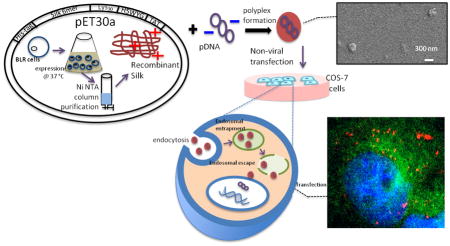
Abstract
Gene delivery research has gained momentum with the use of lipophilic vectors that mimic viral systems to increase transfection efficiency. However, maintaining cell viability with these systems remains a major challenge. Therefore biocompatible and nontoxic biopolymers that are designed by combining non-immunological viral mimicking components with suitable carriers have been explored to address these limitations. In the present study recombinant DNA technology was used to design a multi-functional gene delivery system for nuclear targeting, while also supporting cell viability. Spider dragline silk recombinant proteins were modified with DNA condensing units and the proton sponge endosomal escape pathway was utilized for enhanced delivery. Short-term transfection efficiency in a COS-7 cell line (adherent kidney cells isolated from African green monkey) was enhanced compared to lipofectamine and polyethyleneimine (PEI), as was cell viability with these recombinant bio-polyplexes. Endosomal escape and consequent nuclear targeting were shown with fluorescence microscopy.
Keywords: Gene delivery, Bioengineered silk, Nuclear localization, Endosomal escape
1. Introduction
Increased interest in gene delivery for therapeutic applications has occurred over the last decade resulting in different approaches to replace or knock-down particular genes. [1– 2] Viral gene delivery agents are used for their high transfection efficiency; [2–3] however, they also can cause immunogenic responses resulting in toxicity and low cell viability. [4] Therefore non-viral gene carriers have become important to pursue in order to increase cell viability. [5–6] Non-viral gene delivery agents are derived from natural or synthetic polymers with either lipophilic or hydrophilic characteristics, resulting in different assemblies in vitro and in vivo. [7–8] Biosynthetic methods for synthesizing polymers, rather than chemical synthesis and modification strategies, can avoid chemical residues or impurities, which can negatively impact biological outcomes, while also providing tight control of polymer dispersity. Different amenities can be incorporated into these bioengineered polymers by selecting from a wide range of natural polymer libraries or motifs, or by synthesizing modified recombinant polymers in situ. Some of the common benefits of this approach include increased cell viability, biodegradability, biocompatibility and enhanced functionality. [9]
These bioengineered polymers have to assemble in solution to meet gene delivery and then stay soluble with hydrophilic interactions within the solution interphase. In addition, the polymer chains need to assemble in order to prevent excess penetration of media, which would result in rapid degradation. Silk proteins are biocompatible and biodegradable, and support cell viability. [10] Highly ordered secondary structure in solution stems from beta sheet formation, via inter-chain molecular hydrogen bonding between alanine/glycine residues. [11] Highly ordered structure of silk has been used to construct solid, scaffold-like materials for tissue engineering and extracellular matrix (ECM) mimicking applications, and also for delivering therapeutics and genes. [12–17] Biodegradability of the whole structure that has aminoacids as the building blocks facilitates the clearance of the cargo resulting in excellent biocompatibility. The repetitive peptide units of spider silk act as an excellent polymeric support for gene delivery, mimicking synthetic polymeric functions toward similar goals. [18–19]
Recombinant DNA techniques allows for facile modification of spider silk sequences with endosomal escape chemistries to address endosomal degradation challenges upon endocytosis, a major challenge in the intracellular delivery process. Cells defense mechanisms can disrupt gene delivery processes by degradation of the carrier before it reaches its destination. [20] As with bacteria using membrane destabilizing proteins or viruses using membrane-disrupting molecules, non-viral agents can possess similar abilities. [21] For endosomal escape sequences, the histidine-rich peptide, H5WYG (GLFHAIAHFIHGGWHGLIHGWYG) is expression friendly due to the elimination of repetitive amino acids. The histidines can respond to the drop in pH between the cytoplasm and endosomes to act as a proton sponge by absorbing excess protons in the endosomes. [22] This process prevents proton bombardment of the cargo in the endosomes and results in disruption of endosomal membrane. [23]
For the attachment of oligonucleotides to the large peptidic carrier, non-covalent, ionic attraction is a useful method where the negatively charged phosphate backbone of oligonucleotides interacts with positively charged proteins to form electrostatic bonds. Synthetic polymers such as polyethyleneimine (PEI) have been used for their cationic characteristics and natural cationic polymers such as chitosan have been tried with the same purpose of condensing the negatively charged DNA. However, these highly cationic polymers cause toxicity due to non-selective attachment to the negatively charged membrane phospholipids or to serum proteins. [24–26] Recombinant DNA technology has been useful for obtaining cationic, non-toxic proteins for gene delivery. [9, 18] Lysine has been used successfully in condensing DNA into particles along with the polymer, without toxicity to cells [27, 28, 39] with an optimal number of 30 lysines to bind to medium range sized DNA (5 kbp). [29] Also, inspired by nature, clusters of lysine residues can help to condense DNA, as histones have repetitive lysine residues organized in clusters. High positive charged particles after condensing DNA is not desired due to cytotoxicity caused by non-specific interactions. [30–31]
Nuclear targeting is necessary for successful gene delivery, not only to facilitate transfection, but also to prevent premature degradation in the cytoplasm. [32] For nuclear targeting, the Transactivating Regulatory Protein (TAT) sequence, with an origin from an 86 amino acid long protein from the Human Immunodeficiency Virus (HIV) virus has been pursued. [33] This protein has a nine amino acid long, arginine rich sequence, with nuclear directing ability either as a short peptide or as part of a long chain. [34]
The goal of the current study was to design and study the effects of different functional groups and their placement in recombinant silk proteins for nuclear targeting and gene delivery. We demonstrated with intracellular tracking assays that the endosomal escape unit postponed early degradation of the cargo by preventing endosomal degradation. A nuclear localization signal increased the transfection efficiency as a result of nuclear targeting and in its absence; endosomal escape prolonged the degradation of the cargo.
2. Experimental Section
2.1. Construction and Transformation of Plasmids
The spider silk repeating unit (SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT) was selected from the consensus repeat of the dragline silk spidroin (MaSp-I) sequence from the spider Nephila clavipes. Six contiguous copies of this repeated sequence (silk 6mer) were previously inserted into the pET-30-a plasmid, [29] which contains a linker with SpeI restriction site. pET30-a was linearized with SpeI restriction enzyme to obtain NheI and SpeI flanking ends at C- and N-termini, respectively. Calf intestine alkaline phosphatase was used to dephosphorylate the 5′ end of the linear plasmid. DNA sequences were designed to encode 6XT [(K30)(GLFHAIAHFIHGGWHGLIHGWYG)(RKKRRQRRR)] and 6XK [(K15)(GLFHAIAHFIHGGWHGLIHGWYG)(K15)]. The sequences were purchased as synthetic genes, which were cloned into pUC57 from GenScript (Piscataway, NJ). Ends were designed to be flanked by SpeI and NheI restriction sites, respectively. pUC57 derivatives were digested by SpeI and NheI to liberate the sequences, isolated with gel electrophoresis and purified with Gel Extraction Kits (QIAGEN, Gaithersburg, MD). Purified sequences were ligated with alkaline phosphatase-treated pET30-a-6mer plasmid with T4 DNA ligase for 16 hours at 10°C. The ligation mixture was used to transform chemically competent DH5α E. coli cells (Invitrogen, Carlsbad, CA). Transformed plasmids were confirmed by dideoxy sequencing with both forward and reverse T7 promoter and terminator sequences (Tufts Core Facility, Boston, MA).
2.2. Protein Expression and Purification
6XT and 6XK containing pET30-a plasmids were used to transform chemically competent BLR E. coli strain (EMD Millipore, Darmstadt, Germany). The recombinant strains were grown in Luria-Bertani medium in a shaking incubator at 250 rpm and 37°C. Cells were induced with 1 mM isopropyl-β-D- thiogalactopyranoside when the optical density was between 0.8–1.0 at 600 nm. At 4 hours after induction, cells were collected by centrifugation at 8,000 rpm for 20 minutes at 4°C. Harvested pellets were lysed with denaturing phosphate lysis buffer overnight (100 mM NaH2PO4, 10 mM Trisbase, 8M urea, pH 8.0). The supernatant was collected by centrifugation at 8,700 rpm for 10 minutes at 10°C and loaded onto Ni chelating columns packed with Ni-NTA agarose (Novex, Grand Island, NY) at pH 8.0. The column was washed and eluted with phosphate buffers at pH 6.3, 5.9 and 4.3, respectively. The elution at pH 4.3 was dialyzed (MWCO 3.4kDa) against deionized water for 3 days and lyophilized. The purity of the proteins was monitored by SDS-PAGE using 12% NuPage Bis-Tris gels (Invitrogen, Carlsbad, CA). The molecular weight determinations were confirmed by matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry.
2.3. Preparation of polyplexes
pDNA encoding gLuc luciferase (pCMV-GLuc 5764 bp) was amplified in chemically competent DH5α E. coli. Purification was performed by QIAGEN mini plasmid kit and DNA concentration was obtained with NanoDrop 2000c (Thermo Fisher Scientific, Waltham, MA). Polyplex preparations were prepared by mixing the recombinant proteins with gLuc plasmid with various amine/phosphate (N/P) ratios in deionized water with a final volume of 50 μL. N/P ratio was calculated from the moles of phosphate groups in the gLuc pDNA. The total moles of phosphate groups found in 100ng of gluc pDNA (5,764 bp) was calculated. To balance the charges for phosphate and free amine groups, the molarity of phosphate was tripled. Amount of protein needed was calculated by dividing the amine molarity by total free amine groups and adjusted for previously determined N/P ratios. When changing the N/P ratio, the pDNA amount was kept constant and protein amount was changed accordingly. The mixture of the polplexes was incubated overnight at room temperature.
2.4. Characterization of the polyplexes
Electrophoretic mobility shift assay was performed by loading the polyplex samples with different N/P ratios onto Tris-Acetate-EDTA (TAE) agarose gels (0.8% by mass, 1%EtBr by volume) for 25 minutes. Gels were analyzed under UV. Prior to zeta potential analysis, samples were diluted to a final concentration of 1 mL with deionized water and DMEM, respectively. Zeta potential of the samples was obtained using a Zeta NanoSizer (Nano ZS90, Malvern Instruments, UK), averaging 3 consecutive measurements at room temperature and at 37°C. Scanning Electron Microscopy (SEM) images were taken with Zeiss Ultra Field Emission Scanning Electron Microscope (Thornwood, NY) Both concentrated (1 mg/mL) and diluted samples (0.1 mg/mL) that were previously prepared in deionized water and then vacuum dried were processed. Images were taken in high vacuum with 4 keV energy on carbon-coated surfaces after 5 nm thick of Au coating with Sputter Coater (Cressington HR208). Dynamic Light Scattering was used to detect the particle size with NanoZS90 (Malvern Instruments, UK) equipped with 633nm He-Ne laser at 25 °C with a 90° scattering angle. Polyplexes were prepared in deionized water with previously stated N/P ratios and measurements were taken with 50 μL samples at pH 7.4 and 5.5. Endosomal escape ability was characterized by pH response by simple acid/base titration. Polymer samples with 1 mg/mL concentration in PBS (pH 7.4) were prepared from polyethyleneimine (PEI, 25kDa, branched, Sigma Aldrich, Haverhill, MA), 6XK and 6XT. pH was measured by adding 5 μL increments of 3mM HCl solution. All recordings were processed into a pH response curve.
2.5. Intracellular Tracking Assay
Previously amplified gLuc pDNA was labeled with fluorescein using Label IT Nucleic Acid Labeling Kits (Mirus, Madison, WI). Polyplexes were formed according to the previously described techniques with labeled pDNA. COS-7 cells (ATCC, Manassas, VA) derived from the kidney of African Green Monkey, Cercopithecus aethiops, were seeded onto 96 well glass bottom plates with 5,000 cells/well at the time of transfection. Fresh media was added prior to the transfection and polyplexes were added with maximum DNA amount of 1 ng/μL. Cells were fixed after different time intervals (4 and 10 hours). Cell fixation was done according to standard protocols. Briefly, cells were washed with PBS, 10% formalin and %0.05 Triton, respectively. Then 10% Blocking Solution was used for washing prior to Primary Antibody labeling. LAMP-1 (H4A3) 1ry antibody (Abcam, Cambridge, MA) was diluted by blocking solution (1:200) and added to wells for a 4 hour incubation. After excess washing with PBS, 2ry antibody (Goat Polyclonal Secondary Antibody to Mouse IgG2b- heavy chain FITC, Abcam, Cambridge, MA) solution diluted with blocking solution (1:50, v:v) was added onto wells for 1 hour incubation. Cells were washed with excess PBS prior to the addition of 4′, 6-diamino-2-phenylindole (DAPI, Invitrogen, Carlsbad, CA) solution (1:1000, v:v) and incubated for 10 minutes. Intracellular distributions of the polyplexes and labeled endosomes, along with the nucleus were observed by confocal microscopy. Confocal images (1024×1024 pixels, 12 bit depth) were acquired by PMTs from fixed COS-7 cells, stained for 1ry and 2ry antibodies as indicated above and DAPI, using a commercial Laser Scanning Spectral Leica TCS SP2 confocal microscope (Wetzlar, Germany) equipped with a Ti:sapphire laser (Spectra Physics, Mountain View, CA). 96 wells glass bottom plates were imaged using a 63x/1.2 NA water immersion objective.
2.6. Cell culture, Transfection and Viability
COS-7 cells were grown to 80% confluence using media consisting of Dulbecco’s Modified Eagle Medium (DMEM), 10% FBS, 5% Glutamine. Cells were trypsinized by 0.25% Trypsin (Invitrogen, Carlsbad, CA) and replated on 96-well plates with a density of 10,000 cells/cm2 at the time of transfection after 24 hours of incubation at 37°C. Before transfection, 200 μL of fresh media was added to each well. Polyplexes with varying N/P ratios were added into each well with a pDNA amount of 100ng/well. Lipofectamine LTX (Invitrogen, Carlsbad, CA), branched polyethyleneimine (MW~25kDa, Sigma Aldrich, Haverhill, MA), recombinant silk 6mer and naked pDNA were used as controls and prepared with the same varying N/P ratios. Two sets of incubation times were chosen for the transfections. After 15 and 48 hours, the supernatant was collected and kept at 4°C for luciferase expression analysis with a BioLux Gaussia Luciferase Assay Kit (New England Biolabs, Ipswich, MA). Briefly 50 μL of the supernatant was mixed with 20 μL of GLuc assay solution and monitored after 5–15 seconds incubation at room temperature in the dark. Luminescence activity was monitored by microplate reader (SpectraMax Paradigm Multi-mode Microplate Detection Platform, Sunnyvale, CA). Expression levels were calculated relative to Lipofectamine LTX levels, which were in RLU/mg units. Cell toxicity was monitored by a standard 3-(4,5-dimethylthiazol-2-yl) 2,5-diphenyltetrazolium bromide (MTT) assay (Vybrant MTT Cell Proliferation Assay, Invitrogen, Carlsbad, CA) according to standard protocols. Briefly, COS-7 cells were seeded into 96 well plates with a density of 7,000 cells/well at the time of transfection. Polyplexes with varying N/P ratios were added into each well along with Lipofectamine LTX, PEI, 6mer and naked pDNA as controls. After 15 and 24 hours of incubation at 37°C, pre-mixed MTT solution was added onto each well for 4 hours post-incubation. MTT was solubilized with dimethyl sulfoxide (DMSO) and characterized by obtaining absorbance values.
3. Results and Discussion
The recombinant silk 6mer protein modified with poly-lysine, an endosomal escape unit and the HIV derived TAT sequence was successfully designed, synthesized and purified. Recombinant spider silk with 6 repeating units was synthesized and purified to be used as control in determination of N/P ratio and cytotoxicity.
The yield of purified recombinant silk proteins was around 5 mg/L. Both 6XT and 6XK gave major bands around 27 kDa and 26 kDa, respectively, on SDS-PAGE gels that were higher than the actual molecular weights due to the hydrophobic nature of silk. [28] The molecular weight obtained by MALDI-TOF was 22,502.62 Da for 6XK and 24,125.61 Da for 6XT, which correlated with theoretical calculations. Samples were lyophilized and stored at 4°C for polyplex formation and further characterization. Lyophilized protein samples (6mer, 6XK, 6XT) and PEI (branched, 25 kDa), lipofectamine LTX as controls were weighed and solubilized in 1 mL of DI water. Total pDNA was held constant at 1 ng/μL when calculating the N/P ratios, thus the protein amount was changed accordingly. Polyplexes with varying N/P ratios from 0.02 to 2 were used to analyze the electrostatic interactions by electrophoretic mobility shift assay. Free DNA migrated towards the anode as expected whereas polyplexes with the N/P ratio of 2 migrated towards the cathode. As the amount of protein was increased, migration towards the cathode also increased as seen in Figure 1. This result indicated that using the maximum electrostatic interactions possible, condensing the pDNA within the protein, formed polyplexes. Migration towards the cathode for the polyplexes with N/P=2 suggested that excess positively charge protein was present in a non-condensed form.
Figure 1. Electromobility assay of polyplexes and corresponding zetapotential analysis.

a) Agarose gel showing the migration of the polyplexes towards the cathode. b) Zeta potential of the polyplexes before and after complex formation to verify the polyplex formation with N/P ratio of 1.
Silk protein consisting of six repeating units was negatively charged as reported previously for dragline silk MaSp-I. [28] Zeta potential measurements (Figure 1) indicated that the 6mer was negative and modified 6XT and 6XK proteins were positive, as expected, due to the added lysine and arginine amino acids. The addition of lysine units brings the zeta potential of the silk proteins from negative to positive, indicating that lysine residues may assemble on the surface of the particles due to ionic and hydrophilic interactions in solution. Zeta potential of the polyplexes with N/P =1 dropped below 5 mV, confirming complex formation between pDNA and the cationic protein. Polyplexes with the N/P ratio above 1 gave increasing zeta potential values due to excess cationic charge (data not shown).
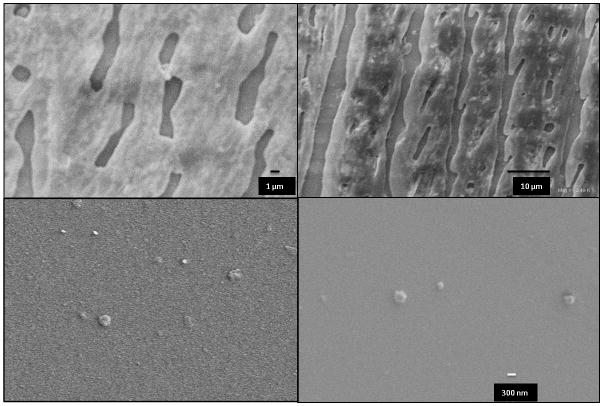
SEM images showed polyplex formation upon addition of pDNA based on the change in morphology of the materials (Figure 2). Before complexing the pDNA, naked 6XK and 6XT protein showed film-like structures resembling the previously stated 6mer film-like morphology whereas the polyplexes resulted in the formation of globular structures when N/P ratio of 1 was used. The sizes of the features were between 300–350 nm for diluted samples. As the concentration of the sample increased, particles tend to show aggregates (data not shown). The sizes of the polyplexes were measured with DLS and 6XK and 6XT polyplexes gave lower size readings, 133 nm and 116 nm, respectively (Table 1). This size is suitable for clathrin mediated endocytosis where a particle size below 500 nm needs to be sustained. [34–35] Differences in size measurements between SEM images and DLS measurements were attributed to the dried state of the polyplexes for SEM measurements vs. hydrated particles in solution for DLS. To test the endosomal escape ability of the polyplexes, samples prepared with deionized water with pH 7.40 were used and analyzed with DLS. After dilute HCl addition to the samples, pH was lowered to 5.50 and analyzed with DLS. Size comparison of the two samples with different pH values revealed the inverse proportionality of size and pH change as expected. Polyplex size for 6XK and 6XT was raised to 170 nm and 144 nm from 133 nm and 116 nm, respectively. Proton sponge ability of the polyplexes was verified by the increase in size acidic medium. Upon lowering the pH from 7.4 to 5.5, mimicking the endosomal conditions, polyplex size was increased due to swelling of the polyplexes caused by protonation of the histidine residues, facilitating the endosomal membrane rupture and endosomal escape.
Figure 2. Scaning Electron Microscopy analysis for size and conformation decetion.
Left and right columns belong to 6XK and 6XT, respectively. Top row of SEM images taken before the polyplex formation showing film-like structures. Protein concentration was 1 mg/mL. Bottom row shows polyplex formation upon addition of pDNA with a ratio of N/P 1. Protein concentration was 0.1 mg/mL. Scale bar is 300 nm.
Table 1.
DLS measurements for recombinant proteins at pH 5.5 and pH 7.4 Measurements were taken at RT
| pH=7 | w/o DNA(nm) | polyplex form (N/P=1) (nm) |
|---|---|---|
| 6XK | 70 ± 2 | 133 ± 6 |
| 6XT | 60 ± 1 | 116 ± 4 |
| pH=5.5 | ||
| 6XK | - | 170 ± 6 |
| 6XT | - | 144 ± 5 |
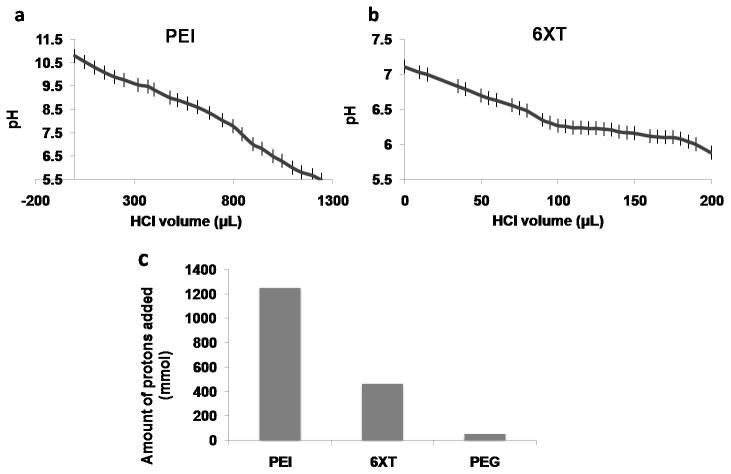
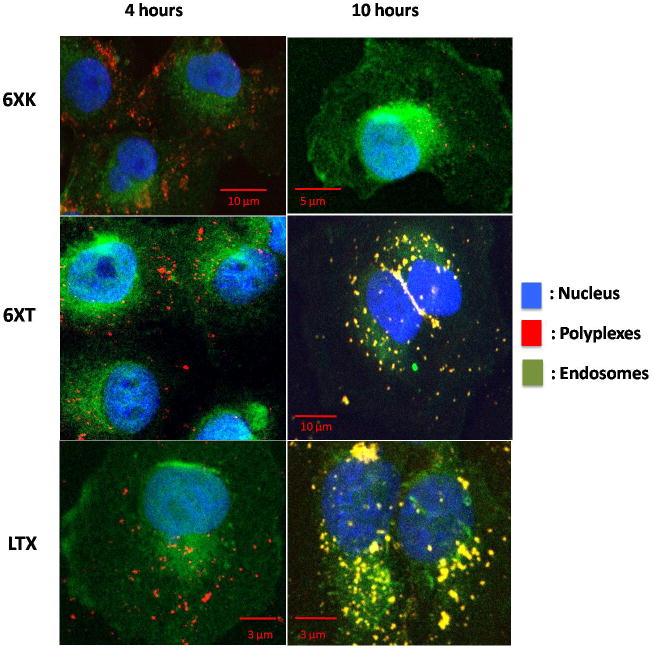
Buffering ability of the polyplexes were tested with pH response curves and showed the slow decrease in pH in the case of endosomal escape units containing constructs. Polyethyleneimine is known to show high buffering capacity, therefore it is used as a control. As the pH is lowered, PEI gave a plateau around pH 8.59 – 8.03 (Figure 3-b). The gold standard PEI has a high buffering capacity above neutral pH, due to the abundance of secondary amines, which protonates the polymer rendering a positive charge before entering the endosomes. [37] The 6XK and 6XT silk constructs showed a plateau-like phase between pH 6.27 – 6.08. This response was attributed to the proton sponge ability of histidines, which is prominent at pH 6.15, which is the pKa value of imidazole ring on the amino acid (Figure 3-a). Amount of protons added during titration were calculated by the change in pH values and compared with Polyethylene glycol, PEI and 6XT (Figure 3-c). Unlike PEI, having a buffering capacity below pH 7 and above endosomal pH 5.5 makes the proposed recombinant biopolymers excellent endosomal escape candidates. The amount of proton added into the titration media containing the polymer in question is a proof of the buffering ability of the polymer. The comparison of quantity of the added protons into the media shows that recombinant polymer 6XT has a buffering capacity lower than PEI and higher than Polyethylene glycol (PEG). Buffering capacity as high as PEI may cause toxicity and difficulty in releasing the pDNA when upon transfection. [38] Recombinant polymers having a buffering capacity lower than PEI, still proves the endosomal escape ability by acting as a proton sponge. Intracellular tracking showed that the particles were internalized and able to target the nucleus by the TAT modification of the recombinant silk proteins. Endosomal and nuclear tracking were carried out by seeding COS-7 cells onto glass bottom 96 well plates. After transfection of the cells with the gene carrier, immunofluorescence labeling of the samples were performed. Confocal images were merged and adjusted by using ImageJ. (US, NIH, Maryland) Nuclear localization ability was verified and compared with Lipofectamine LTX as it is widely used for gene delivery. A comparison between the 6XK and 6XT polyplexes after 4 hours of transfection showed enhanced localization of the latter around the nucleus and the polyplexes were still in the cytoplasm and intact, (Figure 4), whereas free DNA signal in control cells (with only DNA added) was lost (data not shown). The endosomal escape ability of the polyplexes is thought to play a major role in preventing early degradation of the polyplexes. [22– 23] It should also be noted that 6XT transfected cells showed nuclear localization even after 10 hours as well as the lipofectamine LTX transfected cells.
Figure 3. Proton Buffer Analysis.
pH response curve for PEI (a) and 6XT (b) by acid-base titration. Buffering capacity of the recombinant polymer was prominent between pH 6.27 and 6.08. Amount of proton required to change the pH of the media was calculated and compared for PEI, PEG and recombinant polymer (c).
Figure 4. Intracellular Tracking Assay.
Fate of polyplexes traced for 4 and 10 hours post transfection. Comparison between lipofectamine polyplexes and nuclear targeting polyplexes (6XT) shows similarity and separates from 6XK polyplexes, which contained only endosomal escape functions. 6XK are the polylpexes without nuclear targeting domain, 6XT includes the nuclear targeting domain (TAT) and LTX is commercially available from Invitrogen (Carlsbad, CA).
Transfection of COS-7 cells with luciferase expression plasmid was performed in 96 well plates. The pDNA content in each well was kept constant and the N/P ratio was changed by adjusting the protein ratio. Luciferase expression levels were detected as RLU units and then converted to percentages with comparison to lipofectamine transfection.(Figure 5) Degradation of the polyplexes simultaneously released the pDNA which was reflected in the transfection efficiency values. The bioengineered silk proteins showed similar transfection values compared to LTX in short term incubations. After 15 hours of transfection, with the release of luciferase coding pDNA, 6XT showed up to 85% of the transfection efficiency of lipofectamine (5.0 e^4 RLU). Nuclear localization ability and endosomal escape units improved nuclear transfection by delaying the degradation of the polyplex and facilitating the nucleus targeting. Silk-based carriers had lower transfection values after 48 hours of incubation compared to the lipophilic carrier LTX due to the biodegradability of the recombinant silk. Loosening of the polyplex structure due to hydrolysis and subsequent degradation of the polyplexes released the pDNA into the cytosol near the nucleus [40]. The biodegradability of the silk allows the particle to degrade without causing accumulation in the cells, thereby lowering cytotoxicity.
Figure 5. Transfection Efficiency Assay.
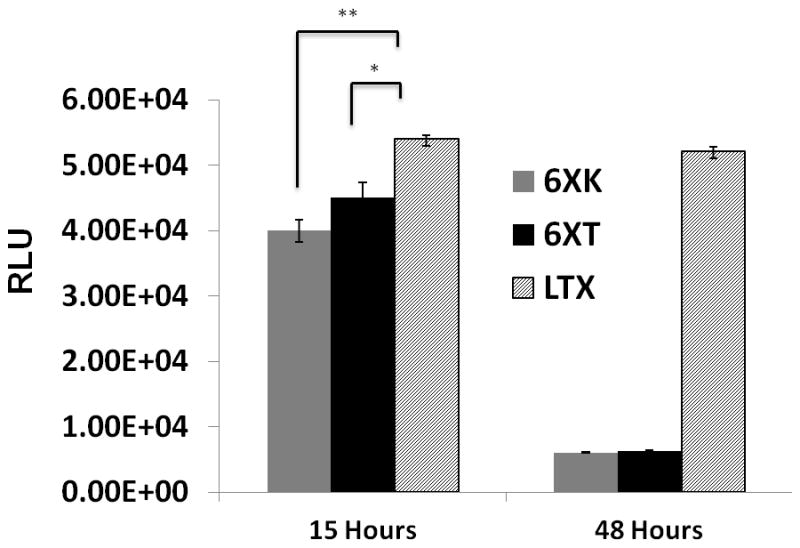
Luciferase expression levels of recombinant 6XK (without nuclear localization unit), 6XT (with nuclear localization unit) and lipofectamine (LTX) polyplexes were compared. Data collected after 15 and 48 hours post transfection (*= p<0.05, **= p<0.01).
Cytotoxicity assays showed that the bioengineered silk proteins were biocompatible and non-cytotoxic compared with controls. The 6mer, 6XT, 6XK showed no cytotoxicity and compared well with the positive control of naked pDNA and cells cultured without additives. (Figure 6) Accepted as the ‘golden standard’ in gene delivery due to high buffering capacity, PEI showed cytotoxic behavior after 24 hours of transfection. Lipophilic LTX also had low cell proliferation after 24 hours. Lipofectamine LTX showed 40% cytotoxicity after 24 hours of cell culture and PEI exhibited 90% cytotoxicity. The addition of 30 poly-lysine residues based on the previous transfection efficiency studies was enough to generate an effective and non-cytotoxic cationic carrier. Biosynthetic and biodegradable silk-based proteins showed almost no cytotoxicity, with comparable values to free DNA and non-modified recombinant silk 6mer used as positive controls.
Figure 6. Cell Metabolic Activity Assay.
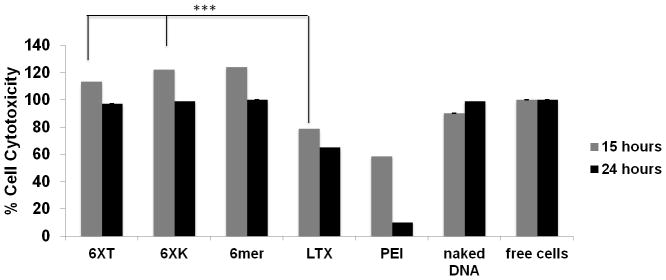
MTT assay for the effect of polyplexes on cell metabolic activity and cytotoxicity. Polyplexes with N/P ratio of 1 were used. Positive controls were cell cultures without additives and cultures with naked DNA. Comparison with lipofectamine (LTX) and polyethyleneimine (PEI) were performed. Cytotoxicity was measured after 15 and 24 hours of post-transfection (*= p<0.05, **= p<0.01, ***= p<0.005)
4. Conclusions
Engineering biopolymers with specific targeting and endosomal escape agents elevated transfection levels and also produced biocompatible, biodegradable, non-toxic and monodisperse carriers. Overall, the non-viral bioengineered polymers with comparable transfection efficiency to lipofectamine LTX, were achieved, yet with higher biocompatibility than lipofectamine LTX and the ‘gold standard’ PEI.
Biosynthetic approaches to the design of gene delivery vectors offers tremendous control over the chemistry, biocompatibity, biodegradability and non-cytotoxic features for gene delivery. Commercially available gene transfection agents show low viability due to their cationic, non-compatible nature. With the present work exploiting virus-derived components, successful non-viral gene carriers were formed with the ability to prevent endosomal degradation and perform nuclear localization. The proposed design of recombinant biopolymers has the virtue of overcoming endosomal degradation and successfully targeting the nucleus without causing cytotoxicity with facilitating the released pDNA for higher transfection results.
Supplementary Material
Acknowledgments
Authors would like to thank Dr. Zhang Ke for providing the use of the NanoZS (Malvern Instruments, UK) and Tessa DesRochers for the cell culture support. This project has been funded by the NIH (EB002520).
Footnotes
Supporting Information is available from the Wiley Online Library or from the author
References
- 1.Yankaskas J, Marshall B, Sufian B, Simon R, Rodman D. Cystic fibrosis adult care: consensus conference report. Chest. 2004;125:1–39. doi: 10.1378/chest.125.1_suppl.1s. [DOI] [PubMed] [Google Scholar]
- 2.Cavazzana-Galvo M, Hacein-Bey S, de Sainte Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanove JL, Bousso P, Deist FL, Fischer A. Gene Therapy of Human Severe Combined Immunodeficiency (SCID)-X1 Disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 3.Kaspar B, Llado J, Sherkat N, Rothstein J, Gage F. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- 4.Miller AD. Retroviral Vectors. Curr Top Microbiol Immunol. 1992;158:1–24. doi: 10.1007/978-3-642-75608-5_1. [DOI] [PubMed] [Google Scholar]
- 5.Luo D, Saltzman M. Synthetic DNA delivery Systems. Nature Biotechnology. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 6.Mintzer M, Simanek E. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 7.Nijdome T, Huang L. Gene Therapy Progress and Prospects: Nonviral Vectors. Gene Therapy. 2002;9:1647–52. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 8.Taira K, Kataoka T, Nijdome T. Non-viral gene therapy: gene design and delivery. New York: Springer; 2005. [Google Scholar]
- 9.Numata K, Kaplan DL. Silk-based delivery systems of bioactive molecules. Adv Drug Delivery Rev. 2010;62:1497–1508. doi: 10.1016/j.addr.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman G, Diaz F, Jakuba C, Calabro T, Horan R, Chen J, Ru H, Richmond J, Kaplan DL. Silk-based Biomaterials. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 11.Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure-function-property-design interplay in biopolymers: Spider Silk. Acta Biomaterialia; 2013 doi: 10.1016/j.actbio.2013.08.020. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tien L, Wu F, Tang-Schomer M, Yoon E, Omenetto F, Kaplan DL. Silk as a Multifunctional Biomaterial Substrate for Reduced Glial Scarring around Brain-Penetrating Electrodes. Adv Funct Mater. 2013:3185–3193. [Google Scholar]
- 13.Yan S, Zhang Q, Wang J, Liu Y, Lu S, Li M. Silk fibroin/chondroitin sulfate/hyaluronic acid ternary scaffolds for dermal tissue reconstruction. Acta Biomaterialia. 2013:6771–6782. doi: 10.1016/j.actbio.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Pritchard E, Hu X, Valentin T, Panilaitis B, Omenetto F, Kaplan DL. Stabilization of vaccines and antibiotics in silk and eliminating the cold chain. PNAS. 2012:11981–11986. doi: 10.1073/pnas.1206210109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Mandal B, Grinberg A, Gil E, Panilaitis B, Kaplan DL. High-strength silk protein scaffolds for bone repair. PNAS. 2012:7699–7704. doi: 10.1073/pnas.1119474109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seib F, Kaplan DL. Doxorubicin-loaded silk films: Drug-silk interactions and in vivo performance in human orthotopic breast cancer. Biomaterials. 2012:8442–8450. doi: 10.1016/j.biomaterials.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das SK, Dey T, Kundu SC. Fabrication of Sericin Nanoparticles for Controlled Gene Delivery. RSC Adv. 2014:2137. [Google Scholar]
- 18.Numata K, Mieszawska-Czajkowska A, Kvenvold L, Kaplan DL. Silk-based Nanocomplexes with Tumor-Homing Peptides for Tumor-Specific Gene Delivery. Macromol Biosci. 2012:75–82. doi: 10.1002/mabi.201100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Numata K, Kaplan DL. Silk-based Gene Carriers with Cell-Membrane Destabilizing Peptides. Biomacromolecules. 2011:3189–3195. doi: 10.1021/bm101055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellman I, Fuchs R, Helenius A. Acidification of the Endocytic and Exocytic Pathways. Annual Reviews Biochemistry. 1986;55:663–70. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 21.Varkouhi A, Scholte M, Storm G, Haisma H. Endosomal Escape Pathway for Delivery of Biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Midoux P, Kichler A, Boutin V, Maurizot J, Monsigny M. Membrane permeabilization and efficient gene transfer by a peptide containing several histidines. Bioconjugate Chemistry. 1998;9:260–67. doi: 10.1021/bc9701611. [DOI] [PubMed] [Google Scholar]
- 23.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjugate Chemistry. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 24.Godbey W, Wu K, Mikos A. Poly(ethyleneimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22:471–80. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 25.Fischer D, Bieber T, Li Y, Elsasser H, Kissel T. A novel nonviral vector for DNA delivery based on low molecular weight branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16:1273–9. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 26.Chang K, Higuchi Y, Kawakami S, Yamashita F, Hashida M. Development of Lysine-Histidine Dendron Modified Chitosan for Improving Transfection Efficiency in HEK293 Cells. Journal of Controlled Release. 2011;156:195–202. doi: 10.1016/j.jconrel.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Zanuer W, Ogris M, Wagner E. Polylysine Based Transfection Systems Utilizing Receptor Mediated Delivery. Adv Drug Del Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 28.Numata K, Hamasaki J, Subramanian B, Kaplan DL. Gene delivery mediated by recombinant silk proteins containing cationic and cell binding motifs. J Control Release. 2010:136–143. doi: 10.1016/j.jconrel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Numata K, Subramanian B, Heather A, Kaplan DL. Bioengineered silk protein-based gene delivery systems. Biomaterials. 2009;30:5775–5784. doi: 10.1016/j.biomaterials.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Nelson CE, Evans B, Duvall C. Delivery of Intracellular-acting Biologicals in Pro-Apoptotic Therapies. Current Pharmeceutical Design. 2011;17:293–319. doi: 10.2174/138161211795049642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ming Yu, Hsu C, Uludag H. Effects of size and topology of DNA molecules on Intracellular Delivery with non-viral Gene Carriers. BMC Biotechnology. 2008;8:23. doi: 10.1186/1472-6750-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langel U. Handbook of Cell-Penetrating Peptides. Boca Raton: Taylor & Francis; 2007. [Google Scholar]
- 33.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R. Structural and Functional Characteristization of Human Immunodefficiency Virus TAT Protein. J Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siomi H, Shida H, Maki M, Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Gersdroff K, Sanders NN, Vandenbroucke R, de Smedt SC, Wagner E, Ogris M. The Internalization Route Resulting in Successful Gene Expression Depends on Both Cell Line and Polyethyleneimine Polyplex Type. Mol Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size Dependent Internalization of Particles via the Pathways of Clathrin and Caveolae Mediated Endocytosis. Biochem J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neu M, Fischer D, Kissel T. Recent Advances in Rational Gene Transfer Vector Design Based on Poly(ethylene imine) and its Derivatives. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 38.Forrest ML, Gabrielson N, Pack DW. Reduction of Polyhethyleneimine Buffer Capacity Enhances In-vitro Gene Delivery Activity. Molecular Therapy. 2004;9:138. [Google Scholar]
- 39.Canine B, Hatefi A. Development of recombinant cationic polymers for gene therapy research. Adv Drug Deliv Rev. 2010;62:1524–1529. doi: 10.1016/j.addr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grigsby CL, Leong KW. Balancing protection and release of DANN: tools to adress a bottleneck of non-viral gene delivery. J R Soc Interface. 2009 doi: 10.1098/rsif.2009.0260. Advance Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.