Abstract
Over several decades, animals have been used as models to investigate the human-specific drug toxicity, but the outcomes are not always reliably extrapolated to the humans in vivo. Appropriate in vitro human-based experimental system that includes in vivo parameters is required for the evaluation of multiple organ interaction, multiple organ/organ-specific toxicity, and metabolism of xenobiotic compounds to avoid the use of animals for toxicity testing. One such versatile in vitro technology in which human primary cells could be used is integrated discrete multiple organ co-culture (IdMOC). IdMOC system adopts wells-within-well concept that facilitates co-culture of cells from different organs in a discrete manner, separately in the respective media in the smaller inner wells which are then interconnected by an overlay of a universal medium in the large containing well. This novel in vitro approach mimics the in vivo situation to a great extent, and employs cells from multiple organs that are physically separated but interconnected by a medium that mimics the systemic circulation and provides for multiple organ interaction. Applications of IdMOC include assessment of multiple organ toxicity, drug distribution, organ-specific toxicity, screening of anticancer drugs, metabolic cytotoxicity, etc.
Keywords: Alternative method, anticancer drug screening, cytotoxicity screening, human drug toxicity, human primary culture, metabolic cytotoxicity
INTRODUCTION
Man is exposed to lots of chemical entities through ingestion, inhalation, or by the agents getting in contact with the skin. The chemical entities can be pharmaceuticals, food preservatives/additives/colorants, agrochemicals, environmental pollutants, cosmetics, etc., All these daily use chemicals produce their own impacts when used at certain concentrations for specific periods of time. In case the concentration or the exposure time of a particular chemical exceeds the permissible limit, it may cause adverse cellular effects including those at the molecular level. Therefore, assessment of toxicological risk and knowledge of side effects about all the chemical substances are essential from the perspective of human welfare. Over a long period of time, animal experimentation has been practiced as a tool to identify the toxicity of chemical entities. Many reports have shown that data obtained from animal experiments are not reliably predictive of the human risks.[1] It is well known that xenobiotic chemicals tested on laboratory animal models do not always reflect the absorption, distribution, metabolism, excretion, and toxicity (ADMET) in humans because in vivo biology and physiology are more complex in humans than in lab animals.[2] Species differences in xenobiotic metabolism, drug–drug interaction, and sensitivity are the major issues which limit the use of animal in vivo test methods for human drug toxicity testing.[3] In the light of Russell and Burch's 3R concept (replacement, reduction, and refinement),[4] the scientific community and regulatory authorities have become conscious about the pain and distress caused to animals during experiments. Both poor relevance of data generated from animal experiments to humans and the ethical considerations of animal experiments have urged attention to in vitro methods for human risk assessment.
In vitro toxicology testing is considered as an essential tool to enrich our understanding about harmful chemicals and also predict their harmful effects on humans. However, the in vitro test methods do provide adequately reliable toxicological data, including mechanistic understanding about the toxicants, but the monoculture of established cell lines in isolation, without any cell-to-cell communication, and deficiency of metabolic enzymes due to altered gene expression give an impression that this approach cannot provide the complexity of human in vivo biology to the same extent as it happens in humans.[5,6,7] Several drawbacks of the conventional cell culture technique warrant a next-generation in vitro technique. Thus, the scientific need for improved methods has brought up a new technique, the Integrated discrete Multiple Organ Co-culture (IdMOC). IdMOC is a patented novel in vitro experimental system which is much advantageous in a way that it allows use of primary cells or organ slices, multiple organ interaction, and analysis of metabolic cytotoxicity. Herein, we review IdMOC in vitro technology to highlight its advantages over the conventional in vitro test system.
PRINCIPLES OF IdMOC TECHNOLOGY AND THE PLATE DESIGN
IdMOC technology has been developed by In Vitro Admet Laboratory (IVAL), USA, and patented by Dr. Albert P. Li as an in vitro experimental system for the assessment of human xenobiotic metabolism, drug distribution and toxicity. According to Dr. Li, IdMOC is based on the concept that in the human body, there are multiple organs that are physically separated but interconnected by the systemic circulation, allowing multiple organ interaction.[8]
The main advantage of the system is design of the culture plate. IdMOC uses wells within well concept which means the plate has chambers/large containing wells, with each having six small inner wells. This kind of plate design allows culture of different cell types from different organs or different cell types from the same organ separately in the small inner wells. In the six inner wells of each chamber, one can culture six different cell types, one in each well, three different cell types each in duplicate or two different cell types each in triplicate. After culturing the two or more cell types in the six small inner wells, in their respective media, they can be interconnected by filling the chamber/large containing well with a universal medium which contains the drug/xenobiotic compound at a certain concentration for a defined incubation period. Then the xenobiotic-treated cells can be subjected to assays and the medium can be used to analyze the metabolite(s) of the parent xenobiotic compound.[9] Figure 1 represents the concept of IdMOC and the schematic design of the IdMOC plate.[10]
Figure 1.
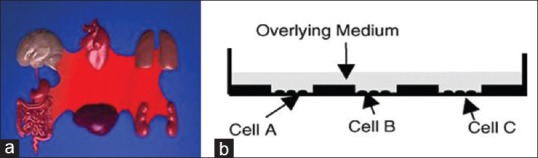
(a) The human body is conceptualized here as multiple organs connected by blood, the principle adopted in the IdMOC system. (b) A cross-section of the IdMOC plate is depicted, demonstrating the culture of multiple cell types (cells A–C) in physically separated cultures in different wells but later interconnected by flooding of a common medium. The IdMOC, thus, mimics the humans in vivo, with multiple organs as physically separated entities connected by a common fluid (blood in vivo; the overlay medium in the IdMOC)[18]
Therefore, the assay plate is designed to interconnect different organ systems to mimic critical in vivo situation under in vitro condition. The IdMOC plates provide equivalents of 6, 24, and 96 well plates for assay. For example, the 96-well assay plate has 16 large containing wells/chambers, each of which has 6 small inner wells [Figure 2]. Therefore, one can test a single xenobiotic compound at eight different concentrations, in duplicates, or 16 different xenobiotic compounds at a particular concentration. Between these two extremes, different numbers of compounds and different combinations of cells are possible. On the other hand, using conventional cell culture plate, one can check the toxicity of only a single compound in different concentrations, but the wells cannot be interconnected for cell-to-cell communication.
Figure 2.

A 96-well IdMOC plate (kind courtesy, Dr. Albert P. Li)
ADVANTAGES AND LIMITATIONS OF IdMOC OVER THE SIMPLE CONVENTIONAL CELL CULTURE TECHNIQUE
In the human body, liver is the major organ involved in the First-phase clearance of ingested xenobiotic compounds and, also, in the control of the systemic levels of drugs and other chemicals. The toxicant gets metabolized in the liver, and the metabolic product(s) is/are transported in circulation to the distal, non-hepatic organs such as kidney, lung, brain, intestine, and so on, and can produce adverse effects, may be in tissue-/organ-specific manner.[10] Therefore, in the body, liver is the first and immediate organ liable for the xenobiotic attack. Drugs such as troglitazone, nefadazone, trovafloxacin, etc., have been withdrawn due to their hepatotoxic effect.[11] Because the parent xenobiotic compound would either directly exert its toxic effect on the target cells or gets metabolized by the liver, the metabolite acts as a toxin to produce an adverse cellular effect either in the very organ that metabolizes the compound (say, liver) or more distal organs (say, kidney and/or lung). Sometimes, after the Phase I metabolism, the parent xenobiotic substance becomes neutralized by liver detoxification and is excreted via kidney. Therefore, the understanding of hepatic detoxification mechanism is very crucial to find the direct hepatotoxicity (liver injury) and the metabolite-induced toxicity of a particular xenobiotic substance. Lack of multiple organ interaction in the conventional cell culture does not allow the metabolites to pass from one cell type to another cell type in the same organ or from one organ to another organ. Thus, the cytotoxic effect produced by the activated metabolite and the differential response of the metabolically competent (e.g., hepatocytes) and incompetent (e.g., fibroblasts) cells toward a particular xenobiotic compound, which cannot be studied adopting the conventional cell culture system, can be conveniently studied adopting IdMOC technology.
Another issue with conventional in vitro testing is the use of continuous, transformed cell lines such as 3T3 cells and HepG2 cells. These cells can be used to detect general toxic agents, but would be of limited use for toxicity that, due to xenobiotic metabolizing enzyme activities or organ-specific biochemical pathways, would lead to organ-specific interaction.[10] Primary cells or organ slices are used in IdMOC because primary hepatocytes are the closest model for the liver in vivo. Especially, human primary cells are important experimental systems for the prediction of human-specific drug properties and risk assessment. Technologies for obtaining, cryopreserving, and culturing primary cells from humans are well established and can be adopted.[12,13] Alternatively, cryopreserved primary cells can be availed from reputed commercial sources also. Organ slices can also be used for multiple organ toxicity testing. However, organ slices prepared from the biopsy samples donated by humans are viable only for 24 h and cannot be used for long-term studies.[10] In such a case, the suspense in the availability of biopsy samples is a lacuna. Also, results from experiment to experiment would vary depending on the genetic makeup, health status, and age of individuals from whom the organ slices are derived. Considering the limitations, IdMOC system using cells, rather than organ slices, is appropriate to screen the chemical entities with respect to liver metabolism.
Identification of pharmacokinetic, pharmacodynamic, and safety profile of a drug is crucial for successful drug development. Failure of drug molecules to testify any one of these properties results in clinical failure.[14] Animal experiments which are adopted to test these properties fail to mimic the in vivo condition of humans. Thus, the testing modality should be very close to humans right from the preliminary screening of drugs to avoid the loss of huge amounts spent in animal experimentation for drug discovery.[15] IdMOC could be the rewarding in vitro model to study the ADMET properties of a drug because the system allows use of appropriate human primary cells or organ slices, co-culture of two to six different cell types from different organs or the same organ, and the introduction of cell–cell communication by flooding with the universal medium in the chamber/containing well which renders it to be a versatile technology for the preliminary screening of potential drugs. However, the information derived using the primary cells that are co-cultured in the IdMOC plate will depend on the quality, particularly with regard to the degree of retention of organ-specific properties.[9,10] One of the major challenges associated with IdMOC plate design is that there is no directed flow of the medium from one inner well to another. Therefore, other in vivo parameters such as concentration and residence of drug/toxin at each organ may not be assessed. Thus, the system cannot model the sequential pharmacokinetic events as present in vivo.[9] This major limitation of the IdMOC technique in the context of pharmacokinetic evaluation can be overcome by the incorporation of microfluidics technique to the inner wells of the IdMOC plate. Such development would potentially broaden the applications of IdMOC technology. But the potential use of cytochrome P450 (CYP) enzyme inducers and inhibitors would help to identify the drug–drug interaction, bio-availability, metabolic pathway, and induction and inhibition of the drug.[16]
IdMOC provides for assessment of acute cytotoxicity by adopting 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay/ATPase assay, apoptotic evaluation using caspase assay, and sub-acute toxicity testing by assessing cell cycle arrest, oxidative stress, DNA damage, etc., Apart from its use for toxicity assessment, IdMOC serves as a potent model to study the end points of hepatic metabolism by determining the gene expression pattern of hepatic function, Phase I, Phase II metabolic enzymes, and efflux and uptake transporters. All these assays can be conducted even by researchers working in small-scale laboratories. In the absence of absolute 3D models, the systemic, repetitive, and quantitative analysis of a biological system in terms of unknown substances studied through the IdMOC system represents human in vivo biology to a better extent than the simple 2D culture of single cell type/primary cells adopting the conventional in vitro approach.
APPLICATIONS OF IdMOC
Multiple organ toxicity screening
Every year the drug regulatory authorities update the details of drugs that are recalled/withdrawn during clinical trials and also at post-marketing stage. One of the main reasons for the withdrawal of a marketed drug is the side effects caused to non-target organs. One such example is Rezulin, a drug that was introduced in the year 1997 for treatment of Type II diabetes but withdrawn in the year 2000 for the reason of hepatotoxicity.[1,16] Every drug is pre-clinically tested on non-human animal models, which is followed by small-scale human clinical trials conducted with the drug. Even then, after marketing, when the drug reaches the heterogeneous large-scale human population, side effects are realized. So, a better approach and effort is necessary to improve the pre-clinical efficacy of the drug to exactly predict human drug toxicity. As discussed earlier, animal models have some drawbacks in the accurate prediction of human drug toxicity. On the other hand, in IdMOC technique, human primary cell types isolated from different organs are cultured in the discrete small inner wells and an overlying universal medium connects all the cell types from different organs. Therefore, the toxic potential of a drug can be evaluated in cells from multiple organs under identical experimental conditions to identify multiple toxic end points at a time.
For example, aflatoxin B1 (AFB1) is a well-known hepatotoxin and its differential and selective toxicity toward hepatocytes has been illustrated using the IdMOC system [Figure 3].[10] Human hepatocytes, human renal proximal tubule cells, and human pulmonary epithelial cells co-cultured in the inner wells of the IdMOC plate and the cells, each in duplicate, were flooded with the universal medium containing AFB1 at different concentrations ranging from 0.27 to 200 μM for 48 h. The percent viability of all the three cells co-cultured in the IdMOC plate was determined adopting ATPase assay. The result showed that even at a low concentration, AFB1 was selectively toxic to hepatocytes. These results were consistent with and also comparable to the in vivo data. Thus, the IdMOC proved to be an effective system for the evaluation of organ-specific toxicity to screen the side effects/efficacy of a drug on the target organ as well as on non-target organs.[9,10,17]
Figure 3.
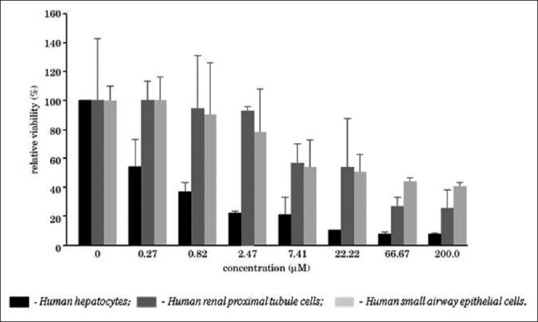
Organ-specific cytotoxicity determination in the IdMOC system, illustrated with aflatoxin B1, a known hepatotoxic agent[10]
Organ-specific toxicity screening
Every organ is made up of different tissues or cell types. Each cell type of the particular organ may have different function and architecture that change the response of the particular cell type toward a particular xenobiotic compound. In such case, a single cell type used in multiple organ toxicity evaluation may not be the complete representative of an organ. Thus, after deriving information for a particular compound from the multiple organ toxicity testing, it is very important to evaluate organ-specific toxicity to check which cell type of the organ is sensitive for a particular xenobiotic compound. For example, liver is made up of three major cell types: hepatocytes, Kupffer cells, and stellate cells. Among these cell types, hepatocytes are mainly involved in the metabolism of xenobiotics and are susceptible to toxic insult. Therefore, using different cell types of the same organ, IdMOC technique can provide hepatotoxicity model, cardiotoxicity model, nephrotoxicity model, etc., The organ-specific toxicity model can be used in paracrine signaling by evaluating the cellular factors secreted by one cell type on a different cell type within the same organ.[9,10,17]
To create a lung model, major cell types of the lung, such as bronchial epithelial cells (bronchus), small airway epithelial cells (alveolus), and micro-vascular endothelial cells (capillary of the lung), each in duplicate, were co-cultured in IdMOC assay plate and the cells were treated with increasing concentrations of nicotine, which is a key ingredient of cigarette smoke, for 24 h [Figure 4]. The cytotoxic concentration of the cigarette smoke was assessed adopting MTT assay and the relative viability against different concentrations of nicotine was plotted. The results revealed that the smoke condensate was cytotoxic to all the three cell types, wherein bronchial epithelial cells were the most susceptible. Thus, the IdMOC lung model proved that cigarette smoke is associated with lung cancer as well as toxicity.[10]
Figure 4.
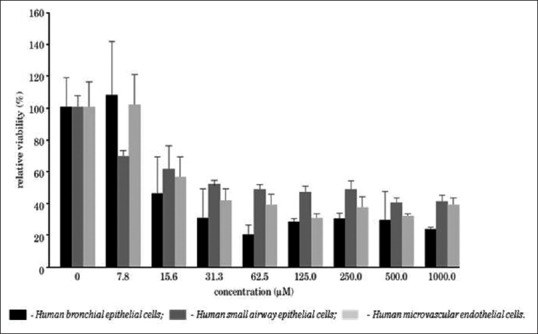
Evaluation of nicotine cytotoxicity toward multiple pulmonary cell types co-cultured in the IdMOC system: A lung model[10]
Anticancer drug screening
Tumor xenograft grown in nude mouse is the most common in vivo model used to test the efficacy of anticancer drugs during pre-clinical screening. But the pre-clinically tested anticancer drug does not always pass through the human trials. The drug may affect the normal cells. At the laboratory screening level, these limitations can be overcome by using a tumor-bearing man model of IdMOC in which one can co-culture the target cancer cells with normal cells from key organs that allows selecting an anticancer drug with acceptable toxic potential toward normal tissues.[8,9,18] The end points such as cell viability, cell proliferation, cell survival, gene expression pattern, DNA repair mechanism, hormone receptor, growth factor receptor, drug metabolism, and drug resistance can be assessed by adopting real-time PCR technique. Thus, the cost factor and number animals used for pre-clinical trial are reduced while adopting IdMOC technique.[18]
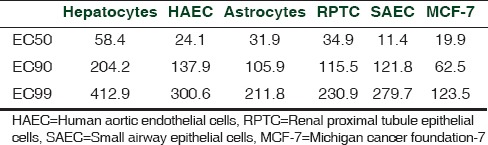
One such study with tamoxifen (TMX) revealed the IdMOC system to perform as an in vitro model of a tumor-bearing man. Since TMX is an anti-mammary cancer drug, it was tested on MCF-7 mammary cancer cell, with human primary hepatocytes, renal proximal tubule cells, pulmonary epithelial cells, aortic endothelial cells, and astrocytes as normal cells. TMX showed cytotoxicity towards the MCF-7 cancer cell at a very low dose (IC50 19.9 μM) compared with the other cell types [Table 1].[18] Thus, the estrogen receptor (ER) antagonist TMX was proved cytotoxic to ER-positive MCF-7 cell, in a dose-dependent manner, with negligible effect on the normal cells.[18] Also, the results revealed that human hepatocytes are highly resistant to TMX treatment compared to the other cell types used in the study. Thus, the study concluded that TMX may have strong metabolic clearance by hepatocytes.[18] IdMOC is an efficient model for discovery of anticancer drugs which are specifically toxic to cancer cells and not toxic or less toxic to normal cells.
Table 1.
EC50, EC90, and EC99 for human hepatocytes (hepatocytes), human aortic endothelial cells, human astrocytes (astrocytes), human renal proximal tubule epithelial cells, human small airway epithelial cells, and human breast adenocarcinoma cells (MCF-7)[18]
Metabolic toxicity
One of the critical limitations of conventional cell culture technique is evaluation of toxicity upon xenobiotic biotransformation. In the liver, Phase I bioactive metabolism and Phase II detoxification process may have several possible effects on xenobiotic compounds. Thus, the xenobiotic compound is either bio-activated or detoxified. The conventional in vitro techniques/methods are inadequate to study the metabolic cytotoxic effect of a xenobiotic compound because monoculture of hepatocytes in isolation with lack of well-to-well interconnection prevents metabolites from reaching their sensitive organs (either hepatic or non-hepatic). IdMOC technology solved this problem concerned with the determination of the metabolic cytotoxicity when the following three model toxicants, each with specific pattern of liver metabolism, were used:
TMX is a non-steroidal antagonist of estrogen receptor used for breast cancer treatment. Since TMX is a direct acting toxicant, it does not need any metabolic activation at liver. So, TMX acts directly on the target cells without liver metabolism, to produce toxic effect.[19,20,21,22]
AFB1 is a mycotoxin that contaminates human food and it is reported to be a hepatotoxic agent.[19] Human CYP isoenzymes 1A2[23,24] and 3A4[25] metabolize AFB1 into a highly reactive (AFB1)-8,9 epoxide that causes toxicity at the site where metabolic activation occurs whereas the other metabolites are detoxified.[26,27,28] Therefore, a non-toxic parent compound, AFB1, requires bio-activation in the liver to form extremely reactive metabolite which causes toxicity to the hepatic cells themselves.[19]
Cyclophosphamide (CPA) is an immune suppressant which is used as an anticancer agent. Mostly, CYP2B6 isoenzyme[29,30] acts on CPA and converts it into a chemotherapeutically active 4-hydroxy-cyclophosphamide metabolite to exert its anticancer activity.[31] Therefore, CPA is a pro-drug because biotransformation of CPA is important to form a toxic diffusible metabolite which is transported via circulatory system to exert its therapeutic effect on the cancer cells and perhaps toxic effect on hepatic or non-hepatic organs.[8,9,19]
To emphasize the importance of liver metabolism and metabolism-dependent cytotoxicity, metabolically competent primary human hepatocytes[32,33] and metabolically incompetent mouse 3T3 fibroblast cells[34,35,36] were chosen and co-cultured in the IdMOC plate to assess the toxic effect of a parent compound/bio-activated metabolite on metabolically competent hepatic cells (where metabolism occurs) and/or metabolically incompetent (target/sensitive organ) cells.
The inner wells of IdMOC 96-well plate were seeded with three wells each of thawed primary human hepatocytes in plating medium and 3T3 cells in Dulbecco's modified Eagle's medium. Cells were incubated for 4 h for attachment. Afterward, the three model toxicants, viz., AFB1, TMX, and CPA, were taken separately at designated concentrations and diluted with hepatocyte induction medium that was flooded into each of the large chamber/well, thereby interconnecting all the inner wells of each chamber. Then, the plates were incubated for 16 h and the cells were subjected to cytotoxicity evaluation by adopting MTT assay.[19]
Table 2 presents the experimental results with the human hepatocytes and 3T3 fibroblast cells adopting IdMOC technology, which classified these model toxins into three distinct profiles as follows:
Table 2.
EC50 values of the model toxicants aflatoxin B1, cyclophosphamide, and tamoxifen for 3T3 cells and human hepatocytes cultured in IdMOC
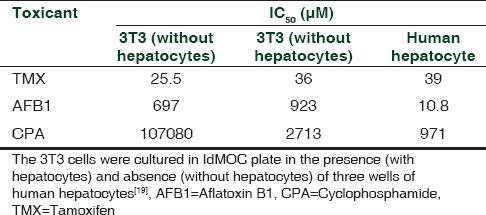
TMX was cytotoxic to both 3T3 cells and hepatic cells directly at low concentrations and the presence of hepatocyte in co-culture system only slightly impinged on 3T3 cells. Thus, TMX was concluded to be a direct acting cytotoxicant with hepatic detoxification.
The hepatotoxicant AFB1 showed greater cytotoxic effect on hepatocytes than 3T3 cells. The presence of hepatocyte promoted the bio-activation of AFB1, but did not increase the cytotoxic potential of AFB1/reactive metabolite to impinge on 3T3 cells, thereby suggesting that AFB1 is a metabolism-dependent selective cytotoxicant to hepatocytes.
The presence and absence of hepatocyte co-cultured with 3T3 cells altered the cytotoxicity of CPA on metabolically incompetent 3T3 cells. In the presence of hepatocytes, the cytotoxic potential of CPA to 3T3 cells was greatly increased. Thus, the presence of metabolically competent hepatocytes facilitated CPA to produce the diffusible active metabolite to exert its cytotoxic effect at a higher rate on metabolically incompetent 3T3 cells. To substantiate this influence, a non-specific P450 inhibitor, 1-aminobenzotraizole, was used to attenuate the cytotoxicity of CPA toward both 3T3 cells and hepatocytes, thus confirming the CYP enzyme activation upon CPA-induced toxicity. Thus, CPA is categorized as a metabolism-dependent cytotoxicant with stable, diffusible toxic metabolites.[19]
Using IdMOC plates, one can model hepatic metabolism with the non-hepatic organs which are prone to be sensitive for a particular drug. For e.g., co-culturing of primary hepatocytes with proximal tubular kidney cells can be an appropriate model to study the metabolism-induced drug toxicity on kidney (nephrotoxicity).
Some of the advantages of such models are as follows:
The complete information about the induction or inhibition of hepatic Phase I and Phase II enzymes, as a result the cellular changes that happened to the target organ, can be obtained simultaneously for a particular test drug.[14,37,38]
The major pathway involved in the drug metabolism can be identified using selective inhibitors or inducers available for the Phase I enzymes. For example, ketoconazole is a selective inhibitor of CYP3A4 enzyme. If the tested drug induces CYP3A4 Phase I enzyme, the inhibitor ketoconazole inhibits the metabolism of the particular drug. As a result, one can infer that the drug is metabolized by CYP3A4 enzyme.[14,15,16,38]
Identification of drug-induced metabolism would permit one to evaluate the drug–drug interaction. In drug–drug interaction, a drug can affect the metabolic stability of another drug. This kind of interaction leads to the failure of the drug in clinical trials. For example, the antifungal ketoconazole, a potent inhibitor of CYP3A4, causes drug–drug interactions with the drugs that are substrates of CYP3A4.[14,15,16,38]
In these cases, the fate of the parent compound and its metabolites can be analyzed by high performance liquid chromatography (HPLC) or liquid chromatography/mass spectrometry (LC/ MS). The outcome of this analysis would provide information regarding the stability and metabolite profile of the tested drug.[9,14]
IdMOC system's application in high content analysis
Use of IdMOC system in high content analysis is a recent development. To increase the quality of information generated by IdMOC system, the high content analysis, a quantitative and multi-parametric tool that is powerful for the evaluation of toxicity in vitro, of cytotoxic endpoints was performed using fluorescent stains calcein-AM (live stain), ethidium homodimer-I (dead stain), and Hoechst 33342 (nuclear stain), which facilitate analysis of live and dead cells and measurement of nuclear area and cell density. As the fluorescence intensity of the live cells decreases, the cytotoxicity increases, whereas the dead cell intensity increases when the cytotoxicity decreases. Two model toxicants that are detoxified and activated by the liver, 4-aminophenol and CPA, respectively, were screened for hepatotoxicity (human primary hepatocytes) and metabolism-dependent toxicity on a non-liver cell type (3T3-L). It was found that 4-aminophenol is less toxic to 3T3 cells in the presence of hepatic cells than in the absence of hepatic cells, which shows the metabolic detoxification of the compound by the hepatic cells. On the other hand, CPA is more toxic to 3T3 cells in the presence of hepatic cells than in the absence of hepatic cells showing that CPA needs metabolic activation by liver. Thus, the conjunction of high content analysis with the IdMOC technology provides quick and high-quality screening of multiple compounds for hepatotoxicity and the effects of hepatic metabolism on non-liver cell types.[39]
POPULARIZATION OF IdMOC TECHNOLOGY
Advancement of 3Rs research is one of the objectives of Mahatma Gandhi-Doerenkamp Center (MGDC).[38,39,40,41] This objective was realized when MGDC and Dr. Albert Li, CEO, AP Sciences, USA, entered into an understanding to distribute IdMOC plates, free of cost, to deserving researchers. Through this arrangement, Dr. Li, the inventor of this technology, provides a gift of a substantial number of IdMOC plates to MGDC for distribution to qualified and competent Indian scientists interested in in vitro toxicology/pharmacology for their research endeavors. At present, research using IdMOC technology is going on at three centers in India in collaboration with MGDC: (i) Narsee Monjee Institute of Management Studies (NMIMS) University, Mumbai; (ii) Sree Chitra Tirunal Institute for Medical Sciences and Technology, Thiruvananthapuram, Kerala; and (iii) Bharathidasan University, Tiruchirappalli, Tamil Nadu.
CONCLUSION
The IdMOC technology is a simple and new-generation in vitro experimental system which does not require any sophisticated laboratory equipment for the evaluation of distribution, metabolism, and toxicity of a xenobiotic. Co-culture of multiple cell types of the same organ or multiple organs in a physically discrete manner allows the system to interact and helps to predict the multiple endpoints. Use of primary human cells and incorporation of metabolic cytotoxicity to the in vitro system provides an insight to the scientific community that IdMOC is a physiologically relevant model for risk assessment. The embodiment of wells-within-well concept in IdMOC technology has promoted in vitro technique from routine two-dimensional cell culture to mimic, to a great extent, the real in vivo conditions. Thus, IdMOC is an innovative and less time-consuming model that could replace animal testing methods perhaps to comply with the changing regulatory needs. In vitro approach has always been an adoptable technique and readily procures many in vivo key features. Thus, the technique could overcome the uncertainty of animal testing and withstand for a long period to reduce and replace the use of animals in scientific research. However, novel inventions and new methodologies will never stop until the in vitro condition matches or supersedes the in vivo condition. The future of cell culture could be the virtual human-on-chip which may simulate a complete human, but in a simple magnitude. IdMOC has a great potential simulating humans in vivo using in vitro conditions and this technique can be adopted by all researchers who are efficiently carrying out conventional in vitro cell culture in the laboratory.
ACKNOWLEDGMENTS
The financial assistance from the Doerenkamp - Zbinden Foundation, Switzerland, is heartily acknowledged. We thank Dr. Albert Li, IVAL/AP Sciences Laboratory, USA, for his support and collaboration with MGDC.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Li AP. Accurate prediction of human drug toxicity: A major challenge in drug development. Chem Biol Interact. 2004;150:3–7. doi: 10.1016/j.cbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Li AP. Human-based in vitro experimental systems for the evaluation of human drug safety. Curr Drug Saf. 2007;2:193–9. doi: 10.2174/157488607781668909. [DOI] [PubMed] [Google Scholar]
- 3.Li AP. Overview: Evaluation of metabolism-based drug toxicity in drug development. Chem Biol Interact. 2009;179:1–3. doi: 10.1016/j.cbi.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Russell WM, Burch RL. London, UK: Methuen; 1959. The Principles of Humane Experimental Technique. [Google Scholar]
- 5.Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line HepG2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–42. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 6.Westerink WM, Schoonen WG. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro. 2007;21:1581–91. doi: 10.1016/j.tiv.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Westerink WM, Schoonen WG. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro. 2007;21:1592–602. doi: 10.1016/j.tiv.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Li AP. In vitro evaluation of human xenobiotic toxicity: Scientific concepts and the novel integrated discrete multiple cell co-culture (IdMOC) technology. ALTEX. 2008;25:43–9. doi: 10.14573/altex.2008.1.43. [DOI] [PubMed] [Google Scholar]
- 9.Uzgare AR, Li AP. New paradigm in toxicity testing: Integrated discrete Multiple Organ Co-cultures (IdMOC) for the evaluation of xenobiotic toxicity. ALTEX Proc. 2013;2:39–46. [Google Scholar]
- 10.Li AP. The use of the integrated discrete multiple organ co-culture (IdMOC) system for the evaluation of multiple organ toxicity. Altern Lab Anim. 2009;37:377–85. doi: 10.1177/026119290903700408. [DOI] [PubMed] [Google Scholar]
- 11.Bale SS, Vernetti L, Senutovitch N, Jindal R, Hegde M, Gough A, et al. In vitro platforms for evaluating liver toxicity. Exp Biol Med (Maywood) 2014;239:1180–91. doi: 10.1177/1535370214531872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li AP, Lu C, Brent JA, Pham C, Fackett A, Ruegg CE, et al. Cryopreserved human hepatocytes: Characterization of drug-metabolizing activities and applications in higher throughput screening assays for hepatotoxicity, metabolic stability, and drug-drug interaction potential. Chem Biol Interact. 1999;121:17–35. doi: 10.1016/s0009-2797(99)00088-5. [DOI] [PubMed] [Google Scholar]
- 13.Li AP. Human hepatocytes: Isolation, cryopreservation and applications in drug development. Chem Biol Interact. 2007;168:16–29. doi: 10.1016/j.cbi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Li AP. Critical human hepatocyte-based in vitro assays for the evaluation of adverse drug effects. In: Kapetanovic IM, editor. Drug Discovery and Development-Present and Future. Croatia: InTech Publisher; 2011. pp. 151–68. [Google Scholar]
- 15.Li AP. Preclinical in vitro screening assays for drug-like properties. Drug Discov Today Technol. 2005;2:179–85. doi: 10.1016/j.ddtec.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Li AP. Screening for human ADME/Tox drug properties in drug discovery. Drug Discov Today. 2001;6:357–66. doi: 10.1016/s1359-6446(01)01712-3. [DOI] [PubMed] [Google Scholar]
- 17.Richter PA, Li AP, Polzin G, Roy SK. Cytotoxicity of eight cigarette smoke condensates in three test systems: Comparisons between assays and condensates. Regul Toxicol Pharmacol. 2010;58:428–36. doi: 10.1016/j.yrtph.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Li AP, Bode C, Sakai Y. A novel in vitro system, the integrated discrete multiple organ cell culture (IdMOC) system, for the evaluation of human drug toxicity: Comparative cytotoxicity of tamoxifen towards normal human cells from five major organs and MCF-7 adenocarcinoma breast cancer cells. Chem Biol Interact. 2004;150:129–36. doi: 10.1016/j.cbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Li AP, Uzgare A, LaForge YS. Definition of metabolism-dependent xenobiotic toxicity with co-cultures of human hepatocytes and mouse 3T3 fibroblasts in the novel integrated discrete multiple organ co-culture (IdMOC) experimental system: Results with model toxicants aflatoxin B1, cyclophosphamide and tamoxifen. Chem Biol Interact. 2012;199:1–8. doi: 10.1016/j.cbi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Al-Akoum M, Dodin S, Akoum A. Synergistic cytotoxic effects of tamoxifen and black cohosh on MCF-7 and MDA-MB-231 human breast cancer cells: An in vitro study. Can J Physiol Pharmacol. 2007;85:1153–9. doi: 10.1139/Y07-111. [DOI] [PubMed] [Google Scholar]
- 21.Benz C, Cadman E, Gwin J, Wu T, Amara J, Eisenfeld A, et al. Tamoxifen and 5-fluorouracil in breast cancer: Cytotoxic synergism in vitro. Cancer Res. 1983;43:5298–303. [PubMed] [Google Scholar]
- 22.Petinari L, Kohn LK, de Carvalho JE, Genari SC. Cytotoxicity of tamoxifen in normal and tumoral cell lines and its ability to induce cellular transformation in vitro. Cell Biol Int. 2004;28:531–9. doi: 10.1016/j.cellbi.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Crespi CL, Penman BW, Steimel DT, Gelboin HV, Gonzalez FJ. The development of a human cell line stably expressing human CYP3A4: Role in the metabolic activation of aflatoxin B1 and comparison to CYP1A2 and CYP2A3. Carcinogenesis. 1991;12:355–9. doi: 10.1093/carcin/12.2.355. [DOI] [PubMed] [Google Scholar]
- 24.Sengstag C, Würgler FE. DNA recombination induced by aflatoxin B1 activated by cytochrome P450 1A enzymes. Mol Carcinog. 1994;11:227–35. doi: 10.1002/mc.2940110408. [DOI] [PubMed] [Google Scholar]
- 25.Guengerich FP, Johnson WW, Shimada T, Ueng YF, Yamazaki H, Langouët S. Activation and detoxication of aflatoxin B1. Mutat Res. 1998;402:121–8. doi: 10.1016/s0027-5107(97)00289-3. [DOI] [PubMed] [Google Scholar]
- 26.Gorelick NJ. Risk assessment for aflatoxin: I. Metabolism of aflatoxin B1 by different species. Risk Anal. 1990;10:539–59. doi: 10.1111/j.1539-6924.1990.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 27.Guengerich FP, Johnson WW, Ueng YF, Yamazaki H, Shimada T. Involvement of cytochrome P450, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B1 and relevance to risk of human liver cancer. Environ Health Perspect. 1996;104(Suppl 3):557–62. doi: 10.1289/ehp.96104s3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CJ, Wang SW, Shiah HS, Lin JK. Effect of ethanol on hepatotoxicity and hepatic DNA-binding of aflatoxin B1 in rats. Biochem Pharmacol. 1990;40:715–21. doi: 10.1016/0006-2952(90)90306-6. [DOI] [PubMed] [Google Scholar]
- 29.Chang TK, Weber GF, Crespi CL, Waxman DJ. Differential activation of cyclophosphamide and ifosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res. 1993;53:5629–37. [PubMed] [Google Scholar]
- 30.McErlane V, Yakkundi A, McCarthy HO, Hughes CM, Patterson LH, Hirst DG, et al. A cytochrome P450 2B6 meditated gene therapy strategy to enhance the effects of radiation or cyclophosphamide when combined with the bioreductive drug AQ4N. J Gene Med. 2005;7:851–9. doi: 10.1002/jgm.728. [DOI] [PubMed] [Google Scholar]
- 31.Huttunen KM, Raunio H, Rautio J. Prodrugs--from serendipity to rational design. Pharmacol Rev. 2011;63:750–71. doi: 10.1124/pr.110.003459. [DOI] [PubMed] [Google Scholar]
- 32.Fabre G, Combalbert J, Berger Y, Cano JP. Human hepatocytes as a key in vitro model to improve preclinical drug development. Eur J Drug Metab Pharmacokinet. 1990;15:165–71. doi: 10.1007/BF03190200. [DOI] [PubMed] [Google Scholar]
- 33.Gómez-Lechón MJ, Donato MT, Castell JV, Jover R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr Drug Metab. 2003;4:292–312. doi: 10.2174/1389200033489424. [DOI] [PubMed] [Google Scholar]
- 34.De Groene EM, Nijmeijer SM, Horbach GJ, Witkamp RF. Tiamulin inhibits human CYP3A4 activity in an NIH/3T3 cell line stably expressing CYP3A4 cDNA. Biochem Pharmacol. 1995;50:771–3. doi: 10.1016/0006-2952(95)00197-8. [DOI] [PubMed] [Google Scholar]
- 35.De Groene EM, Seinen W, Horbach GJ. A NIH/3T3 cell line stably expressing human cytochrome P450-3A4 used in combination with a lacZ’ shuttle vector to study mutagenicity. Eur J Pharmacol. 1995;293:47–53. doi: 10.1016/0926-6917(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 36.Liebsch HM, Spielmann H. Balb/c 3T3 cytotoxicity test. Methods Mol Biol. 1995;43:177–87. doi: 10.1385/0-89603-282-5:177. [DOI] [PubMed] [Google Scholar]
- 37.Oleson FB, Berman CL, Li AP. An evaluation of the P450 inhibition and induction potential of daptomycin in primary human hepatocytes. Chem Biol Interact. 2004;150:137–47. doi: 10.1016/j.cbi.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Li AP. In vitro human hepatocyte-based experimental systems for the evaluation of human drug metabolism, drug-drug interactions, and drug toxicity in drug development. Curr Top Med Chem. 2014;14:1325–38. doi: 10.2174/1568026614666140506114411. [DOI] [PubMed] [Google Scholar]
- 39.Cole SD, Madren-Whalley JS, Li AP, Dorsey R, Salem H. High content analysis of an in vitro model for metabolic toxicity: Results with the model toxicants 4-aminophenol and cyclophosphamide. J Biomol Screen. 2014;19:1402–8. doi: 10.1177/1087057114550399. [DOI] [PubMed] [Google Scholar]
- 40.Gunatilake M, Busquet F, Akbarsha MA. Alternatives initiative in Sri Lanka: Pre- and post-conference workshops at the inaugural scientific conference of the Sri Lanka association for laboratory animal science. ALTEX. 2014;31:224–6. doi: 10.14573/altex.1403121. [DOI] [PubMed] [Google Scholar]
- 41.Akbarsha MA. Alternatives to animal experiments. In: Tandon PN, Muralidhar K, Gupta YK, editors. Use of Animals in Scientific Research and Education. New Delhi: Indian National Science Academy; 2012. pp. 147–68. [Google Scholar]