Abstract
Neural progenitor cells (NPCs) have distinct proliferation capacities at different stages of brain development. Lin28 is an RNA-binding protein with two homologs in mice: Lin28a and Lin28b. Here we show that Lin28a/b are enriched in early NPCs and their expression declines during neural differentiation. Lin28a single-knockout mice show reduced NPC proliferation, enhanced cell cycle exit and a smaller brain, whereas mice lacking both Lin28a alleles and one Lin28b allele display similar but more severe phenotypes. Ectopic expression of Lin28a in mice results in increased NPC proliferation, NPC numbers and brain size. Mechanistically, Lin28a physically and functionally interacts with Imp1 (Igf2bp1) and regulates Igf2-mTOR signaling. The function of Lin28a/b in NPCs could be attributed, at least in part, to the regulation of their mRNA targets that encode Igf1r and Hmga2. Thus, Lin28a and Lin28b have overlapping functions in temporally regulating NPC proliferation during early brain development.
Keywords: Lin28a, Lin28b, Neural progenitor cells, Proliferation, Brain development, Mouse
Summary: The proliferation of neural progenitor cells is regulated by Lin28a/b via their interaction with Imp1 and their regulation of Igf2-mTOR signaling.
INTRODUCTION
Neural progenitor cells (NPCs) use distinct self-renewal and differentiation programs for brain development and homeostasis. At different stages, NPCs exhibit unique morphologies, proliferation capacities and differentiation potentials. Early embryonic NPCs replicate rapidly to expand their population, and their proliferation rate declines later in development to coordinate with the neuronal differentiation programs (He et al., 2009; Salomoni and Calegari, 2010). This temporal change of NPC properties has been partially attributed to intricate gene expression programs. For example, unique sets of transcription factors are sequentially expressed in different populations of NPCs during brain development, including Pax6, Tbr2 (also known as Eomes) and Sox2 (Englund et al., 2005; Graham et al., 2003; Sessa et al., 2008). Epigenetic regulators modulate chromatin states to control NPC self-renewal and neurogenic potentials in a developmental stage-dependent manner (Kishi et al., 2012; Lim et al., 2009; Nishino et al., 2008). Although these studies have provided significant insights into the highly coordinated gene expression program at transcriptional levels for NPC behavior control, little is known about gene regulation at post-transcriptional levels for NPC self-renewal and brain development.
Signals from local and extrinsic sources regulate NPC proliferation and differentiation during brain development (Johansson et al., 2010), including insulin-like growth factor 1/2 (Igf1/2) signaling (Lehtinen et al., 2011; O'Kusky and Ye, 2012). The action of Igf1/2 is mainly mediated by insulin-like growth factor receptor 1 (Igf1r) followed by Akt activation and downstream signaling events, including mTOR pathway activation (Laplante and Sabatini, 2012; O'Kusky and Ye, 2012). In humans, mutations in IGF1 or IGF1R are associated with severe growth retardation and microcephaly (Abuzzahab et al., 2003; Woods et al., 1997). Knockout of Igf1, Igf2 or Igf1r in mice dysregulates progenitor cell division leading to deficiencies in embryonic and postnatal growth, and ultimately results in reduced brain size (Baker et al., 1993; Liu et al., 1993). Although the importance in NPC behaviors is established, how Igf1/2 signaling is temporally regulated to contend with the changes in NPC proliferation that occur during brain development remains largely unknown.
Lin28 is an RNA-binding protein with a cold-shock domain (CSD) and retroviral-type CCHC zinc knuckle RNA-binding domain. Mammals have two Lin28 homologs: Lin28a and Lin28b. Since our original identification of LIN28 as a developmental timing regulator in C. elegans (Moss et al., 1997), substantial efforts have been put into understanding Lin28a/b functions in mammals. Studies from recent years suggest that Lin28a/b function in a wide spectrum of biological processes and diseases, including embryonic stem cell (ESC) self-renewal, induced pluripotent stem cell (iPSC) generation, cancers and diabetes (Shyh-Chang and Daley, 2013; Thornton and Gregory, 2012). Our previous studies, together with those by other groups, suggest that Lin28a regulates cell proliferation and neurogenesis in vitro (Balzer et al., 2010; Cimadamore et al., 2013). However, the physiological functions of Lin28a/b in somatic stem cells remain largely unknown, and their in vivo roles in NPC self-renewal and brain development have not been determined.
Whereas previous research focused on the microRNA (miRNA) let-7 as the major target mediating Lin28 functions (Shyh-Chang and Daley, 2013; Thornton and Gregory, 2012), our recent studies suggest that Lin28a could function in a let-7-independent manner (Balzer et al., 2010). Moreover, recent genome-wide studies suggest that mRNAs are the major targets of Lin28a, whereas miRNA loci represent only 0.07% of Lin28a binding sequence reads (Cho et al., 2012; Hafner et al., 2013). Overall, these studies raise questions as to the identity of mRNA targets of Lin28 and their importance in mediating Lin28 functions during brain development.
Here, we use both gain- and loss-of-function genetic approaches to reveal that Lin28a and Lin28b have overlapping functions in promoting NPC proliferation and brain development in mouse. Lin28a/b function, at least in part, through modulation of Igf2-mTOR signaling activities and the protein expression of the chromatin regulator Hmga2.
RESULTS
Lin28a deletion results in reduced body and brain size in mice
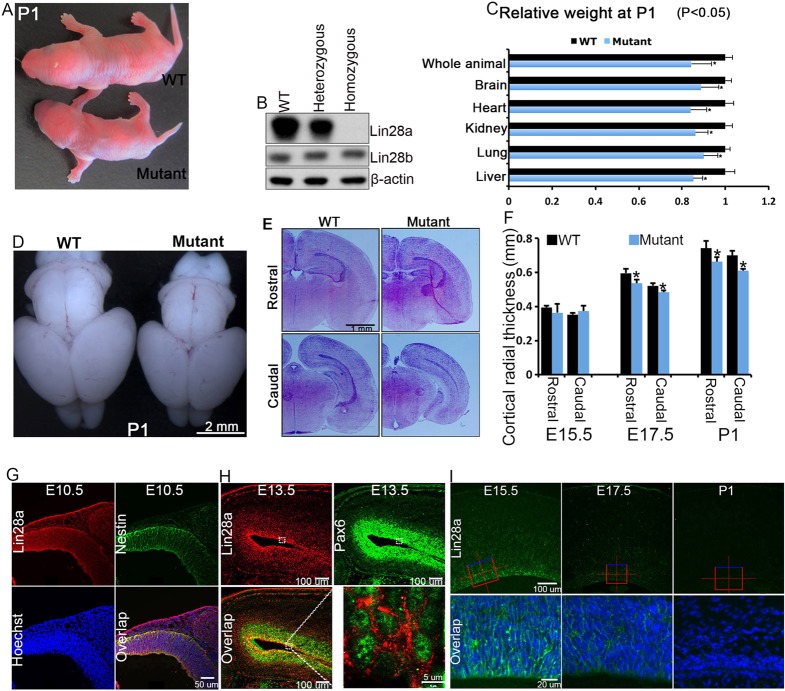
Mammals possess two Lin28 genes, encoding the Lin28a and Lin28b proteins (supplementary material Fig. S1A), which may have arisen by duplication of an ancestral gene (Guo et al., 2006; Moss and Tang, 2003). We and others have previously observed Lin28a expression in early neural development (Balzer et al., 2010; Yokoyama et al., 2008). The purpose of this study was to investigate the in vivo roles of Lin28 with a particular focus on the developing mammalian nervous system. To do so we created Lin28a null mice, which harbor an exon 2 deletion. Heterozygous Lin28a mice were viable and fertile and of normal size, whereas homozygous mutant mice exhibited reduced body size at birth (Fig. 1A). Western blot analysis verified the complete elimination of Lin28a (Fig. 1B, top panel) and not the homolog Lin28b (Fig. 1B, middle panel) protein in the brain of homozygous mutant embryos. Mutant organ mass was proportionally reduced with total body weight at embryonic day (E) 18.5 and postnatal day (P) 1 compared with wild type (Fig. 1C; supplementary material Fig. S1C), including a reduction in brain size (Fig. 1C,D; supplementary material Fig. S1C). No significant difference in morphology or organ size was observed at E15.5 between homozygous mutant, heterozygous and wild-type embryos (supplementary material Fig. S1B). Histological staining and statistical analysis revealed a ∼10% reduction in cerebral cortex thickness in Lin28a mutant brains compared with littermate controls at E17.5 and P1 (Fig. 1E,F). These findings suggest that Lin28a is crucial for murine growth control, including brain development.
Fig. 1.
Loss of Lin28a results in reduced body and brain size. (A) Lin28a mutant and its wild-type (WT) littermate on P1. (B) Western blot analyses of Lin28a and Lin28b expression in the cerebral cortex of E10.5 wild-type, heterozygous and homozygous mutant embryos. β-actin serves as a loading control. (C) Relative weights of different organs isolated from P1 wild-type and mutant mice. Error bars indicate s.e.m. of measurements from three independent experiments. *P<0.05 for each WT versus mutant comparison (Student's t-test). (D) Dorsal views of P1 wild-type and mutant mouse brains. (E) Coronal sections of P1 cerebral cortex stained with H&E. (F) Lin28a-deficient cortex shows decreased radial thickness. Error bars indicate s.e.m. of nine sections at comparable positions along the rostral/caudal axis between wild-type and mutant cerebral cortex from three independent experiments. *P<0.05 (Student's t-test). (G-I) Confocal microscopy images of coronal sections of the developing brain at different stages using the antibodies indicated. Hoechst stains nuclei (blue). Overlap refers to merge. Scale bars: 2 mm in D; 1 mm in E; 50 µm in G; 100 µm in H, except 5 µm bottom right; 100 µm top row and 20 µm bottom row in I.
To guide our mechanistic studies of Lin28a in brain development, we assessed the distribution of Lin28a protein in the cerebral cortex at different stages. These experiments showed that Lin28a is expressed in the NPCs, as indicated by its localization in early neural tube and at the ventricular/subventricular zone (VZ/SVZ) of the developing cerebral cortex (Fig. 1G-I; supplementary material Fig. S1D). At subcellular levels, Lin28a is localized in the cytoplasm of NPCs labeled by Pax6 or nestin (Fig. 1H; supplementary material Fig. S1D). Immunoblots of whole-cell extracts of cortices from different embryonic stages showed that Lin28a/b are predominant at early stages when the neuroepithelium is primarily composed of NPCs and that protein levels decrease as neural differentiation proceeds (supplementary material Fig. S1E). These data, together with our previous studies (Balzer et al., 2010), suggest that Lin28a/b are highly expressed in NPCs in the early developing brain.
Lin28a deletion leads to reduced NPC numbers due to decreased proliferation and enhanced cell cycle exit
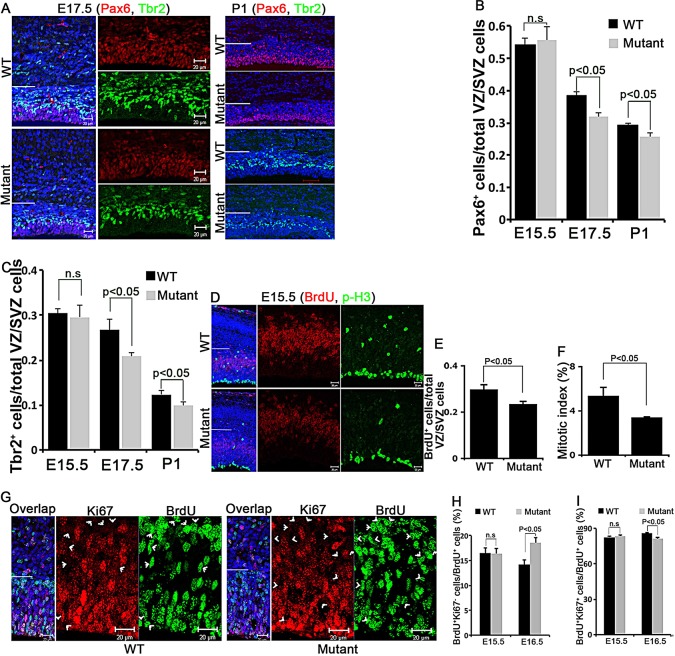
The enriched expression of Lin28a in NPCs (Fig. 1G-I) and reduced brain size of Lin28a mutants (Fig. 1D-F) prompted us to test whether there is a decreased number of NPCs in the mutant brains. We used Pax6 and Tbr2, respectively, to mark apical progenitors (APs) and intermediate neural progenitors (INPs) in the developing cerebral cortex. We focused our analyses on the VZ/SVZ of the cerebral cortex (the area below the white lines in Fig. 2). Examination of comparable cerebral cortical sections from wild-type and mutant littermates showed that the numbers of Pax6+ APs and Tbr2+ INPs were significantly decreased in the mutants at E17.5 and P1, whereas no difference was detected at E15.5 (Fig. 2A-C). Together, these data indicate that Lin28a is required for the maintenance of AP and INP populations during normal development.
Fig. 2.
Lin28a deficiency results in reduced proliferation, enhanced cell cycle exit and depletion of cortical NPCs. (A) Confocal micrographs of coronal sections from E17.5 or P1 wild-type and mutant cortex stained with antibodies against Pax6 (red) or Tbr2 (green). Hoechst stains nuclei (blue). The ventricular/subventricular zone (VZ/SVZ) is the area below the white line. (B,C) Percentage of Pax6+ (B) or Tbr2+ (C) cells among total cells within the VZ/SVZ in the experiment in A at the stages indicated. n.s., not significant (P>0.05) at E15.5; P<0.05 at E17.5 and P1 (Student's t-test). (D) Confocal micrographs of coronal sections from E15.5 wild-type and mutant cortex stained with antibodies against p-H3 (green) and BrdU (red) 1 h after a single pulse label of BrdU. The VZ/SVZ regions (below the white line) are enlarged to the right. (E,F) Percentage of BrdU+ cells (E) and p-H3+ cells (F) among total cells within the VZ/SVZ (below the white line) in the experiment in D. P<0.05 in E and F (Student's t-test). (G) Confocal micrographs of coronal sections from E16.5 mice 24 h after a single pulse label of BrdU; sections were stained with antibodies against BrdU (green) and Ki67 (red). Arrowheads indicate cells that exited the cell cycle (BrdU+/Ki67–). (H,I) The percentage of BrdU+/Ki67− cells (H) or BrdU+/Ki67+ cells (I) among total BrdU+ cells within the VZ/SVZ (below the white line in the experiment in G). n.s., not significant (P>0.05) at E15.5; P<0.05 at E16.5 (Student's t-test). (B,C,E,F,H,I) Error bars indicate s.e.m. of nine sections from three independent experiments. Scale bars: 20 µm.
Programed cell death occurs during normal cerebral cortex development (Blaschke et al., 1998), and reduced brain size could be due to increased cell death in Lin28a mutant mice. TUNEL staining of comparable coronal sections in wild-type and mutant brains showed no significant increase in TUNEL+ cells in mutants compared with wild-type controls, either prior to or during stages of observable brain size reduction (supplementary material Fig. S2A,B). Because TUNEL detects the final step of the apoptosis process, we also examined an early event of apoptosis: the activation of caspase 3. Again, we did not find any significant difference in the expression of activated caspase 3 in the cerebral cortex of the wild type and Lin28a mutants (supplementary material Fig. S2D,E).
Next, we investigated whether the decrease in NPC numbers in Lin28a−/− mutants is due to decreased cell proliferation and/or premature differentiation of NPCs. We addressed this using comparable cerebral cortex sections from wild-type and mutant embryos at E15.5, when no significant morphological or cellular differences were observed (Fig. 2B,C; supplementary material Fig. S1B), examining cells in S phase by performing 1-h bromodeoxyuridine (BrdU) labeling experiments. There was a significant reduction of BrdU+ cells in the VZ/SVZ of Lin28a−/− cerebral cortex compared with wild type (Fig. 2D,E). Since a BrdU pulse experiment only labels cells undergoing S phase at a given time, we used a phospho-histone H3 (p-H3) antibody to label mitotic cells. In E15.5 wild-type cerebral cortex, p-H3+ NPCs were mainly localized at the VZ, with some scattered cells in the SVZ (Fig. 2D, upper panel), whereas the Lin28a−/− brains exhibited a reduction of p-H3+ cells in the VZ/SVZ (Fig. 2D,F). Thus, Lin28a is required to maintain the proper NPC proliferation rate during cerebral cortex development.
To determine whether Lin28a−/− NPCs prematurely exit the cell cycle in addition to reducing their proliferation rate, we performed 24-h BrdU pulse labeling experiments at different stages of cortical development, including E15.5 and E16.5. Using Ki67 to mark cells in all phases of the cell cycle except G0, we assessed the fraction of BrdU+ cells that had exited the cell cycle within 24 h after the incorporation of BrdU (referred to as BrdU+/Ki67− cells) (Fig. 2G). Although no difference was detected at E15.5, we found a significant increase in the proportion of mutant NPCs that had exited the cell cycle compared with the wild-type control at E16.5 (Fig. 2G,H). We also quantified the number of actively proliferating cells within 24 h after BrdU labeling (referred to as BrdU+/Ki67+), and found that actively proliferating cells are slightly reduced in the Lin28a mutants compared with the wild-type control at E16.5 (Fig. 2I). Together, these data indicate that Lin28a is required for NPCs to maintain an appropriate proliferation rate and prevent premature cell cycle exit during cerebral cortex development.
Lin28a and Lin28b are both required for normal NPC proliferation and brain development
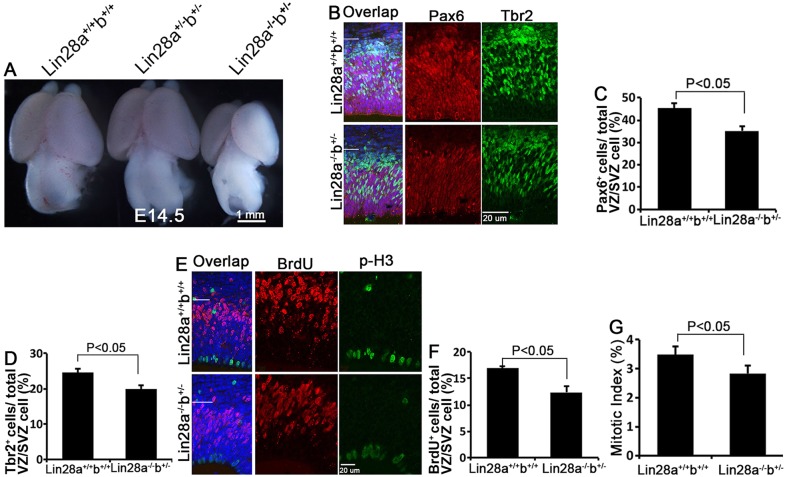
The high protein sequence similarity between Lin28a and Lin28b (supplementary material Fig. S1A) and their similar protein expression patterns during cerebral cortex development (supplementary material Fig. S1E) prompted us to examine whether they play redundant roles in brain development. It has been reported that Lin28b null mice exhibit no detectable developmental phenotypes (Shinoda et al., 2013). We crossed Lin28a+/− with Lin28b+/− mice to produce compound heterozygotes, which were intercrossed to generate various genotypes including double-homozygous knockout (DKO) offspring. No DKO mice were found postnatally or between E14.5 and E18.5 (Table 1), suggesting early lethality of DKO embryos. By contrast, Lin28a−/−; Lin28b+/− (referred to as Lin28a−/−b+/−) compound heterozygotes were recovered at normal Mendelian ratios (12.5%) before birth (Table 1), suggesting that they can survive through development. Intriguingly, Lin28a−/−b+/− compound heterozygotes had visibly smaller brains than littermate controls at E14.5 with more than 90% penetrance (Fig. 3A), and brain size reduction was more severe than with Lin28a−/− embryos, which showed a normal brain size at least a day later in development. Therefore, we focused our NPC behavior analysis on the cerebral cortex of Lin28a−/−b+/− compound mutants.
Table 1.
Genotypes of 91 embryos from E14.5 to E18.5
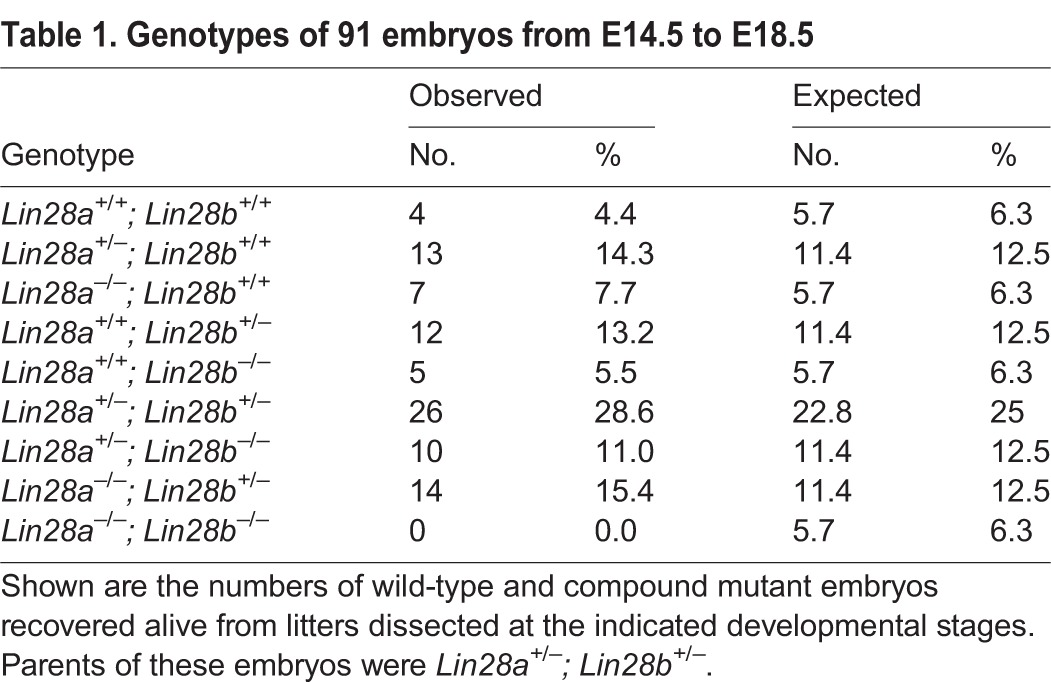
Fig. 3.
Lin28a and Lin28b have overlapping functions in NPC proliferation and brain development. (A) Reduced brain size in Lin28a−/−b+/− compound mutants compared with wild type and Lin28a/b double heterozygotes at E14.5. (B) Confocal micrographs of coronal sections from E14.5 wild-type and Lin28a−/−b+/− cortex stained with antibodies against Pax6 (red) or Tbr2 (green). Hoechst stains nuclei (blue). The VZ/SVZ regions (below the white line) are enlarged to the right. (C,D) Percentage of Pax6+ (C) or Tbr2+ (D) cells among total cells within the VZ/SVZ (below the white line in the experiment in B). (E) Confocal micrographs of coronal sections from E14.5 wild-type and Lin28a−/−b+/− compound mutant cortex stained with antibodies against p-H3 (green) and BrdU (red) 1 h after a single pulse label of BrdU. The VZ/SVZ regions (below the white line) are enlarged to the right. (F,G) Percentage of BrdU+ cells (F) and p-H3+ cells (G) among total cells within the VZ/SVZ (below the white line in the experiment in E). (C,D,F,G) Error bars indicate s.e.m. of nine sections from three independent experiments; P<0.05 (Student's t-test). Scale bars: 1 mm in A; 20 µm in B,E.
The defect in NPC numbers and proliferation was also more severe in Lin28a−/−b+/− compound mutants than in Lin28a−/− embryos. NPCs were significantly decreased at E14.5 in Lin28a−/−b+/− cerebral cortex, as evidenced by reduced numbers of Pax6+ APs and Tbr2+ INPs compared with controls (Fig. 3B-D), whereas Lin28a knockout mice start to exhibit reduced NPC numbers at E17.5 (Fig. 2A-C). Furthermore, 1-h BrdU labeling and p-H3 staining experiments suggest that there is a significant reduction of S-phase and M-phase NPCs in Lin28a−/−b+/− compared with control E14.5 cerebral cortex (Fig. 3E-G). TUNEL and activated caspase 3 assays did not show any significant difference in cell death between wild-type and Lin28a−/−b+/− cerebral cortex at different stages, including E12.5, E13.5 and E14.5 (supplementary material Fig. S2C,F). Together, these results suggest that Lin28a and Lin28b have overlapping functions in regulating NPC proliferation and brain development.
Ectopic expression of Lin28a in NPCs results in NPC expansion and an enlarged brain
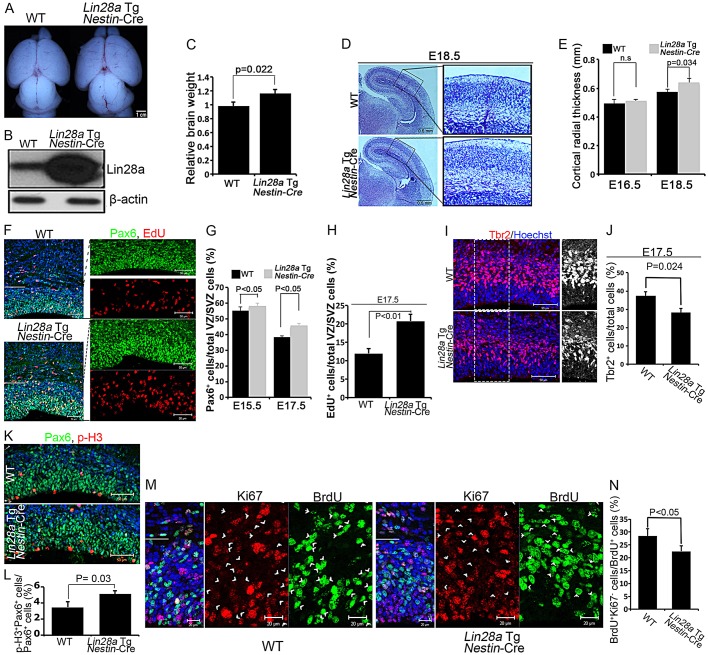
The loss-of-function studies above suggest that Lin28a/b are required for NPC proliferation and brain development. We next performed Lin28a gain-of-function studies. We created an inducible Lin28a transgenic mouse model (Lin28a Tg), in which a STOP codon flanked with loxP sequences is placed downstream of a constitutive promoter and upstream of a copy of the Lin28a gene (supplementary material Fig. S1F); Lin28a expression is induced upon the spatiotemporal expression of Cre. In the absence of Cre, the floxed Lin28a transgenic mice do not overexpress Lin28a, as indicated by our immunohistochemical staining studies at different developmental stages (supplementary material Fig. S1G,H), and there are no noticeable morphological or behavioral abnormalities. The Nestin-Cre driver induces recombination at ∼E11.5 in NPCs of the cerebral cortex (Kim et al., 2009; Tronche et al., 1999). In Lin28a Tg; Nestin-Cre embryos, Lin28a expression is significantly increased in E14.5 NPCs (Fig. 4B); brain size and weight are increased at E18.5 (Fig. 4A,C) and Hematoxylin and Eosin (H&E) staining and statistical analysis revealed slightly increased thickness of the cerebral cortex compared with the control at E18.5, although no difference was detected at E16.5 (Fig. 4D,E). We found no evidence of altered cell death in Lin28a-overexpression cerebral cortex compared with control (supplementary material Fig. S2G-J). These results suggest that Lin28a could play instructive roles in brain development.
Fig. 4.
Ectopic expression of Lin28a results in NPC expansion and enlarged brains. (A) Dorsal views of E18.5 wild-type and Nestin-Cre-directed Lin28a-overexpression mouse brains. (B) Western blot analyses of Lin28a protein expression in NPCs isolated from E14.5 wild-type and Lin28a-overexpression cerebral cortex. β-actin serves as a loading control. (C) Analysis of brain weights of E18.5 wild-type and Lin28a-overexpression brains. Error bars indicate s.e.m. from four independent experiments. P=0.022 (Student's t-test). (D) Coronal sections of E18.5 cerebral cortex stained with H&E. (E) The Lin28a-overexpression cortex shows increased radial thickness. n.s., not significant at E16.5; P=0.034 at E18.5 (Student's t-test). (F) Confocal micrographs of coronal sections from E17.5 wild-type and Lin28a-overexpression mouse cortex labeled for Pax6 (green) and with EdU (red) 1 h after a single pulse label of EdU. The VZ/SVZ is the area below the white line. (G) Percentage of Pax6+ cells among total cells within the VZ/SVZ at E15.5 and E17.5. P<0.05 (Student's t-test). (H) Percentage of EdU+ cells among total cells within the VZ/SVZ at E17.5 in the experiment in F. P<0.01 (Student's t-test). (I) E17.5 cerebral cortex samples from wild-type and Lin28a transgenic mice were immunostained with an antibody against Tbr2 (red). Tbr2+ cells from the boxed area are shown in the right-hand panel. (J) Percentage of Tbr2+ cells among total cells in the experiment in I. P=0.024 (Student's t-test). (K) Confocal micrographs of coronal sections from E17.5 wild-type and Lin28a transgenic mouse cortex stained with antibodies against Pax6 (green) and p-H3 (red). (L) Percentage of p-H3/Pax6 double-positive cells among Pax6+ cells in K. P=0.03 (Student's t-test). (M) Confocal micrographs of coronal sections from E17.5 mice 24 h after a single pulse label of BrdU; sections were stained with antibodies against BrdU (green) and Ki67 (red). Arrowheads indicate cells that exited the cell cycle (BrdU+/Ki67–). (N) Percentage of BrdU+/Ki67– cells among total BrdU+ cells in the experiment in M. P<0.05 (Student's t-test). (E,G,H,J,L,N) Error bars indicate s.e.m. of nine sections from three independent experiments. Scale bars: 1 cm in A; 0.5 mm in D; 50 µm in F,I,K; 20 µm in M.
The enlarged brain size of Lin28a Tg; Nestin-Cre mice prompted us to examine whether there is increased proliferation and numbers of NPCs in the brain. Indeed, Pax6+ APs were significantly increased in Lin28a Tg; Nestin-Cre cerebral cortex compared with the control (Fig. 4F,G) and this was detected as early as E15.5, when the cerebral cortex is similar in size in wild-type and Lin28a-overexpression mice, suggesting that an increased NPC population could be the cause of the enlarged brain. Correspondingly, there is a robust increase in the cell proliferation rate, as indicated by increased numbers of EdU+ cells as well as p-H3+ cells in the VZ/SVZ zone of Lin28a Tg; Nestin-Cre mice compared with the control (Fig. 4F,H,K,L). Interestingly, the numbers of Tbr2+ INPs were significantly reduced in Lin28a Tg; Nestin-Cre cerebral cortex compared with the control (Fig. 4I,J). Together, these data suggest that Lin28a overexpression results in the amplification of Pax6+ APs and in an imbalance in the conversion to INPs, and this could preferentially generate neurons at the expense of INPs.
We assayed whether Lin28a overexpression delays NPC cell cycle exit in the VZ/SVZ by applying a single BrdU pulse 24 h prior to sacrifice and Ki67 labeling. Consistent with the observation that loss of Lin28a results in premature cell cycle exit (Fig. 2G-I), we found that Lin28a overexpression significantly inhibits the cell cycle exit of NPCs compared with the control (Fig. 4M,N). Together, these results indicate that Lin28a plays regulatory roles in NPC proliferation and brain development.
Lin28a interacts with Imp1 to regulate NPC proliferation
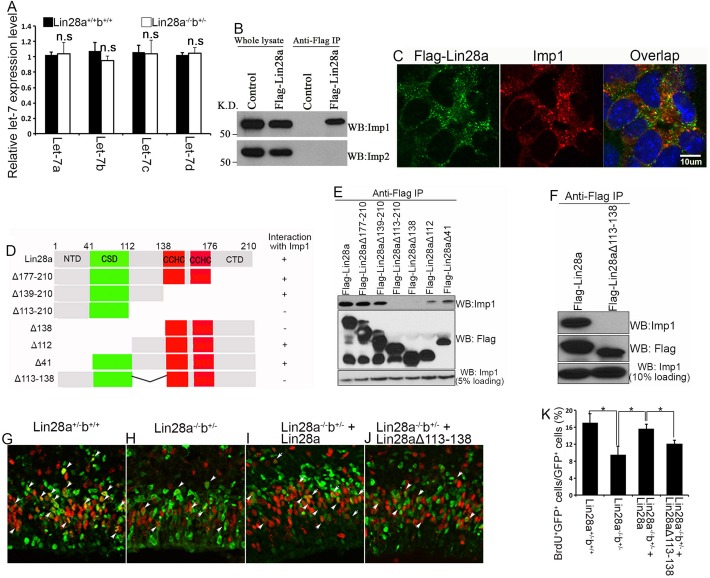
Lin28 could function in NPCs through miRNA let-7-dependent and/or -independent mechanisms (Balzer et al., 2010; Thornton and Gregory, 2012; Zhu et al., 2010). We examined let-7 (let7a-d) expression in NPCs isolated from the cerebral cortex of E14.5 wild-type and Lin28a−/−b+/− embryos, when NPC proliferation, numbers and brain size are reduced (Fig. 3). However, we did not detect any significant difference in let-7 expression in NPCs between wild-type and Lin28a−/−b+/− NPCs (Fig. 5A).
Fig. 5.
Lin28a interacts with Imp1. (A) miRNA RT-PCR analysis of let-7 expression levels in NPCs isolated from E14.5 cerebral cortex of wild type and Lin28a−/−b+/− compound mutants. n.s., not significant (P>0.05) (Student's t-test). Error bars indicate s.e.m. from three independent experiments. (B) Lin28a BAC with Flag tag was used to generate the NE-4C stable cell line. Lin28a protein was immunoprecipitated (IP) with m2 beads (anti-Flag) followed by western blot (WB) analysis using an antibody against Imp1 or Imp2. K.D. indicates kDa. (C) Confocal micrographs of Flag-Lin28a stable NE-4C cells stained with antibodies against Flag (Lin28a, green) and Imp1 (red). Hoechst stains nuclei (blue). Scale bar: 10 µm. (D) Schematic representation of Lin28a full-length and deletion constructs. The interval between the RNA-binding CSD and CCHC domains of Lin28a is required for its interaction with Imp1. (E,F) Total lysates from HEK 293T cells transfected with Flag-Lin28a or various deletion constructs were immunoprecipitated with anti-Flag antibody and western blot analysis was performed using the antibodies indicated. (G-J) Twenty-four hours after in utero electroporation of the indicated plasmids, together with pCAG-H2BGFP constructs, into E14.5 brains, the animals were administered a single pulse label of BrdU for 1 h followed by immunostaining of cerebral cortex for GFP (green) and BrdU (red). Arrowheads indicate GFP/BrdU double-positive cells. (K) Quantification of GFP/BrdU double-positive cells among total GFP+ cells from the experiment in G-J. Data are expressed as s.e.m. of six sections from two independent experiments. *P<0.05 (Student's t-test).
To broaden our viewpoint of Lin28a/b functional mechanisms, we sought to identify Lin28a-associated proteins in NE-4C cells, a cell line established from the cerebral vesicle of E9.0 mouse embryos and which mimics NPC proliferation and differentiation in vitro (Varga et al., 2008). We generated a genetically modified Lin28a bacterial artificial chromosome (BAC) vector that encodes a Lin28a fusion protein containing two N-terminal tags: EGFP and Flag (supplementary material Fig. S3A). An NE-4C stable cell line with the genetically modified Lin28a BAC was used for affinity purification (supplementary material Fig. S3B). Of ∼40 proteins pulled down with Lin28a (supplementary material Fig. S3C,D), Imp1 (also known as Igf2bp1) was of particular interest because our previous studies have shown that Lin28a is associated with Imp1-3 in skeletal muscle (Polesskaya et al., 2007). In addition, recent studies suggest that Imp1 plays essential roles in NPC self-renewal during brain development (Nishino et al., 2013). We confirmed that Lin28a specifically interacts with Imp1, and not its homolog Imp2 (Igf2bp2), in NE-4C cells (Fig. 5B). Furthermore, Lin28a colocalizes with Imp1 in the cytoplasm of NE-4C cells (Fig. 5C). Deletion analysis indicates that the interaction between Lin28a and Imp1 is specific and requires the region between the RNA-binding CSD and CCHC domains of Lin28a (Fig. 5D-F). These data suggest that Lin28a is associated with Imp1 in NE-4C cells.
To examine the functional importance of the domain that interacts with Imp1, we used in utero electroporation to label individual NPCs and to introduce full-length Lin28a or the Lin28a deletion mutant (Lin28aΔ113-138) into NPCs of Lin28a−/−b+/− compound mutants. BrdU labeling experiments show that wild-type Lin28a almost fully rescues the proliferation deficiency of NPCs seen in Lin28a−/−b+/− mutants (Fig. 5G-K). By contrast, Lin28aΔ113-138 exhibits significantly reduced rescue effects (Fig. 5G-K). These studies suggest that the region of Lin28a that interacts with Imp1 is functionally important in regulating NPC proliferation during brain development.
Lin28 regulates Igf2-mTOR signaling in NPCs
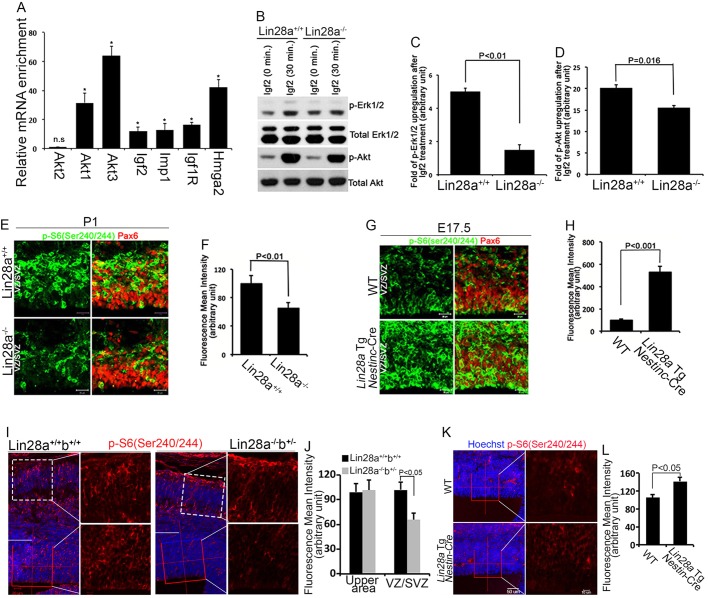
Recent genome-wide studies have identified Lin28a and Imp1 targets in HEK 293T cells (Hafner et al., 2013, 2010). Our finding that Lin28a and Imp1 interact in NE-4C cells prompted us to examine their common targets following RNA immunoprecipitation (RIP) to isolate Lin28a-associated RNAs in NE-4C cells. This showed that Lin28a is associated with mRNAs encoding Hmga2, a regulator of NPC self-renewal (Nishino et al., 2008), and several components of the Igf2-mTOR signaling pathway, including Igf2, Igf1r, Akt1/3 and Imp1 (Fig. 6A). Therefore, we next tested the hypothesis that Lin28a/b regulates Igf2-mTOR signaling by examining the responses of wild-type and Lin28a−/− cerebral cortex tissues to Igf2 stimulation. Cortex tissues were dissected from the lateral pallium of mouse brains at E12.5, when more than 85% of the cells are NPCs, as evidenced by Pax6 expression (data not shown), and cultured the tissue for 30 min with Igf2. We used phosphorylation of Erk1/2 (Mapk3/1) at Thr202/204 (p-Erk1/2) and of Akt at Ser473 (p-Akt) as readouts of Igf2 signaling activity (Lehtinen et al., 2011). Western blot results showed that Igf2 treatment significantly increased p-Erk1/2 and p-Akt levels in wild-type cortex tissue, whereas this response was significantly blunted in Lin28a mutant tissue (Fig. 6B-D). These studies indicate that the response of Lin28a mutant cerebral cortex to Igf2 stimulation is compromised.
Fig. 6.
Lin28a/b regulate Igf2-mTOR signaling in NPCs. (A) Using the NE-4C stable cell line with Flag-tagged Lin28a, mRNAs associated with Lin28a were pulled down using m2 (Lin28a) antibodies followed by RT-PCR analyses with the probes indicated. Error bars indicate s.e.m. of three independent experiments. *P<0.05; n.s., not significant (P>0.05) (Student's t-test). (B) Western blot analyses of the levels of p-Erk1/2 (Thr202/204) and p-Akt (Ser473) using embryonic cortex tissues dissected from the lateral pallium of E12.5 mouse brains. The embryonic cortices were grown in DMEM in the presence or absence of 20 ng/ml Igf2 for 30 min. Total Erk1/2 and total Akt serve as controls. (C,D) Quantification of western blot data in experiment B using three independent blots. Densitometry of blot signals was quantified with ImageJ software (NIH); levels of p-Erk1/2 or p-Akt in samples with Igf2 treatment were normalized to respective controls without Igf2 treatment. Error bars indicate s.e.m. from three independent experiments. P<0.01 (C) and P=0.016 (D) (Student's t-test). (E,G) Confocal micrographs of coronal sections of cerebral cortex stained with antibodies against Pax6 (red) and p-S6 (Ser240/244) (green). Hoechst stains nuclei (blue). The erebral cortex tissues are from P1 wild-type and Lin28a−/− mutant mice (E) or from E17.5 wild-type and Nestin-Cre-directed Lin28a-overexpression mice (G). (F,H) Statistical analysis of fluorescence intensity of p-S6 (Ser240/244) in the VZ/SVZ of the cerebral cortex. P<0.01 (F) and P<0.001 (H) (Student's t-test). (I,K) Confocal micrographs of coronal sections from E14.5 Lin28a−/−b+/− compound mutant (I) or Lin28a-overexpression (K) cortex stained with antibodies against p-S6 (Ser240/244). Hoechst stains nuclei (blue). The VZ/SVZ is the area below the white line. (J,L) Statistical analysis of fluorescence intensity of p-S6 (Ser240/244) at the VZ/SVZ in experiments in I or K. P<0.05 (Student's t-test). (F,H,J,L) Error bars indicate s.e.m. of nine sections from three independent experiments. Scale bars: 20 µm in E,G and I left; 50 µm in K left; 10 µm in I,K right.
To examine whether Igf2-mTOR signaling is disrupted in vivo, we used phosphorylation of the S6 ribosome protein (Ser240/244 or Ser235/236; p-S6) as a downstream readout of Igf2-mTOR signaling activity (Ferrari et al., 1991; Lehtinen et al., 2011; Magri et al., 2011). mTOR activity was reduced in NPCs (labeled by Pax6) of the Lin28a knockout, as evidenced by a significant decrease of p-S6 levels compared with controls at P1 (Fig. 6E,F; supplementary material Fig. S4A,B) and at E14.5 in Lin28a−/−b+/− brains (Fig. 6I,J), at a time when let-7 expression levels are unchanged in the mutant NPCs (Fig. 5A). These data suggest that the integrity of Igf2-mTOR signaling is compromised in NPCs of Lin28 loss-of-function mutants. Consistent with the idea that Lin28a positively regulates Igf2-mTOR signaling, the overexpression of Lin28a significantly increased p-S6 staining in the Lin28a Tg; Nestin-Cre mouse NPCs compared with controls (Fig. 6G,H; supplementary material Fig. S4C,D) and specifically in the VZ/SVZ of the cerebral cortex as early as E14.5 (Fig. 6K,L). Together, these data suggest that Lin28a regulates Igf2-mTOR signaling in NPCs in vivo.
Igf1r and Hmga2 are functionally important targets of Lin28 in regulating NPC proliferation
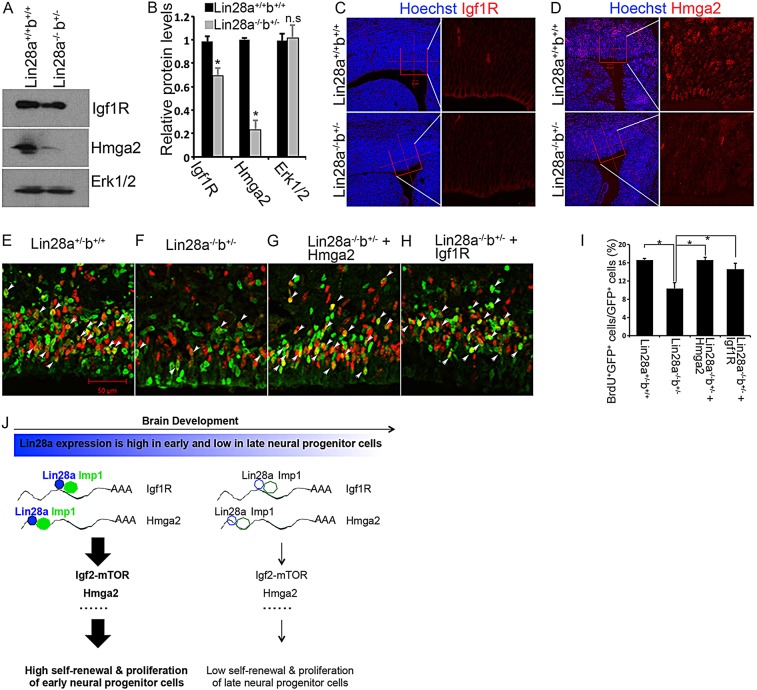
To further understand the molecular mechanisms by which Lin28 regulates Igf2-mTOR signaling and NPC proliferation, we examined the expression of proteins encoded by Lin28a/b-associated mRNAs as confirmed in our RIP studies in NE-4C cells (Fig. 6A). The protein levels of Igf1r, Akt3 and Hmga2 were significantly reduced in NPCs isolated from Lin28a−/−b+/− cerebral cortex (Fig. 7A,B; supplementary material Fig. 4G,H). By contrast, Akt1, Akt2 and Imp1 protein levels were not significantly changed in Lin28a−/− or Lin28a−/−b+/− NPCs compared with the control (supplementary material Fig. 4E-H). Immunohistochemical staining confirmed the reduction in Igf1r and Hmga2 in the VZ of the Lin28a−/−b+/− cerebral cortex (Fig. 7C,D). These studies suggest that Lin28a/b regulate the protein expression of Igf1r and Hmga2 in vivo.
Fig. 7.
Igf1r and Hmga2 are functionally important targets of Lin28a/b. (A) Western blot analysis of Igf1r and Hmga2 expression using lysates from NPCs of wild-type and Lin28a−/−b+/− compound mutant cerebral cortex. (B) Quantification of western blot data in experiment A using three independent blots. n.s., not significant; *P<0.05 (Student's t-test). Error bars indicate s.e.m. from three independent experiments. (C,D) Confocal micrographs of coronal sections from E14.5 wild-type and Lin28a−/−b+/− cerebral cortex stained with antibodies against Igf1r (red in C) and Hmga2 (red in D). Hoechst stains nuclei (blue). (E-H) Twenty-four hours after in utero electroporation of the indicated plasmids, together with pCAG-H2BGFP constructs, into E14.5 brains, the animals were administered with a single pulse label of BrdU for 1 h followed by immunostaining of cerebral cortex for GFP (green) and BrdU (red). Arrowheads indicate GFP/BrdU double-positive cells. (I) Quantification of GFP/BrdU double-positive cells among total GFP+ cells from the experiment in E-H. Data are expressed as s.e.m. of six sections from two independent experiments. *P<0.05 (Student's t-test). (J) In the working model, Lin28a interacts with Imp1 to regulate the levels of Igf1r and Hmga2 protein, therefore providing early NPCs with higher self-renewal capacity as compared with late NPCs in the developing brain. Scale bars: 20 µm in C,D left; 10 µm in C,D right; 50 µm in E-H.
Previous studies suggest that Igf2-mTOR signaling and Hmga2 are crucial regulators of NPC self-renewal and brain development (Lehtinen et al., 2011; Magri et al., 2011; Nishino et al., 2008). To examine the functional importance of Igf1r/Hmga2 downregulation in mediating Lin28a/b functions in NPC proliferation, we performed rescue experiments using the in utero electroporation method. BrdU labeling studies showed that ectopic expression of Hmga2 or Igf1r significantly rescued the proliferation deficiency of Lin28a−/−b+/− NPCs (Fig. 7E-I). Together, these data suggest that Lin28a/b regulate NPC proliferation, at least in part, through the regulation of Igf1r and Hmga2 protein expression in the developing brain.
DISCUSSION
Lin28a/b play essential roles in a variety of biological processes and diseases, including ESC self-renewal, iPSC formation, cancers and type II diabetes (Peng et al., 2011; Shyh-Chang and Daley, 2013; Thornton and Gregory, 2012; Yu et al., 2007). However, the roles of Lin28a/b in somatic progenitor cells remain largely unknown, and their potential in vivo functions in neural stem/progenitor cells and brain development have not been examined. Here, our gain- and loss-of-function studies have identified Lin28a/b as crucial regulators for the temporal control of NPC proliferation during mouse brain development (Fig. 7J).
Our previous studies suggest that Lin28 regulates neurogliogenesis in vitro (Vadla et al., 2012). Recent studies of human ESC-differentiated NPCs suggest that LIN28 is capable of rescuing NPC proliferation and neurogenic deficits in the absence of SOX2 (Cimadamore et al., 2013). Our current studies provide substantial new information about Lin28a/b functions in vivo. First, we found that Lin28a is not only necessary but also sufficient for promoting NPC proliferation in mice. Second, this Lin28a-mediated NPC regulation is functionally important for brain development because Lin28a loss- and gain-of-function mice exhibit smaller and enlarged brains, respectively. Third, whereas it is unclear if Lin28a and Lin28b play distinct or redundant roles, our studies suggest that they have overlapping functions in regulating NPC proliferation and brain development. The RNA-binding proteins Lin28 and Lin41 are founding members of the heterochronic pathway, which was originally characterized in C. elegans, and are required for the progression of specific developmental stages in the seam cells (Ambros, 2011; Blaschke et al., 1998; Nimmo and Slack, 2009; Rougvie and Moss, 2013). Previously, we showed that mammalian Lin41 (Trim71 in mouse) regulates the balance of NPC self-renewal and differentiation during neural development, and that loss of Lin41 results in precocious NPC differentiation (Chen et al., 2012); our current study suggests that Lin28a/b promote the temporal proliferation capacities of NPCs and brain development in mice. Thus, these heterochronic genes play evolutionarily conserved roles to regulate progenitor cell behaviors across different species.
Lin28a/b are highly expressed in early NPCs and their expression is almost absent in the cerebral cortex after E14.5, based on western blot analysis (supplementary material Fig. S1E), whereas E15.5 is the earliest stage at which the cellular phenotypes can be detected in the Lin28a−/− cerebral cortex (Fig. 2D-F). There are several possible explanations for these observations. First, the early molecular changes, such as the compromised Igf2 signaling in Lin28a−/− NPCs at E12.5 (Fig. 6B-D), are not sufficient to cause detectable cellular/morphological phenotypes until later developmental stages, such as E15.5. Second, Lin28a/b have redundant functions, as suggested by our analysis of Lin28a−/−b+/− compound mutants (Fig. 3), which might lead to the late manifestation of cellular phenotypes in the Lin28a single-knockout cerebral cortex. Lastly, Lin28a is locally restricted to the VZ/SVZ of the cerebral cortex at late developmental stages, based on immunohistochemical staining (Fig. 1I). The near absence of Lin28a in western blot assays (supplementary material Fig. S1E) could be due to the fact that the protein lysate is from a mixture of NPCs and differentiated neurons.
Mechanistic studies of Lin28a/b functions have mainly focused on inhibition of miRNA let-7 biogenesis (Shyh-Chang and Daley, 2013; Thornton and Gregory, 2012). Our previous in vitro studies suggest that Lin28a regulates neurogliogenesis through let-7-independent mechanisms (Balzer et al., 2010). Here, our in vivo evidence further suggests that Lin28a/b may function through let-7-independent mechanisms to regulate NPC proliferation. First, let-7 expression levels remain unchanged in NPCs isolated from E14.5 Lin28a−/−b+/− compound mutants (Fig. 5A), which already exhibit smaller brains and reduced NPC numbers compared with controls (Fig. 3A-D). Second, Lin28a is associated with mRNAs encoding Hmga2 and components of the Igf2-mTOR pathway, including Igf2, Igf1r, Akt1/3 and Imp1 (Fig. 6A). In the E14.5 NPCs of Lin28a−/−b+/− mutants, the protein levels of Hmga2 and Igf1r are reduced compared with controls (Fig. 7A-D). Importantly, ectopic expression of Hmga2 or Igf1r can partially rescue the proliferation defects of Lin28a−/−b+/− NPCs, suggesting that these downstream targets are functionally important in mediating Lin28a/b regulation of NPCs. Together, these results suggest that Lin28a/b regulate NPC proliferation, at least in part, through modulation of Igf2-mTOR signaling activities and the production of Hmga2 protein (Fig. 7J).
A recent study showed that Imp1 temporally regulates NPC self-renewal in the developing brain (Nishino et al., 2013). Interestingly, we found that Lin28a specifically associates with Imp1 and not its homolog Imp2 (Fig. 5B), and that the Imp1 protein level is unchanged in Lin28a−/−b+/− NPCs. Thus, epistasis analysis will be informative in defining the genetic interactions between Lin28a and Imp1 in NPC regulation. It has been shown that Imp1 increases protein production by enhancing the stability of its target mRNA Hmga2 (Nishino et al., 2013), whereas Lin28a regulates mRNA translation, based on studies in cultured cells (Cho et al., 2012; Hafner et al., 2013). Therefore, examining the mRNA expression levels of Lin28 candidate targets, including Hmga2 and Igf1r, in Lin28a/b genetically modified mouse brains will further advance our understanding of Lin28a/b functional mechanisms.
MATERIALS AND METHODS
Lin28a/b mutant and Lin28a transgenic mice
The generation of Lin28a knockout and inducible Lin28a transgenic mice is described in the supplementary Materials and Methods. Lin28b knockout mice were kindly provided by the Dr George Daley laboratory, and have been described elsewhere (Shinoda et al., 2013).
Mutant phenotypic and BrdU/EdU labeling analyses
Histological processing, TUNEL assays, BrdU/EdU labeling and immunohistochemical labeling of cryosections were performed as described previously (Chen et al., 2014); details are provided in the supplementary Materials and Methods and antibodies are listed in Table S1.
RNA immunoprecipitation (RIP)
To examine Lin28a-associated RNAs, we grew Flag-tagged Lin28a stable NE-4C cells in 6×100 mm plates to 100% confluence. Cells were then cross-linked for 4 min using 245 nm UV, and collected using 1 ml of lysis buffer [50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP40, and one tablet of protease inhibitor (Roche) per 10 ml]. Cell lysate was incubated on ice for 15 min, vortexed three times during incubation, and centrifuged at 13,000 rpm (15,700 g) for 15 min. Supernatant was collected in fresh tubes, and myc beads as a control (50 µl for three plates; Sigma) and Flag beads (50 µl for three plates; Sigma) were added. The lysate was rotated with beads at 4°C for 3 h followed by centrifugation at 6000 rpm (3300 g) for 2 min. Supernatant was removed and the beads washed four times with 1 ml wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP40) for 10 min each. Trizol (Invitrogen) was used to extract RNAs followed by RT-PCR analyses.
Affinity purification of Lin28a-associated proteins
Lin28a-interacting proteins were purified from genetically modified Lin28a BAC-transfected NE-4C mouse cells as detailed in the supplementary Materials and Methods.
Quantitative RT-PCR
For the mRNA RT-PCR analyses in Fig. 6A, RNA samples were from RIP experiments as described above and the probes listed were obtained from Life Technologies. For the miRNA let-7 RT-PCR in Fig. 5A, E14.5 wild-type and Lin28a−/−b+/− dorsal telencephalon tissue was microdissected, and isolated NPCs were cultured in adherent culture medium overnight or in self-renewal medium for 5 days before RNA collection (for NPC isolation and culture see the supplementary Materials and Methods). RNAs were extracted using Trizol and converted into cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Stem-loop reverse transcription primers for individual let-7 members were used for the reverse transcription reaction, followed by the real-time PCR analysis, which provides quantitation of mature let-7 miRNA expression.
In utero electroporation
Mouse cortical NPCs were electroporated with various plasmids as described previously (Chen et al., 2014). Briefly, DNA constructs were prepared using the EndoFree Plasmid Maxi Kit (Qiagen), including pCAG-H2BGFP, pCAG-Lin28a, pCAG-Lin28aΔ113-138, pCAG-Igf1r and pCAG-Hmga2. Individual plasmids or combinations, as indicated (Fig. 5G-J and Fig. 7E-H), plus 0.5% Fast Green (Sigma) were injected into the lateral ventricle of E14.5 brains followed by electroporation using an ECM 830 electroporator (BTX) with four 100 ms pulses separated by 100 ms intervals at 32 V. To improve retention of the pregnancy, we avoided plasmid injection and electroporation of the two embryos next to the upper vagina. In utero development was allowed to continue for 24 h (or as indicated) followed by intraperitoneal injection of BrdU at 100 mg/kg body weight for 1 h before embryo dissection. E15.5 brains were dissected and processed for immunohistochemical staining analysis as described above.
Supplementary Material
Acknowledgements
We thank Lori Bulwith for technical assistance and our laboratory colleagues for stimulating discussions; Dr F. C. Nielsen for Imp1, Imp2 and Imp3 antibodies; Dr Anthony A. Hyman for the R6Kamp-hNGFP plasmid used for the generation of tagged Lin28a BAC; and the Dr George Daley laboratory for sharing Lin28b knockout mice.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
We are grateful for the support of The National Institutes of Health (NIH)/NICHD [K99HD073269 to J.-F.C.] and the work was supported in part by the University of Georgia start-up fund, NIH [HD081562 and NS085749 to L.N.], NIH/NCATS Colorado CTSI [UL1 TR000154 to L.N.], the Neuroscience Program [NS48154 to L.N.], the Colorado IDDRC (Animal Models Core; to L.N.) and the National Science Foundation (NSF) [IOS-0924497 to E.G.M.]. Deposited in PMC for release after 12 months.
Author contributions
M.Y., S.-L.Y., C.L. and J.-F.C. conceived and performed all experiments. S.H., E.G.M. and L.N. helped with the manuscript writing. R.D. and A.N. generated Lin28a transgenic mice. M.D. and K.C.H. performed mass spectrophotometry. J.-F.C. designed and interpreted the experiments and wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.120543/-/DC1
References
- Abuzzahab M. J., Schneider A., Goddard A., Grigorescu F., Lautier C., Keller E., Kiess W., Klammt J., Kratzsch J., Osgood D. et al. (2003). IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. N. Engl. J. Med. 349, 2211-2222 10.1056/NEJMoa010107 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2011). MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21, 511-517 10.1016/j.gde.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J., Liu J. P., Robertson E. J. and Efstratiadis A. (1993). Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75, 73-82 10.1016/0092-8674(93)90680-O [DOI] [PubMed] [Google Scholar]
- Balzer E., Heine C., Jiang Q., Lee V. M. and Moss E. G. (2010). LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 137, 891-900 10.1242/dev.042895 [DOI] [PubMed] [Google Scholar]
- Blaschke A. J., Weiner J. A. and Chun J. (1998). Programmed cell death is a universal feature of embryonic and postnatal neuroproliferative regions throughout the central nervous system. J. Comp. Neurol. 396, 39-50 [DOI] [PubMed] [Google Scholar]
- Chen J., Lai F. and Niswander L. (2012). The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 26, 803-815 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-F., Zhang Y., Wilde J., Hansen K. C., Lai F. and Niswander L. (2014). Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5, 3885 10.1038/ncomms4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J., Chang H., Kwon S. C., Kim B., Kim Y., Choe J., Ha M., Kim Y. K. and Kim V. N. (2012). LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765-777 10.1016/j.cell.2012.10.019 [DOI] [PubMed] [Google Scholar]
- Cimadamore F., Amador-Arjona A., Chen C., Huang C.-T. and Terskikh A. V. (2013). SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. USA 110, E3017-E3026 10.1073/pnas.1220176110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R. A. M., Bulfone A., Kowalczyk T. and Hevner R. F. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247-251 10.1523/JNEUROSCI.2899-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Bandi H. R., Hofsteenge J., Bussian B. M. and Thomas G. (1991). Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J. Biol. Chem. 266, 22770-22775. [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P. and Pevny L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749-765 10.1016/S0896-6273(03)00497-5 [DOI] [PubMed] [Google Scholar]
- Guo Y., Chen Y., Ito H., Watanabe A., Ge X., Kodama T. and Aburatani H. (2006). Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384, 51-61 10.1016/j.gene.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129-141 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Max K. E. A., Bandaru P., Morozov P., Gerstberger S., Brown M., Molina H. and Tuschl T. (2013). Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 19, 613-626 10.1261/rna.036491.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Nakada D. and Morrison S. J. (2009). Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol. 25, 377-406 10.1146/annurev.cellbio.042308.113248 [DOI] [PubMed] [Google Scholar]
- Johansson P. A., Cappello S. and Götz M. (2010). Stem cells niches during development—lessons from the cerebral cortex. Curr. Opin. Neurobiol. 20, 400-407 10.1016/j.conb.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Kim W.-Y., Wang X., Wu Y., Doble B. W., Patel S., Woodgett J. R. and Snider W. D. (2009). GSK-3 is a master regulator of neural progenitor homeostasis. Nat. Neurosci. 12, 1390-1397 10.1038/nn.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi Y., Fujii Y., Hirabayashi Y. and Gotoh Y. (2012). HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat. Neurosci. 15, 1127-1133 10.1038/nn.3165 [DOI] [PubMed] [Google Scholar]
- Laplante M. and Sabatini D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274-293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M. K., Zappaterra M. W., Chen X., Yang Y. J., Hill A. D., Lun M., Maynard T., Gonzalez D., Kim S., Ye P. et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893-905 10.1016/j.neuron.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D. A., Huang Y.-C., Swigut T., Mirick A. L., Garcia-Verdugo J. M., Wysocka J., Ernst P. and Alvarez-Buylla A. (2009). Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529-533 10.1038/nature07726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-P., Baker J., Perkins A. S., Robertson E. J. and Efstratiadis A. (1993). Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59-72 10.1016/S0092-8674(05)80084-4 [DOI] [PubMed] [Google Scholar]
- Magri L., Cambiaghi M., Cominelli M., Alfaro-Cervello C., Cursi M., Pala M., Bulfone A., Garcia-Verdugo J. M., Leocani L., Minicucci F. et al. (2011). Sustained activation of mTOR pathway in embryonic neural stem cells leads to development of tuberous sclerosis complex-associated lesions. Cell Stem Cell 9, 447-462 10.1016/j.stem.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Moss E. G. and Tang L. (2003). Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258, 432-442 10.1016/S0012-1606(03)00126-X [DOI] [PubMed] [Google Scholar]
- Moss E. G., Lee R. C. and Ambros V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637-646 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- Nimmo R. A. and Slack F. J. (2009). An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma 118, 405-418 10.1007/s00412-009-0210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K. and Morrison S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227-239 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim S., Zhu Y., Zhu H. and Morrison S. J. (2013). A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. Elife 2, e00924 10.7554/eLife.00924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kusky J. and Ye P. (2012). Neurodevelopmental effects of insulin-like growth factor signaling. Front. Neuroendocrinol. 33, 230-251 10.1016/j.yfrne.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Chen L.-L., Lei X.-X., Yang L., Lin H., Carmichael G. G. and Huang Y. (2011). Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 29, 496-504 10.1002/stem.591 [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E. G. and Harel-Bellan A. (2007). Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21, 1125-1138 10.1101/gad.415007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E. and Moss E. G. (2013). Developmental transitions in C. elegans larval stages. Curr. Top. Dev. Biol. 105, 153-180 10.1016/B978-0-12-396968-2.00006-3 [DOI] [PubMed] [Google Scholar]
- Salomoni P. and Calegari F. (2010). Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 20, 233-243 10.1016/j.tcb.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Sessa A., Mao C.-a., Hadjantonakis A.-K., Klein W. H. and Broccoli V. (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56-69 10.1016/j.neuron.2008.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda G., Shyh-Chang N., de Soysa T. Y., Zhu H., Seligson M. T., Shah S. P., Abo-Sido N., Yabuuchi A., Hagan J. P., Gregory R. I. et al. (2013). Fetal deficiency of Lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 31, 1563-1573 10.1002/stem.1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N. and Daley G. Q. (2013). Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395-406 10.1016/j.stem.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J. E. and Gregory R. I. (2012). How does Lin28 let-7 control development and disease? Trends Cell Biol. 22, 474-482 10.1016/j.tcb.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R. and Schütz G. (1999). Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99-103 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Vadla B., Kemper K., Alaimo J., Heine C. and Moss E. G. (2012). lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 8, e1002588 10.1371/journal.pgen.1002588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga B. V., Hádinger N., Gócza E., Dulberg V., Demeter K., Madarász E. and Herberth B. (2008). Generation of diverse neuronal subtypes in cloned populations of stem-like cells. BMC Dev. Biol. 8, 89 10.1186/1471-213X-8-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K. A., Camacho-Hübner C., Barter D., Clark A. J. L. and Savage M. O. (1997). Insulin-like growth factor I gene deletion causing intrauterine growth retardation and severe short stature. Acta Paediatr. 86Suppl. 423, 39-45 10.1111/j.1651-2227.1997.tb18367.x [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Hashimoto M., Shimizu H., Ueno-Kudoh H., Uchibe K., Kimura I. and Asahara H. (2008). Dynamic gene expression of Lin-28 during embryonic development in mouse and chicken. Gene Expr. Patterns 8, 155-160 10.1016/j.gep.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R. et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917-1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zhu H., Shah S., Shyh-Chang N., Shinoda G., Einhorn W. S., Viswanathan S. R., Takeuchi A., Grasemann C., Rinn J. L., Lopez M. F. et al. (2010). Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 42, 626-630 10.1038/ng.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.