoparticles. Analytical Chemistry. 2013;85:915–22. doi: 10.1021/ac302403e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Li L. Mass Spectral Imaging and Profiling of Neuropeptides at the Organ and Cellular Domains. Analytical and Bioanalytical Chemistry. 2010;397:3185–93. doi: 10.1007/s00216-010-3723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye H, Greer T, Li L. Probing Neuropeptide Signaling at the Organ and Cellular Domains via Imaging Mass Spectrometry. Journal of Proteomics. 2012;75:5014–26. doi: 10.1016/j.jprot.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy RT. Emerging Trends in In Vivo Neurochemical Monitoring by Microdialysis. Current Opinion in Chemical Biology. 2013;17:860–7. doi: 10.1016/j.cbpa.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christie AE. Crustacean Neuroendocrine Systems and Their Signaling Agents. Cell and tissue research. 2011;345:41–67. doi: 10.1007/s00441-011-1183-9. [DOI] [PubMed] [Google Scholar]
- 19.Hummon AB, Amare A, Sweedler JV. Discovering New Invertebrate Neuropeptides Using Mass Spectrometry. Mass Spectrometry Reviews. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 20.Bendena W, Donly B, Tobe S. Allatostatins: a growing family of neuropeptides with structural and functional diversity. Annals of the New York Academy of Sciences. 1999;897:311–29. doi: 10.1111/j.1749-6632.1999.tb07902.x. [DOI] [PubMed] [Google Scholar]
- 21.Skiebe P, Schneider H. Allatostatin peptides in the crab stomatogastric nervous system: inhibition of the pyloric motor pattern and distribution of allatostatin-like immunoreactivity. The Journal of Experimental Biology. 1994;194:195–208. doi: 10.1242/jeb.194.1.195. [DOI] [PubMed] [Google Scholar]
- 22.Jorge-Rivera J, Marder E. Allatostatin decreases stomatogastric neuromuscular transmission in the crab Cancer borealis. The Journal of Experimental Biology. 1997;200:2937–46. doi: 10.1242/jeb.200.23.2937. [DOI] [PubMed] [Google Scholar]
- 23.Dircksen H, Skiebe P, Abel B, Agricola H, Buchner K, Muren JE, Nässel DR. Structure, distribution, and biological activity of novel members of the allatostatin family in the crayfish Orconectes limosus. Peptides. 1999;20:695–712. doi: 10.1016/s0196-9781(99)00052-2. [DOI] [PubMed] [Google Scholar]
- 24.Kreissl S, Weiss T, Djokaj S, Balezina O, Rathmayer W. Allatostatin modulates skeletal muscle performance in crustaceans through pre- and postsynaptic effects. European Journal of Neuroscience. 1999;11:2519–30. doi: 10.1046/j.1460-9568.1999.00674.x. [DOI] [PubMed] [Google Scholar]
- 25.Cruz-Bermúdez ND, Marder E. Multiple modulators act on the cardiac ganglion of the crab, Cancer borealis. The Journal of Experimental Biology. 2007;210:2873–84. doi: 10.1242/jeb.002949. [DOI] [PubMed] [Google Scholar]
- 26.Fu Q, Tang LS, Marder E, Li L. Mass spectrometric characterization and physiological actions of VPNDWAHFRGSWamide, a novel B type allatostatin in the crab, Cancer borealis. Journal of Neurochemistry. 2007;101:1099–107. doi: 10.1111/j.1471-4159.2007.04482.x. [DOI] [PubMed] [Google Scholar]
- 27.Tobe SS, Bendena WG. Chapter 29 - Allatostatins. In: Kastin AJ, editor. Handbook of Biologically Active Peptides. 2. Boston: Academic Press; 2013. pp. 191–6. [Google Scholar]
- 28.Ma M, Szabo TM, Jia C, Marder E, Li L. Mass spectrometric characterization and physiological actions of novel crustacean C-type allatostatins. Peptides. 2009;30:1660–8. doi: 10.1016/j.peptides.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stangier J, Hilbich C, Beyreuther K, Keller R. Unusual cardioactive peptide (CCAP) from pericardial organs of the shore crab Carcinus maenas. Proceedings of the National Academy of Sciences. 1987;84:575–9. doi: 10.1073/pnas.84.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fort TJ, García-Crescioni K, Brezina V, Miller MW. Regulation of the crab heartbeat by crustacean cardioactive peptide (CCAP): central and peripheral actions. Journal of Neurophysiology. 2007;97:3407–20. doi: 10.1152/jn.00939.2006. [DOI] [PubMed] [Google Scholar]
- 31.Mercier A, Lee J. Differential effects of neuropeptides on circular and longitudinal muscles of the crayfish hindgut. Peptides. 2002;23:1751–7. doi: 10.1016/s0196-9781(02)00151-1. [DOI] [PubMed] [Google Scholar]
- 32.Jorge-Rivera J, Marder E. TNRNFLRFamide and SDRNFLRFamide modulate muscles of the stomatogastric system of the crab Cancer borealis. Journal of Comparative Physiology A. 1996;179:741–51. doi: 10.1007/BF00207353. [DOI] [PubMed] [Google Scholar]
- 33.Wilkens J, Shinozaki T, Yazawa T, ter Keurs H. Sites and modes of action of proctolin and the FLP F2 on lobster cardiac muscle. The Journal of Experimental Biology. 2005;208:737. doi: 10.1242/jeb.01430. [DOI] [PubMed] [Google Scholar]
- 34.Swensen AM, Marder E. Multiple peptides converge to activate the same voltage-dependent current in a central pattern-generating circuit. The Journal of Neuroscience. 2000;20:6752–9. doi: 10.1523/JNEUROSCI.20-18-06752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stangier J, Hilbich C, Burdzik S, Keller R. Orcokinin: a novel myotropic peptide from the nervous system of the crayfish, Orconectes limosus. Peptides. 1992;13:859. doi: 10.1016/0196-9781(92)90041-z. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E. Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. The Journal of Comparative Neurology. 2002;444:227–44. doi: 10.1002/cne.10139. [DOI] [PubMed] [Google Scholar]
- 37.Skiebe P, Dreger M, Meseke M, Evers JF, Hucho F. Identification of orcokinins in single neurons in the stomatogastric nervous system of the crayfish, Cherax destructor. The Journal of Comparative Neurology. 2002;444:245–59. doi: 10.1002/cne.10145. [DOI] [PubMed] [Google Scholar]
- 38.Saideman S, Ma M, Kutz-Naber K, Cook A, Torfs P, Schoofs L, Li L, Nusbaum M. Modulation of rhythmic motor activity by pyrokinin peptides. Journal of Neurophysiology. 2007;97:579–95. doi: 10.1152/jn.00772.2006. [DOI] [PubMed] [Google Scholar]
- 39.Vazquez-Acevedo N, Rivera NM, Torres-Gonzalez AM, Rullan-Matheu Y, Ruiz-Rodriguez EA, Sosa MA. GYRKPPFNGSIFamide (Gly-SIFamide) modulates aggression in the freshwater prawn Macrobrachium rosenbergii. The Biological bulletin. 2009;217:313–26. doi: 10.1086/BBLv217n3p313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nässel DR. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Progress in Neurobiology. 2002;68:1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 41.Chen R, Hui L, Sturm RM, Li L. Three Dimensional Mapping of Neuropeptides and Lipids in Crustacean Brain by Mass Spectral Imaging. Journal of the American Society for Mass Spectrometry. 2009;20:1068–77. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higuchi H. Molecular Analysis of Central Feeding Regulation by Neuropeptide Y (NPY) Neurons with NPY Receptor Small Interfering RNAs (siRNAs) Neurochemistry International. 2012;61:936–41. doi: 10.1016/j.neuint.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 43.Kanai A, Zabbarova I, Oefelein M, Radziszewski P, Ikeda Y, Andersson KE. Mechanisms of Action of Botulinum Neurotoxins, Beta3-adrenergic Receptor Agonists, and PDE5 Inhibitors in Modulating Detrusor Function in Overactive Bladders: ICI-RS 2011. Neurourology and Urodynamics. 2012;31:300–8. doi: 10.1002/nau.21246. [DOI] [PubMed] [Google Scholar]
- 44.Do Rego JL, Seong JY, Burel D, Luu-The V, Larhammar D, Tsutsui K, Pelletier G, Tonon MC, Vaudry H. Steroid Biosynthesis within the Frog Brain: A Model of Neuroendocrine Regulation. Annals of the New York Academy of Sciences. 2009;1163:83–92. doi: 10.1111/j.1749-6632.2008.03664.x. [DOI] [PubMed] [Google Scholar]
- 45.Verleyen P, Huybrechts J, Schoofs L. SIFamide Illustrates the Rapid Evolution in Arthropod Neuropeptide Research. General and Comparative Endocrinology. 2009;162:27–35. doi: 10.1016/j.ygcen.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 46.Szabo TM, Chen R, Goeritz ML, Maloney RT, Tang LS, Li L, Marder E. Distribution and Physiological Effects of B-type Allatostatins (Myoinhibitory Peptides, MIPs) in the Stomatogastric Nervous System of the Crab Cancer borealis. The Journal of Comparative Neurology. 2011;519:2658–76. doi: 10.1002/cne.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caprioli RM, Farmer TB, Gile J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Analytical Chemistry. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 48.Chughtai K, Heeren RM. Mass Spectrometric Imaging for Biomedical Tissue Analysis. Chemical Reviews. 2010;110:3237–77. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye H, Greer T, Li L. From Pixel to Voxel: a Deeper View of Biological Tissue by 3D Mass Spectral Imaging. Bioanalysis. 2011;3:313–32. doi: 10.4155/bio.10.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lietz CB, Gemperline E, Li L. Qualitative and Quantitative Mass Spectrometry Imaging of Drugs and Metabolites. Advanced Drug Delivery Reviews. 2013;65:1074–85. doi: 10.1016/j.addr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye H, Gemperline E, Li L. A Vision for Better Health: Mass Spectrometry Imaging for Clinical Diagnostics. Clinica chimica acta; international journal of clinical chemistry. 2013;420:11–22. doi: 10.1016/j.cca.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erin G, Bingming C, Lingjun L. Challenges and Recent Advances in Mass Spectrometric Imaging of Neurotransmitters. Bioanalysis. 2014 doi: 10.4155/bio.13.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. Imaging Mass Spectrometry of Neuropeptides in Decapod Crustacean Neuronal Tissues. Journal of Proteome Research. 2007;6:1782–91. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen R, Hui L, Cape SS, Wang J, Li L. Comparative Neuropeptidomic Analysis of Food Intake via a Multifaceted Mass Spectrometric Approach. ACS Chemical Neuroscience. 2010;1:204–14. doi: 10.1021/cn900028s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korte AR, Lee YJ. Multiplex Mass Spectrometric Imaging with Polarity Switching for Concurrent Acquisition of Positive and Negative Ion Images. Journal of the American Society for Mass Spectrometry. 2013;24:949–55. doi: 10.1007/s13361-013-0613-1. [DOI] [PubMed] [Google Scholar]
- 56.Perdian DC, Lee YJ. Imaging MS Methodology for More Chemical Information in Less Data Acquisition Time Utilizing a Hybrid Linear Ion Trap-orbitrap Mass Spectrometer. Analytical Chemistry. 2010;82:9393–400. doi: 10.1021/ac102017q. [DOI] [PubMed] [Google Scholar]
- 57.Yagnik GB, Korte AR, Lee YJ. Multiplex Mass Spectrometry Imaging for Latent Fingerprints. Journal of Mass Spectrometry : JMS. 2013;48:100–4. doi: 10.1002/jms.3134. [DOI] [PubMed] [Google Scholar]
- 58.Jimenez CR, Li KW, Dreisewerd K, Spijker S, Kingston R, Bateman RH, Burlingame AL, Smit AB, van Minnen J, Geraerts WP. Direct Mass Spectrometric Peptide Profiling and Sequencing of Single Neurons Reveals Differential Peptide Patterns in a Small Neuronal Network. Biochemistry. 1998;37:2070–6. doi: 10.1021/bi971848b. [DOI] [PubMed] [Google Scholar]
- 59.Jimenez CR, Li KW, Dreisewerd K, Mansvelder HD, Brussaard AB, Reinhold BB, Van der Schors RC, Karas M, Hillenkamp F, Burbach JP, Costello CE, Geraerts WP. Pattern changes of pituitary peptides in rat after salt-loading as detected by means of direct, semiquantitative mass spectrometric profiling. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9481–6. doi: 10.1073/pnas.94.17.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jimenez CR, ter Maat A, Pieneman A, Burlingame AL, Smit AB, Li KW. Spatio-temporal Dynamics of the Egg-laying-inducing Peptides During an Egg-laying Cycle: A Semiquantitative Matrix-assisted Laser Desorption/Ionization Mass Spectrometry Approach. Journal of Neurochemistry. 2004;89:865–75. doi: 10.1111/j.1471-4159.2004.02353.x. [DOI] [PubMed] [Google Scholar]
- 61.Jimenez CR, Li KW, Smit AB, Janse C. Auto-Inhibitory Control of Peptidergic Molluscan Neurons and Reproductive Senescence. Neurobiology of Aging. 2006;27:763–9. doi: 10.1016/j.neurobiolaging.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 62.Ellis SR, Bruinen AL, Heeren RM. A Critical Evaluation of the Current State-of-the-art in Quantitative Imaging Mass Spectrometry. Analytical and Bioanalytical Chemistry. 2014;406:1275–89. doi: 10.1007/s00216-013-7478-9. [DOI] [PubMed] [Google Scholar]
- 63.Chen R, Jiang X, Conaway MC, Mohtashemi I, Hui L, Viner R, Li L. Mass Spectral Analysis of Neuropeptide Expression and Distribution in the Nervous System of the Lobster Homarus americanus. Journal of Proteome Research. 2010;9:818–32. doi: 10.1021/pr900736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kutz KK, Schmidt JJ, Li L. In Situ Tissue Analysis of Neuropeptides by MALDI FTMS In-Cell Accumulation. Analytical Chemistry. 2004;76:5630–40. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- 65.Jia X, Hollung K, Therkildsen M, Hildrum KI, Bendixen E. Proteome Analysis of Early Post-mortem Changes in Two Bovine Muscle Types: M. longissimus dorsi and M. semitendinosis. Proteomics. 2006;6:936–44. doi: 10.1002/pmic.200500249. [DOI] [PubMed] [Google Scholar]
- 66.Svensson M, Skold K, Nilsson A, Falth M, Nydahl K, Svenningsson P, Andren PE. Neuropeptidomics: MS Applied to the Discovery of Novel Peptides from the Brain. Analytical Chemistry. 2007;79:15–6. 8–21. doi: 10.1021/ac071856q. [DOI] [PubMed] [Google Scholar]
- 67.Svensson M, Skold K, Svenningsson P, Andren PE. Peptidomics-based Discovery of Novel Neuropeptides. Journal of Proteome Research. 2003;2:213–9. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 68.O’Callaghan JP, Sriram K. Focused Microwave Irradiation of the Brain Preserves In Vivo Protein Phosphorylation: Comparison with Other Methods of Sacrifice and Analysis of Multiple Phosphoproteins. Journal of Neuroscience Methods. 2004;135:159–68. doi: 10.1016/j.jneumeth.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 69.Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M, Svenningsson P, Andren PE. Heat Stabilization of the Tissue Proteome: A New Technology for Improved Proteomics. Journal of Proteome Research. 2009;8:974–81. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 70.Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. The Significance of Biochemical and Molecular Sample Integrity in Brain Proteomics and Peptidomics: Stathmin 2-20 and Peptides as Sample Quality Indicators. Proteomics. 2007;7:4445–56. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 71.Sturm RM, Greer T, Woodards N, Gemperline E, Li L. Mass Spectrometric Evaluation of Neuropeptidomic Profiles upon Heat Stabilization Treatment of Neuroendocrine Tissues in Crustaceans. Journal of Proteome Research. 2013;12:743–52. doi: 10.1021/pr300805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. Journal of Proteome Research. 2007;6:1295–305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 73.Groseclose MR, Massion PP, Chaurand P, Caprioli RM. High-throughput Proteomic Analysis of Formalin-fixed Paraffin-embedded Tissue Microarrays Using MALDI Imaging Mass Spectrometry. Proteomics. 2008;8:3715–24. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ronci M, Bonanno E, Colantoni A, Pieroni L, Di Ilio C, Spagnoli LG, Federici G, Urbani A. Protein Unlocking Procedures of Formalin-fixed Paraffin-embedded Tissues: Application to MALDI-TOF Imaging MS Investigations. Proteomics. 2008;8:3702–14. doi: 10.1002/pmic.200701143. [DOI] [PubMed] [Google Scholar]
- 75.Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. MALDI Imaging of Formalin-fixed Paraffin-embedded Tissues: Application to Model Animals of Parkinson Disease for Biomarker Hunting. Journal of Proteome Research. 2008;7:969–78. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- 76.Djidja M-CC, Francese S, Loadman PM, Sutton CW, Scriven P, Claude E, Snel MF, Franck J, Salzet M, Clench MR. Detergent Addition to Tryptic Digests and Ion Mobility Separation Prior to MS/MS Improves Peptide Yield and Protein Identification for In Situ Proteomic Investigation of Frozen and Formalin-fixed Paraffin-embedded Adenocarcinoma Tissue Sections. Proteomics. 2009;9:2750–63. doi: 10.1002/pmic.200800624. [DOI] [PubMed] [Google Scholar]
- 77.Crecelius A, Gotz A, Arzberger T, Frohlich T, Arnold GJ, Ferrer I, Kretzschmar HA. Assessing Quantitative Post-mortem Changes in the Gray Matter of the Human Frontal Cortex Proteome by 2-D DIGE. Proteomics. 2008;8:1276–91. doi: 10.1002/pmic.200700728. [DOI] [PubMed] [Google Scholar]
- 78.Schwartz SA, Reyzer ML, Caprioli RM. Direct Tissue Analysis Using Matrix-assisted Laser Desorption/Ionization Mass Spectrometry: Practical Aspects of Sample Preparation. Journal of Mass Spectrometry : JMS. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 79.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-Dimensional Visualization of Protein Expression in Mouse Brain Structures Using Imaging Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2005;16:1093–9. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 80.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct Molecular Analysis of Whole-Body Animal Tissue Sections by Imaging MALDI Mass Spectrometry. Analytical Chemistry. 2006;78:6448–56. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 81.Crossman L, McHugh NA, Hsieh Y, Korfmacher WA, Chen J. Investigation of the Profiling Depth in Matrix-assisted Laser Desorption/Ionization Imaging Mass Spectrometry. Rapid Communications Mass Spectrometry. 2006;20:284–90. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 82.Woo HK, Northen TR, Yanes O, Siuzdak G. Nanostructure-initiator Mass Spectrometry: a Protocol for Preparing and Applying NIMS Surfaces for High-sensitivity Mass Analysis. Nature Protocols. 2008;3:1341–9. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- 83.Greving MP, Patti GJ, Siuzdak G. Nanostructure-initiator Mass Spectrometry Metabolite Analysis and Imaging. Analytical Chemistry. 2011;83:2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sturm RM, Greer T, Chen R, Hensen B, Li L. Comparison of NIMS and MALDI Platforms for Neuropeptide and Lipid Mass Spectrometric Imaging in C. borealis Brain Tissue. Analytical Methods. 2013;5:1623. doi: 10.1039/C3AY26067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawamoto T. Use of a New Adhesive Film for the Preparation of Multi-purpose Fresh-frozen Sections from Hard Tissues, Whole-animals, Insects and Plants. Archives of Histology and Cytology. 2003;66:123–43. doi: 10.1679/aohc.66.123. [DOI] [PubMed] [Google Scholar]
- 86.Ye H, Hui L, Kellersberger K, Li L. Mapping of Neuropeptides in the Crustacean Stomatogastric Nervous System by Imaging Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2013;24:134–47. doi: 10.1007/s13361-012-0502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lemaire R, Wisztorski M, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. MALDI-MS Direct Tissue Analysis of Proteins: Improving Signal Sensitivity Using Organic Treatments. Analytical Chemistry. 2006;78:7145–53. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 88.Goodwin RJ, Pennington SR, Pitt AR. Protein and Peptides in Pictures: Imaging with MALDI Mass Spectrometry. Proteomics. 2008;8:3785–800. doi: 10.1002/pmic.200800320. [DOI] [PubMed] [Google Scholar]
- 89.Kaletas BK, van der Wiel IM, Stauber J, Guzel C, Kros JM, Luider TM, Heeren RM. Sample Preparation Issues for Tissue Imaging by Imaging MS. Proteomics. 2009;9:2622–33. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 90.MacAleese L, Stauber J, Heeren RM. Perspectives for Imaging Mass Spectrometry in the Proteomics Landscape. Proteomics. 2009;9:819–34. doi: 10.1002/pmic.200800363. [DOI] [PubMed] [Google Scholar]
- 91.Shanta SR, Zhou L-H, Park YS, Kim YH, Kim Y, Kim KP. Binary Matrix for MALDI Imaging Mass Spectrometry of Phospholipids in Both Ion Modes. Analytical Chemistry. 2011;83:1252–9. doi: 10.1021/ac1029659. [DOI] [PubMed] [Google Scholar]
- 92.Hui L, Zhang Y, Wang J, Cook A, Ye H, Nusbaum MP, Li L. Discovery and Functional Study of a Novel Crustacean Tachykinin Neuropeptide. ACS Chemical Neuroscience. 2011;2:711–22. doi: 10.1021/cn200042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jia C, Hui L, Cao W, Lietz CB, Jiang X, Chen R, Catherman AD, Thomas PM, Ge Y, Kelleher NL, Li L. High-definition De Novo Sequencing of Crustacean Hyperglycemic Hormone (CHH)-family Neuropeptides. Molecular and Cellular Proteomics : MCP. 2012;11:1951–64. doi: 10.1074/mcp.M112.020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen B, Lietz CB, Li L. In Situ Characterization of Proteins Using Laserspray Ionization on a High-Performance MALDI-LTQ-Orbitrap Mass Spectrometer. Journal of The American Society for Mass Spectrometry. 2014 doi: 10.1007/s13361-014-0986-9. published online Oct 2, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass Spectrometry Sampling under Ambient Conditions with Desorption Electrospray Ionization. Science (New York, NY) 2004;306:471–3. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 96.Demian RI, Justin MW, Qingyu S, Cooks RG. Development of Capabilities for Imaging Mass Spectrometry Under Ambient Conditions with Desorption Electrospray Ionization (DESI) International Journal of Mass Spectrometry. 2007:259. [Google Scholar]
- 97.Trimpin S, Ren Y, Wang B, Lietz CB, Richards AL, Marshall DD, Inutan ED. Extending the Laserspray Ionization Concept to Produce Highly Charged Ions at High Vacuum on a Time-of-flight Mass Analyzer. Analytical Chemistry. 2011;83:5469–75. doi: 10.1021/ac2007976. [DOI] [PubMed] [Google Scholar]
- 98.Chansela P, Goto-Inoue N, Zaima N, Sroyraya M, Sobhon P, Setou M. Visualization of Neuropeptides in Paraffin-Embedded Tissue Sections of the Central Nervous System in the Decapod Crustacean, Penaeus Monodon, by Imaging Mass Spectrometry. Peptides. 2012;34:10–8. doi: 10.1016/j.peptides.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 99.Seeley EH, Caprioli RM. 3D Imaging by Mass Spectrometry: A New Frontier. Analytical Chemistry. 2012;84:2105–10. doi: 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiong X, Xu W, Eberlin LS, Wiseman JM, Fang X, Jiang Y, Huang Z, Zhang Y, Cooks RG, Ouyang Z. Data Processing for 3D Mass Spectrometry Imaging. Journal of the American Society for Mass Spectrometry. 2012;23:1147–56. doi: 10.1007/s13361-012-0361-7. [DOI] [PubMed] [Google Scholar]
- 101.Zhu ZJ, Rotello VM, Vachet RW. Engineered Nanoparticle Surfaces for Improved Mass Spectrometric Analyses. Analyst. 2009;134:2183–8. doi: 10.1039/b910428c. [DOI] [PubMed] [Google Scholar]
- 102.Peterson DS. Matrix-free Methods for Laser Desorption/Ionization Mass Spectrometry. Mass Spectrometry Reviews. 2007;26:19–34. doi: 10.1002/mas.20104. [DOI] [PubMed] [Google Scholar]
- 103.Chiang CK, Chen WT, Chang HT. Nanoparticle-based Mass Spectrometry for the Analysis of Biomolecules. Chemical Society Reviews. 2011;40:1269–81. doi: 10.1039/c0cs00050g. [DOI] [PubMed] [Google Scholar]
- 104.Sudhir P-R, Wu H-F, Zhou Z-C. Identification of Peptides Using Gold Nanoparticle-Assisted Single-Drop Microextraction Coupled with AP-MALDI Mass Spectrometry. Analytical Chemistry. 2005;77:7380–5. doi: 10.1021/ac051162m. [DOI] [PubMed] [Google Scholar]
- 105.Moyer SC, Cotter RJ. Peer Reviewed: Atmospheric Pressure MALDI. Analytical Chemistry. 2002;74:468 A–76 A. doi: 10.1021/ac022091j. [DOI] [PubMed] [Google Scholar]
- 106.Dickinson PS, Stevens JS, Rus S, Brennan HR, Goiney CC, Smith CM, Li L, Towle DW, Christie AE. Identification and Cardiotropic Actions of Sulfakinin Peptides in the American Lobster Homarus americanus. The Journal of Experimental Biology. 2007;210:2278–89. doi: 10.1242/jeb.004770. [DOI] [PubMed] [Google Scholar]
- 107.Bito L, Davson H, Levin E, Murray M, Snider N. The Concentrations of Free Amino Acids and Other Electrolytes in Cerebrospinal Fluid, In Vivo Dialysate of Brain, and Blood Plasma of the Dog. Journal of Neurochemistry. 1966;13:1057–67. doi: 10.1111/j.1471-4159.1966.tb04265.x. [DOI] [PubMed] [Google Scholar]
- 108.Nandi P, Lunte SM. Recent Trends in Microdialysis Sampling Integrated with Conventional and Microanalytical Systems for Monitoring Biological Events: a Review. Analytica Chimica Acta. 2009;651:1–14. doi: 10.1016/j.aca.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shippenberg TS, Thompson AC. Overview of Microdialysis. Current Protocols in Neuroscience. 2001;Chapter 7(Unit 7):1. doi: 10.1002/0471142301.ns0701s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chung YT, Ling YC, Yang CS, Sun YC, Lee PL, Lin CY, Hong CC, Yang MH. In Vivo Monitoring of Multiple Trace Metals in the Brain Extracellular Fluid of Anesthetized Rats by Microdialysis-membrane Desalter-ICPMS. Analytical Chemistry. 2007;79:8900–10. doi: 10.1021/ac070981z. [DOI] [PubMed] [Google Scholar]
- 111.Cebada J, Alvarado-Alvarez R, Becerra E, Neri-Bazan L, Rocha L, Garcia U. An Improved Method for Long-term Measuring of Hemolymph Fluctuations of Non-essential Amino Acids, GABA and Histamine from Freely Moving Crayfish. Journal of Neuroscience Methods. 2006;153:1–7. doi: 10.1016/j.jneumeth.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 112.Watson CJ, Venton BJ, Kennedy RT. In Vivo Measurements of Neurotransmitters by Microdialysis Sampling. Analytical Chemistry. 2006;78:1391–9. doi: 10.1021/ac0693722. [DOI] [PubMed] [Google Scholar]
- 113.Lada MW, Kennedy RT. Quantitative In Vivo Monitoring of Primary Amines in Rat Caudate Nucleus Using Microdialysis Coupled by a Flow-Gated Interface to Capillary Electrophoresis with Laser-Induced Fluorescence Detection. Analytical Chemistry. 1996;68:2790–7. doi: 10.1021/ac960178x. [DOI] [PubMed] [Google Scholar]
- 114.Cremers TI, de Vries MG, Huinink KD, van Loon JP, vd Hart M, Ebert B, Westerink BH, De Lange EC. Quantitative Microdialysis Using Modified Ultraslow Microdialysis: Direct Rapid and Reliable Determination of Free Brain Concentrations with the MetaQuant Technique. Journal of Neuroscience Methods. 2009;178:249–54. doi: 10.1016/j.jneumeth.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 115.Lonnroth P, Jansson PA, Smith U. A Microdialysis Method Allowing Characterization of Intercellular Water Space in Humans. American Journal of Physiology. 1987;253:E228–31. doi: 10.1152/ajpendo.1987.253.2.E228. [DOI] [PubMed] [Google Scholar]
- 116.Hooks MS, Colvin AC, Juncos JL, Justice JB., Jr Individual Differences in Basal and Cocaine-stimulated Extracellular Dopamine in the Nucleus Accumbens Using Quantitative Microdialysis. Brain Research. 1992;587:306–12. doi: 10.1016/0006-8993(92)91012-4. [DOI] [PubMed] [Google Scholar]
- 117.Martin-Fardon R, Sandillon F, Thibault J, Privat A, Vignon J. Long-term Monitoring of Extracellular Dopamine Concentration in the Rat Striatum by a Repeated Microdialysis Procedure. Journal of Neuroscience Methods. 1997;72:123–35. doi: 10.1016/s0165-0270(96)02170-x. [DOI] [PubMed] [Google Scholar]
- 118.Krebs-Kraft DL, Rauw G, Baker GB, Parent MB. Zero Net Flux Estimates Of Septal Extracellular Glucose Levels and the Effects of Glucose on Septal Extracellular GABA Levels. European Journal of Pharmacology. 2009;611:44–52. doi: 10.1016/j.ejphar.2009.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Larsson CI. The Use of an “Internal Standard” for Control of the Recovery in Microdialysis. Life Sciences. 1991;49:PL73–8. doi: 10.1016/0024-3205(91)90082-m. [DOI] [PubMed] [Google Scholar]
- 120.Scheller D, Kolb J. The Internal Reference Technique in Microdialysis: A Practical Approach to Monitoring Dialysis Efficiency and to Calculating Tissue Concentration from Dialysate Samples. Journal of Neuroscience Methods. 1991;40:31–8. doi: 10.1016/0165-0270(91)90114-f. [DOI] [PubMed] [Google Scholar]
- 121.Roy MC, Ikimura K, Nishino H, Naito T. A High Recovery Microsampling Device Based on a Microdialysis Probe for Peptide Sampling. Analytical Biochemistry. 2010;399:305–7. doi: 10.1016/j.ab.2009.12.036. [DOI] [PubMed] [Google Scholar]
- 122.Duo J, Fletcher H, Stenken JA. Natural and Synthetic Affinity Agents as Microdialysis Sampling Mass Transport Enhancers: Current Progress and Future Perspectives. Biosensors and Bioelectronics. 2006;22:449–57. doi: 10.1016/j.bios.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 123.Herbaugh AW, Stenken JA. Antibody-enhanced Microdialysis Collection of CCL2 from Rat Brain. Journal of Neuroscience Methods. 2011;202:124–7. doi: 10.1016/j.jneumeth.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pettersson A, Markides K, Bergquist J. Enhanced Microdialysis of Neuropeptides. Acta Biochimica Polonica. 2001;48:1117–20. [PubMed] [Google Scholar]
- 125.Lada MW, Vickroy TW, Kennedy RT. High Temporal Resolution Monitoring of Glutamate and Aspartate In Vivo Using Microdialysis On-line with Capillary Electrophoresis with Laser-Induced Fluorescence Detection. Analytical Chemistry. 1997;69:4560–5. doi: 10.1021/ac970518u. [DOI] [PubMed] [Google Scholar]
- 126.Bert L, Robert F, Denoroy L, Stoppini L, Renaud B. Enhanced Temporal Resolution for the Microdialysis Monitoring of Catecholamines and Excitatory Amino Acids Using Capillary Electrophoresis with Laser-induced Fluorescence Detection. Analytical Developments and In Vitro Validations. Journal of Chromatography A. 1996;755:99–111. doi: 10.1016/s0021-9673(96)00595-x. [DOI] [PubMed] [Google Scholar]
- 127.Hogan BL, Lunte SM, Stobaugh JF, Lunte CE. On-line Coupling of In Vivo Microdialysis Sampling with Capillary Electrophoresis. Analytical Chemistry. 1994;66:596–602. doi: 10.1021/ac00077a004. [DOI] [PubMed] [Google Scholar]
- 128.Wang M, Roman GT, Schultz K, Jennings C, Kennedy RT. Improved Temporal Resolution for In Vivo Microdialysis by Using Segmented Flow. Analytical Chemistry. 2008;80:5607–15. doi: 10.1021/ac800622s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deeba S, Corcoles EP, Hanna GB, Pareskevas P, Aziz O, Boutelle MG, Darzi A. Use of Rapid Sampling Microdialysis for Intraoperative Monitoring of Bowel Ischemia. Diseases of the Colon & Rectum. 2008;51:1408–13. doi: 10.1007/s10350-008-9375-4. [DOI] [PubMed] [Google Scholar]
- 130.Wang M, Roman GT, Perry ML, Kennedy RT. Microfluidic Chip for High Efficiency Electrophoretic Analysis of Segmented Flow from a Microdialysis Probe and In Vivo Chemical Monitoring. Analytical Chemistry. 2009;81:9072–8. doi: 10.1021/ac901731v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang M, Slaney T, Mabrouk O, Kennedy RT. Collection of Nanoliter Microdialysate Fractions in Plugs for Off-line In Vivo Chemical Monitoring with Up To 2 s Temporal Resolution. Journal of Neuroscience Methods. 2010;190:39–48. doi: 10.1016/j.jneumeth.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Slaney TR, Nie J, Hershey ND, Thwar PK, Linderman J, Burns MA, Kennedy RT. Push-Pull Perfusion Sampling with Segmented Flow for High Temporal and Spatial Resolution In Vivo Chemical Monitoring. Analytical Chemistry. 2011;83:5207–13. doi: 10.1021/ac2003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rogers M, Leong C, Niu X, de Mello A, Parker KH, Boutelle MG. Optimisation of a Microfluidic Analysis Chamber for the Placement of Microelectrodes. Physical Chemistry Chemical Physics. 2011;13:5298–303. doi: 10.1039/c0cp02810j. [DOI] [PubMed] [Google Scholar]
- 134.Song P, Hershey ND, Mabrouk OS, Slaney TR, Kennedy RT. Mass Spectrometry “Sensor” for In Vivo Acetylcholine Monitoring. Analytical Chemistry. 2012;84:4659–64. doi: 10.1021/ac301203m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fastner S, Predel R, Kahnt J, Schachtner J, Wegener C. A Simple Purification Protocol for the Detection of Hormones in the Hemolymph of Individual Insects by Matrix-assisted Laser Desorption/Ionization Time-of-flight Mass Spectrometry. Rapid Communications in Mass Spectrum. 2007;21:23–8. doi: 10.1002/rcm.2800. [DOI] [PubMed] [Google Scholar]
- 136.Ebner K, Rjabokon A, Pape HC, Singewald N. Increased In Vivo Release of Neuropeptide S in the Amygdala of Freely Moving Rats after Local Depolarisation and Emotional Stress. Amino Acids. 2011;41:991–6. doi: 10.1007/s00726-011-1058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Frost SI, Keen KL, Levine JE, Terasawa E. Microdialysis Methods for In Vivo Neuropeptide Measurement in the Stalk-median Eminence in the Rhesus Monkey. Journal of Neuroscience Methods. 2008;168:26–34. doi: 10.1016/j.jneumeth.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Perry M, Li Q, Kennedy RT. Review of Recent Advances in Analytical Techniques for the Determination of Neurotransmitters. Analytica Chimica Acta. 2009;653:1–22. doi: 10.1016/j.aca.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Emmett MR, Andren PE, Caprioli RM. Specific Molecular Mass Detection of Endogenously Released Neuropeptides Using In Vivo Microdialysis/Mass Spectrometry. Journal of Neuroscience Methods. 1995;62:141–7. doi: 10.1016/0165-0270(95)00070-4. [DOI] [PubMed] [Google Scholar]
- 140.Buck K, Voehringer P, Ferger B. Rapid Analysis of GABA and Glutamate in Microdialysis Samples Using High Performance Liquid Chromatography and Tandem Mass Spectrometry. Journal of Neuroscience Methods. 2009;182:78–84. doi: 10.1016/j.jneumeth.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 141.Carrozzo MM, Cannazza G, Pinetti D, Di Viesti V, Battisti U, Braghiroli D, Parenti C, Baraldi M. Quantitative Analysis of Acetylcholine in Rat Brain Microdialysates by Liquid Chromatography Coupled with Electrospray Ionization Tandem Mass Spectrometry. Journal of Neuroscience Methods. 2010;194:87–93. doi: 10.1016/j.jneumeth.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 142.Uutela P, Ketola RA, Piepponen P, Kostiainen R. Comparison of Different Amino Acid Derivatives and Analysis of Rat Brain Microdialysates by Liquid Chromatography Tandem Mass Spectrometry. Analytica Chimica Acta. 2009;633:223–31. doi: 10.1016/j.aca.2008.11.055. [DOI] [PubMed] [Google Scholar]
- 143.Song P, Mabrouk OS, Hershey ND, Kennedy RT. In Vivo Neurochemical Monitoring Using Benzoyl Chloride Derivatization and Liquid Chromatography-Mass Spectrometry. Analytical Chemistry. 2012;84:412–9. doi: 10.1021/ac202794q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reed B, Bidlack JM, Chait BT, Kreek MJ. Extracellular Biotransformation of Beta-endorphin in Rat Striatum and Cerebrospinal Fluid. Journal of Neuroendocrinology. 2008;20:606–16. doi: 10.1111/j.1365-2826.2008.01705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bernay B, Gaillard MC, Guryca V, Emadali A, Kuhn L, Bertrand A, Detraz I, Carcenac C, Savasta M, Brouillet E, Garin J, Elalouf JM. Discovering New Bioactive Neuropeptides in the Striatum Secretome Using In Vivo Microdialysis and Versatile Proteomics. Molecular and Cell Proteomics. 2009;8:946–58. doi: 10.1074/mcp.M800501-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang X, Rauch A, Xiao H, Rainer G, Logothetis NK. Mass Spectrometry-based Neurochemical Analysis: Perspectives for Primate Research. Expert Rev Proteomics. 2008;5:641–52. doi: 10.1586/14789450.5.5.641. [DOI] [PubMed] [Google Scholar]
- 147.DiFeliceantonio AG, Mabrouk OS, Kennedy RT, Berridge KC. Enkephalin Surges in Dorsal Neostriatum as a Signal to Eat. Current Biology. 2012;22:1918–24. doi: 10.1016/j.cub.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]