Abstract
Cell fate decisions are fundamental to the development of multicellular organisms. In mammals the first cell fate decision involves segregation of the pluripotent inner cell mass and the trophectoderm, a process regulated by cell polarity proteins, HIPPO signaling and lineage-specific transcription factors such as CDX2. However, the regulatory mechanisms that operate upstream to specify the trophectoderm lineage have not been established. Here we report that transcription factor AP-2γ (TFAP2C) functions as a novel upstream regulator of Cdx2 expression and position-dependent HIPPO signaling in mice. Loss- and gain-of-function studies and promoter analysis revealed that TFAP2C binding to an intronic enhancer is required for activation of Cdx2 expression during early development. During the 8-cell to morula transition TFAP2C potentiates cell polarity to suppress HIPPO signaling in the outside blastomeres. TFAP2C depletion triggered downregulation of PARD6B, loss of apical cell polarity, disorganization of F-actin, and activation of HIPPO signaling in the outside blastomeres. Rescue experiments using Pard6b mRNA restored cell polarity but only partially corrected position-dependent HIPPO signaling, suggesting that TFAP2C negatively regulates HIPPO signaling via multiple pathways. Several genes involved in regulation of the actin cytoskeleton (including Rock1, Rock2) were downregulated in TFAP2C-depleted embryos. Inhibition of ROCK1 and ROCK2 activity during the 8-cell to morula transition phenocopied TFAP2C knockdown, triggering a loss of position-dependent HIPPO signaling and decrease in Cdx2 expression. Altogether, these results demonstrate that TFAP2C facilitates trophectoderm lineage specification by functioning as a key regulator of Cdx2 transcription, cell polarity and position-dependent HIPPO signaling.
Keywords: TFAP2C, Cell polarity, HIPPO signaling, CDX2, Mouse
Summary: Mouse trophectoderm specification is regulated by AP-2γ/TFAP2C, which activates Cdx2 expression, controls actin dynamics and suppresses HIPPO signaling via multiple pathways, including the potentiation of cell polarity.
INTRODUCTION
The development of multicellular organisms is dependent on proper cell fate choices that result in different cell types. In mammals the divergence of the inner cell mass (ICM) and trophectoderm denotes the first cell fate choice during preimplantation embryo development. This process is highly regulated, being controlled by a combination of cell-cell interactions, cell polarity proteins, position-dependent HIPPO signaling and lineage-specific transcription factors (Wennekamp et al., 2013). For example, after fertilization the totipotent zygote undergoes a series of symmetrical cell divisions between the 1- and 8-cell stages. At the 8-cell stage the adherens junction proteins E-cadherin and β-catenin assemble on cell membranes, facilitating embryo compaction (Ducibella et al., 1977; Riethmacher et al., 1995; De Vries et al., 2004). Concomitant with compaction, core cell polarity complexes consisting of partitioning-defective protein 6B (PARD6B), PARD3 and atypical PKC (e.g. PKCζ or PKCλ/ι) assemble on the apical membranes (i.e. the outer region) of each compacted blastomere (Plusa et al., 2005; Vinot et al., 2005; Dard et al., 2009). Next, the compacted 8-cell embryo undergoes two rounds of symmetrical and asymmetrical cell divisions that generate two populations of cells: polar outside cells and apolar inside cells (Tarkowski and Wroblewska, 1967; Ziomek and Johnson, 1980, 1982; Johnson and Ziomek, 1981). These two cell populations will serve as precursors for trophectoderm and ICM, respectively.
Based on the formation of outside and inside cell populations, the activity of the HIPPO signaling pathway becomes position dependent (Nishioka et al., 2009; Hirate et al., 2013). For instance, on the outside of the embryo where there is low cell contact, cell polarity suppresses HIPPO signaling by sequestering angiomotin (AMOT), a tight junction-associated scaffold protein, preventing it from interacting with large tumor suppressor 1/2 (LATS1/2) kinase (Hirate et al., 2013). Consequently, YES-associated protein (YAP) enters the nucleus (Nishioka et al., 2009) and interacts with TEA domain family member 4 (TEAD4), which directly and selectively upregulates Cdx2 transcription in the outside cells (Nishioka et al., 2009). By contrast, on the inside of the embryo where there is high cell contact and an absence of apical cell polarity, AMOT is recruited to adherens junctions where it interacts with neurofibromatosis 2 (NF2) and LATS1/2 to activate the HIPPO pathway (Cockburn et al., 2013; Hirate et al., 2013). Subsequently, LATS1/2 phosphorylates YAP, preventing it from entering the nucleus and interacting with TEAD4. It has also been reported that the nuclear localization of TEAD4 itself may be impaired within the inside cells (Home et al., 2012), and the lack of a functional TEAD4-YAP complex causes downregulation of Cdx2 expression and upregulation of pluripotency genes. Future studies are warranted to delineate alternative mechanisms that may specify early cell lineages.
Despite our current understanding of the mechanisms that underlie position-dependent HIPPO signaling and upregulation of Cdx2 expression in the outside cells, the transcriptional mechanisms that function upstream of cell polarity and HIPPO signaling have not been elucidated. For example, which transcriptional mechanisms are responsible for the initiation of Cdx2 expression and cell polarization in the cleavage stage embryo? At the morula and early blastocyst stages, which transcriptional mechanisms negatively regulate HIPPO signaling in the outside cells to specify the trophectoderm lineage?
Recently, we identified a novel and crucial role for transcription factor AP-2γ (TFAP2C) in the regulation of core processes that underlie blastocyst formation (Choi et al., 2012). We found that TFAP2C is initially widely expressed in the preimplantation embryo, and then, during the morula to blastocyst transition, it becomes enriched in the mural trophectoderm and downregulated in the ICM. Notably, this expression pattern was largely consistent with the role of TFAP2C in tight junction biogenesis and blastocoel formation (Choi et al., 2012), suggesting that TFAP2C is a key trophectoderm-specific transcription factor in the preimplantation embryo. Here we report that TFAP2C plays a much wider regulatory role in early lineage formation. During the 2- to 8-cell stages TFAP2C functions as an upstream initiator of Cdx2 expression and cell polarization. At the 8-cell to morula transition, TFAP2C negatively regulates HIPPO signaling in the outside cells via transcriptional regulation of Pard6b and Rock1/2. Depletion of TFAP2C or inhibition of ROCK kinase activity triggered a loss of position-dependent HIPPO signaling and downregulation of Cdx2 expression. Our findings demonstrate that TFAP2C acts as a master regulator of trophectoderm formation in mouse preimplantation embryos.
RESULTS
TFAP2C is required for the initiation of zygotic Cdx2 expression in preimplantation embryos
We recently established that TFAP2C is a key regulator of genes required for blastocyst formation in mice (Choi et al., 2012). TFAP2C-depleted embryos arrest around the morula stage and fail to establish the trophectoderm epithelium. However, the biological role of TFAP2C in trophectoderm lineage specification was not investigated. A previous study showed that forced expression of Tfap2c in embryonic stem cells (ESCs) could induce a trophectoderm cell fate (Kuckenberg et al., 2010). Thus, we hypothesized that TFAP2C is required for the onset of Cdx2 transcription during preimplantation development. To test this we first evaluated the developmental expression of TFAP2C and CDX2 in oocytes and preimplantation embryos (supplementary material Fig. S1). TFAP2C protein was first detected at the 1-cell stage and was expressed up until the blastocyst stage, when it became enriched in the trophectoderm epithelium. CDX2 protein was first detected at the 8-cell stage and its expression continued up to the blastocyst stage, when it became restricted to the trophectoderm. These analyses revealed that the expression of TFAP2C preceded that of CDX2.
To determine whether TFAP2C regulates Cdx2 transcription during the window of preimplantation development, real-time qPCR analysis was used to evaluate Cdx2 transcripts in TFAP2C knockdown (KD) and control embryos at different developmental stages. Maternally derived and zygotic Tfap2c transcripts were depleted by microinjection of Tfap2c siRNA at the 1-cell stage. Control embryos were microinjected with a non-specific siRNA. Using this approach we were able to significantly downregulate TFAP2C protein as early as the 4-cell stage (supplementary material Fig. S2). Moreover, the specificity and efficacy of the Tfap2c siRNA sequences were previously tested via an elaborate series of control and rescue experiments (Choi et al., 2012). Cdx2 transcripts were reduced by 77%, 87%, 76% at the 4-cell, 8-cell and morula stage, respectively (Fig. 1A). Because CDX2 protein is first detectable around the 8-cell stage, we used immunofluorescence analysis to evaluate the expression of CDX2 at the morula stage. Consistent with the mRNA expression profile, CDX2 protein was substantially downregulated in TFAP2C KD morulae (Fig. 1B). Evaluation of cell nuclei revealed that total cell numbers were similar between control and TFAP2C KD morulae (27.2±3.6 versus 23.6±5.6), ruling out any treatment effects on proliferation that could influence the timing of CDX2 expression. The expression of GATA3, a co-activator of Cdx2 (Home et al., 2009), was also evaluated in TFAP2C KD morulae (supplementary material Fig. S2). This analysis revealed that GATA3 expression was undisturbed in TFAP2C KD embryos, suggesting that activation of Cdx2 expression depends on TFAP2C.
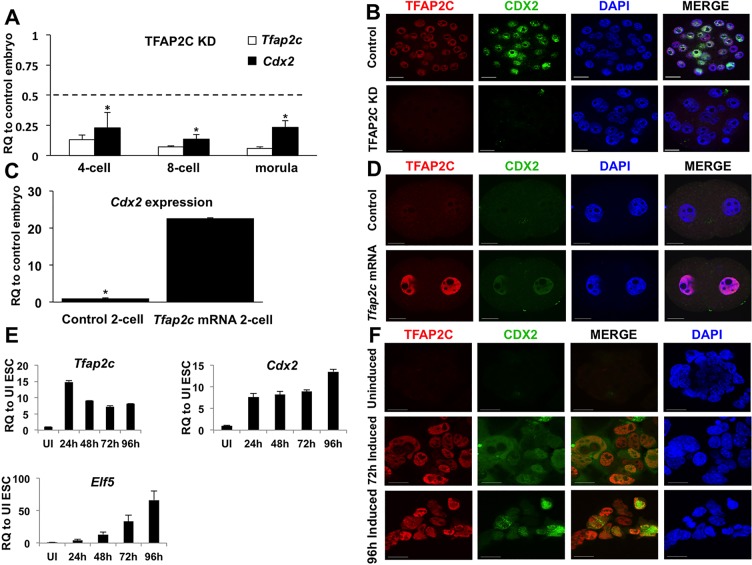
Fig. 1.
TFAP2C regulates the onset of Cdx2 expression in preimplantation mouse embryos. (A) Real-time qPCR analysis of Tfap2c and Cdx2 transcripts in TFAP2C KD and control preimplantation embryos at different developmental stages. Cdx2 transcripts were downregulated at the 4-cell, 8-cell and morula stages. A total of three biological replicates were performed. RQ, relative quantification. The dashed line corresponds to a 50% reduction in specific gene transcripts. (B) Immunofluorescence analysis of TFAP2C and CDX2 in TFAP2C KD and control morulae. A total of three biological replicates were performed using 28 control and 30 TFAP2C KD embryos. All control embryos (28/28) stained positive for CDX2. By contrast, 90% of TFAP2C KD embryos (27/30) exhibited a strong reduction in CDX2 protein. (C) Real-time qPCR analysis of Cdx2 transcripts in 2-cell embryos from zygotes microinjected with 25 ng/μl Tfap2c mRNA or that were uninjected (control). (D) Immunofluorescence analysis of TFAP2C and CDX2 in 2-cell embryos from zygotes microinjected with 25 ng/μl Tfap2c mRNA or that were uninjected (control). All Tfap2c mRNA-injected 2-cell embryos (27/27) exhibited an increase in CDX2 protein. CDX2 protein was not visible in any of the control 2-cell embryos (0/27). (E) Real-time qPCR analysis of TFAP2C target genes in Tfap2c-inducible ESCs at 24, 48, 72 and 96 h following induction. (F) Immunofluorescence analysis of CDX2 in Tfap2c-inducible ESCs at 72 and 96 h following induction. Embryo and ESC nuclei were counterstained with DAPI. UI, uninduced. (A,C,E) Error bars indicate mean±s.e.m. *P<0.05. Scale bars: 20 μm.
To determine whether TFAP2C is both necessary and sufficient for Cdx2 transcription, Tfap2c was overexpressed in fertilized 1-cell embryos and mouse ESCs. Microinjection of Tfap2c mRNA into 1-cell embryos triggered a significant increase in Cdx2 transcripts and protein at the 2-cell stage (Fig. 1C,D). Treatment of Tfap2c mRNA-injected 1-cell embryos with alpha-amanitin, a potent RNA polymerase II inhibitor, blocked transcriptional activation of Cdx2, confirming that increased expression is via zygotic transcription and not from stored maternal Cdx2 transcripts (data not shown). Forced expression of TFAP2C in ESCs via transgene expression triggered a substantial increase in Cdx2 transcripts and protein in a temporal manner (Fig. 1E,F). Moreover, other TFAP2C target genes, such as Elf5, were induced in conjunction with Cdx2. Collectively, these results demonstrate that TFAP2C is required for the activation of Cdx2 expression during early embryonic development and demonstrate that TFAP2C plays an instructive role in trophectoderm specification via regulation of Cdx2.
TFAP2C directly regulates Cdx2 expression via a regulatory element in intron 1
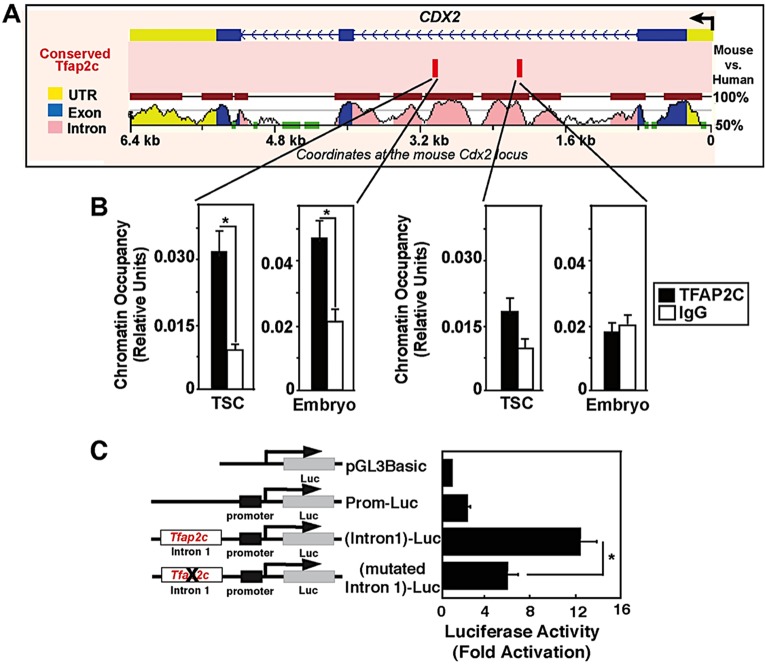
The Cdx2 gene contains several regulatory elements (Wang and Shashikant, 2007; Benahmed et al., 2008) and is directly regulated by several transcription factors including CDX2, GATA3 and TEAD4 (Home et al., 2009; Barros et al., 2011; Rayon et al., 2014). To ascertain whether TFAP2C is a direct regulator of Cdx2 transcription, we performed motif analysis of TFAP2C binding sites in the Cdx2 gene. This analysis revealed that there are two conserved TFAP2C binding motifs located within an enhancer in intron 1 (Fig. 2A). This intronic enhancer contains binding motifs for both GATA3 and TEAD4, which are crucial for trophectoderm lineage specification (Home et al., 2009; Rayon et al., 2014). Chromatin immunoprecipitation (ChIP) analysis in trophoblast stem cells (TSCs) confirmed that TFAP2C occupancy was enriched on the distal motif (Fig. 2B). Furthermore, to confirm that TFAP2C binds to this Cdx2 enhancer in preimplantation embryos, microChIP analysis was carried out in pools of morulae. This analysis showed that TFAP2C occupies the Cdx2 enhancer in preimplantation embryos (Fig. 2B). To test whether this motif is functional, a luciferase reporter was fused to the Cdx2 promoter along with the intronic enhancer containing a wild-type or mutant TFAP2C motif. These constructs were transiently transfected into TSCs and luciferase activity was measured. Disruption of the distal TFAP2C motif caused a >50% reduction in luciferase activity as compared with the wild-type construct (Fig. 2C). Altogether, these results, in combination with the loss-of-function and gain-of-function experiments above, demonstrate that TFAP2C is a direct regulator of Cdx2 transcription during early development.
Fig. 2.
TFAP2C directly regulates Cdx2 transcription. (A) VISTA regulatory sequence alignment of mouse and human Cdx2 loci. The location of two conserved TFAP2C motifs (red bars) within mouse Cdx2 intron 1 is indicated. (B) ChIP analyses confirming occupancy of TFAP2C at the distal conserved motif within intron 1 of the Cdx2 locus in mouse TSCs (mean±s.e., three independent experiments) and in mouse morulae (Embryo; mean±s.e., two independent experiments). (C) Transient transfection analyses with luciferase reporters fused to the mouse Cdx2 promoter and intron 1 region with or without the distal TFAP2C motif. The bar chart shows that Cdx2 promoter activity is positively regulated by the TFAP2C motif in TSCs. This experiment was performed four times. Error bars indicate s.e. *P<0.05.
TFAP2C regulates apical cell polarity via PARD6B
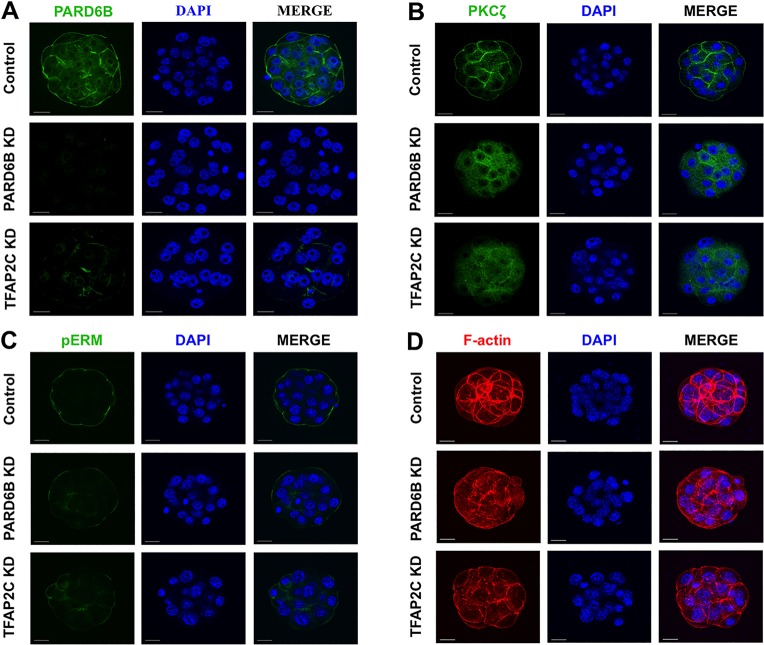
Trophectoderm-specific expression of Cdx2 is mediated by the establishment of apical cell polarity and position-dependent HIPPO signaling during the 8-cell to morula and morula to blastocyst transitions, respectively (Alarcon, 2010; Hirate et al., 2013). We previously showed that Pard6b is a direct target gene of TFAP2C in preimplantation embryos and TS-like cells (Choi et al., 2012). However, the biological relevance of this putative TFAP2C-PARD6B pathway in apical cell polarity was not explored. To address this, the expression and localization of PARD6B and PKCζ were evaluated in TFAP2C KD and PARD6B KD embryos. In control embryos PARD6B and PKCζ were localized to the apical region of the outside cells (Fig. 3A,B). By contrast, in TFAP2C KD and PARD6B KD embryos PARD6B was significantly reduced and PKCζ was mislocalized to the cytoplasm (Fig. 3A,B). To test whether TFAP2C can induce Pard6b expression, Tfap2c was overexpressed in fertilized 1-cell embryos and ESCs (supplementary material Fig. S3). Microinjection of Tfap2c mRNA triggered a significant increase in Pard6b transcripts in 2-cell embryos and ESCs. Tfap2c overexpression in 1-cell embryos caused an increase in PARD6B protein at the 2-cell stage. In ESCs Tfap2c overexpression induced a trophectoderm-like morphology, with PARD6B enriched at the cell membranes.
Fig. 3.
TFAP2C positively regulates apical cell polarity via PARD6B. (A) Immunofluorescence analysis of PARD6B expression and localization in TFAP2C KD and control morulae. PARD6B KD embryos were used as a control. 80% of control embryos (24/30) exhibited strong PARD6B staining and apical localization. By contrast, PARD6B was significantly downregulated in PARD6B KD and TFAP2C KD morulae. The apical localization of PARD6B was lost in 14/20 TFAP2C KD embryos (70%) and 32/32 PARD6B KD embryos (100%). (B) Immunofluorescence analysis of PKCζ sublocalization in PARD6B KD, TFAP2C KD and control morulae. PKCζ was mislocalized to the cytoplasm in PARD6B KD and TFAP2C KD morulae. (C) Immunofluorescence analysis revealed that pERM (microvilli marker) was decreased in PARD6B KD and TFAP2C KD morulae. (D) Immunofluorescence analysis showing that F-actin was disorganized in PARD6B KD and TFAP2C KD morulae. Nuclei were counterstained with DAPI. (A-D) A total of two to three biological replicates were performed using 20-32 embryos per group. Scale bars: 20 μm.
To assess whether other markers of apical polarity are disrupted in TFAP2C KD morulae, we evaluated the localization of phosphorylated forms of ezrin/radixin/moesin (collectively pERM). These proteins are involved in the formation of microvilli of the apical domain (Sato et al., 1992; Louvet et al., 1996). Furthermore, because establishment of the apical domain involves reorganization of the actin cytoskeleton, F-actin localization was evaluated using phalloidin. In control embryos pERM and F-actin were enriched at the apical regions (Fig. 3C,D). Conversely, in TFAP2C KD and PARD6B KD embryos pERM was reduced and F-actin was disorganized (Fig. 3C,D). Altogether, these results demonstrate that TFAP2C is a key regulator of Pard6b expression and is required for the establishment of apical cell polarity.
TFAP2C negatively regulates HIPPO signaling in the outside cells
Because TFAP2C-deficient morulae exhibit defects in apical cell polarity and contain reduced levels of CDX2, we hypothesized that TFAP2C modulates position-dependent HIPPO signaling to upregulate Cdx2 expression in the outside cells. To test this we first evaluated the expression of several HIPPO signaling pathway members in TFAP2C KD and control embryos (supplementary material Fig. S4). This analysis revealed that the expression of Lats1, Lats2 and Yap (Yap1) mRNA was unaffected in TFAP2C KD embryos. Moreover, the expression of Tead4 transcripts and of TEAD4 protein were similar in TFAP2C KD and control embryos (supplementary material Fig. S4). These results imply that TFAP2C does not directly regulate the expression of individual HIPPO signaling genes.
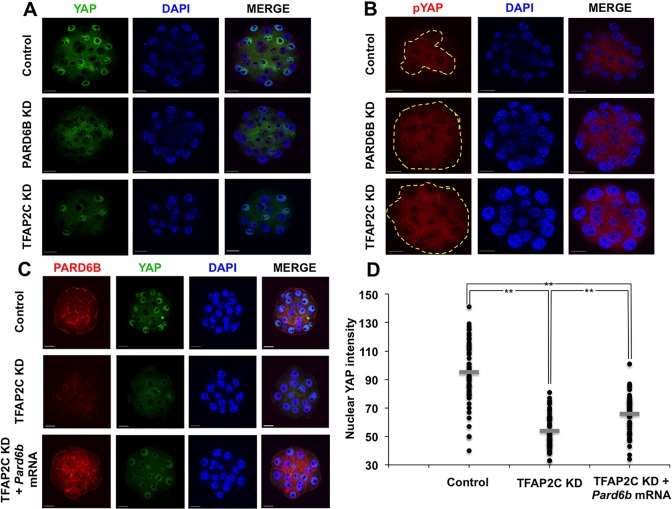
Next we examined whether TFAP2C can regulate the activity of the HIPPO signaling pathway by evaluating the expression and localization of YAP. PARD6B KD embryos were used as a positive control because PARD6B functions downstream of TFAP2C and is required for position-dependent HIPPO signaling (Hirate et al., 2013). Nuclear YAP (nYAP) and cytoplasmic YAP (cYAP) were evaluated using YAP and phospho-YAP (pYAP) antibodies (Hirate et al., 2013; Lorthongpanich et al., 2013). In control embryos YAP was localized in the nuclei of outside cells (Fig. 4A). By contrast, in TFAP2C KD and PARD6B KD embryos there was a significant decrease in nYAP and an increase in cYAP in the outside cells (Fig. 4A,D). Consistent with this, pYAP was present in the cytoplasm of both inside and outside cells (Fig. 4B). Combined, these results demonstrate that position-dependent HIPPO signaling is dysregulated in TFAP2C KD embryos.
Fig. 4.
TFAP2C negatively regulates HIPPO signaling in the outside cells. (A) Immunofluorescence analysis of nuclear and cytoplasmic YAP in PARD6B KD and TFAP2C KD morulae. In control embryos YAP was present in the nuclei of the outside cells. In PARD6B KD and TFAP2C KD embryos, by contrast, YAP was largely absent from the nuclei and was instead present in the cytoplasm. (B) Immunofluorescence analysis of pYAP in TFAP2C KD and control morulae. PARD6B KD embryos were used as a further control. In control embryos, pYAP localized predominantly to the cytoplasm of the inside cells; only 16% of embryos exhibited abnormal inside-outside localization (4/25). By contrast, in PARD6B KD (96%) and TFAP2C KD morulae (86%) pYAP was present in the cytoplasm of both the inside and outside cells (24/25 and 19/22, respectively). The dashed line delineates the region of the embryo containing cytoplasmic pYAP. (C) Rescue of PARD6B expression in TFAP2C KD embryos partially restored nuclear YAP in the outside cells. (D) Quantitative analysis of YAP fluorescence intensity indicated that nuclear YAP was significantly reduced in TFAP2C KD morulae and could be partially restored by rescue of PARD6B. Nuclei were counterstained with DAPI. A total of two to three biological replicates were performed using 22-37 embryos per group. **P<0.01. Scale bars: 20 μm.
Since TFAP2C-dependent regulation of Pard6b is required for apical cell polarity, we hypothesized that TFAP2C negatively regulates HIPPO signaling in the outside cells via the PARD6B-PARD3-PKCζ complex. To test this we rescued Pard6b expression by microinjection of Pard6b mRNA into TFAP2C KD embryos. Injection of Pard6b mRNA rescued PARD6B and restored PKCζ localization to the apical region in TFAP2C KD embryos (Fig. 4C; supplementary material Fig. S5). However, analysis of YAP localization revealed that nYAP was only partially restored in the outside cells (Fig. 4C,D). The levels of nYAP were higher in PARD6B rescued embryos than in TFAP2C KD embryos, yet were still lower than in control embryos. The expression of Cdx2 transcripts and of CDX2 protein were not restored in PARD6B rescued TFAP2C KD embryos (Fig. 5A,B). These results demonstrate that TFAP2C regulates position-dependent HIPPO signaling in part through PARD6B. However, PARD6B rescue alone is not sufficient to restore HIPPO signaling and Cdx2 expression, suggesting that TFAP2C might function via multiple pathways.
Fig. 5.
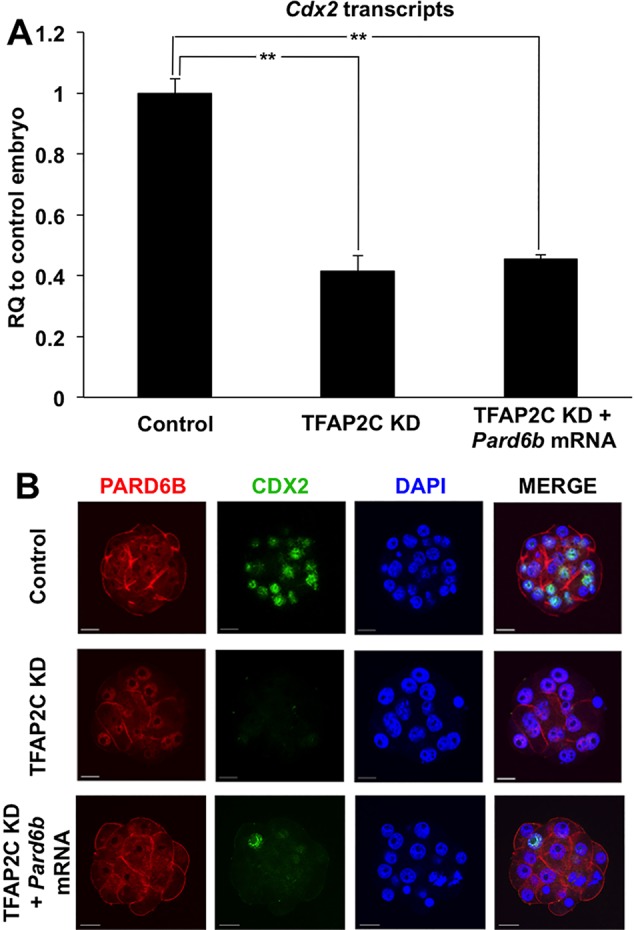
Re-establishment of apical polarity does not rescue Cdx2 expression in TFAP2C KD morulae. (A) Real-time qPCR analysis of Cdx2 transcripts in TFAP2C KD and PARD6B-rescued TFAP2C KD morulae. Cdx2 expression could not be restored by rescuing PARD6B expression in TFAP2C KD morulae. Control group was set to 1. Error bars indicate mean±s.e.m. **P<0.01. (B) Immunofluorescence analysis of CDX2 in TFAP2C KD and PARD6B-rescued TFAP2C KD morulae. CDX2 protein was not rescued by re-expression of PARD6B. Nuclei were counterstained with DAPI. A total of two biological replicates were performed using 24-26 embryos per group. Scale bars: 20 μm.
ROCK acts downstream of TFAP2C to repress HIPPO signaling
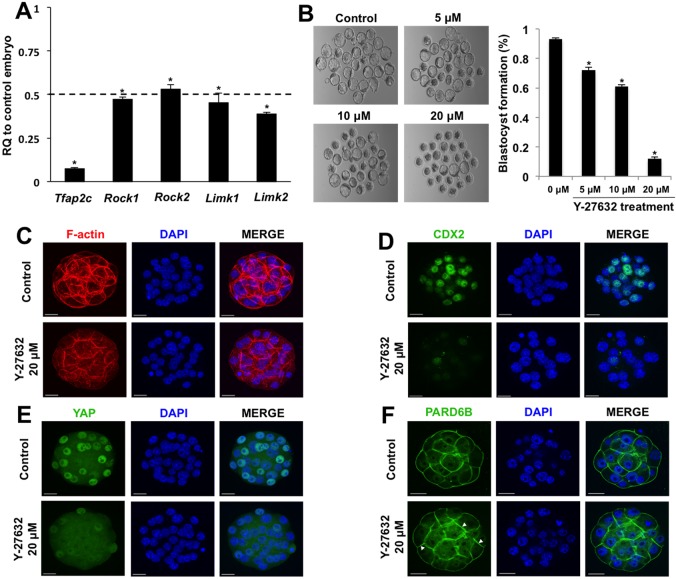
Recently, it was reported that Rho-associated protein kinase (ROCK) regulates position-dependent HIPPO signaling through cell polarity and inhibition of LATS activity in the outside cells (Kono et al., 2014). Previously, we showed that Rock2 is a direct target gene of TFAP2C in early embryos and TS-like cells (Choi et al., 2012). Thus, we reasoned that TFAP2C suppresses HIPPO signaling by promoting the expression of key genes required for inhibition of LATS activity. To address this we evaluated the expression of several putative TFAP2C target genes involved in ROCK signaling (Choi et al., 2012). The expression of Rock1, Rock2, Limk1 and Limk2 transcripts was evaluated in TFAP2C KD morulae. These transcripts were found to be reduced by >50% in TFAP2C KD embryos as compared with control embryos (Fig. 6A). Moreover, immunofluorescence analysis of ROCK1 protein revealed that it was reduced in TFAP2C KD embryos (supplementary material Fig. S6).
Fig. 6.
TFAP2C regulates position-dependent HIPPO signaling in part via ROCK signaling. (A) Real-time qPCR analysis of Rock1, Rock2, Limk1 and Limk2 transcripts in TFAP2C KD morulae. There was a significant decrease in Rock1, Rock2, Limk1 and Limk2 transcripts in TFAP2C KD morulae. A total of three biological replicates were performed. (B) Treatment of wild-type 8-cell embryos with Y-27632 blocks blastocyst formation in a dose-dependent manner. A total of three biological replicates were performed using 42 embryos in each group. (A,B) *P<0.05. Error bars indicate mean±s.e.m. (C) F-actin organization was disrupted in a subset of embryos treated with 20 μM Y-27632. (D) Cdx2 expression was severely reduced in embryos treated with 20 μM Y-27632. (E) Treatment of embryos with 20 μM Y-27632 activates HIPPO signaling in the outside cells causing loss of YAP in the nuclei. (F) In embryos treated with 20 μM Y-27632, PARD6B was enriched at both the apical and basolateral regions. Nuclei were counterstained with DAPI. Immunofluorescence experiments were performed using a total of two to three biological replicates with 16-30 embryos per group. In several of the experimental replicates, control and treated embryos were co-stained with phalloidin to visualize F-actin in conjunction with CDX2 and YAP. Scale bars: 20 μm.
To determine whether inhibition of ROCK1 and ROCK2 could phenocopy TFAP2C KD, we cultured 8-cell embryos in the presence of Y-27632, a specific inhibitor of ROCK1 and ROCK2. Embryos treated with 0, 5, 10 and 20 μM Y-27632 for 48 h exhibited a dose-dependent reduction in blastocyst formation and arrested at the morula stage (Fig. 6B). Phalloidin staining of F-actin revealed that 22/54 (41%) treated embryos had a disorganized cytoskeleton (Fig. 6C). Immunofluorescence analysis revealed that CDX2 and YAP were dramatically reduced in embryos treated with Y-27632 (Fig. 6D,E). To test whether apical cell polarity was altered in Y-27632-treated embryos, we evaluated the expression and localization of PARD6B. Consistent with a previous report (Kono et al., 2014), PARD6B was no longer largely confined to the apical region, but was localized to both the apical and basal region of the embryos (Fig. 6F). Altogether, our results demonstrate that TFAP2C regulates position-dependent HIPPO signaling via ROCK and PARD6B.
DISCUSSION
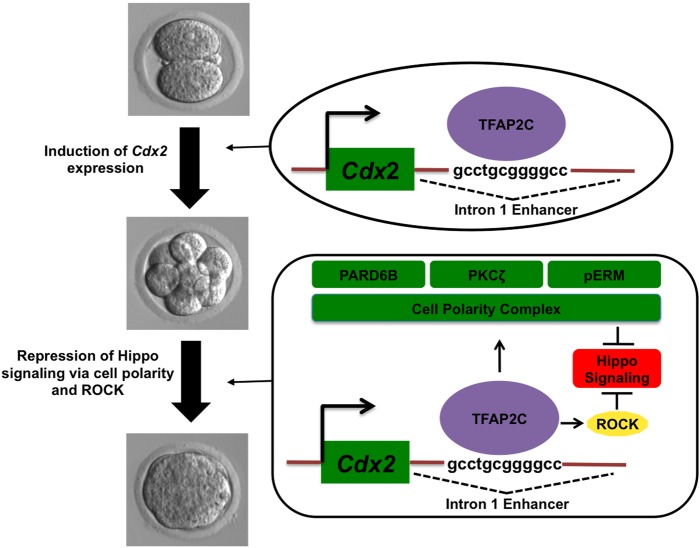
We previously showed that TFAP2C functions as a key regulator of the morula to blastocyst transition in mouse preimplantation embryos (Choi et al., 2012). Bioinformatics, gene expression and functional analyses revealed that TFAP2C regulates a large number of genes involved in tight junction biogenesis, blastocoel formation and cell cycle control. In the present study we extend these findings and show that TFAP2C plays a major role in trophectoderm lineage specification. Our results in preimplantation embryos, Tfap2c-inducible ESCs and TSCs show that TFAP2C: (1) regulates early Cdx2 expression via binding to an intronic enhancer; (2) facilitates apical cell polarity through regulation of Pard6b expression; and (3) negatively regulates HIPPO signaling in the outside blastomeres through PARD6B and ROCK signaling. We propose a working model in which TFAP2C functions as an upstream regulator of trophectoderm specification (Fig. 7).
Fig. 7.
TFAP2C-dependent model for regulation of trophectoderm lineage specification. TFAP2C promotes trophectoderm lineage specification via two major mechanisms. The first involves activation of Cdx2 expression during cleavage stages. The second mechanism is through negative regulation of HIPPO signaling in the outside cells of morulae to restrict Cdx2 expression to the emerging trophectoderm lineage. TFAP2C positively regulates apical cell polarity and ROCK signaling to repress HIPPO signaling.
The regulation of Cdx2 expression in early embryos and other developmental contexts is mediated by transcriptional and epigenetic mechanisms (Chen et al., 2009; Home et al., 2009, 2012; Yeap et al., 2009; Yuan et al., 2009; Saha et al., 2013; Rayon et al., 2014). Several key regulatory regions are present in Cdx2 (Wang and Shashikant, 2007; Benahmed et al., 2008). In this study we identified two conserved TFAP2C binding motifs in intron 1. We demonstrated that TFAP2C can positively regulate Cdx2 expression by binding to this enhancer. Interestingly, this intronic enhancer also contains a binding motif for GATA3 (Home et al., 2009). Previously, it was shown that GATA3 positively regulates Cdx2 around the morula to blastocyst transition to specify the trophectoderm lineage (Home et al., 2009). We found that TFAP2C positively regulates zygotic Cdx2 transcription during an earlier period, between the 1-cell and 4-cell stages. This implies that TFAP2C acts earlier than GATA3 and may induce the stochastic Cdx2 expression pattern that is observed in preimplantation embryos between the 8-cell and morula stages (Dietrich and Hiiragi, 2007).
Importantly, our data demonstrate for the first time that TFAP2C is a direct activator of Cdx2 expression during the early stages of preimplantation development. One could postulate that TFAP2C-dependent activation of Cdx2 expression is indirect and is mediated by downregulation of OCT4 (POU5F1); however, this is unlikely to be the case. For example, TFAP2C overexpression in ESCs induces Cdx2 expression even in the presence of constitutive OCT4 expression (Adachi et al., 2013). Moreover, during the early to middle stages of preimplantation development when Cdx2 is initially expressed, CDX2 and OCT4 are co-expressed in the same blastomeres (Dietrich and Hiiragi, 2007). The documented reciprocal negative regulation of OCT4 and CDX2 occurs later in preimplantation development when the ICM and trophectoderm lineages are established during blastocyst formation (Strumpf et al., 2005; Wang et al., 2010). Thus, our experimental data, as obtained by TFAP2C KD and overexpression, ChIP analysis and Cdx2 reporter assays, strongly support that TFAP2C is a direct activator of Cdx2 expression in preimplantation embryos. Combined, our results and those of others show that during the window of preimplantation development the Cdx2 gene is regulated by multiple inputs that culminate in trophectoderm-specific expression at the blastocyst stage. This mode of regulation is analogous to that of Oct4, which is under tight control in preimplantation embryos and ESCs (Yeom et al., 1996; Niwa et al., 2005; Wang et al., 2010).
Trophectoderm and ICM specification is controlled by differential HIPPO signaling in the outside and inside of the embryo (Hirate et al., 2013). In this study we show that TFAP2C negatively regulates HIPPO signaling in the outside blastomeres to specify the trophectoderm lineage. Depletion of TFAP2C triggered activation of LATS kinase (as inferred by elevated pYAP), a loss of position-dependent HIPPO signaling and downregulation of Cdx2. We further show that TFAP2C regulates HIPPO signaling through apical cell polarity. Previously, we established that Pard6b is a direct target gene of TFAP2C (Choi et al., 2012). In the current study we show that TFAP2C is required for Pard6b expression; depletion of TFAP2C triggered downregulation of Pard6b and loss of apical polarity, whereas Tfap2c overexpression induced elevated Pard6b transcript and protein levels in early embryos and ESCs. Consistent with our findings, two recent studies showed that PARD6B is required for position-dependent HIPPO signaling and Cdx2 expression in preimplantation embryos (Alarcon, 2010; Hirate et al., 2013). Interestingly, in the current study PARD6B rescue restored apical cell polarity, but only partially re-established position-dependent HIPPO signaling and did not rescue Cdx2 expression. Our results demonstrate that TFAP2C controls position-dependent HIPPO signaling in part through PARD6B. Importantly, these experimental findings offer new insight into the underlying transcriptional mechanisms that regulate apical cell polarity in the preimplantation embryo.
Multiple mechanisms can negatively regulate HIPPO signaling (Salah et al., 2011; Yu et al., 2012; Hirate et al., 2013). In some developmental contexts, F-actin negatively regulates HIPPO signaling (Matsui and Lai, 2013). Interestingly, we and others have observed that F-actin is disorganized in PARD6B KD and TFAP2C KD morulae (Alarcon, 2010; Choi et al., 2012). This prompted us to evaluate the expression and function of putative TFAP2C target genes involved in the regulation of F-actin. Accordingly, this analysis revealed that Rock and Limk family members were downregulated in TFAP2C KD embryos. Incubation of 8-cell embryos in the presence of Y-27632, a specific inhibitor of ROCK1 and ROCK2, blocked blastocyst formation and resulted in a loss of position-dependent HIPPO signaling. These findings are largely consistent with those of a recent study which showed that RHO-ROCK signaling is required for trophectoderm specification (Kono et al., 2014). Specifically, the authors showed that ROCK controls position-dependent HIPPO signaling by repressing LATS activity in the outside cells. However, the mechanism by which ROCK represses LATS kinase was not determined. In the current study, phalloidin staining revealed that 41% of treated embryos had a disorganized F-actin cytoskeleton, suggesting that ROCK signaling might repress HIPPO signaling via F-actin. However, closer examination of F-actin and nYAP staining in Y-27632-treated embryos revealed no association, as embryos with normal and abnormal F-actin staining exhibited reduced levels of YAP (data not shown). Thus, it is possible that ROCK negatively regulates HIPPO signaling independently of F-actin. Future studies are necessary to determine the precise mechanism by which ROCK negatively regulates HIPPO signaling in the preimplantation embryo.
To our knowledge, TFAP2C is the first transcription factor shown to act upstream of the HIPPO signaling pathway in the preimplantation embryo. Other transcription factors, such as GATA3 and TEAD4, function downstream of cell polarity and HIPPO signaling to selectively upregulate Cdx2 expression in the emerging trophectoderm lineage (Yagi et al., 2007; Nishioka et al., 2008; Home et al., 2009). This implies that TFAP2C might sit at the top of a hierarchy of signaling pathways and transcription factors that program the early trophectoderm lineage. Interestingly, previous studies showed that zygotic TFAP2C is required for placental development after implantation (Auman et al., 2002; Werling and Schorle, 2002). This suggests that TFAP2C regulates trophoblast lineage development during two critical periods of embryonic development. In the preimplantation embryo, maternal and zygotic TFAP2C mediates trophectoderm lineage specification via regulation of position-dependent HIPPO signaling and Cdx2 (this study), whereas in the post-implantation embryo TFAP2C promotes placental development by controlling the proliferation and differentiation of trophoblast cells (Werling and Schorle, 2002). Future ChIP-Seq and RNA-Seq studies are warranted to delineate the overarching transcriptional networks in which TFAP2C, GATA3, TEAD4 and CDX2 may intersect to program the trophoblast lineage.
In summary, the results reported here demonstrate that TFAP2C orchestrates trophectoderm lineage specification by functioning as an upstream regulator of Cdx2 transcription and position-dependent HIPPO signaling. These findings have implications in understanding the molecular basis of clinical reproductive disorders and congenital defects that may originate during early embryonic development. From a broader viewpoint, such information might contribute to our understanding of the mechanisms that control HIPPO signaling and cellular differentiation in other developmental contexts.
MATERIALS AND METHODS
Ethics statement
All experiments involving animals were conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines under current approved protocols at Michigan State University.
Embryo collection and microinjection
Mouse embryos were isolated from superovulated CF1 females mated with B6D2F1 males (Charles River Laboratories). Fertilized 1-cell embryos were collected at 16-18 h post-human chorionic gonadotropin (hCG) treatment and then cultured in modified KSOM medium (EMD Millipore) under mineral oil at 37°C in a humidified atmosphere of 5% O2, 5% CO2, 90% N2. Microinjection was carried out as described previously (Choi et al., 2012). Briefly, 5-10 pl 100 μM Tfap2c siRNA, 100 μM control siRNA, 100 μM Pard6b siRNA, Pard6b mRNA (1.0 μg/μl) or Tfap2c mRNA (25 ng/μl) was injected into the cytoplasm of 1-cell embryos using a PL100 picoinjector (Harvard Apparatus) at 19-21 h post-hCG. Following microinjection, embryos were placed back into KSOM and cultured for 2 to 5 days.
Treatment of embryos with the ROCK inhibitor Y-27632
A stock of Y-27632 dissolved in H2O was purchased (EMD Millipore) and stored at −20°C until use. Working solutions of 0, 5, 10 and 20 μM Y-27632 were prepared in KSOM. To evaluate the effects of Y-27632 on the 8-cell to morula and morula to blastocyst transitions, 8-cell embryos were cultured in the presence and absence of the inhibitor for 24 and 48 h, respectively.
Tfap2c-inducible ESC and TSC culture
Mouse Tfap2c-inducible ESCs were obtained from Corriel Cell Repositories (#AG23567). This engineered ESC line was originally generated by Dr Minoru Ko at the National Institute of Aging (Correa-Cerro et al., 2011). Cells were grown on a feeder layer of mitomycin-treated puromycin-resistant mouse embryonic fibroblasts (MEFs) in standard ESC medium, supplemented with 0.2 mg/ml doxycycline and 1.0 mg/ml puromycin. Prior to Tfap2c induction, cells were switched onto gelatin and cultured with 3.0 mg/ml puromycin for 3 days. Tfap2c expression was induced by removal of doxycycline. At 0, 24, 48, 72 and 96 h following Tfap2c induction, cells were isolated for real-time PCR and immunofluorescence analysis.
Mouse TSCs were cultured as described previously (Home et al., 2009). In brief, TSCs were initially grown on a layer of mitotically inactivated MEFs in the presence of 25 ng/ml FGF4 (Sigma-Aldrich) and heparin (1 μg/ml) in TSC medium. Prior to ChIP and reporter assays, TSCs were switched onto gelatin-coated dishes to remove MEFs.
siRNA sequences
SMART pool siRNA was purchased from Dharmacon (Thermo Fisher). The Tfap2c siRNA was described previously (Choi et al., 2012). siRNA sequences were (5′-3′): Tfap2c AAGCUGAGUUCCCUAGUAA, GCACGGGACUUCGCCUAUG, AGCGGUGGCUGACUAUUUA and CCGCAGUGCAGAAUUAUAU; Pard6b GACCUGCCGCCUAUAAAUA, GGAACAACGUGGUGCGCAA, GUUAGAAGUCAACGGUAUA and GAAGACGGUACCAUCAUCA. Control UAAGGCUAUGAAGAGAUAC, AUGUAUUGGCCUGUAUUAG, AUGAACGUGAAUUGCUCAA and UGGUUUACAUGUCGACUAA.
Gene expression analysis by real-time quantitative PCR (qPCR)
Total RNA was isolated using the PicoPure RNA Isolation Kit (Arcturus) or RNeasy Mini Kit (Qiagen). cDNA synthesis was carried out using SuperScript II reverse transcriptase (Invitrogen). Real-time qPCR analysis was conducted utilizing TaqMan probes (Applied Biosystems) or gene-specific designed primers (SYBR Green detection) and a StepOnePlus real-time PCR system (Applied Biosystems). Ubtf was used as an endogenous control for embryos as described previously (Wang et al., 2010). Real-time qPCR primers are listed in supplementary material Table S1.
Quantitative ChIP assay
Real-time PCR-based quantitative ChIP analysis was employed to assess TFAP2C occupancy at the Cdx2 intronic enhancer. TSCs were trypsinized, spun down, and resuspended in PBS containing 1% formaldehyde for 10 min. The reaction was quenched by adding 0.125 M glycine. Fixed cells were sonicated and the chromatin extract was snap frozen and stored at −80°C. Anti-TFAP2C was used to immunoprecipitate TFAP2C-bound DNA fragments; as a negative control, a species-matched IgG was used (antibodies are described in supplementary material Table S2). Immunoprecipitated DNA was decrosslinked, digested with proteinase K and purified. Primers are listed in supplementary material Table S1.
microChIP in embryos
For microChIP analysis, 100 morula were crosslinked with 1% formaldehyde, and ChIP analysis was performed following a previously described protocol (Home et al., 2009) with some modifications. In brief, sonicated, crosslinked chromatin fragments were immunoprecipitated with a rabbit anti-TFAP2C antibody; rabbit IgG was used as control antibody (antibodies are described in supplementary material Table S2). Immunoprecipitated chromatin was decrosslinked, digested with proteinase K and purified. Immunoprecipitated DNA fragments were amplified using the whole-genome amplification procedure [Genomeplex Single-Cell Genome Amplification (WGA4) kit, Sigma], and were analyzed by qPCR for TFAP2C occupancy. Primers are listed in supplementary material Table S1.
In vitro transcription
Pard6b and Tfap2c mRNA used for microinjection was synthesized in vitro. The Pard6b open reading frame (ORF) was amplified from mouse embryo cDNA with gene-specific primers containing T7 promoter sequence (supplementary material Table S1). PCR products were used as templates for in vitro transcription. The Tfap2c ORF was amplified from TSC cDNA. PCR amplification was performed using a nested PCR approach with the following primers (5′-3′): reaction #1 forward, CCTGGATTTAACTGGCGACTAT; reaction #1 reverse, CTCCTACCAGAAGGTGTGATTG; reaction #2 forward, GGACGCCATGTTGTGGAAAATAAC; reaction #2 reverse, TACTTCCTGTGCTTTTCCATTTTC. The PCR product from the second reaction was then TA-cloned into pCR2.1 vector (Life Technologies). The 1.4 kb BamHI-EcoRV fragment containing the Tfap2c ORF was then subcloned into the pIVT vector containing 5′ and 3′ Xenopus β-globin untranslated regions (UTRs) for enhanced translation (Igarashi et al., 2007). This was accomplished using a blunt site created at the SacI restriction site that was treated with Klenow large fragment. pIVT was then digested with BamHI to create complementary sites that could facilitate directional subcloning to correctly orient the T7 promoter. Tfap2c-pIVT was then linearized in preparation for in vitro transcription by digestion with NdeI. In vitro transcription of Pard6b and Tfap2c mRNA was performed using the mMESSAGE mMACHINE T7 kit (Ambion) according to the manufacturer's manual. After in vitro transcription, mRNA was treated with DNase I to remove the DNA template. Purified mRNA was dissolved in RNase-free water and the concentration determined by A260. Synthesis and quality of mRNA were confirmed using an RNA bioanalyzer (Agilent).
Motif analysis and luciferase reporter assays
To identify conserved TFAP2C motifs, VISTA regulatory sequence alignment of Cdx2 loci was performed as described previously (Home et al., 2009). For transient transfection analysis, RP23–355P12 BAC DNA was used to amplify regulatory elements of the mouse Cdx2 locus. The promoter region (−663 to −18 bp) of the mouse Cdx2 gene was cloned into BglII/HindIII sites of the pGL3 Basic vector (Promega) to generate Cdx2(pro)Luc. A region of Cdx2 intron 1 (+2601-3469 bp) containing the conserved TFAP2C motif (GCCTGCGGGGC) was cloned into KpnI/XhoI sites upstream of the Cdx2 promoter in Cdx2(pro)Luc. To delete the conserved TFAP2C motif, the regions upstream and downstream of the TFAP2C motif were PCR amplified with 5′ KpnI/3′ EcoRV and 5′ EcoRV/3′XhoI primer sets, respectively. Finally, the TFAP2C motif-deleted fragment was cloned into KpnI/XhoI sites upstream of the Cdx2 promoter in Cdx2(pro)Luc.
Embryo staging, immunofluorescence and confocal microscopy
Control and treated embryos (siRNA, mRNA, ROCK inhibitor) were collected at either the 2-cell stage (48 h post-hCG) or morula stage (90 h post-hCG). This allowed us to precisely compare control and treatment groups at similar stages. Zona pellucidae of preimplantation embryos were removed by tyrode acid treatment. Embryos were then fixed with 3.7% paraformaldehyde for 20 min, permeabilized with PBS containing 0.1% Tween 20 for 15 min, blocked with PBS containing 0.1% BSA for 1 h at room temperature, and incubated with primary antibodies in blocking solution overnight at 4°C, followed by incubation with Alexa Fluor 488 and 594 secondary antibodies. Antibody information is provided in supplementary material Table S2. Embryos were mounted in Vectashield containing DAPI (4,6-diamidino-2-phenylindole; Vector Laboratories) and imaged using an Olympus FluoView 1000 confocal microscope (CARV; Atto Bioscience) with FluoView Viewer 3.0 software (Molecular Devices). Quantification of nYAP was accomplished using ImageJ software (NIH). In some experiments, F-actin was stained using Alexa Fluor 586-phalloidin (Molecular Probes). The vast majority of images presented in Figs 1-5 are z-sections representing the middle region of the embryos. The only exceptions are found in Fig. 3D, Fig. 6 and supplementary material Fig. S6, where z-stacks were obtained to highlight the 3D localization of F-actin, PARD6B and ROCK1.
Statistical analysis
Data were analyzed by analysis of variance (ANOVA) using SPSS (IBM) and Student's t-test. Data are presented as mean±s.e.m. P<0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Dr Chen Chen (Michigan State University) for critically reading the manuscript and Dr Keith Latham (Michigan State University) for providing alpha-amanitin.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This research was supported by a grant from the National Institute of General Medical Sciences to J.G.K. [GM095347] and grants from the National Institute of Child Health and Human Development to S.P. [HD079363, HD062546 and HD075233]. Deposited in PMC for release after 12 months.
Author contributions
J.G.K. and Z.C. designed research; Z.C., T.S.C., A.G. and C.A.W. performed research; S.P. and J.G.K. contributed reagents/analytical tools; Z.C., J.G.K., T.S.C., A.G. and S.P. analyzed data; and J.G.K. and Z.C. wrote the paper.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.120238/-/DC1
References
- Adachi K., Nikaido I., Ohta H., Ohtsuka S., Ura H., Kadota M., Wakayama T., Ueda H. R. and Niwa H. (2013). Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell 52, 380-392 10.1016/j.molcel.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Alarcon V. B. (2010). Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347-358 10.1095/biolreprod.110.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman H. J., Nottoli T., Lakiza O., Winger Q., Donaldson S. and Williams T. (2002). Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 129, 2733-2747. [DOI] [PubMed] [Google Scholar]
- Barros R., da Costa L. T., Pinto-de-Sousa J., Duluc I., Freund J.-N., David L. and Almeida R. (2011). CDX2 autoregulation in human intestinal metaplasia of the stomach: impact on the stability of the phenotype. Gut 60, 290-298 10.1136/gut.2010.222323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benahmed F., Gross I., Gaunt S. J., Beck F., Jehan F., Domon-Dell C., Martin E., Kedinger M., Freund J.-N. and Duluc I. (2008). Multiple regulatory regions control the complex expression pattern of the mouse Cdx2 homeobox gene. Gastroenterology 135, 1238-1247.e3 10.1053/j.gastro.2008.06.045 [DOI] [PubMed] [Google Scholar]
- Chen L., Yabuuchi A., Eminli S., Takeuchi A., Lu C.-W., Hochedlinger K. and Daley G. Q. (2009). Cross-regulation of the Nanog and Cdx2 promoters. Cell Res. 19, 1052-1061 10.1038/cr.2009.79 [DOI] [PubMed] [Google Scholar]
- Choi I., Carey T. S., Wilson C. A. and Knott J. G. (2012). Transcription factor AP-2gamma is a core regulator of tight junction biogenesis and cavity formation during mouse early embryogenesis. Development 139, 4623-4632 10.1242/dev.086645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn K., Biechele S., Garner J. and Rossant J. (2013). The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195-1201 10.1016/j.cub.2013.05.044 [DOI] [PubMed] [Google Scholar]
- Correa-Cerro L. S., Piao Y., Sharov A. A., Nishiyama A., Cadet J. S., Yu H., Sharova L. V., Xin L., Hoang H. G., Thomas M. et al. (2011). Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Sci. Rep. 1, 167 10.1038/srep00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard N., Le T., Maro B. and Louvet-Vallée S. (2009). Inactivation of aPKClambda reveals a context dependent allocation of cell lineages in preimplantation mouse embryos. PLoS ONE 4, e7117 10.1371/journal.pone.0007117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries W. N., Evsikov A. V., Haac B. E., Fancher K. S., Holbrook A. E., Kemler R., Solter D. and Knowles B. B. (2004). Maternal beta-catenin and E-cadherin in mouse development. Development 131, 4435-4445 10.1242/dev.01316 [DOI] [PubMed] [Google Scholar]
- Dietrich J.-E. and Hiiragi T. (2007). Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219-4231 10.1242/dev.003798 [DOI] [PubMed] [Google Scholar]
- Ducibella T., Ukena T., Karnovsky M. and Anderson E. (1977). Changes in cell surface and cortical cytoplasmic organization during early embryogenesis in the preimplantation mouse embryo. J. Cell Biol. 74, 153-167 10.1083/jcb.74.1.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y., Hirahara S., Inoue K.-i., Suzuki A., Alarcon V. B., Akimoto K., Hirai T., Hara T., Adachi M., Chida K. et al. (2013). Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23, 1181-1194 10.1016/j.cub.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P., Ray S., Dutta D., Bronshteyn I., Larson M. and Paul S. (2009). GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 284, 28729-28737 10.1074/jbc.M109.016840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P., Saha B., Ray S., Dutta D., Gunewardena S., Yoo B., Pal A., Vivian J. L., Larson M., Petroff M. et al. (2012). Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl. Acad. Sci. USA 109, 7362-7367 10.1073/pnas.1201595109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H., Knott J. G., Schultz R. M. and Williams C. J. (2007). Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev. Biol. 312, 321-330 10.1016/j.ydbio.2007.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. H. and Ziomek C. A. (1981). The foundation of two distinct cell lineages within the mouse morula. Cell 24, 71-80 10.1016/0092-8674(81)90502-X [DOI] [PubMed] [Google Scholar]
- Kono K., Tamashiro D. A. A. and Alarcon V. B. (2014). Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev. Biol. 394, 142-155 10.1016/j.ydbio.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckenberg P., Buhl S., Woynecki T., van Furden B., Tolkunova E., Seiffe F., Moser M., Tomilin A., Winterhager E. and Schorle H. (2010). The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell. Biol. 30, 3310-3320 10.1128/MCB.01215-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C., Messerschmidt D. M., Chan S. W., Hong W., Knowles B. B. and Solter D. (2013). Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 27, 1441-1446 10.1101/gad.219618.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet S., Aghion J., Santa-Maria A., Mangeat P. and Maro B. (1996). Ezrin becomes restricted to outer cells following asymmetrical division in the preimplantation mouse embryo. Dev. Biol. 177, 568-579 10.1006/dbio.1996.0186 [DOI] [PubMed] [Google Scholar]
- Matsui Y. and Lai Z.-C. (2013). Mutual regulation between Hippo signaling and actin cytoskeleton. Protein Cell 4, 904-910 10.1007/s13238-013-3084-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K. and Sasaki H. (2008). Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270-283 10.1016/j.mod.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K.-i., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N. et al. (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398-410 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R. and Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917-929 10.1016/j.cell.2005.08.040 [DOI] [PubMed] [Google Scholar]
- Plusa B., Frankenberg S., Chalmers A., Hadjantonakis A.-K., Moore C. A., Papalopulu N., Papaioannou V. E., Glover D. M. and Zernicka-Goetz M. (2005). Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 118, 505-515 10.1242/jcs.01666 [DOI] [PubMed] [Google Scholar]
- Rayon T., Menchero S., Nieto A., Xenopoulos P., Crespo M., Cockburn K., Cañon S., Sasaki H., Hadjantonakis A.-K., de la Pompa J. L. et al. (2014). Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell 30, 410-422 10.1016/j.devcel.2014.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethmacher D., Brinkmann V. and Birchmeier C. (1995). A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. USA 92, 855-859 10.1073/pnas.92.3.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B., Home P., Ray S., Larson M., Paul A., Rajendran G., Behr B. and Paul S. (2013). EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol. Cell. Biol. 33, 2691-2705 10.1128/MCB.00069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Z., Melino G. and Aqeilan R. I. (2011). Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 71, 2010-2020 10.1158/0008-5472.CAN-10-3516 [DOI] [PubMed] [Google Scholar]
- Sato N., Funayama N., Nagafuchi A., Yonemura S. and Tsukita S. (1992). A gene family consisting of ezrin, radixin and moesin. Its specific localization at actin filament/plasma membrane association sites. J. Cell Sci. 103, 131-143. [DOI] [PubMed] [Google Scholar]
- Strumpf D., Mao C.-A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F. and Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093-2102 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Tarkowski A. K. and Wroblewska J. (1967). Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J. Embryol. Exp. Morphol. 18, 155-180. [PubMed] [Google Scholar]
- Vinot S., Le T., Ohno S., Pawson T., Maro B. and Louvet-Vallée S. (2005). Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev. Biol. 282, 307-319 10.1016/j.ydbio.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Wang W. C. H. and Shashikant C. S. (2007). Evidence for positive and negative regulation of the mouse Cdx2 gene. J. Exp. Zool. B Mol. Dev. Evol. 308B, 308-321 10.1002/jez.b.21154 [DOI] [PubMed] [Google Scholar]
- Wang K., Sengupta S., Magnani L., Wilson C. A., Henry R. W. and Knott J. G. (2010). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS ONE 5, e10622 10.1371/journal.pone.0010622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennekamp S., Mesecke S., Nédélec F. and Hiiragi T. (2013). A self-organization framework for symmetry breaking in the mammalian embryo. Nat. Rev. Mol. Cell Biol. 14, 454-461 10.1038/nrm3602 [DOI] [PubMed] [Google Scholar]
- Werling U. and Schorle H. (2002). Transcription factor gene AP-2 gamma essential for early murine development. Mol. Cell. Biol. 22, 3149-3156 10.1128/MCB.22.9.3149-3156.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Kohn M. J., Karavanova I., Kaneko K. J., Vullhorst D., DePamphilis M. L. and Buonanno A. (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827-3836 10.1242/dev.010223 [DOI] [PubMed] [Google Scholar]
- Yeap L.-S., Hayashi K. and Surani M. A. (2009). ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12 10.1186/1756-8935-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom Y. I., Fuhrmann G., Ovitt C. E., Brehm A., Ohbo K., Gross M., Hubner K. and Scholer H. R. (1996). Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122, 881-894. [DOI] [PubMed] [Google Scholar]
- Yu F.-X., Zhao B., Panupinthu N., Jewell J. L., Lian I., Wang L. H., Zhao J., Yuan H., Tumaneng K., Li H. et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780-791 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Han J., Guo G., Orlov Y. L., Huss M., Loh Y.-H., Yaw L.-P., Robson P., Lim B. and Ng H.-H. (2009). Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507-2520 10.1101/gad.1831909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziomek C. A. and Johnson M. H. (1980). Cell surface interaction induces polarization of mouse 8-cell blastomeres at compaction. Cell 21, 935-942 10.1016/0092-8674(80)90457-2 [DOI] [PubMed] [Google Scholar]
- Ziomek C. A. and Johnson M. H. (1982). The roles of phenotype and position in guiding the fate of 16-cell mouse blastomeres. Dev. Biol. 91, 440-447 10.1016/0012-1606(82)90050-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.