Abstract
As a deficient virus due to the lack of envelope proteins, hepatitis D virus (HDV) causes chronic or fulminant “delta hepatitis” only in people with simultaneous hepatitis B virus (HBV) infection. HBV encodes three types of surface proteins known as small (S), medium (M) and large (L) envelope proteins. All three types of HBV surface antigens (HBsAgs) are present on HDV virions. The envelopment process of HDV occurs through interactions between the HDV ribonucleoprotein (RNP) complex and HBV HBsAgs. While HBsAg is the only protein required by HDV, the exact interaction sites between the S protein and pre-mature HDV are not well defined yet. In fact, these sites are distributed along the S protein with some hot spots for the envelopment process. Moreover, in most clinically studied samples, HDV infection is associated with a dramatically reduced HBV viral load, temporarily or permanently, while HBsAg resources are available for HDV packaging. Thus, beyond interacting with HBV envelope proteins, controlling mechanisms exist by which HDV inhibits HBV-DNA replication while allowing a selective transcription of HBV proteins. Here we discuss the molecular interaction sites between HBsAg and the HDV-RNP complex and address the proposed indirect mechanisms, which are employed by HBV and HDV to facilitate or inhibit each other’s viral replication. Understanding molecular interactions between HBV and HDV may help to design novel therapeutic strategies for delta hepatitis.
Keywords: Viral hepatitis, Hepatitis B virus, Hepatitis D virus, Hepatitis B virus surface antigens, Hepatitis D virus antigen, Ag loop, Liver cirrhosis
Core tip: Hepatitis D virus (HDV) causes accelerated liver disease in form of fulminant or chronic hepatitis in patients with hepatitis B virus (HBV) infection. HBV supports HDV replication by sharing its surface proteins. Even without overt HBV-DNA replication, transcription of HBV surface proteins (HBsAgs) remains stable in HDV infected cells, which is essential for assembly of HDV virions containing HBsAg proteins. HDV replication is oftentimes associated with a suppression of HBV-DNA levels, and several mechanisms have been suggested how HBV or HDV may influence each other’s replication. Understanding molecular interactions between HBV and HDV may help to design novel therapeutic strategies.
INTRODUCTION
Globally, about 350 million people are chronically infected with hepatitis B virus (HBV), of which 15 million are positive for hepatitis D virus (HDV) antibodies[1]. HDV causes chronic or fulminant “delta hepatitis”, in form of co- or super-infection in HBV infected patients[2]. Delta virus is considered as a very deleterious pathogen since its infection commonly leads to progression of hepatic fibrosis, cirrhosis and increased risk of hepatocellular carcinoma[2,3]. There are eight known genotypes of HDV (from 1 to 8), of which genotype 1 has a worldwide distribution and genotype 3 has been associated with the most severe outcome of liver disease[4,5]. With a virion size of 36 nm and a 1.7 Kb genomic circular RNA, HDV is the smallest known human virus. Its genome encodes only two structural proteins termed small- and large-HD-antigens (S- and L-HDAg). The proteins are transcribed from the same open reading frame (ORF) and are identical except for a 19 amino acid extension in C-terminal domain of L-HDAg[2].
HDV requires the function of a helper virus as an envelope source for virion envelopment and propagation. This function can be provided through HBV (all genotypes from A to H) or other Orthohepadnaviridae members, such as Woodchuck hepatitis virus (WHV), by sharing the surface proteins[2]. The 19 amino acid extension of L-HDAg, which is called the “packaging signal”, is responsible for this interaction[6]. While HBV thereby provides an essential basis for HDV viremia and infectivity, most clinical studies reported that HBV replication is diminished in HBV-HDV-infected patients and that HDV co-infection is associated with lower HBV viremia than HBV mono-infection[7]. However, HBV-DNA, HDV-RNA and HBsAg apparently fluctuate in longitudinally studied patients indicating ongoing and dynamic interactions between HBV and HDV in infected cells[8].
Although the direct contact between HBsAg and HDAg for HDV virion envelopment can be considered the main interaction, other less well understood mechanisms may also interfere with the replication of both viruses in infected cells[9]. Here we describe possible mechanisms for HBV/HDV interactions and their probable molecular cross-talks in infected cells. These mechanisms include HBsAg-HDAg interactions and HDV-trans-controlling of HBV genome replication/transcription, cellular transcriptional pathways and RNA polymerase activity in dually infected hepatocytes.
HBsAg-HDAg INTERACTIONS
HBV encodes three surface proteins with different initiation-of-replication sites from one ORF. These proteins are large, medium and small HBsAgs (L-, M- and S-HBsAg)[10]. As an integral protein, S-HBsAg (226 amino acids) is anchored in the lipid bilayer of the endoplasmic reticulum (ER) through its N-terminal (residues 4-28 and 80-100) and C-terminal (residues 165-226) transmembrane domains (TMDs). It also includes an antigenic loop (Ag loop, residues 101-164) with immunodominant epitopes, facing the ER lumen. The rest of residues located between TMDs face the cytoplasm and are called cytosolic loops (CYLs). These are expected to be residues 29 to 79 (CYL-I) and 194 to 201 (CYL-II)[11]. The M-HBsAg (281 amino acids), contains the whole S-HBsAg plus an N-terminal preS2 region facing the ER lumen. The L-HBsAg (389-400 amino acids), contains preS1, preS2 (preS) and S domains[12]. This protein has two conformations based on the positioning of preS in ER membrane towards the cytoplasm (for virion formation) or ER lumen (for receptor binding)[11]. All three types of HBsAgs are found on the surface of mature HDV particles[2]. The schematic features of HBsAg proteins and their localization in ER membrane are shown in Figure 1.
Figure 1.
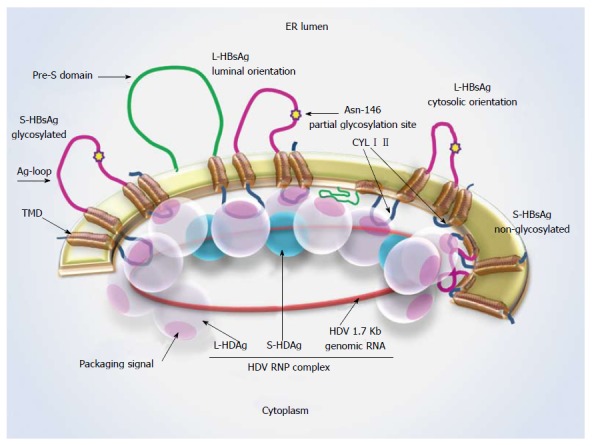
Hepatitis D virus-ribonucleoprotein complex interaction with S-hepatitis B virus surface antigen. Schematic representation of L- and S-HBsAg locations in the ER membrane and the interaction sites with the HDV-RNP complex. The Asn-146 glycosylation site is highlighted by a star. About half of the residues remain non-glycosylated at this site. A cytosolic orientation has been suggested for the non-glycosylated Ag loop, which may interfere with HBsAg-HDAg interaction. L-HDAg mediates HDV assembly at the late stage of viral replication through forming connections with HDV-RNA, S-HDAg and HBsAg. Ag loop: Antigenic loop; CYL I, II: Cytosolic loop I, II; ER: Endoplasmic reticulum; L-/ S-HBsAg: Large and small hepatitis B virus surface antigens; L-/ S-HDAg: Large and small hepatitis D virus proteins; RNP: Ribonucleoprotein; TMD: Transmembrane domain.
Both HDV small and large proteins form connections to one another as well as to HDV RNA through RNA binding domains to assemble the HDV ribonucleoprotein (RNP) complex[6]. The L-HDAg is responsible for RNP localization in the ER membrane through a CXXX farnesylation signal (C stands for cytosine, and X for any amino acid) and also interactions with HBV surface proteins through its packaging signal (Figure 1)[2]. The packaging signal is very genotype specific in HDV (74% divergence between genotypes 1 and 2) and plays an important role in the envelopment. Although an association between HDV-1/HBV-A and -D and HDV-3/HBV-F and -A has been observed, independent investigations suggest that the co-infections are mainly representative of common genotypes of each of the viruses in certain geographical areas and not specific for distinct HBV or HDV genotypes[13-15]. Studies indicate that HDV genotype 2 is associated with a less aggressive disease compared to genotype 1 which has been attributed to the higher packaging efficiency of genotype 1 than that of genotype 2[16,17]. Moreover, a variation in the packaging efficiency has been observed among different isolates of the same genotype, which reflects the critical role of this length in HDAg interactions with HBsAg. It has been reported that the hydrophobic nature of the L-HDAg C-terminal domain provided by C211-farnesylation as well as the number of hydrophobic residues of the packaging signal (which differs among HDV genotypes) enhance HDV interactions with surface proteins of HBV and therefore the packaging efficiency[17].
While HBV requires both S- and L-HBAgs for viral assembly, HDV needs S-HBsAg for its in vitro virion packaging and L-HBsAg for infectivity[18,19]. Therefore, it is very likely that most of the HDAg binding sites are located on the S domain of HBsAg[20]. While an intact HBsAg is not able to interact with L-HDAg, a denatured form of HBsAg is competent for such interactions suggesting that L-HDAg has no connection to the external domains of HBsAg but rather to the domains inside the particles[21]. The cytoplasmic orientation of the CYLs of the S protein, provides a reasonable condition for these sites to interact with pre-mature HDV virions in the cytosol (Figure 1)[22].
The importance of S-HBsAg residues 24 to 28 and 56 to 80 for HDV secretion has been shown in previous studies[23,24]. Also, a C-terminal truncation of HBsAg by 50 amino acids inhibits HDV envelopment and secretion[25]. Based on these data and also from our recent observation indicating a high rate of amino acid selection at CYLs of S-HBsAg in HBV isolates from HBV/HDV infected patients (own unpublished observations), these domains are expected to make a significant contribution in HDV packaging. Of special importance are tryptophan residues at positions 196, 199 and 201 at the C-terminal domain of S-HBsAg, which are suggested to have a central localization in binding interface with HDV-RNP complex[26].
Mutational studies revealed that in addition to the receptor binding site on the pre-S1 domain of L-HBsAg, the Ag loop is also responsible for HDV virion infectivity[27]. On the other hand, in vitro experiments using mutant HBsAg with deletions in Ag loop resulted in the lack of subviral particles as well as HDV virion secretion[27]. In more detailed studies it was shown that N-glycosylation of S-HBsAg, which is mediated by the C-terminal domain of S protein and occurs partially on Asn-146, affects HDV envelopment and secretion[12]. Based on these studies, HDV secretion is delayed or reduced (about ten folds) in the presence of non-glycosylated HBsAg, while HBV and HBsAg formation is not affected[21]. Although a weakened interaction between HBsAg and other components of HDV-RNP complex (rather than L-HDAg) has been suggested for this reduction, based on the luminal positioning of Ag loop in the ER membrane, a direct interaction of this domain and HDV components is unlikely[21,27]. Different mechanisms have been suggested to explain the effects of the antigenic domain, especially in its non-glycosylated form, on HDV packaging and secretion. One is a modified maturation and trafficking process for non-glycosylated HBsAg, which in turn will affect the rate of interactions with HDV[21]. The association of HBsAg with calnexin (a molecular chaperon in ER membrane) is also affected by the glycosylation process[28]. Therefore, a non-glycosylated HBsAg is more prone to misfolding and late maturation[29]. Furthermore, it has been suggested that a non-glycosylated Ag loop faces the cytoplasm, which possibly masks the cytosolic interaction sites with HDV or hinders appropriate connections between HBsAg and HDV (Figure 1)[11]. Due to deferent propagation responses of HBV and HDV to non-glycosylated Ag loop, it is possible that these viruses apply different mechanisms to interact with HBsAg[21]. Likewise, the lateral S-S interactions between S protein carbohydrates, which play a critical role in virion stability, are suggested to occur differently for HBV and HDV due to their particle sizes[12].
INDIRECT INTERACTIONS BETWEEN HBV AND HDV AFFECTING VIRAL REPLICATION
There are several indications of low HBV replication levels in patients co-infected with HDV[7,9,30]. On the other hand, longitudinal analyses of HBV/HDV co-infected individuals demonstrated a fluctuating pattern of HBV and HDV replication over time[8]. In case that HDV is temporarily or permanently the dominant virus during dual infection with HBV, there should be a molecular scenario for these viruses to control each other’s replication. Most of the studies so far, indicate a controlling role of HDV over HBV replication or its protein expression in infected cells[7,9,31].
Previous investigations showed that HBV DNA in the host cell genome can produce enough surface antigen molecules for HDV virion assembly even in the absence of precore and pregenomic RNAs and regardless of an active HBV replication[32,33]. These cells, which still produce some of the viral products, may be selected through immune responses, appear as a result of a resolved infection or just due to the support of the infected cells for parts of the viral proteins such as envelope antigens but not the complete replication of the virus[33-35]. Nonetheless, regarding the role of HBV as an envelope provider, HDV nucleoproteins can be considered as competitors with HBV for HBsAg. Therefore, they may induce a selective suppression on HBV replication associated with an increase in PreS/S RNAs and HBsAg levels in co-infected patients[9]. Investigations on the effects of S- and L-HDAgs on HBV replication have shown that these proteins inhibit HBV replication through a strong suppression of HBV enhancers (EnhI and II) and also trans-activation of the IFN-alpha-inducible MxA gene[31]. The inhibitory effects of L-HDAg on RNA polymerase II, which is involved in replication of both HBV and HDV viruses, might be another reason for reduced HBV replication in the presence of co-infection with HDV[36].
Another indication of HBV controlled replication/gene expression in HDV infected cells is the presence of basal core promoter (BCP) and precore (PC) mutations in the HBV genome of patients co-infected with HDV. Occurrence of HBV BCP and PC mutations is associated with lower levels of HBV DNA in both serum and liver without affecting HDV replication and clinical manifestations in patients[9,37]. In contrast, PC/BCP point mutations with HBeAg negative phenotype can significantly increase HBV viremia and replication of polymerase mutated strains in HDV-negative patients[38].
Other instances of indirect effects of HBV and HDV on each other’s replication include the synergistic activation of serum response element (SRE)-dependent pathways by HBxAg and L-HDAg, thus affecting factors which are involved in transcription regulation mediated by SRE[39]. From the cross-talks between HBV and HDV we can also refer to the NF-κB activation, which results from ER stress (induced by HBsAgs) or TNFα secretion from immune cells (in response to HBV infection) and correlates with L-HDAg nuclear export and HDV secretion[40].
CONCLUSION
The clinical observation of aggravated liver disease in patients with HBV/HDV co- or super-infection has prompted intense research on molecular interactions between both viruses. A major interaction between HBV and HDV is that they share a surface protein supply; this fact is currently being translated into novel therapeutic approaches using entry inhibitors in clinical trials for delta hepatitis[41]. However, in spite of the direct interaction sites between HBsAg and HDV-RNP, it seems that the key interference between HBV and HDV cannot be devoted to a certain domain or residue of the S protein but connection spots are rather distributed along the HBsAg. Moreover, besides the main reason for HBV/HDV interactions to share a surface protein supply, this is not the only interface between the two viruses in infected cells. Further investigations are required to unravel yet unknown molecular interactions that are employed by HBV or HDV to dominate in dual infections.
Footnotes
P- Reviewer: Rodriguez-Frias F, Rizzetto M S- Editor: Ji FF L- Editor: A E- Editor: Jiao XK
Supported by The German Research Foundation (DFG Ta434/2-1 and SFB/TRR57); and by the Interdisciplinary Center for Clinical Research (IZKF) Aachen.
Conflict-of-interest: The authors declare no conflict of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: January 15, 2015
First decision: February 7, 2015
Article in press: March 9, 2015
References
- 1.Price J. An update on hepatitis B, D, and E viruses. Top Antivir Med. 2014;21:157–163. [PMC free article] [PubMed] [Google Scholar]
- 2.Dastgerdi ES, Herbers U, Tacke F. Molecular and clinical aspects of hepatitis D virus infections. World J Virol. 2012;1:71–78. doi: 10.5501/wjv.v1.i3.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wedemeyer H. Re-emerging interest in hepatitis delta: new insights into the dynamic interplay between HBV and HDV. J Hepatol. 2010;52:627–629. doi: 10.1016/j.jhep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Le Gal F, Gault E, Ripault MP, Serpaggi J, Trinchet JC, Gordien E, Dény P. Eighth major clade for hepatitis delta virus. Emerg Infect Dis. 2006;12:1447–1450. doi: 10.3201/eid1209.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexopoulou A, Dourakis SP. Genetic heterogeneity of hepatitis viruses and its clinical significance. Curr Drug Targets Inflamm Allergy. 2005;4:47–55. doi: 10.2174/1568010053622867. [DOI] [PubMed] [Google Scholar]
- 6.Shirvani-Dastgerdi E, Amini-Bavil-Olyaee S, Alavian SM, Trautwein C, Tacke F. Comprehensive analysis of mutations in the hepatitis delta virus genome based on full-length sequencing in a nationwide cohort study and evolutionary pattern during disease progression. Clin Microbiol Infect. 2014:in press. doi: 10.1016/j.cmi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Jardi R, Rodriguez F, Buti M, Costa X, Cotrina M, Galimany R, Esteban R, Guardia J. Role of hepatitis B, C, and D viruses in dual and triple infection: influence of viral genotypes and hepatitis B precore and basal core promoter mutations on viral replicative interference. Hepatology. 2001;34:404–410. doi: 10.1053/jhep.2001.26511. [DOI] [PubMed] [Google Scholar]
- 8.Schaper M, Rodriguez-Frias F, Jardi R, Tabernero D, Homs M, Ruiz G, Quer J, Esteban R, Buti M. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J Hepatol. 2010;52:658–664. doi: 10.1016/j.jhep.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Pollicino T, Raffa G, Santantonio T, Gaeta GB, Iannello G, Alibrandi A, Squadrito G, Cacciola I, Calvi C, Colucci G, et al. Replicative and transcriptional activities of hepatitis B virus in patients coinfected with hepatitis B and hepatitis delta viruses. J Virol. 2011;85:432–439. doi: 10.1128/JVI.01609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prange R, Streeck RE. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sureau C, Fournier-Wirth C, Maurel P. Role of N glycosylation of hepatitis B virus envelope proteins in morphogenesis and infectivity of hepatitis delta virus. J Virol. 2003;77:5519–5523. doi: 10.1128/JVI.77.9.5519-5523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes-Gouvêa MS, Pereira Soares Mdo C, Guedes de Carvalho Mello IM, Brito EM, Pereira Moia Lde J, Bensabath G, Nunes HM, Carrilho FJ, Pinho JR. Hepatitis D and B virus genotypes in chronically infected patients from the Eastern Amazon Basin. Acta Trop. 2008;106:149–155. doi: 10.1016/j.actatropica.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutierrez MF, Carrilho FJ, Pinho JR. Dynamics of hepatitis D (delta) virus genotype 3 in the Amazon region of South America. Infect Genet Evol. 2011;11:1462–1468. doi: 10.1016/j.meegid.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Amini N, Alavian SM, Kabir A, Aalaei-Andabili SH, Saiedi Hosseini SY, Rizzetto M. Prevalence of hepatitis d in the eastern mediterranean region: systematic review and meta analysis. Hepat Mon. 2013;13:e8210. doi: 10.5812/hepatmon.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu SC, Wu JC, Sheen IJ, Syu WJ. Interaction and replication activation of genotype I and II hepatitis delta antigens. J Virol. 2004;78:2693–2700. doi: 10.1128/JVI.78.6.2693-2700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SC, Syu WJ, Sheen IJ, Liu HT, Jeng KS, Wu JC. Varied assembly and RNA editing efficiencies between genotypes I and II hepatitis D virus and their implications. Hepatology. 2002;35:665–672. doi: 10.1053/jhep.2002.31777. [DOI] [PubMed] [Google Scholar]
- 18.Sureau C, Guerra B, Lanford RE. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J Virol. 1993;67:366–372. doi: 10.1128/jvi.67.1.366-372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudima S, Meier A, Dunbrack R, Taylor J, Bruss V. Two potentially important elements of the hepatitis B virus large envelope protein are dispensable for the infectivity of hepatitis delta virus. J Virol. 2007;81:4343–4347. doi: 10.1128/JVI.02478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang CJ, Sung SY, Chen DS, Chen PJ. N-linked glycosylation of hepatitis B surface antigens is involved but not essential in the assembly of hepatitis delta virus. Virology. 1996;220:28–36. doi: 10.1006/viro.1996.0282. [DOI] [PubMed] [Google Scholar]
- 22.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 23.Jenna S, Sureau C. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology. 1998;251:176–186. doi: 10.1006/viro.1998.9391. [DOI] [PubMed] [Google Scholar]
- 24.Hourioux C, Sureau C, Poisson F, Brand D, Goudeau A, Roingeard P. Interaction between hepatitis delta virus-encoded proteins and hepatitis B virus envelope protein domains. J Gen Virol. 1998;79(Pt 5):1115–1119. doi: 10.1099/0022-1317-79-5-1115. [DOI] [PubMed] [Google Scholar]
- 25.Chen PJ, Lai WJ, Wang CJ, Chen DS. Hepatitis B surface antigen and large-form hepatitis delta antigen in HDV assembly: a further study. Prog Clin Biol Res. 1993;382:29–34. [PubMed] [Google Scholar]
- 26.Komla-Soukha I, Sureau C. A tryptophan-rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J Virol. 2006;80:4648–4655. doi: 10.1128/JVI.80.10.4648-4655.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaoudé GA, Sureau C. Role of the antigenic loop of the hepatitis B virus envelope proteins in infectivity of hepatitis delta virus. J Virol. 2005;79:10460–10466. doi: 10.1128/JVI.79.16.10460-10466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- 29.Ou WJ, Cameron PH, Thomas DY, Bergeron JJ. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 30.Sakugawa H, Nakasone H, Nakayoshi T, Kawakami Y, Yamashiro T, Maeshiro T, Kinjo F, Saito A, Zukeran H. Hepatitis B virus concentrations in serum determined by sensitive quantitative assays in patients with established chronic hepatitis delta virus infection. J Med Virol. 2001;65:478–484. [PubMed] [Google Scholar]
- 31.Williams V, Brichler S, Radjef N, Lebon P, Goffard A, Hober D, Fagard R, Kremsdorf D, Dény P, Gordien E. Hepatitis delta virus proteins repress hepatitis B virus enhancers and activate the alpha/beta interferon-inducible MxA gene. J Gen Virol. 2009;90:2759–2767. doi: 10.1099/vir.0.011239-0. [DOI] [PubMed] [Google Scholar]
- 32.Freitas N, Cunha C, Menne S, Gudima SO. Envelope proteins derived from naturally integrated hepatitis B virus DNA support assembly and release of infectious hepatitis delta virus particles. J Virol. 2014;88:5742–5754. doi: 10.1128/JVI.00430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol. 2010;84:8308–8315. doi: 10.1128/JVI.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason WS, Litwin S, Jilbert AR. Immune selection during chronic hepadnavirus infection. Hepatol Int. 2008;2:3–16. doi: 10.1007/s12072-007-9024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason WS, Litwin S, Xu C, Jilbert AR. Hepatocyte turnover in transient and chronic hepadnavirus infections. J Viral Hepat. 2007;14 Suppl 1:22–28. doi: 10.1111/j.1365-2893.2007.00911.x. [DOI] [PubMed] [Google Scholar]
- 36.Modahl LE, Lai MM. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J Virol. 2000;74:7375–7380. doi: 10.1128/jvi.74.16.7375-7380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JC, Chen CM, Chen TZ, Lee SD, Yen FS, Choo KB. Prevalence and type of precore hepatitis B virus mutants in hepatitis D virus superinfection and its clinical implications. J Infect Dis. 1996;173:457–459. doi: 10.1093/infdis/173.2.457. [DOI] [PubMed] [Google Scholar]
- 38.Tacke F, Gehrke C, Luedde T, Heim A, Manns MP, Trautwein C. Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of Lamivudine-resistant mutants. J Virol. 2004;78:8524–8535. doi: 10.1128/JVI.78.16.8524-8535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto T, Kato N, Yoshida H, Otsuka M, Moriyama M, Shiratori Y, Koike K, Matsumura M, Omata M. Synergistic activation of the serum response element-dependent pathway by hepatitis B virus x protein and large-isoform hepatitis delta antigen. J Infect Dis. 2003;187:820–828. doi: 10.1086/368389. [DOI] [PubMed] [Google Scholar]
- 40.Huang CR, Lo SJ. Hepatitis D virus infection, replication and cross-talk with the hepatitis B virus. World J Gastroenterol. 2014;20:14589–14597. doi: 10.3748/wjg.v20.i40.14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urban S, Bartenschlager R, Kubitz R, Zoulim F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147:48–64. doi: 10.1053/j.gastro.2014.04.030. [DOI] [PubMed] [Google Scholar]


