Abstract
Molecular genetic analyses in Schizosaccharomyces pombe are greatly enhanced by our ability to delete chromosomal genes via homologous recombination and to introduce genes expressed from autonomous plasmids. In this paper, we describe a novel approach to generating marked deletion cassettes that bypasses the need for the long, PAGE-purified oligonucleotides required in the currently used PCR-based deletion approach. We also describe additional uses of this two-step PCR method for constructing chromosomal insertion cassettes. Finally, we describe how gap repair in S. pombe can facilitate plasmid constructions in a manner that circumvents the reliance on compatible restriction sites in the DNA molecules that are being joined. Several applications of this gap repair plasmid construction strategy are discussed.
Keywords: Schizosacharomyces pombe, Fission yeast, Gap repair, Selectable marker, Gene disruption, Two-step PCR
1. Introduction to gene deletions in Schizosacharomyces pombe
Homologous recombination, both in the budding yeast Saccharomyces cerevisiae and in the fission yeast S. pombe, has been widely exploited to construct chromosomal gene deletions using one-step gene replacement protocols [1,2]. In their earliest form, these techniques required the replacement of a cloned open reading frame (ORF) by a selectable marker on a plasmid, followed by transformation of an appropriate host strain with the linearized construct to allow for selection of transformants carrying the marked deletion at the targeted genomic locus. Due to limitations of available restriction sites in the target gene, many of these constructions are more correctly described as gene disruptions, rather than deletions. This is due to the fact that the selectable marker was simply inserted into the ORF leaving the possibility that a portion of the disrupted gene could be transcribed and translated. The advent of polymerase chain reaction (PCR; [3]) and advances in yeast transformation protocols have dramatically changed the ease with which deletions are constructed. Gene deletions are now carried out by using oligonucleotides whose 5′ ends are homologous to sequences flanking the target gene to be deleted, and whose 3′ ends can prime the amplification of the desired selectable marker [4,5]. The resulting PCR products are designed to possess approximately 40 bp of targeting sequence for S. cerevisiae deletions, and 60–80 bp of targeting sequence for S. pombe deletions.
It is generally believed that longer flanking sequences are required on deletion cassettes to promote homologous recombination in S. pombe than in S. cerevisiae. This perception may be due in part to the early use of poorly expressed selectable markers that led to the isolation of transformants which received multiple copies of the transforming DNA fragment [6]. It is also apparent that the transformation protocol used affects the efficiency of homologous recombination with a high efficiency protocol described by Keeney and Boeke [7] and by Bäahler et al. [4]. The use of these transformation protocols has made the PCR-based approach to gene deletions the preferred method. However, we and others have encountered problems with certain gene deletions due to a combination of the generation of a low number of candidate transformants and a low efficiency of homologous recombination. Since this technique requires very long (80- to 100-mer) oligonucleotides that require PAGE purification, these deletions can become expensive and time-consuming to carry out.
2. Generation of a gene deletion cassette by two-step PCR and cloning
We describe here a method for creating a deletion cassette in a way that bypasses the need for the synthesis of large primers that require PAGE purification, yet creates a deletion fragment with long flanking sequences to produce a high efficiency of homologous recombination (Fig. 1). This approach is a variation of a technique used to create a plasmid to facilitate gap repair cloning of budding yeast genes [8]. Four oligonucleo-tides are used to amplify sequences flanking the target gene. The outer pair of primers (labeled 5′for and 3′rev; Fig. 1) has relatively short complementary sequences at their 5′ ends (see underlined sequences in Table 1) which facilitates the joining of the two “first-round” PCR products into a single product during a second round of PCR. This junction region also contains a unique restriction site to allow for later linearization of the deletion cassette. The orientation of the joined products is inverted relative to the genomic sequence. Key to production of the second-round PCR product is a five-cycle amplification of the first-round products in the absence of additional primers that allows annealing of the lower strand of the 5′ flanking region product (labeled AB in Fig. 1) with the upper strand of the 3′ flanking region product (labeled CD in Fig. 1). We pool equivalent amounts of the first-round PCR products (as determined by electrophoresis), make 10- and 100-fold dilutions, and use 2 ll of this DNA in a 50 μl PCR. The five-cycle reaction is carried out with a 3 min 45 °C annealing step, a 1 min 72 °C polymerization step, and a 1 min 94 °C denaturation step [8]. This is followed by a standard, 30 cycle, PCR amplification in the presence of the two primers that do not carry the complementary 5′ sequences (labeled 5′rev and 3′for; Fig. 1). We use the Epicentre Failsafe PCR System (primarily using buffers B and F), although we expect that this protocol will work with any PCR system.
Fig. 1.
Schematic for construction of gene deletion cassettes via two-step PCR. The deletion cassette is made through the use of two rounds of PCR and the cloning of the resulting PCR product. The first round of PCR involves two reactions to amplify chromosomal sequences (labeled AB and CD to define the relative orientation of the sequences) that flank the target gene. The PCR primers that are distal to the target gene (labeled 5′for and 3′rev) carry complementary sequences at their 5′ ends (see underlined sequences in Table 1) that allow the joining of the first-round PCR products to create the second-round PCR product. The second-round of PCR consists of a five-cycle amplification step to join the two first-round PCR products, carried out in the absence of additional primers, followed by a standard PCR using primers 5′rev and 3′for to amplify the combined PCR product. The second-round PCR product is then cloned into a vector that supplies an S. pombe selectable marker to mark the eventual gene deletion. Digestion of the plasmid with a restriction enzyme that cuts in the junction between the first-round PCR sequences produces a DNA molecule with long sequences at each end that are identical to the flanking regions of the target gene.
Table 1.
Oligonucleotides used to construct git2 deletion cassettesa
| git2Δ-5′for-BamHI |
| 5′-ACGGGATCCAGGCGCAGTATTCGAACGCGC-3′ |
| git2Δ-3′rev-BamHI |
| 5′-CCTGGATCCCGTGTTTTTTTATACCGGGCTG-3′ |
| git2Δ-5′for-SapI |
| 5′-GAAGAGCGCTCTTCGCAGTATTCGAACGCGC-3′ |
| git2Δ-3′rev-SapI |
| 5′-GAAGAGCGCTCTTCGTTTTTTTATACCGGGCTG-3′ |
| git2Δ-5′rev |
| 5′-TTTCAAAAATACCACTATACGCG-3′ |
| git2Δ-3′for |
| 5′-AAACTGAACAACGGAACGAC-3′ |
Three pairs of oligonucleotide sequences are presented. The first pair represents the distal oligonucleotides (see Fig. 1) that possess a unique BamHI restriction site in the complementary 5′ ends (underlined). The second pair represents the distal oligonucleotides (see Fig. 1) that possess a pair of inverted SapI restriction sites in the complementary 5′ ends (underlined). The third pair represents the proximal oligonucleotides (see Fig. 1) used in conjunction with either of the first two sets of oligonucleotides. The lengths of the git2 flanking sequences amplified in the first-round PCR products are 302 bp for the 5′ targeting region and 334 bp for the 3′ targeting region.
To construct a deletion cassette, the second-round PCR product is cloned into a vector possessing a selectable marker that works in single copy in S. pombe (Fig. 1). We have used the SpECTRA TOPO cloning vectors from Invitrogen, however traditional cloning vectors could also be used for this procedure. These vectors utilize the S. cerevisiae LEU2 selectable marker, which has been problematic in the past in single copy due to weak suppression of the S. pombe leu1-32 allele. However, transcription of LEU2 in the SpECTRA vectors is driven from the SV40 promoter, which allows for sufficient expression to produce Leu+ transformants carrying single copy integrations. By linearizing the vector with a restriction enzyme that cuts at the unique restriction site within the complementary sequences used to join the first-round PCR products, one produces a DNA molecule that can be used to transform an S. pombe strain to carry out a one-step gene deletion. Construction of this cassette requires more time and manipulation relative to the single PCR performed when using 80-mer to 100-mer primers [4]. However, this protocol utilizes four short oligonucleo-tides that do not require purification reducing the time of production and purification, and the expense associated with use of the longer PCR primers. Thus, the time from design of deletion oligonucleotides to transformation of yeast is similar for the two approaches.
We used this protocol to delete the git2/cyr1 aden-ylate cyclase gene. In the course of this work, we constructed three related cassettes to evaluate two variables with respect to this approach. The SpECTRA vector used to clone the second-round PCR product contains an S. pombe ars element that improves transformation efficiency and mitotic stability, but which is normally not present in integration plasmids. After constructing a deletion cassette that possesses a unique BamHI site for linearization, we deleted the ars element by dropping out a 1 kb BstZ17I (Bst1107I) fragment in an effort to reduce the number of nonintegration events that were observed in our earliest efforts using these cassettes. We also tested two different restriction sites in the junction sequence used to join the first-round PCR products to examine the importance of maintaining homology to the genomic DNA at the very ends of the deletion cassette. Digestion of our original cassette with BamHI leaves a four bp nonhomologous sequence, along with a 4 base 5′ overhang, at the ends of the linearized plasmid (see Table 1 for oligonucleotide sequences). To examine the effect of having nonhomologous sequences at the end of the cassette, we constructed a cassette possessing a pair of inverted SapI sites in the fusion junction. As SapI cuts outside of recognition sequence, SapI digestion of this plasmid removes the entire fusion junction so that the ends of the digested plasmid are homologous to the target region in the chromosome.
The transformation protocol, based on the procedure described by Bäahler et al. [4], has been modified to enhance the identification of homologous recombination events (Table 2). The 48 h growout step serves to increase the ratio of stable integrants to unstable transformants among the Leu+ colonies. Obviously, the selective medium used to detect integrants depends upon the marker in the vector used to create the disruption cassette.
Table 2.
DMSO transformation of S. pombe for gene deletions
| 1. Grow host strain in YE liquid medium (0.5% yeast extract, 3% glucose) to approximately 107 cells/ml. |
| 2. For each transformation, wash 2× 108 cells once with an equal volume of water and then resuspend in 1 ml sterile water. Transfer to an Eppendorf tube. |
| 3. Wash once with 1 ml of 1× LiOAc/TE solution and bring to 0.1 ml in 1× LiOAc/TE (made from 10× filter-sterilized stocks). |
| 4. Add 2 μl boiled salmon testes DNA (Sigma, 10mg/ml) and 10 μl transforming DNA. |
| 5. Incubate 10 min at room temperature and then add 260 μl of 40% PEG/LiOAc/TE solution. Mix gently and incubate 30–60 min at 30°C. |
| 6. Add 43 μl filter-sterilized DMSO. Mix gently and heat shock for 5 min at 42 °C (do not exceed five minutes). |
| 7. Transfer cells to a 10 ml YE liquid culture and grow at 30 °C for 48 h. |
| 8. Plate 0.2 ml undiluted cells, as well as of 10–1 and 10–2 dilutions of the culture, onto PM selective medium lacking leucine [17]. Colonies should appear in three to five days. |
| Stock solutions |
| 10× LiOAc |
| 1 M lithium acetate, pH 7.5 (adjusted with diluted acetic acid) |
| 10× TE |
| 0.1 M Tris–HCl/0.01 M EDTA, pH 7.5 |
| 40% PEG/LiOAc/TE (can be stored for one month) |
| 8 g PEG 4000 |
| 2 ml of 10× LiOAc |
| 2 ml of 10× TE |
| 9.75 ml water |
| dissolve PEG completely and filter sterilize. |
As the git2 gene is not essential, these tests were carried out in a haploid strain, although we have also successfully used this protocol to delete an essential gene in a diploid host strain (L. Wang, unpublished data). Deletion of the ars from the BamHI site-containing plasmid enhanced the percent of transformants carrying the deletion (Table 3), but this additional step in construction of the deletion cassette was by no means required. In addition, we saw no benefit in using inverted SapI sites to remove nonhomologous sequences from the very ends of the deletion cassettes. In fact, we observed a lower percent of git2 deletion strains when using the SapI cassette, which may reflect poorer digestion by SapI as compared to BamHI. These results demonstrate the ease with which one can delete an S. pombe gene without depending upon long oligonucleotides that require purification.
Table 3.
Analysis of Leu+ colonies from git2 deletion transformations
| Deletion cassette | Leu+ git2+ coloniesa | Leu+ git2Δ colonies |
|---|---|---|
| BamHI site/ars+ | 152 | 103 |
| BamHI site/arsΔ | 59 | 77 |
| SapI site/ars+ | 144 | 33 |
Deletion of the git2 gene was scored by replica plating colonies to media designed to detect defects in glucose regulation of an fbp1-ura4 reporter in the host FWP72 (h– leu1-32 fbp1-ura4+ ura4::fbp1-lacZ) strain [22]. Presence of the deletion in several strains was confirmed by PCR.
Most, if not all, of these colonies appear to be unstable transformants rather than nonhomologous integrants, since the Leu+ can be lost by passaging on rich medium and is poorly transmitted to progeny through meiosis.
3. Additional uses of two-step PCR cassette replacements
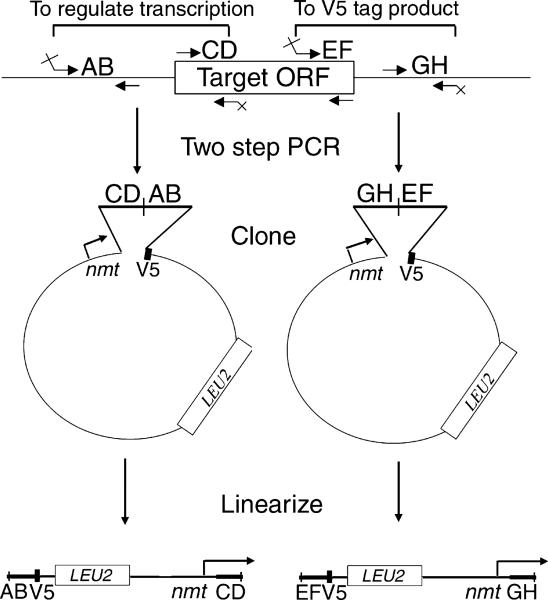
The two-step PCR-based method for deleting S. pombe genes described above can be adapted to carry out alternative manipulations of a gene at its chromosomal locus (Fig. 2). One goal would be to place the transcription of the target gene under the control of one of the nmt-based regulated promoters. This might be done to achieve high levels of expression using the nmt1 promoter or to have the ability to significantly repress expression using the nmt41 or nmt81 promoters. As shown in Fig. 2, one could clone a two-step PCR product such that the 5′ portion of the ORF (labeled CD in Fig. 2) is placed next to the nmt promoter in one of the three SpECTRA vectors, followed by a segment of the DNA from the promoter region of the target gene. Linearization of this plasmid would produce a cassette that replaces the endogenous promoter of the target gene with the nmt promoter from the vector. A second application of this method would be to tag the C-terminus of the target gene's product with the V5-6his dual epitope (Fig. 2). The V5 epitope [9] allows for detection of proteins in Westerns, while the 6his tag facilitates protein purification [10]. For this, one would fuse the 3′ end of the target ORF (labeled EF in Fig. 2) to the SpECTRA vector-encoded V5-6his coding region, preceded by a segment of the chromosomal DNA from downstream of the target gene. Linearization of this plasmid would produce a cassette that replaces the STOP codon of the gene with the V5-6his coding region. (One could also construct a C-terminal truncation of the target ORF by amplifying an internal portion of the ORF rather than the very 3′ end.) While we have not carried out either procedure yet, these constructions should be as straightforward to perform as were the gene deletions described above.
Fig. 2.
Alternative constructions using two-step PCR. The two-step PCR method for making chromosomal insertion cassettes in the SpECTRA cloning vectors can be used for purposes other than gene replacement. The three SpECTRA vectors differ only with regard to the nmt promoter, carrying either the full-strength nmt1 promoter, the intermediate-strength nmt41 promoter, or the low-strength nmt81 promoter [21]. Two alternative types of integration cassettes are described. First, a chromosomal gene's promoter can be replaced with the plasmid-borne nmt-based promoter to regulate the target gene's expression (left side of figure). Second, the V5-6his dual epitope tag encoded by the plasmid can be translationally fused to the 3′ end of the target gene at its chromosomal locus such that the gene remains under the control of its endogenous promoter (right side of figure). This could also be combined with a C-terminal truncation of the target gene by amplifying an internal region of the open reading frame rather than the very 3′ end of the open reading frame during the first round of PCR (the region designated EF). It is critical to note that, unlike the protocol for constructing a gene deletion cassette, the second-round PCR product must be inserted in a particular orientation with respect to the cloning vector as shown in the figure.
4. Plasmid construction via gap repair in S. pombe
The process of gap repair was originally used in S. cerevisiae to rescue mutant alleles from the host chromosome onto plasmids that had been gapped in the region of interest by restriction enzyme digestion [11]. Since then, gap repair has been used to localize sites of mutations in genes and in the construction of libraries that carry random mutations in a targeted region of the plasmid [12,13]. Plasmid constructions in S. cerevisiae have been carried out by gap repair using restriction fragments [14] or PCR products [15] to replace a portion of the original construct. We have recently used gap repair in S. pombe to replace the S. cerevisiae LEU2+ selectable marker in SpECTRA-derived plasmids with PCR products carrying either the S. pombe his3+ or ura4+ gene [16]. The oligonucleotides used for these constructions carry approximately 50 nucleotides of sequence homologous to the vector at their 5′ ends and 20 nucleotides of sequence homologous to the target selectable marker to be amplified at their 3′ ends. The gap-repaired plasmids carrying the swapped markers were easily obtained by co-transforming yeast with the PCR product and plasmid DNA that had been linearized within the LEU2 gene to promote an exchange of the LEU2 sequence with the PCR product that contains the desired selectable marker.
We show here that gap repair transformation can be used to exchange components of a plasmid other than the selectable marker in S. pombe. For a given construct, one might want to exchange the promoter to alter the level of expression or a portion of an ORF to create a chimeric protein (Fig. 3). Alternatively, one might want to make a translational fusion by adding an epitope tag for immune detection, GFP for localization in live cells, or GST for purification. As the selectable marker is the same for the starting plasmid and the gap-repaired plasmid, it was not clear that these constructions would be as easily carried out as our marker-swap constructions which benefited from a selection in S. pombe for cells that receive the new marker.
Fig. 3.
Schematic of a typical clone in a SpECTRA vector backbone with examples of targets for gap repair plasmid constructions. The SpECTRA plasmids are TOPO cloning expression vectors that utilize an SV40 promoter-driven LEU2 selectable marker, carry either the nmt1, nmt41, or nmt81 promoters, and possess a dual V5-6his tag for C-terminal tagging of the cloned ORF. Possible targets for replacement by gap repair include (A) the selectable marker, (B) the promoter, (C) a portion of the open reading frame, and (D) a carboxy-terminal tag (in this example, the addition of a GFP tag is shown).
We have carried out three independent constructions to test the feasibility of gap repair-based plasmid constructions involving exchanges of sequences other than the selectable marker. In two constructions, we replaced the 6his sequence that follows the V5 epitope [9] on pNMT1-derivatives with GFP, to create a C-terminal V5-GFP tag (Fig. 3, example D). The PCR primers used, V5-GFP-forward (5′-GTAAGCCTATCCCTAACCCTCT CCTCGGTCTCGATTCTACGGGTGTCGACCGGA TCCCCGGGTTAATTAA- 3′) and V5-GFP-reverse (5−GCCTAGGAAAACAAACGCA AACAAGGCATCG ACTTTTTCAATAACCAACCTCGCTTATTTAGAA GTGGCGCG-3′), carry 5′ sequences from the SpECTRA vectors and 20 nucleotides of 3′ sequence (see underlined sequences) that amplify the GFP coding region from plasmid pFA6a-GFP [4]. As such, these two oligo-nucleotides can be used to add a GFP tag to the C-terminus of any protein expressed as a V5-tagged protein from any SpECTRA vector. The one limitation is that the cloned ORF cannot possess a SalI restriction site since SalI is the only enzyme that cuts uniquely in this region of the SpECTRA vector without removing a portion of the S. pombe ars. The PCR product was introduced into two different constructs by co-transforming strain FWP5 (h+ leu1-32) cells to Leu+ with approximately 100–200 ng each of PCR product and SalI-linearized vector. The transformation protocol of Bäahler et al. [4] was used; however, unlike the deletion protocol described above, we plated the transformation mix directly to PM-leucine plates [17] without a growout step. Leu+ colonies (we typically observe thousands of colonies on the yeast transformation plates) were pooled and the plasmids were rescued into Escherichia coli by “Smash and Grab” (Table 4; [18]). For these two constructions, 50 and 80% of E. coli transformants carried plasmids containing the GFP fusion.
Table 4.
Smash and Grab rescue of plasmids from yeast to E. coli
| 1. Allow colonies to form on the S. pombe transformation plate (4–5 days after plating). Collect the cells by gentle scraping of the plate in 1 ml sterile water using a glass spreader. Transfer cells to a microfuge tube and concentrate by a brief centrifugation. |
| 2. Resuspend the cell pellet in an equal volume of Smash and Grab buffer (1% SDS; 2% Triton X-100; 100 mM NaCl; 10mM Tris, pH 8.0; and 1 mM EDTA). Transfer 200 μl of cells to a new tube and add 200 μl phenol–chloroform and 300 μl acid washed glass beads. Vortex for 5 min to lyse >50% of the cells as determined by microscopy. |
| 3. Pellet cell debris by microfugation for 5 min. Remove 50 μl of the top (aqueous) layer to a new microfuge tube. Avoid material close to the interface that may be contaminated with phenol–chloroform. |
| 4. Add 50 μl isopropanol, precipitate on ice for 10 min, and pellet DNA by microfugation for 10 min. Remove the liquid and allow pellet to air-dry. Resuspend in 5 μl sterile water. |
| 5. Transform electroporation-competent E. coli strain XL1-Blue (Stratagene) with 1 μl DNA according to manufacturer's instructions (in a slight modification we use 10 μl competent cells in 89 μl sterile water for electroporation and plate the entire transformation onto a single LB Amp plate). |
A third test construction was carried out in which we replaced the nmt promoter with the endogenous promoter for the cloned gpa2+ gene (Fig. 3 example B). We used one long primer containing 50 bases homologous to the SpECTRA vector plus 20 bases that anneal to the gpa2+ promoter and one short reverse primer that anneals to the gpa2+ open reading frame [19] to generate the PCR replacement product. The plasmid was linearized within the nmt promoter region with either a BsgI or PshAI digestion and co-transformed with the PCR product into S. pombe cells as described for the GFP tagging transformations. Nine E. coli transformants from each of the two plasmid rescues from S. pombe were shown to carry the original nmt-containing plasmid. However, we were able to enrich for the desired plasmid by pooling the remaining E. coli transformants, isolating plasmid, and treating this pooled plasmid preparation with either a PshAI digestion for the construction initiated with the BsgI digestion, or a BsgI digestion for the construction initiated with the PshAI digestion. Upon retransforming E. coli with this DNA, we found that nine of nine transformants carried the desired plasmid after PshAI enrichment while four of nine transformants carried the desired plasmid after BsgI enrichment. These gap repair transformations had been carried out using more input vector DNA than was used in the GFP constructions described above. Thus, we assume that the failure to obtain the desired plasmid among the initial transformants was due to incomplete plasmid digestion prior to the S. pombe co-transformation. However, the enrichment by digesting the pooled plasmid prep with a second enzyme that cuts the parental plasmid, but not the gap-repaired plasmid, effectively overcame this problem. These constructions clearly demonstrate that gap repair can be used for plasmid constructions in S. pombe even for constructions that do not involve plasmid marker exchange.
5. Conclusions
We have shown here two distinct molecular approaches that should enhance our ability to manipulate S. pombe genes either at their chromosomal location or on autonomously replicating plasmids. The two-step PCR method for generating insertion cassettes may at first appear too involved to justify the effort relative to the single PCR needed when using long PCR primers. However, we have found that the ease with which we recover the desired deletion strains more than compensates for the added manipulations. In addition, the actual time from conception to completion of the deletion protocol is similar to that when using the long PCR primers as this procedure utilizes short oligonucleotides that do not require additional purification. This protocol can also be modified by the addition of sequences into the PCR primers that would act as barcodes to mark individual gene deletions in the course of construction of a deletion strain collection as has been done with S. cerevisiae [20]. However, it is not clear at the present time whether this additional sequence would cause the need for oligonucleotide purification that might dramatically increase the cost of construction of such a strain collection.
While we have described this deletion protocol in the context of cloning the second-round PCR product into the Invitrogen SpECTRA cloning vectors, this choice of vector is not essential to the success of the deletion. The additional uses described for these insertion cassettes are specific to features found at the cloning site for the SpECTRA vectors (the nmt promoter and the V5-6his dual tag), however, a similar strategy could be used to take advantage of other vectors' features for manipulating genes at their endogenous loci.
The gap repair method for plasmid constructions described here provides considerable flexibility in creating novel constructions. The only limitation is that there must be a restriction site specific to the region of the plasmid to be replaced by the PCR product used to co-transform the yeast. From our experience, it is important to keep the vector concentration low to reduce the amount of uncut plasmid present during the yeast transformation step. We have not tried to optimize this procedure, but we expect that one could use considerably less input vector than the recommended 100– 200 ng. While additional optimization is not required, it should be possible to further increase the efficiency with which we are able to manipulate plasmid sequences in S. pombe.
Acknowledgments
We thank Andrea Giokas for expert technical assistance and Jan Paluh for critical reading of this manuscript. This work was supported by Grant GM46226 from the National Institutes of Health to C.S.H.
References
- 1.Grimm C, Kohli J, Murray J, Maundrell K. Mol. Gen. Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein RJ. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 3.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 4.B€ahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 5.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 6.Russell P. In: Molecular Biology of the Fission Yeast. Nasim A, Young P, Johnson BF, editors. Academic Press; San Diego: 1989. pp. 243–271. [Google Scholar]
- 7.Keeney JB, Boeke JD. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson BM, Hernando Y, Schweizer M. Yeast. 1998;14:391–399. doi: 10.1002/(SICI)1097-0061(19980315)14:4<391::AID-YEA235>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 9.Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. J. Gen. Virol. 1991;72:1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 10.Hochuli E, Chromatogr J. 1988;444:293–302. doi: 10.1016/s0021-9673(01)94032-4. [DOI] [PubMed] [Google Scholar]
- 11.Orr-Weaver TL, Szostak JW, Rothstein RJ. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 12.Muhlrad D, Hunter R, Parker R. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 13.Kostrub CF, Lei EP, Enoch T. Nucleic Acids Res. 1998;26:4783–4784. doi: 10.1093/nar/26.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H, Kunes S, Schatz PJ, Botstein D. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 15.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly DA, Hoffman CS. Biotechniques. 2002;33:978–982. doi: 10.2144/02335bm02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe Y, Lino Y, Furuhata K, Shimoda C, Yamamoto M. EMBO J. 1988;7:761–767. doi: 10.1002/j.1460-2075.1988.tb02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman CS, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 19.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 20.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Nat. Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 21.Basi G, Schmid E, Maundrell K. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman CS, Winston F. Genetics. 1990;124:807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]