Hao et al. report that a distant anti-silencer element interacts with the Rag1 and Rag2 gene promoters in double-positive thymocytes and that SATB1 is a regulator of Rag locus organization in these cells.
Abstract
Rag1 and Rag2 gene expression in CD4+CD8+ double-positive (DP) thymocytes depends on the activity of a distant anti-silencer element (ASE) that counteracts the activity of an intergenic silencer. However, the mechanistic basis for ASE activity is unknown. Here, we show that the ASE physically interacts with the distant Rag1 and Rag2 gene promoters in DP thymocytes, bringing the two promoters together to form an active chromatin hub. Moreover, we show that the ASE functions as a classical enhancer that can potently activate these promoters in the absence of the silencer or other locus elements. In thymocytes lacking the chromatin organizer SATB1, we identified a partial defect in Tcra gene rearrangement that was associated with reduced expression of Rag1 and Rag2 at the DP stage. SATB1 binds to the ASE and Rag promoters, facilitating inclusion of Rag2 in the chromatin hub and the loading of RNA polymerase II to both the Rag1 and Rag2 promoters. Our results provide a novel framework for understanding ASE function and demonstrate a novel role for SATB1 as a regulator of Rag locus organization and gene expression in DP thymocytes.
The diverse antigen receptor repertoires of T and B lymphocytes are generated by a site-specific DNA recombination process that assembles antigen receptor variable (V), diversity (D), and joining (J) gene segments in developing lymphocytes. This process, known as V(D)J recombination, is initiated by a protein complex composed of recombination activating genes 1 and 2 (RAG1 and RAG2), which can recognize and cleave at recombination signal sequences (RSSs) that flank TCR and immunoglobulin V, D, and J gene segments (Schatz and Swanson, 2011). The Rag1 and Rag2 genes display a distinctive, tightly linked genomic organization with stringently and coordinately regulated expression during T and B lymphocyte development (Kuo and Schlissel, 2009).
There are two developmental windows of Rag1 and Rag2 (hereafter, Rag) gene expression during T and B lymphocyte development (Kuo and Schlissel, 2009). In developing thymocytes, the Rag genes are first expressed at the CD4−CD8− double-negative (DN) stage to promote recombination of the Tcrb, Tcrg, and Tcrd genes. Productive Tcrb recombination causes Rag gene down-regulation, cellular proliferation, and differentiation to the CD4+CD8+ double-positive (DP) stage. Rag genes are then reexpressed in DP thymocytes to promote recombination of Tcra genes. After productive Tcra gene assembly and positive selection of TCR-expressing DP thymocytes, Rag genes are silenced during differentiation to the CD4+CD8− or CD4−CD8+ single-positive (SP) stage. In similar fashion, an initial phase of Rag gene expression in prepro– and pro–B cells mediates recombination of Igh genes, whereas a subsequent phase of Rag gene expression in small pre–B cells mediates recombination of Igk and Igl genes.
Transcriptional regulation of the Rag genes is complex, involving distinct sets of lineage- and stage-specific cis-elements that cooperate with the Rag1 and Rag2 promoters (Kuo and Schlissel, 2009). Rag expression in developing B cells depends on sequences upstream of the Rag2 promoter, including a proximal enhancer at −2.6 kb, a distal enhancer at −8 kb, and the well-studied Erag enhancer at −23 kb (Hsu et al., 2003; Kuo and Schlissel, 2009). Sequences within 10 kb of Rag2 appear capable of supporting Rag gene expression in DN thymocytes (Monroe et al., 1999; Yu et al., 1999); however, in DP cells, Rag gene expression is critically dependent on a more distant element located between 71 and 79 kb upstream of Rag2 (Yannoutsos et al., 2004). Elimination of this so-called anti-silencer element (ASE) by gene targeting reduced Rag1 and Rag2 expression by two orders of magnitude in DP thymocytes and prevented differentiation into SP cells, but had only modest effects on Rag gene expression in DN thymocytes (Yannoutsos et al., 2004). Studies with transgenic mice also revealed the presence of a silencer element between the Rag1 and Rag2 genes, which can extinguish Rag expression in DP thymocytes and partially suppress expression in DN thymocytes. Importantly, ASE activity was shown to be essential to counteract the suppressive effects of the intergenic silencer in DP thymocytes (Yannoutsos et al., 2004). Silencer activity depends on a binding site for Runx transcription factors, but further mechanistic information about ASE or silencer function has been lacking.
Gene regulation by distal elements generally depends on long range interactions that are facilitated by chromatin architectural proteins (Gibcus and Dekker, 2013; Merkenschlager and Odom, 2013). Special AT-rich binding protein 1 (SATB1) is a nuclear matrix/scaffold-associated DNA-binding protein that participates in the maintenance of chromatin architecture and regulates the expression of a large number of genes (Alvarez et al., 2000; Kumar et al., 2007; Han et al., 2008; Ahlfors et al., 2010). SATB1 binds ATC-rich DNA sequences (also referred to as base unpairing regions) and anchors these sequences to the nuclear matrix to form loops (Cai et al., 2003, 2006; Kumar et al., 2007). In addition, SATB1 can tether genes to the nuclear matrix by protein–protein interactions (Skowronska-Krawczyk et al., 2014). SATB1 can also recruit chromatin remodeling complexes that either promote or inhibit gene expression (Kumar et al., 2006). Given these activities, it is not surprising that SATB1 regulates gene expression programs in a wide variety of cells, including embryonal stem cells (Savarese et al., 2009), neuronal cells (Balamotis et al., 2012), epithelial progenitor cells (Fessing et al., 2011), pituitary cells (Skowronska-Krawczyk et al., 2014), and many tumors (Kohwi-Shigematsu et al., 2013). Within the hematopoietic compartment, SATB1 is expressed in stem cells and is then up-regulated during their commitment to lymphoid lineages (Satoh et al., 2013; Will et al., 2013). In hematopoietic stem cells, SATB1 maintains quiescence and the potential for long-term self-renewal (Will et al., 2013). In hematopoietic progenitor cells, SATB1 supports the expression of genes critical for lymphocyte development and plays an important role in lymphopoiesis (Satoh et al., 2013). SATB1 is expressed at unusually high levels in the thymus (Dickinson et al., 1992), and Satb1-null mice display inefficient T cell development with the major block at the DP stage, resulting in dramatically reduced numbers of SP thymocytes and peripheral T cells (Alvarez et al., 2000; Satoh et al., 2013). SATB1 is also regulated in the context of peripheral T cell activation and differentiation (Cai et al., 2006; Lund et al., 2005), with increased expression required for Th2 differentiation and cytokine gene expression (Cai et al., 2006; Ahlfors et al., 2010; Notani et al., 2010). Conversely, reductions in SATB1 expression are critical for regulatory T cell function (Beyer et al., 2011).
Given the well-established role of chromosome architecture in regulating gene expression and assembly of antigen receptor loci (Jhunjhunwala et al., 2009; Shih and Krangel, 2013), we investigated whether SATB1 functions to regulate V(D)J recombination in DP thymocytes. We found that Tcra gene rearrangement is partially impaired in SATB1-deficient thymocytes, a defect that was associated with substantially reduced expression of Rag1 and Rag2 at the DP stage. Our analysis of this expression defect revealed that the ASE and Rag promoters interact over long-distances in DP thymocytes and that SATB1 is important to bring Rag2 into this complex and to load RNA polymerase II (RNA pol II) to the Rag1 and Rag2 promoters. Our results provide a novel framework for understanding ASE function and the mechanistic basis for Rag gene expression in DP thymocytes.
RESULTS
Impaired Tcra rearrangement in SATB1-deficient DP thymocytes
To investigate its role in V(D)J recombination, we disrupted the gene encoding SATB1 in long-term hematopoietic stem cells of Satb1f/f mice (unpublished data) using a Vav-Cre transgene. Unlike Satb1-null mice (Alvarez et al., 2000), Satb1f/fVav-Cre mice displayed normal growth and survival. Total thymocyte numbers were reduced ∼60% in Satb1f/fVav-Cre as compared with Satb1f/f (also referred to as WT) mice, with similar reductions in the numbers of DN and DP thymocytes (Table 1 and Fig. S1). However, there were 85–90% reductions in SP thymocytes. Thus, Satb1-deficient thymocytes undergo a normal DN to DP transition, but an impaired transition from DP to SP. Satb1 mRNA expression was reduced by ∼95% in DN3 thymocytes from Satb1f/fVav-Cre mice and its expression was essentially undetectable in DP thymocytes (Table 2).
Table 1.
Quantification of thymocyte subsets in WT (Satb1fl/fl) and Satb1−/− (Satb1f/fVav-Cre) micea
| Cell population | WT (×10−5) | Satb1−/− (×10−5) |
| Total thymocytes (n = 6-7) | 1,730 ± 610 | 670 ± 300b |
| DN1 (n = 3-4) | 0.45 ± 0.30 | 0.20 ± 0.17 |
| DN2 (n = 3-4) | 0.89 ± 0.10 | 0.11 ± 0.10c |
| DN3 (n = 3-4) | 17.9 ± 10.3 | 5.1 ± 3.4 |
| CD8 ISP (n = 5-6) | 22.0 ± 11.3 | 12.4 ± 9.1 |
| DP (n = 6-7) | 1,460 ± 500 | 570 ± 250b |
| CD4 SP (n = 6-7) | 109 ± 49 | 17 ± 7c |
| CD8 SP (n = 6-7) | 28.2 ± 11.6 | 2.7 ± 2.1c |
Vav-Cre transgenic mice have no intrinsic thymic phenotype (de Boer et al., 2003).
P < 0.01 when compared with WT counterparts (Student’s t test).
P < 0.001 when compared with WT counterparts (Student’s t test).
Table 2.
Satb1 mRNA expression (relative to B2m) in thymocyte subsets of WT (Satb1fl/fl) and Satb1−/− (Satb1f/fVav-Cre) micea
| SATB1/β2-microglobulin | WT (×102) | Satb1−/− (×102) |
| DN3 (n = 2) | 3.28 ± 0.31 | 0.19 ± 0.02 |
| DN4 (n = 2) | 36.5 ± 2.68 | 0.13 ± 0.05 |
| CD8 ISP (n = 2) | 90.9 ± 40.8 | NDb |
| DP (n = 2) | 241 ± 51.5 | 0.0046 ± 0.0008 |
| CD4 SP (n = 2) | 26.4 ± 3.49 | 0.093 ± 0.041 |
| CD8 SP (n = 2) | 5.15 ± 0.30 | 0.0049 ± 0.0029 |
RNA and cDNA were prepared from 30,000 cells.
ND, not detected.
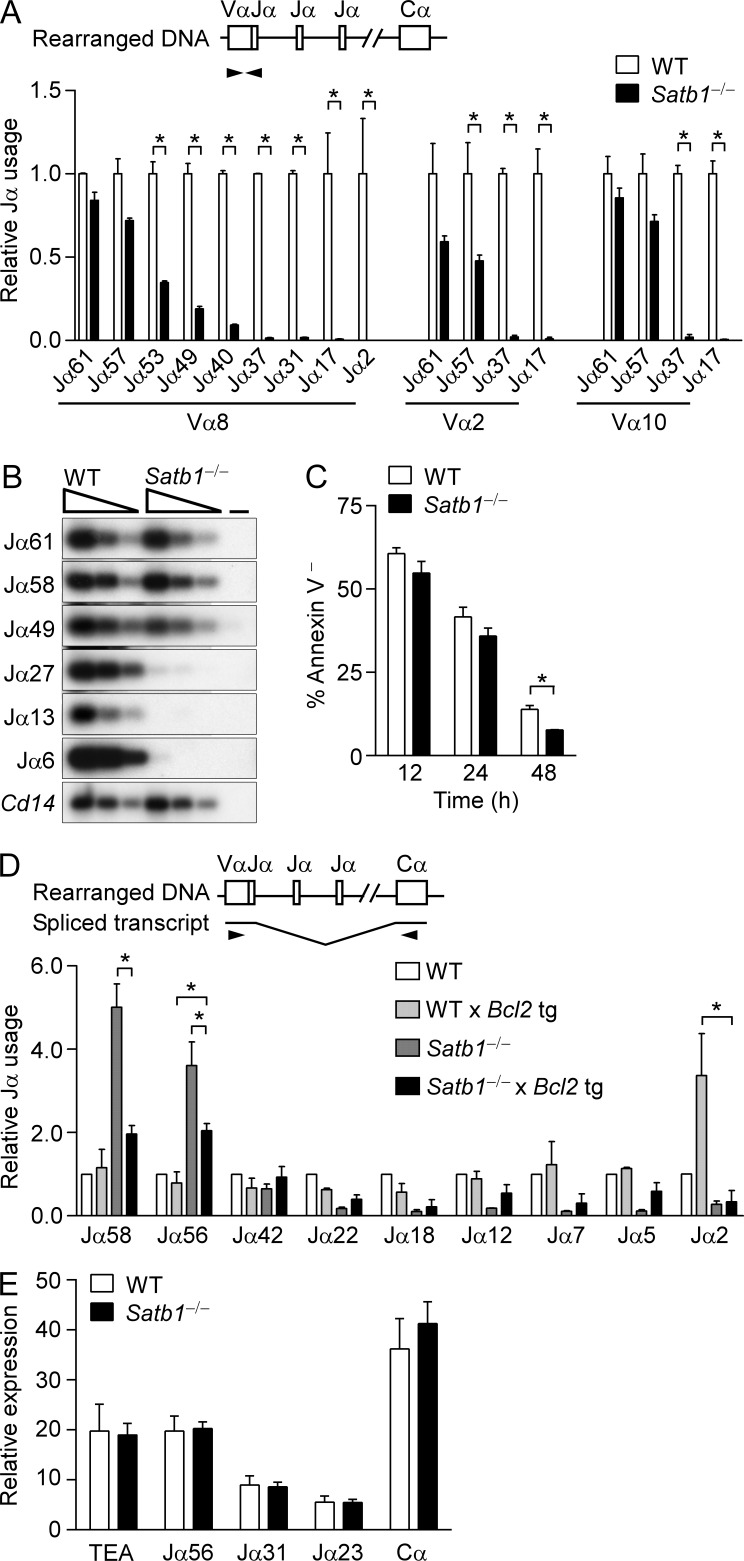
We tested for V(D)J recombination defects using PCR to quantify Tcra locus Vα-to-Jα rearrangement in genomic DNA from WT and Satb1f/fVav-Cre thymocytes. The murine Tcra/Tcrd locus contains ∼100 Vα and 61 Jα gene segments which can undergo several rounds of recombination in DP thymocytes (Krangel, 2009). Newly generated DP thymocytes preferentially rearrange Vα segments to the most 5′ Jα segments, whereas more mature DP thymocytes may replace these initial rearrangements by joining upstream Vα to more 3′ Jα segments. In Satb1f/fVav-Cre thymocytes, quantitative PCR revealed relatively normal Vα rearrangement to 5′ Jα segments, but impaired rearrangement to 3′ Jα segments (Fig. 1 A). This represented a deficiency in RAG-mediated cleavage at 3′ Jα gene segments rather than a deficiency in double-strand break repair, because similar reductions in signal end recombination intermediates at 3′ Jα gene segments were detected by ligation-mediated PCR (Fig. 1 B).
Figure 1.
Defective Tcra rearrangement in SATB1-deficient thymocytes. (A) Tcra coding joints. Thymocyte genomic DNA samples were amplified by quantitative PCR using primers specific for the Vα8 (TRAV12), Vα2 (TRAV14), or Vα10 (TRAV13) families in conjunction with different Jα primers. The data are plotted as mean ± SEM of two experiments, each with one mouse per genotype, with values for SATB1-deficient thymocytes (Satb1−/−, Satb1f/fVav-Cre) normalized to those for WT littermates (Satb1f/f). *, P ≤ 0.05 by two-way ANOVA with Sidak’s multiple comparisons test. (B) Jα signal ends. Three-fold serial dilutions of linker-ligated thymocyte genomic DNA samples were analyzed by PCR using Jα-specific and linker-primers. PCR products were visualized on Southern blots using radiolabeled Jα-specific oligonucleotide probes. The data are representative of two experiments, each with one mouse per genotype. −, no DNA. (C) DP thymocyte survival. WT and SATB1-deficient thymocytes were cultured in vitro for the indicated times and the percentage of Annexin V− DP thymocytes was determined. The data represent the mean ± SEM of two experiments, each with one mouse per genotype. *, P ≤ 0.05 by two-tailed Student’s t test. (D) Relative Jα usage as a function of DP thymocyte lifespan. Vα8-Cα RT-PCR products were analyzed by Southern blot using radiolabeled Jα- and Cα-specific oligonucleotide probes. Relative Jα usage was evaluated by Phosphorimager as (Jα signal for [genotype]/Cα signal for [genotype])/(Jα signal for WT/Cα signal for WT). The data represent the mean ± SEM of two experiments, each with one mouse per genotype. *, P ≤ 0.05 by two-way ANOVA with Tukey’s multiple comparisons test, comparing Satb1−/− Bcl2 tg to WT Bcl2 tg and Satb1−/−. (E) Tcra germline transcription in SATB1-deficient thymocytes. WT (Rag2−/−Satb1f/f) and Satb1−/− (Rag2−/−Satb1f/fVav-Cre) mice were injected with anti-CD3ε and total thymocytes were harvested 10 d later. Germline Tcra transcription was evaluated by quantitative RT-PCR. The data represent mean ± SEM of three experiments with WT and four experiments with Satb1−/− (one mouse per experiment), with normalization to values for hprt.
Defective 3′ Jα usage can result from impaired thymocyte survival (Guo et al., 2002). However, the survival of purified Satb1f/fVav-Cre DP thymocytes in culture was reduced only slightly as compared with WT thymocytes (Fig. 1 C). To further investigate a role for DP thymocyte viability in the Jα rearrangement defect, we introduced a Bcl2 transgene into the Satb1f/fVav-Cre and WT backgrounds to extend thymocyte lifespan (Fig. 1 D). We then analyzed equal amounts of Tcra cDNA for 5′ and 3′ Jα segment usage. Consistent with the rearrangement defect noted above (Fig. 1, A and B), we observed a strong bias toward 5′ Jα usage in Satb1f/fVav-Cre as compared with WT thymocytes (Fig. 1 D; note that 5′ Jα usage in Satb1f/fVav-Cre thymocytes appears to be elevated over WT because Jα signals were normalized to total Tcra transcripts in this assay). The Bcl2 transgene had minimal effect on the pattern of Jα usage in WT thymocytes; the notable exception was Jα2, where Bcl2 expression appeared to protect from cell death those thymocytes that had exhausted the entire Jα array but failed to be positively selected. Although the Bcl2 transgene partially ameliorated the strong bias toward 5′ Jα usage in Satb1f/fVav-Cre thymocytes, the 5′ bias nevertheless persisted and Jα usage in Satb1f/fVav-Cre Bcl2 tg and Satb1f/f Bcl2 tg thymocytes remained distinct. Thus, differential lifespan of DP thymocytes cannot account for differential Jα usage in SATB1-deficient and WT thymocytes. We also detected no change in germline transcription across the Jα-Cα region, suggesting that differential Jα usage does not reflect a change in Jα locus accessibility (Fig. 1 E).
Reduced Rag1 and Rag2 gene expression in SATB1-deficient DP thymocytes
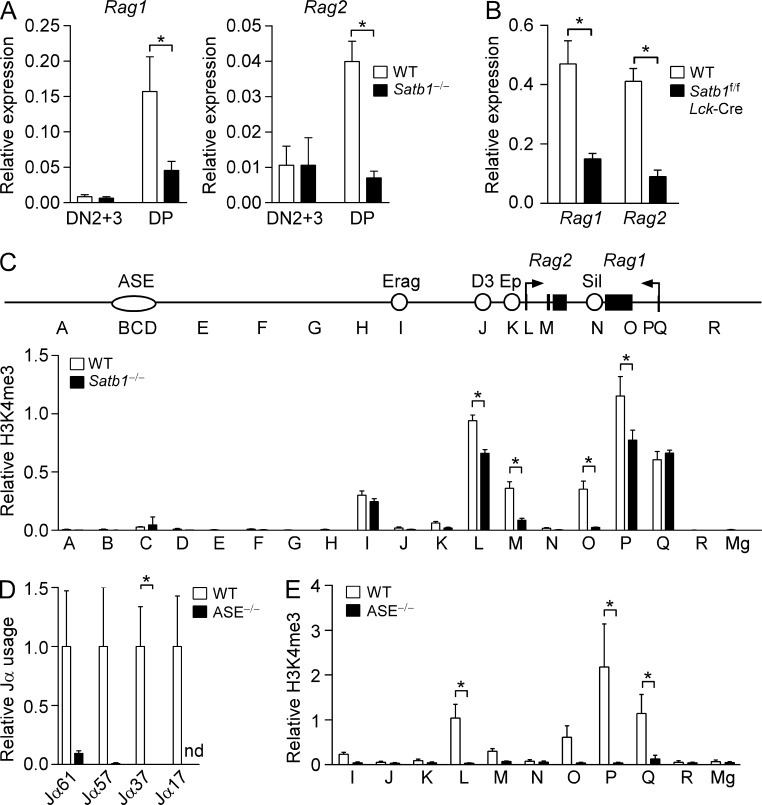
Previous work had shown that impaired 3′ Jα usage can result from defective Rag gene expression in DP thymocytes (Yannoutsos et al., 2001). We therefore assessed Rag1 and Rag2 mRNA expression in DN and DP thymocytes from Satb1f/fVav-Cre and WT mice using real-time PCR (Fig. 2 A). Consistent with previous results (Yannoutsos et al., 2004) Rag1 and Rag2 expression were higher in DP thymocytes than in DN3 thymocytes from WT mice. SATB1-deficient thymocytes displayed normal Rag1 and Rag2 expression at the DN stage but displayed ∼70 and 80% reductions in Rag1 and Rag2 expression, respectively, in DP thymocytes. Similar reductions in Rag1 and Rag2 transcripts were observed in DP thymocytes from Satb1f/fLck-Cre mice; thus, the Rag expression defect is T cell intrinsic (Fig. 2 B). Consistent with the reduced expression phenotype, chromatin immunoprecipitation (ChIP) revealed substantial reductions in the transcription-associated modification histone H3 lysine 4 trimethylation (H3K4me3) at the Rag1 and Rag2 promoters and gene bodies in Satb1f/fVav-Cre DP thymocytes (sites L, M, O and P, Fig. 2 C). We conclude that biased 5′ Jα usage in Satb1f/fVav-Cre thymocytes reflects inefficient rearrangement due to reduced Rag gene expression, and that this 5′ bias may be partially ameliorated by the Bcl2 tg (Fig. 1 D) because thymocytes have additional time for rearrangements to occur.
Figure 2.
Defective Rag1 and Rag2 gene expression in SATB1-deficient and ASE-deleted DP thymocytes. (A) The abundance of Rag1 (left) and Rag2 (right) transcripts in CD25+ (DN2+3) and DP thymocytes was evaluated by quantitative RT-PCR. The data represent the mean ± SEM of four experiments for DN2+3 and five experiments for DP (WT, Satb1f/f; Satb1−/−, Satb1f/fVav-Cre; one mouse per genotype per experiment) with normalization to values for Actb. *, P ≤ 0.05 by two-tailed Student’s t test. (B) A T cell–intrinsic effect of SATB1 was evaluated by measuring transcript abundance in DP thymocytes of WT (Satb1f/f) and Satb1f/fLck-Cre mice by quantitative RT-PCR. The data represent mean ± SEM of three experiments for WT and four experiments for Satb1f/fLck-Cre cDNA (one mouse per experiment), with normalization to values for hprt. *, P ≤ 0.05 by two-tailed Student’s t test. (C) ChIP analysis of Rag locus histone H3K4me3. A map of the Rag locus depicts the convergently transcribed Rag1 and Rag2 genes and known cis-regulatory elements ASE, Erag (enhancer of Rag), D3 (distal enhancer), Ep (enhancer proximal), and Sil (silencer; Kuo and Schlissel, 2009). Sorted DP thymocytes of WT and SATB1-deficient mice were analyzed by ChIP followed by quantitative PCR. Sites A–R in the Rag locus were analyzed by ChIP. The MageA2 promoter (Mg) served as a negative control. The data represent the mean ± SEM of three experiments for each genotype (one mouse per experiment), with values of bound/input expressed relative to those for the B2m promoter (normalized to 1) in each sample. *, P ≤ 0.05 by two-way ANOVA with Sidak’s multiple comparisons test. (D) Tcra coding joints in thymocytes of ASE-deleted mice. Thymocyte genomic DNA samples were amplified by quantitative PCR using a Vα8 family primer in conjunction with different Jα primers, with values for ASE−/− thymocytes normalized to those for WT littermates. The data represent the mean ± SEM of 2–3 independent preparations for each genotype. *, P ≤ 0.05 by two-tailed Student’s t test. nd, not detected. (E) ChIP analysis of Rag locus histone H3K4me3 in DP thymocytes of ASE−/− mice. The data represent the mean ± SEM of four experiments for WT (ASE+/+; one mouse per experiment) and five experiments for ASE−/− (one mouse per experiment). P ≤ 0.05 by two-way ANOVA with Sidak’s multiple comparisons test.
Stage-specific and SATB1-dependent conformations of the Rag locus
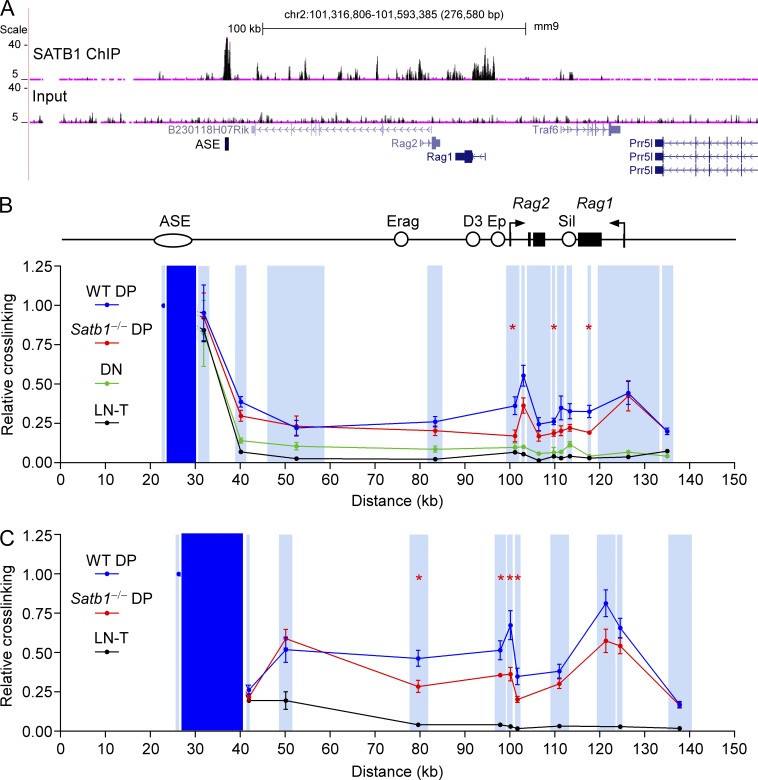
Because Satb1f/fVav-Cre mice displayed a DP stage-specific defect in Rag gene expression, we hypothesized that SATB1 might play a role in ASE function, perhaps by facilitating interactions between the ASE, 71-kb upstream of Rag2, and more proximal cis-acting elements. Prior analysis of ASE−/− mice revealed Rag1 and Rag2 gene expression to be <1% of the level in WT DP thymocytes (Yannoutsos et al., 2004), suggesting a more dramatic expression defect than in Satb1f/fVav-Cre mice. In accord with this, we found that ASE−/− DP thymocytes displayed more dramatic reductions in Vα-to-Jα recombination (Fig. 2 D) and in histone H3 lysine 4 trimethylation (H3K4me3) (Fig. 2 E) than in Satb1f/fVav-Cre mice. On this basis, we predicted that ASE functions and interactions might be partially diminished in Satb1f/fVav-Cre mice. Consistent with such a role, ChIP-sequencing (ChIP-seq) analysis revealed prominent SATB1 binding to the ASE, as well as to the Rag1 and Rag2 promoter regions in DP thymocytes (Fig. 3 A).
Figure 3.
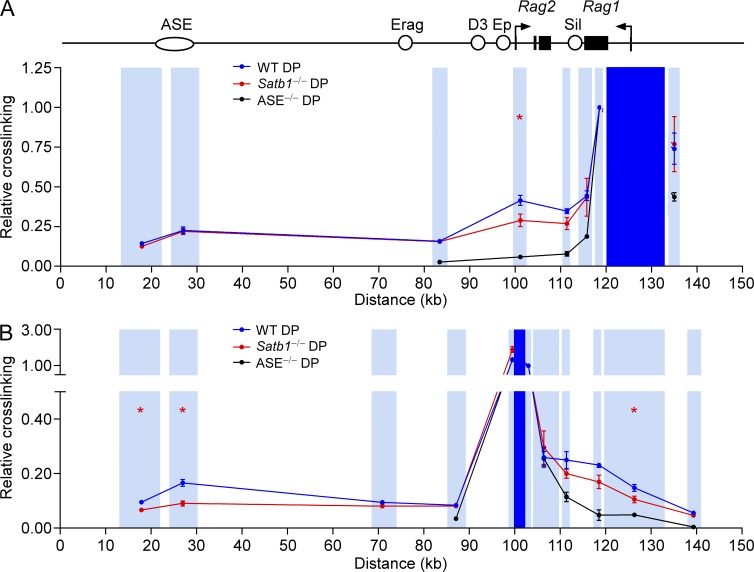
Developmentally regulated and SATB1-dependent interactions between the ASE and Rag promoters. (A) SATB1 ChIP-seq. Sequencing reads for SATB1 ChIP and input DNA are plotted. The ASE region marked corresponds to fragment AB (see Fig. 6). Long-distance interactions of the ASE-containing HindIII (B) or BglII (C) fragments were analyzed by 3C, followed by quantitative PCR. ASE viewpoint restriction fragments are shaded dark blue and target restriction fragments are shaded light blue. Primers were all reverse orientation and were positioned at the left end of each restriction fragment. Relative cross-linking data for each restriction fragment were plotted in the center of the fragment. Data in B represent mean ± SEM of two experiments with lymph node T cells (LN-T) cells (one mouse per experiment), three experiments with DN (Lat−/−) thymocytes (one litter of five to eight mice per experiment), four experiments with WT (Satb1f/f) DP thymocytes (one mouse per experiment), and four experiments with Satb1−/− (Satb1f/fVav-Cre) DP thymocytes (one mouse per experiment), all normalized to results for a nearest neighbor fragment (=1). *, P ≤ 0.05 by two-tailed Student’s t test comparing WT to Satb1−/− DP. Data in C represent mean ± SEM of two experiments with LN-T cells (one mouse per experiment), eight experiments with WT DP thymocytes (one mouse per experiment), and six experiments with Satb1−/− DP thymocytes (one mouse per experiment), normalized as in B. *, P ≤ 0.05 by two-tailed Student’s t test comparing WT to Satb1−/− DP.
Previous studies showed that the ASE is required to counteract functions of the Rag silencer; however, the mechanistic basis for this process remained uncertain (Yannoutsos et al., 2004). In this regard, the ASE could be envisioned to promote Rag gene expression as a consequence of direct physical interaction with the silencer in DP thymocytes. Alternatively, the ASE might counteract the influence of the silencer by interacting directly with the Rag gene promoters.
To investigate Rag locus architecture, we explored interactions between the ASE and Rag genes using chromosome conformation capture (3C; Dekker et al., 2002). In this approach, long-distance interactions in nuclei are initially captured by formaldehyde cross-linking; after restriction enzyme digestion, dilution, and intermolecular ligation, interacting DNA fragments are detected by real-time PCR. Using the ASE-containing HindIII fragment as a viewpoint, in WT DP thymocytes we detected interactions with HindIII fragments carrying the Rag1 and Rag2 promoters 70–100 kb away (Fig. 3 B). Similar ASE-Rag promoter interactions were detected between the relevant BglII fragments as well (Fig. 3 C). These interactions were specific, because they were not detected in LN T cells, which do not express the Rag genes. We evaluated chromatin interactions in the DN compartment by analyzing thymocytes from LAT-deficient mice, which are blocked at the DN stage of development (Zhang et al., 1999). Interaction frequencies were only slightly higher than in LN T cells (Fig. 3 B), consistent with a limited role for the ASE in Rag gene expression in DN thymocytes (Yannoutsos et al., 2004).
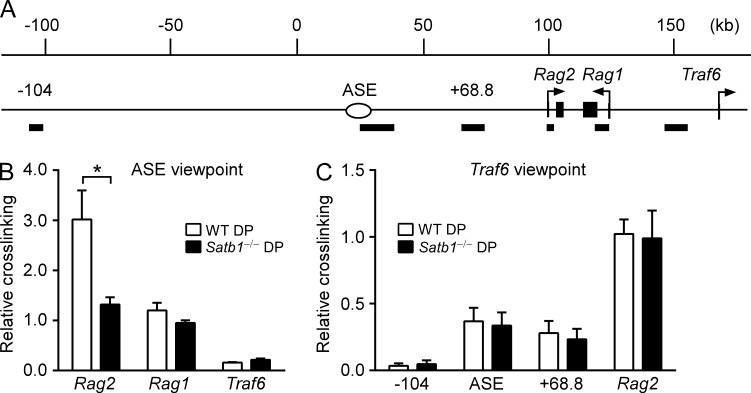
Notably, ASE interactions with the Rag2 promoter were significantly attenuated in Satb1f/fVav-Cre DP thymocytes (Fig. 3, B and C). However, ASE-Rag1 promoter contacts were either maintained (Fig. 3 B) or marginally reduced in a manner that did not reach statistical significance (Fig. 3 C). To address this point further, we analyzed an independent set of BglII-digested 3C samples and confirmed reduced ASE–Rag2 promoter interaction but normal ASE–Rag1 promoter interaction in Satb1f/fVav-Cre DP thymocytes (Fig. 4, A and B). Moreover, the requirement for SATB1 to facilitate ASE-Rag2 promoter interaction was highly specific, because Rag2 promoter contacts with the Traf6 promoter were maintained in SATB1-deficient mice (Fig. 4, A and C). Although the Rag2–Traf6 interaction serves as a useful control, it is of unknown biological significance.
Figure 4.
SATB1 specifically influences the ASE-Rag2 promoter interaction. (A) Rag locus map including flanking regions, with numbering concordant with Fig. 3. -104, ASE (+33.6), +68.8, Rag2 (+100), Rag1 (+121.2), and Traf6 (+151) identify the midpoints of BglII fragments (solid bars) chosen for 3C. Long-distance interactions of the ASE (B) or Traf6 promoter (C) fragments were analyzed by 3C followed by quantitative PCR. Data represent mean ± SEM of four experiments with WT (Satb1f/f) DP thymocytes (one mouse per experiment) and five experiments with Satb1−/− (Satb1f/fVav-Cre) DP thymocytes (one mouse per experiment), all normalized to results for the Traf6 nearest neighbor fragment (=1). *, P ≤ 0.0001 by two-way ANOVA with Sidak’s multiple comparisons test.
Our 3C data indicated that, in addition to the Rag promoters, the ASE interacts broadly across the Rag locus (above the background in LN T cells), including sites between the ASE and Rag2, and within the Rag2 and Rag1 gene bodies (Fig. 3, B and C). Although the ASE contacts the region that includes the intergenic Rag silencer, these interactions were not elevated above the general levels observed across the Rag locus. Collectively, these data support a model in which the ASE indirectly counteracts silencer activity via direct interactions with the Rag1 and Rag2 promoters.
The ASE organizes the Rag locus
To further explore the role of the ASE in Rag locus conformation, we used 3C to assess long distance interactions in DP thymocytes from ASE−/− mice. Using the Rag1 promoter fragment as a viewpoint, we detected an interaction between the Rag1 and Rag2 promoters in WT DP thymocytes and found this interaction to be substantially diminished in ASE−/− DP thymocytes (Fig. 5 A). Similar results were obtained using a Rag2 promoter-containing fragment as the viewpoint (Fig. 5 B). Moreover, Rag1–Rag2 promoter interactions were reduced partially in Satb1f/fVav-Cre DP thymocytes (Fig. 5, A and B). Using these new viewpoints, we also confirmed reduced interactions of the Rag2 but not the Rag1 promoter with the ASE in Satb1f/fVav-Cre DP thymocytes (Fig. 5, A and B), a finding consistent with 3C analyses from the ASE viewpoint (Figs. 3 and 4). We conclude that the ASE organizes the Rag locus by tethering the Rag1 and Rag2 promoters in DP thymocytes, and that efficient tethering of the Rag2 promoter depends on SATB1.
Figure 5.
ASE- and SATB1-dependent interactions between the Rag1 and Rag2 promoters. Long-distance interactions of Rag1 promoter-containing (A) or Rag2 promoter-containing (B) HindIII fragments were analyzed by 3C followed by quantitative PCR. The Rag1 promoter viewpoint (A) and Rag2 promoter viewpoint (B) restrictions fragments are shaded dark blue and target restriction fragments are shaded light blue. Primers for (A) were all reverse orientation and were positioned at the left end of each restriction fragment. Those for B were all forward orientation and were positioned at the right end of each restriction fragment. Relative cross-linking data for each restriction fragment were plotted in the center of the fragment. Data represent mean ± SEM of four experiments each for WT, Satb1−/− (Satb1f/f Vav-Cre), and ASE−/− DP thymocytes (one mouse per genotype per experiment), all normalized to results for a nearest neighbor fragment (=1). *, P ≤ 0.05 by two-tailed Student’s t test comparing WT to Satb1−/− DP.
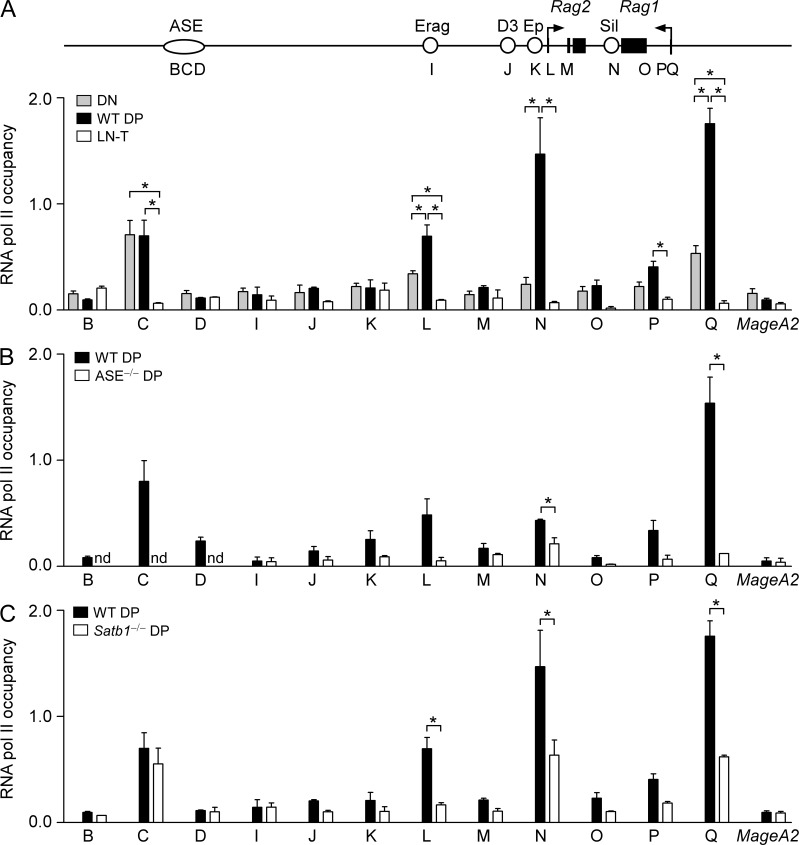
The ASE is required for RNA pol II occupancy at the Rag promoters
To explore the functional significance of the DP-specific chromatin conformation of the Rag locus, we analyzed the RNA pol II distribution using ChIP. As anticipated, RNA pol II occupancy was high at the Rag1 and Rag2 promoters in WT DP thymocytes (sites L and Q, Fig. 6 A). However, RNA pol II was prominent at the ASE and silencer as well (sites C and N, Fig. 6 A). In DN thymocytes from Lat−/− mice, lower levels of RNA pol II were present at the Rag promoters and silencer, but at the ASE, RNA pol II was as abundant as in DP thymocytes. In contrast, RNA pol II was minimal at all sites in LN T cells (Fig. 6 A) and in ASE−/− DP thymocytes (Fig. 6 B). Thus, RNA pol II occupancy at the Rag promoters and silencer is developmentally regulated in a manner that correlates with transcription and depends on the ASE. A previous study also identified substantial RNA pol Il binding to the Rag promoters, ASE, and silencer in DP thymocytes, although other stages of T cell development were not examined (Koch et al., 2011).
Figure 6.
Locus conformation-dependent RNA pol II occupancy of the Rag1 and Rag2 promoters. RNA pol II occupancy was assessed by ChIP and quantitative PCR at sites spanning the Rag locus. Values of bound/input were expressed relative to those for B2m (normalized to 1) in each sample. MageA2 served as a negative control. (A) Data represent mean ± SEM of three experiments each for DN (Lat−/−) thymocytes (one litter of five to eight mice per experiment), WT DP (Satb1f/f) thymocytes (one mouse per experiment), and lymph node T cells (LN-T; one mouse per experiment). Data for these three genotypes and Satb1−/− (Satb1f/fVav-Cre) DP thymocytes (three experiments, one mouse per experiment) were analyzed together by two way ANOVA with Tukey’s multiple comparisons test, with the data for WT DP and Satb1−/− DP plotted separately in C. *, P ≤ 0.05. (B) Data represent the mean ± SEM of three experiments for WT (ASE+/+) DP thymocytes (one mouse per experiment) and two experiments for ASE−/− DP thymocytes (one mouse per experiment). *, P ≤ 0.05 by two-tailed Student’s t test. (C) Data represent the mean ± SEM of three experiments each for WT (Satb1f/f; identical to A) DP thymocytes (one mouse per experiment) and Satb1−/− (Satb1f/fVav-Cre) DP thymocytes (one mouse per experiment). *, P ≤ 0.05 by two-way ANOVA with Tukey’s multiple comparisons test, as noted above. Samples in A and C were all analyzed in the same series of experiments; samples in B were analyzed separately.
Notably, RNA pol II occupancy at the Rag promoters and silencer was partially reduced in DP thymocytes from Satb1f/fVav-Cre mice, whereas occupancy at the ASE was unaffected (Fig. 6 C). Thus, RNA pol II occupancy at the ASE occurs independent of ASE–promoter interactions, whereas RNA pol II occupancy at the promoters and silencer correlates with assembly of an intact complex containing the ASE and the Rag1 and Rag2 promoters. Similarly, DN thymocytes display RNA pol II at the ASE but, in the absence of ASE–promoter interactions, only low levels at the Rag genes themselves (Fig. 6 A). We suggest that the Rag1 and Rag2 promoters acquire high levels of RNA pol II when they are tethered to each other and to the ASE. Whether this reflects delivery of RNA pol II previously bound to the ASE or newly recruited RNA pol II remains an open question.
The ASE directly activates the Rag1 and Rag2 promoters
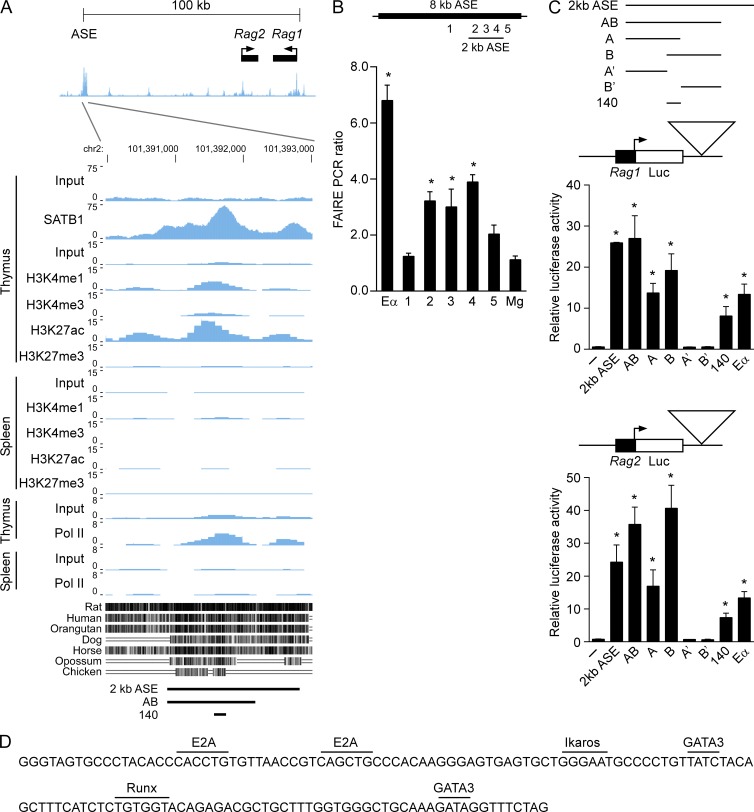
The above data imply that the ASE stimulates Rag gene transcription through direct long distance interactions with the Rag1 and Rag2 promoters, perhaps functioning as a classical enhancer. To test this possibility, we sought to define an ASE fragment that activates Rag1 and Rag2 promoter-driven luciferase reporter constructs in transiently transfected cells. Because previous work had only ascribed ASE activity to a broad 8-kb region (Yannoutsos et al., 2004), we asked whether a smaller core ASE possessed functional activity. Initially, we leveraged chromatin data from the mouse ENCODE database (Stamatoyannopoulos et al., 2012; Rosenbloom et al., 2013) to narrow our search. DNase-Seq data from unsorted thymocytes reveals a tightly linked pair of strong DNase I hypersensitive sites that span ∼2-kb and correspond to a previously identified region notable for substantial interspecies sequence conservation (Fig. 7 A; Yannoutsos et al., 2004). We further mapped open chromatin in the Rag locus in purified DP thymocytes using formaldehyde-assisted isolation of regulatory elements (FAIRE; Giresi et al., 2007), which similarly detected a 2-kb open chromatin region (Fig. 7 B). ENCODE data show that the same region harbors the characteristic chromatin signature of active enhancers in thymus but not in spleen: high histone H3 lysine 4 monomethylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac) coupled with low H3K4me3 and histone H3 lysine 27 trimethylation (Fig. 7 A; Bulger and Groudine, 2011; Natoli and Andrau, 2012; Calo and Wysocka, 2013). Moreover, our ChIP-seq data mapped SATB1 binding to the same region and, consistent with our ChIP data (Fig. 6 A), the region displayed high RNA pol II occupancy as well (Fig. 7 A).
Figure 7.
The ASE directly activates the Rag1 and Rag2 promoters. (A) ENCODE data tracks in the Mouse July 2007 (NCBI37/mm9) assembly are shown. (top) DNase-seq data for the Rag locus in unfractionated thymocytes. (bottom) ChIP-Seq data and sequence conservation for the DNase hypersensitive region in unfractionated thymocytes and splenocytes. Note that primer sets B and D (Fig. 6) map upstream and downstream, respectively, of the 2-kb open chromatin region whereas primer set C maps within the 140-bp minimal enhancer region. (B) FAIRE samples were prepared to evaluate open chromatin at the ASE in WT DP thymocytes. Quantitative PCR was used to evaluate enrichment of sequences in DNA purified from formaldehyde-cross-linked as compared with uncrosslinked samples. The Tcra enhancer (Eα) and MageA2 (Mg) served as positive and negative controls, respectively. Positions of test amplicons relative to the 8-kb ASE region are indicated above the graph. The data are presented as mean ± SEM of values of cross-linked/uncrosslinked in three experiments (one mouse per experiment). *, P ≤ 0.05 by two-tailed Student’s t test comparing Eα and test amplicons to MageA2. (C) ASE activation of the Rag1 and Rag2 promoters. Test ASE fragments were cloned downstream of either a Rag1 promoter-driven or a Rag2 promoter-driven luciferase gene and were assayed by transient transfection into VL3-3M2 DP thymocytes. The data represent the mean ± SEM of two to four independent experiments. *, P ≤ 0.05 by two-tailed Student’s t test comparing results for test ASE fragments to promoter-only controls (−). (D) Nucleotide sequence of the 140-bp minimal enhancer with predicted binding sites for transcription factors marked.
We tested enhancer activity of the 2.0-kb ASE fragment that corresponded to the region of DNase I hypersensitivity and sequence conservation, as well as a 1.3-kb subfragment (AB) corresponding to the region of highest H3K4me1, H3K27ac, and RNA pol II. These test fragments were introduced into promoter-driven luciferase reporters that were transiently transfected into the murine DP thymocyte cell line VL3-3M2 (Fig. 7 C). By themselves, the Rag1 and Rag2 promoters drove minimal luciferase expression compared with positive control plasmids containing the promoters plus the Tcra enhancer (Eα), a potent enhancer in DP thymocytes. Notably, the 2-kb ASE fragment and subfragment AB were more potent than Eα as activators of both promoters. To determine the minimal region responsible for enhancer activity, we tested two partially overlapping subfragments of AB (fragments A and B). Both displayed enhancer activity, with the activity of the 2-kb ASE or fragment AB only marginally better than either A or B. This suggested that the 140-bp region shared by fragments A and B, which is centered within the strong peaks of H3K4me1, H3K27ac, RNA pol II, and SATB1, might play a dominant role in enhancer activity. Indeed, fragments A’ and B’, truncated versions of A and B lacking the 140-bp overlap region, displayed no detectable enhancer activity, whereas the 140-bp overlap region exhibited enhancer function comparable to that of Eα. We conclude that the 140-bp ASE fragment, which contains evolutionarily conserved binding sites for E2A-, Runx-, GATA-, and Ikaros-family transcription factors (Fig. 7 D), is a core enhancer element that augments the function of the Rag promoters. However, because activity of the 140-bp fragment was substantially reduced relative to larger ASE fragments, flanking sequences are likely to boost enhancer activity even further.
DISCUSSION
The Rag silencer and ASE were previously shown to coordinate Rag gene expression in DP thymocytes (Yannoutsos et al., 2004). The silencer was defined using bacterial artificial chromosome (BAC) reporter transgenes lacking the ASE. In such constructs, silencer deletion caused increases in Rag gene expression in DN and DP thymocytes but not in developing B cells. Moreover, the silencer could suppress the expression of a heterologous reporter transgene in pre–B cells, DN and DP thymocytes, and splenic T cells. Thus, the silencer appeared to be capable of broadly suppressing the activities of linked promoters in lymphoid cells. In BAC constructs containing the intergenic silencer, the ASE was shown to be required for Rag expression in DP thymocytes and to increase Rag expression in DN3 thymocytes. However, it had no apparent effect on Rag expression in constructs lacking the silencer. Consistent with these observations, gene-targeted deletion of the ASE reduced Rag expression in DN thymocytes, abrogated Rag expression in DP thymocytes, and caused a developmental block at the DP stage that was likely secondary to impaired Tcra gene recombination. Thus, ASE activity was strongest in DP thymocytes, but the ASE was judged to be distinct from a classical enhancer because it appeared to function by counteracting the activity of an intergenic silencer (Yannoutsos et al., 2004).
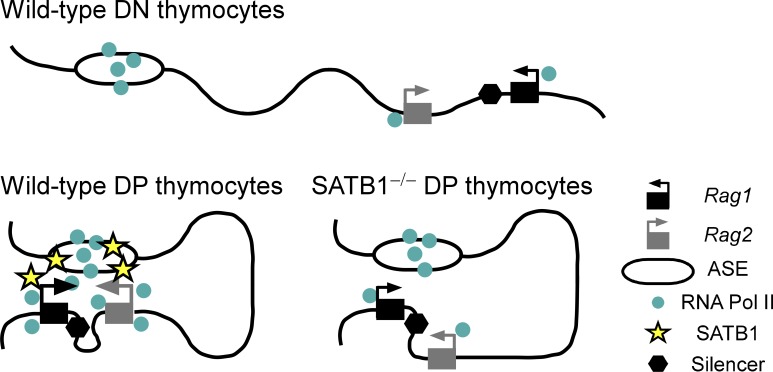
These foundational studies left the mechanistic basis for ASE activity unclear. As one possibility, the ASE could functionally interact with the silencer to neutralize its ability to suppress the Rag promoters. Alternatively, the ASE could functionally interact with the Rag promoters to overcome the suppressive effects of the silencer. Our data strongly support the latter scenario and argue that the ASE functions as a classical enhancer: it displays a chromatin signature typical of active enhancers, it interacts physically with the distant Rag1 and Rag2 promoters and brings these promoters together to form a chromatin hub in DP thymocytes (Fig. 8), and it can directly and potently activate the Rag1 and Rag2 promoters in the absence of the silencer or any other locus elements.
Figure 8.
Model depicting ASE- and SATB1-dependent Rag1 and Rag2 gene expression in DP thymocytes. The Rag1 and Rag2 promoters have minimal contact with the RNA polII-loaded ASE in DN thymocytes and are transcribed at modest levels. SATB1 is up-regulated in DP thymocytes, binds to the ASE and Rag promoters, and facilitates formation of a chromatin hub that promotes RNA Pol II recruitment and trans-activation of the promoters. In the absence of SATB1, the Rag2 promoter is preferentially excluded from the chromatin hub, leading to reduced RNA Pol II recruitment and trans-activation of both promoters.
Our data yield a picture that diverges in two respects from the initial description of ASE activity. First, the chromosomal BAC reporter studies identified no ASE activity in constructs lacking the silencer (Yannoutsos et al., 2004), whereas we show that the ASE can function as a direct activator of the Rag1 and Rag2 promoters in extrachromosomal luciferase reporters that lack the silencer. One explanation for this difference could be that the analysis of a small number of BAC integrants did not provide an accurate picture of ASE activity. However, we favor an alternate possibility: in the environment of a chromosomally integrated locus, the silencer may be essential to create a repressive environment at the promoters that would enforce the need for ASE activity, whereas extrachromosomal reporters may be intrinsically suppressed at the promoters even in the absence of the silencer. A second point of divergence is that the BAC reporter studies showed the ASE to be active but not essential for Rag gene expression in DN thymocytes, whereas we see no clear evidence for long-distance interaction between the ASE and the Rag promoters in that compartment. This difference may simply reflect insensitivity of our 3C analysis to low frequency ASE–promoter interactions that may occur in DN thymocytes. Nevertheless, the BAC transgene and our current work are concordant in many regards, especially the greater reliance on ASE activity and long-distance interactions in DP compared with DN thymocytes.
The intriguing distribution of RNA pol II at the Rag locus suggests potential mechanisms by which the ASE might regulate Rag gene expression. At the ASE, we found RNA pol II to be present at high levels in DN and DP thymocytes and in SATB1-deficient DP thymocytes, but not in LN T cells. However, high level RNA pol II occupancy at the Rag1 and Rag2 promoters was only detected in SATB1-sufficient DP thymocytes, thereby correlating with looping between the ASE and the two Rag promoters. This suggests that an important function of these regulatory loops is to promote RNA pol II loading to the promoters (Fig. 8). RNA pol II binding is a shared property of many enhancers and is often associated with the transcription of enhancer RNAs (Kim et al., 2010; Koch et al., 2011; Natoli and Andrau, 2012). Recent data suggest that enhancer RNAs may play a direct role in enhancer-promoter looping and promoter activation via a mechanism that may involve cohesin (Li et al., 2013). Consistent with these possibilities, the ASE binds TATA-binding protein and general transcription factors in addition to RNA pol II (Koch et al., 2011), and there is low-level ASE transcription in DP thymocytes. RNA pol II initially recruited to the ASE could be delivered to the Rag promoters as a consequence of ASE–promoter interactions. Alternatively, ASE–promoter interactions could stabilize the binding of newly recruited RNA pol II at the Rag promoters. Viewed in another way, the ASE may frequently localize to a transcription factor (Cisse et al., 2013; Cook, 1999; Ghamari et al., 2013) in DN and DP thymocytes, and ASE–promoter interactions and elevated promoter RNA pol II occupancy may reflect high frequency recruitment of the Rag promoters into the same transcription factory in DP thymocytes. The substantially reduced Rag transcription that occurs in DN thymocytes may then be interpreted as a reduced frequency of promoter recruitment to ASE-containing transcription factories in these cells.
RNA pol II was also detected in the Rag silencer region. This binding had a profile similar to that of the Rag promoters, in that it was elevated in DP thymocytes in an ASE- and SATB1-dependent manner, thereby correlating with long-distance looping and high levels of Rag gene transcription. Of note, RNA pol II occupancy maps to the very 3′ end of the Rag1 transcription unit (Rosenbloom et al., 2013; Stamatoyannopoulos et al., 2012), ∼700 bp away from the Runx binding-site that was shown to be essential for silencer activity (Yannoutsos et al., 2004). Moreover, this RNA pol II is primarily in the serine 2 phosphorylated form (Koch et al., 2011). Based on this, we suspect that the accumulation of RNA pol II at this site is related to the termination of Rag1 transcription and not to silencer function.
Finally, our data suggest important roles for chromatin organizer SATB1 in Rag gene expression and Tcra recombination in DP thymocytes. SATB1 expression is dramatically up-regulated in DP thymocytes (Table 2), correlating with the substantial increases in Rag gene expression in this compartment. Further, SATB1 binds to multiple sites across the Rag locus, most prominently at the ASE and Rag1 and Rag2 promoters. We suspect that SATB1 binding to the Rag locus is up-regulated in parallel with its increased expression in DP thymocytes, although our experiments do not directly address this point. However, loss of SATB1 led to significant reductions in Rag gene expression, in Rag2 promoter interactions with the ASE and Rag1 promoter, and in RNA pol II occupancy at the Rag promoters and silencer in DP thymocytes (Fig. 8). Thus, our results for the Rag locus are fully consistent with prior examples in which SATB1 was shown to regulate genomic loci by tethering the bases of chromatin loops and recruiting critical chromatin remodeling complexes and RNA Pol II (Cai et al., 2003, 2006; Kumar et al., 2007). Our results indicate that ASE–Rag1 promoter interactions can be assembled in a SATB1-independent fashion, but that the Rag2 promoter depends on SATB1 to effectively join this complex (Fig. 8). Reduced RNA pol II loading and transcription of Rag2 in the absence of SATB1 may be a straightforward consequence of exclusion of this promoter from the ASE-Rag1 complex. However, RNA pol II loading and transcription of Rag1 are also reduced in the absence of SATB1 even though the Rag1 promoter maintains contact with the ASE. This suggests that Rag1 promoter activity may depend on the formation of a holocomplex containing the ASE and both promoters. Alternatively, SATB1 could play a more direct role in ASE or promoter activity in addition to mediating their long-distance interactions. Consistent with this possibility, SATB1 binding is centered within the 140-bp enhancer core of the ASE.
In summary, we demonstrate that Rag gene expression in DP thymocytes depends on the assembly of a multi-component chromatin complex, or hub, containing the ASE and both Rag gene promoters. We show, too, that chromatin organizer SATB1 plays an important role in the assembly of this complex and in Rag gene transcription, functioning at least in part by stimulating RNA pol II loading to the Rag promoters. Whether SATB1 serves purely in an architectural capacity to facilitate long-distance looping, or also functions in a more direct manner to effect promoter activation, will be an important issue for future studies.
MATERIALS AND METHODS
Mice.
ASE−/− mice (Yannoutsos et al., 2004) and Vav-Cre mice (de Boer et al., 2003) were obtained from The Jackson Laboratory. Bcl2 transgenic mice (Domen et al., 1998) and Lat−/− mice (Zhang et al., 1999) were provided by J. Domen (Duke University, Durham, NC) and W. Zhang (Duke University, Durham, NC), respectively. Mice were maintained on a C57BL/6 background and were housed in specific pathogen-free conditions. All mice were used in accordance with protocols approved by the Duke University Institutional Animal Care and Use Committee.
Flow cytometry and cell sorting.
For surface staining, erythrocyte-lysed single-cell suspensions were prepared in staining medium (DMEM + 2% FCS) with 1 mg/ml normal rat IgG (Sigma-Aldrich) and were stained on ice for 20 min using the following antibodies: FITC-CD8α (53–6.7), PE-CD3ε (145-2C11), PE/Cy5-B220 (RA3-6B2), Mac-1 (M1/70), Gr-1 (RB6-8C5), TER119 (TER-119), PE/Cy7-CD4 (GK1.5), APC-CD44 (IM7), TCRβ (H57-597), and APC/Cy7-CD25 (PC61.5) from eBioscience, as well as PE/Cy5-CD11c (HL3) from BD. FACS analyses were performed on a FACSCanto II (BD) and data were analyzed with FlowJo software (Tree Star).
For cell sorting, single-cell suspensions were prepared following erythrocyte lysis. DN3 thymocytes (CD4−CD8−CD25+CD44−/lo) and a combined DN2/3 thymocyte population (CD4−CD8−CD25+) were obtained after initial depletion of CD4+ and CD8+ cells using biotinylated-CD4 and CD8 antibodies (GK1.5 and 53–6.7, respectively [eBioscience] and streptavidin MACS beads (Miltenyi Biotec). DP thymocytes (CD4+CD8+) were obtained by sorting of whole thymocytes using FITC-CD8 (53–6.7) and PE/Cy7-CD4 (GK1.5) antibodies (eBioscience) or PE-CD4 (GK1.5) and FITC-CD8 (53–6.7) antibodies (BioLegend). Sorting was performed on a FACSVantage (BD) and the purity of cells after double sorting was >95%. Lymph node T cells were isolated as previously described (Jackson and Krangel, 2005).
In vitro survival assay.
Sorted DP thymocytes (105) were cultured in 200 µl RPMI 1640 medium containing 10% FBS, 55 µM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin for varying times in the presence of 10 ng/ml mouse recombinant IL-7 (R&D Systems). Apoptosis was measured by Annexin V and 7-aminoactinomycin D staining using the Annexin V-PE kit (BD) according to the manufacturer’s instructions.
ChIP.
For immunoprecipitation using anti-trimethylated H3K4 (Millipore 04–745) or control rabbit IgG (ab-105-c; R&D Systems), chromatin was prepared without formaldehyde cross-linking and was immunoprecipitated exactly as previously described (Hao and Krangel, 2011). Immunoprecipitated and input samples were quantified by real-time PCR using a Roche LightCycler and a FastStart DNA Master Syber Green I kit (Roche). PCR conditions were as follows: 5 min at 95°C followed by 45 cycles of 1 s at 95°C, 5 s at 62°C, 7 s at 72°C. Analysis of B2m was used to normalize ratios of bound/input in different samples. Primers sequences are provided in Table S1.
For immunoprecipitation using anti-RNA pol II (Millipore; 05–623) or control rabbit IgG (R&D Systems; ab-105-c), 1 × 107 cells were subjected to cross-linking by incubation for 10 min on ice in 10 ml of RPMI 1640 containing 10% FBS and 1% formaldehyde. The reaction was stopped by addition of glycine to 0.125 M and incubation for 5 min at 23°C. Cells were then washed in PBS and lysed by incubation for 10 min on ice in 1 ml of 5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40, 0.1 mM PMSF, and 0.1 mM benzamidine. Nuclei were pelleted and lysed by resuspension in 0.3 ml 50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS. Chromatin was then pelleted through 8 M urea by centrifugation at 30,000 rpm for 16 h at 10°C in a Beckman SW 40Ti rotor. After centrifugation, pelleted chromatin was resuspended in 10 mM Tris, pH 8.0, 1 mM EDTA, 5% glycerol, was dialyzed overnight at 4°C against the same buffer. The volume was then adjusted to 1 ml and the suspension was sonicated using a Model 550 Sonic Dismembrator (Thermo Fisher Scientific), alternating 15 s on and 20 s off for 10 cycles with the sample immersed in an ice/water bath. Chromosomal DNA was reduced to an average size of 300–500 bp, as determined by agarose gel electrophoresis. Sonicated chromatin was precleared with Protein A-Sepharose/salmon sperm DNA slurry (Millipore), incubated overnight at 4°C with anti-RNA pol II or control rabbit IgG, and was subsequently incubated for 1 h with Protein A–Sepharose/salmon sperm DNA slurry. Immunoprecipitated DNA was then purified after vigorous washing of immunoprecipitates and reversal of cross-links by overnight incubation at 60°C. Immunoprecipitated and input samples were quantified by real-time PCR using a Roche LightCycler 480 and a QuantiFast SYBRgreen kit (QIAGEN). PCR conditions were as follows: 5 min at 95°C followed by 45 cycles of 10 s at 95°C, 30 s at 62°C.
ChIP-seq.
For ChIP-seq using anti-SATB1 (Abcam; ab109122), 4 × 107 C57BL/6 thymocytes were freshly prepared and washed with PBS containing 0.5 mM PMSF and were subjected to cross-linking by incubation for 10 min in 5 mM Hepes, pH 7.5, 10 mM NaCl, 0.1 mM EDTA, 0.05 mM EGTA, 1% formaldehyde at 23°C. The reaction was stopped by addition of glycine to 0.125 M. Cells were then immediately washed in ice-cold PBS containing 0.5 mM PMSF and lysed by incubation for 10 min on ice in 1 ml of 50 mM Hepes, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP-40, 0.25% TritonX-100 containing protease inhibitor (Roche). Nuclei were pelleted and lysed by resuspending in 10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA containing protease inhibitor (Roche). Pelleted chromatin was resuspended in 400 µl of 10 mM Tris, pH 8.0, 300 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% TritonX-100, 0.1% sodium deoxycholate and 0.5% N-laurylsarcosine, and was sonicated using a model XL2000 ultrasonic cell disruptor (MICROSON) so that relatively large fragments (1–10 kbp) were included. Sonicated chromatin was incubated overnight at 4°C with anti-SATB1 antibody that was preconjugated with Magnet beads (Dynabeads). Immunoprecipitated DNA was then purified after vigorous washing of immunoprecipitates and reversal of cross-links by overnight incubation at 65°C in 50 mM Tris, pH 8.0, 10 mM EDTA, and 1% SDS. Input DNA and ChIP DNA (10–20 ng) were subjected to sonication (Bioruptor; UCD-200) at medium power for 20 min, alternating 30 s on and 30 s off. Library construction was conducted as follows: (1) End repair by T4 DNA polymerase, Klenow DNA polymerase and T4 kinase; (2) A-tailing by Klenow exo-; (3) Adaptor ligation using Illimina TruSeq adaptors (Illumina) and T4 DNA ligase; (4) 15 cycles of PCR amplification by AccuPrime DNA Taq polymerase (Invitrogen) and TruSeq primers; (5) purification of final products by size selection (200–1,000 bp) using SPRI beads (Thermo Fisher Scientific). The ChIP-sequencing was performed by the UC Berkeley sequence facility. The Gene Expression Omnibus accession no. for the SATB1 ChIP-seq dataset is GSE66248.
Quantitative RT-PCR.
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions, and cDNA was synthesized using oligo dT primers and SuperScript III (Invitrogen). PCR was performed using an iCycler MyiQ Real-Time PCR Detection System (Bio-Rad Laboratories) and SYBR Green PCR core reagents (Applied Biosystems). PCR conditions (for Rag1, Rag2, and Actb) were as follows: 95°C for 10 min, followed by 35 cycles of 95°C for 15 s and 60°C for 45 s. Relative expression levels of target genes were normalized to the value for Actb in each sample. Primers sequences are provided in Table S1.
Tcra recombination.
To quantify Tcra coding joints, thymocytes were lysed by overnight incubation at 37°C in 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM EDTA, 0.4% SDS, 0.1 mg/ml proteinase K, and genomic DNA was prepared by phenol/chloroform extraction and ethanol precipitation. Quantitative real-time PCR with Vα and Jα-specific primers was performed using a Roche LightCycler and a FastStart DNA Master Syber Green I kit (Roche). PCR conditions were as described above.
To quantify Jα signal ends, genomic DNA was linker-ligated as described (McMurry et al., 1997) and linker-ligated DNA was used to amplify signal ends by ‘touchdown’ PCR (Jackson et al., 2005). Conditions for Cd14 amplification were: 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 62°C for 30 s and 72°C for 1 min, and a 10-min extension at 72°C. Amplicons were separated by agarose gel electrophoresis and analyzed by Southern blot with 32P-labeled oligonucleotide probes. Primers and probes were described previously (Seitan et al., 2011).
To evaluate relative Jα usage in Tcra transcripts, total RNA was extracted from whole thymus using TRIzol (Invitrogen) according to the manufacturer’s instructions. Contaminating genomic DNA was removed by incubation with 1 U DNaseI (New England Biolabs) for 10 min at 37°C. cDNA was synthesized using SuperScript reverse transcriptase and oligo(dT)20 according to the manufacturer’s instructions. Jα usage was determined in Vα8 Tcra transcripts as previously described (Hawwari et al., 2005).
3C.
This technique was performed with some modifications to a previously described protocol (Hagège et al., 2007). 1 × 107 cells were subjected to cross-linking by incubation for 10 min on ice in 10 ml of RPMI-1640 containing 10% FBS and 2% formaldehyde. The reaction was stopped by addition of glycine to 0.125 M and incubation for 5 min at 23°C. Cells were then washed in PBS and lysed by incubation for 10 min on ice in 5 ml of 10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP-40, 0.1 mM benzamidine, and 0.1 mM PMSF. Nuclei were pelleted, washed once with PBS, and lysed by incubation for 1 h at 37°C in 0.5 ml of 1× NEB II or NEB III digestion buffer (New England Biolabs) containing 0.3% SDS. Triton-X100 was then added to a final concentration of 2% and incubation was continued for an additional 1 h at 37°C. Chromatin was then digested by addition of 200 U BglII or HindIII for overnight incubation at 37°C, followed by a second addition of 200 U of enzyme for an additional 8 h incubation at 37°C. Digested chromatin was then sedimented through 8 M urea by centrifugation at 35,000 rpm for 16 h at 10°C in a Beckman SW 40Ti rotor. After centrifugation, the digested chromatin (0.5 ml of gel and liquid) was diluted and resuspended in 2 ml H2O, dialyzed overnight at 4°C against 30 mM Tris, pH 7.4, 10 mM MgCl2, diluted to 7 ml in 30 mM Tris, pH 7.4, 10 mM MgCl2, 1 mM DTT, and 0.1 mM ATP, and was ligated by addition of 4,000 U T4 DNA ligase for overnight incubation at 16°C. Ligated chromatin was incubated overnight at 60°C to reverse cross-links and DNA was purified by extraction with phenol/chloroform and ethanol precipitation.
Ligation products were quantified by TaqMan quantitative real-time PCR (Roche) using a Roche LightCycler 480 and PCR conditions as follows: 5 min at 95°C followed by 45 cycles of 10 s at 95°C and 30 s at 62°C. To generate ligation product standards, 10 µg of BAC RP23-325I3 was digested at 37°C overnight with 50 U HindIII or BglII, after which DNA was purified by phenol/chloroform extraction and ethanol precipitation, and then ligated overnight at 16°C in a 200 µl reaction containing 40 U T4 DNA ligase. Purified, ligated DNA was serially diluted in 10-fold increments from a 2 ng/µl stock to generate the standard curve.
Digestion efficiencies of different experimental samples were 90–94% as determined by quantitative real-time PCR in which yields of several amplicons that span HindIII or BglII sites were compared with yields of neighboring amplicons that were not disrupted by digestion. Normalization of 3C PCR signals from different samples was accomplished by setting the nonspecific interaction of the bait fragment with one of its nearest neighbor fragments equal to one in each sample. Primer and probe sequences are provided in Table S1.
FAIRE.
FAIRE was performed essentially as previously described (Giresi et al., 2007) except that quantitative real-time PCR was used to evaluate enrichment of sequences in DNA purified from formaldehyde-cross-linked as compared with uncrosslinked samples. Primer sequences are provided in Table S1.
Luciferase.
VL3-3M2 cells were provided by S. Sarafova (Davidson College, Davidson, NC) and were cultured in RPMI-1640 supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin, 55 µM 2-mercaptoethanol, 2 mM l-glutamine, and 25 mM Hepes, pH 7.0. Approximately 3 × 105 cells were transfected with versions of the pXPG firefly luciferase reporter plasmid (Bert et al., 2000) containing the Rag1 or Rag2 promoter and different anti-silencer region or control DNA fragments. In brief, 1 µg of each construct was cotransfected with 100 ng of plasmid expressing Renilla luciferase using the Superfect transfection reagent (QIAGEN). Cells were cultured for 48 h in 0.5 ml of medium in a 24-well plate and were then harvested to assay for luciferase activity using an Infinite F200 Tecan plate luminometer and a Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer’s instructions.
Online supplemental material.
Fig. S1 shows normal DN-to-DP transition in SATB1-deficient thymocytes. Table S1 lists oligonucleotides used as PCR primers and probes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20142207/DC1.
Supplementary Material
Acknowledgments
We thank Lynn Martinek, Nancy Martin, and Mike Cook of the Duke Cancer Institute Flow Cytometry Facility for help with cell sorting and analysis, Kingshuk Roy Choudhury for advice on statistical analysis, and Eugene Oltz for critical reading of the manuscript.
This work was supported by National Institutes of Health grants R37 GM10452 (to M.S.K.) and R37 CA39681 (to T.K.S.), by a Grant-in-Aid for Scientific Research (S) and for Scientific Research on Priority Areas (to I.T.), and by grant JSPS KAKENHI 24390121 and a Leukemia and Lymphoma Society Scholar Award (to M.K.).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 3C
- chromosome conformation capture
- ASE
- anti-silencer element
- BAC
- bacterial artificial chromosome
- ChIP
- chromatin immunoprecipitation
- ChIP-seq
- ChIP-sequencing
- DP
- double positive
- FAIRE
- formaldehyde-assisted isolation of regulatory elements
- H3K4me1
- histone H3 lysine 4 monomethylation
- H3K4me3
- histone H3 lysine 4 trimethylation
- H3K27ac
- histone H3 lysine 27 acetylation
- H3K27me3
- histone H3 lysine 27 trimethylation
- RAG
- recombination activating gene
- RNA Pol II
- RNA polymerase II
- RSS
- recombination signal sequence
- SATB1
- special AT-rich binding protein 1
- SP
- single positive
- V(D)J
- variable, diversity, and joining gene segment
References
- Ahlfors H., Limaye A., Elo L.L., Tuomela S., Burute M., Gottimukkala K.V., Notani D., Rasool O., Galande S., and Lahesmaa R.. 2010. SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood. 116:1443–1453 10.1182/blood-2009-11-252205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J.D., Yasui D.H., Niida H., Joh T., Loh D.Y., and Kohwi-Shigematsu T.. 2000. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Balamotis M.A., Tamberg N., Woo Y.J., Li J., Davy B., Kohwi-Shigematsu T., and Kohwi Y.. 2012. Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol. Cell. Biol. 32:333–347 10.1128/MCB.05917-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert A.G., Burrows J., Osborne C.S., and Cockerill P.N.. 2000. Generation of an improved luciferase reporter gene plasmid that employs a novel mechanism for high-copy replication. Plasmid. 44:173–182 10.1006/plas.2000.1474 [DOI] [PubMed] [Google Scholar]
- Beyer M., Thabet Y., Müller R.U., Sadlon T., Classen S., Lahl K., Basu S., Zhou X., Bailey-Bucktrout S.L., Krebs W., et al. . 2011. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 12:898–907 10.1038/ni.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., and Groudine M.. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell. 144:327–339 10.1016/j.cell.2011.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Han H.J., and Kohwi-Shigematsu T.. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34:42–51 10.1038/ng1146 [DOI] [PubMed] [Google Scholar]
- Cai S., Lee C.C., and Kohwi-Shigematsu T.. 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38:1278–1288 10.1038/ng1913 [DOI] [PubMed] [Google Scholar]
- Calo E., and Wysocka J.. 2013. Modification of enhancer chromatin: what, how, and why? Mol. Cell. 49:825–837 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse I.I., Izeddin I., Causse S.Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., and Darzacq X.. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 341:664–667 10.1126/science.1239053 [DOI] [PubMed] [Google Scholar]
- Cook P.R.1999. The organization of replication and transcription. Science. 284:1790–1795 10.1126/science.284.5421.1790 [DOI] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., and Kioussis D.. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314–325 10.1002/immu.200310005 [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., and Kleckner N.. 2002. Capturing chromosome conformation. Science. 295:1306–1311 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Dickinson L.A., Joh T., Kohwi Y., and Kohwi-Shigematsu T.. 1992. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 70:631–645 10.1016/0092-8674(92)90432-C [DOI] [PubMed] [Google Scholar]
- Domen J., Gandy K.L., and Weissman I.L.. 1998. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood. 91:2272–2282. [PubMed] [Google Scholar]
- Fessing M.Y., Mardaryev A.N., Gdula M.R., Sharov A.A., Sharova T.Y., Rapisarda V., Gordon K.B., Smorodchenko A.D., Poterlowicz K., Ferone G., et al. . 2011. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 194:825–839 10.1083/jcb.201101148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamari A., van de Corput M.P., Thongjuea S., van Cappellen W.A., van Ijcken W., van Haren J., Soler E., Eick D., Lenhard B., and Grosveld F.G.. 2013. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 27:767–777 10.1101/gad.216200.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus J.H., and Dekker J.. 2013. The hierarchy of the 3D genome. Mol. Cell. 49:773–782 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., and Lieb J.D.. 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17:877–885 10.1101/gr.5533506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hawwari A., Li H., Sun Z., Mahanta S.K., Littman D.R., Krangel M.S., and He Y.W.. 2002. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 3:469–476 10.1038/ni791 [DOI] [PubMed] [Google Scholar]
- Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., and Forné T.. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2:1722–1733 10.1038/nprot.2007.243 [DOI] [PubMed] [Google Scholar]
- Han H.J., Russo J., Kohwi Y., and Kohwi-Shigematsu T.. 2008. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 452:187–193 10.1038/nature06781 [DOI] [PubMed] [Google Scholar]
- Hao B., and Krangel M.S.. 2011. Long-distance regulation of fetal V(δ) gene segment TRDV4 by the Tcrd enhancer. J. Immunol. 187:2484–2491 10.4049/jimmunol.1100468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Bock C., and Krangel M.S.. 2005. Regulation of T cell receptor α gene assembly by a complex hierarchy of germline Jalpha promoters. Nat. Immunol. 6:481–489 10.1038/ni1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L.Y., Lauring J., Liang H.E., Greenbaum S., Cado D., Zhuang Y., and Schlissel M.S.. 2003. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 19:105–117 10.1016/S1074-7613(03)00181-X [DOI] [PubMed] [Google Scholar]
- Jackson A.M., and Krangel M.S.. 2005. Allele-specific regulation of TCR β variable gene segment chromatin structure. J. Immunol. 175:5186–5191 10.4049/jimmunol.175.8.5186 [DOI] [PubMed] [Google Scholar]
- Jackson A., Kondilis H.D., Khor B., Sleckman B.P., and Krangel M.S.. 2005. Regulation of T cell receptor β allelic exclusion at a level beyond accessibility. Nat. Immunol. 6:189–197 10.1038/ni1157 [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., and Murre C.. 2009. Chromatin architecture and the generation of antigen receptor diversity. Cell. 138:435–448 10.1016/j.cell.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. . 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature. 465:182–187 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F., Fenouil R., Gut M., Cauchy P., Albert T.K., Zacarias-Cabeza J., Spicuglia S., de la Chapelle A.L., Heidemann M., Hintermair C., et al. . 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18:956–963 10.1038/nsmb.2085 [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Poterlowicz K., Ordinario E., Han H.J., Botchkarev V.A., and Kohwi Y.. 2013. Genome organizing function of SATB1 in tumor progression. Semin. Cancer Biol. 23:72–79 10.1016/j.semcancer.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel M.S.2009. Mechanics of T cell receptor gene rearrangement. Curr. Opin. Immunol. 21:133–139 10.1016/j.coi.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.P., Purbey P.K., Sinha C.K., Notani D., Limaye A., Jayani R.S., and Galande S.. 2006. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell. 22:231–243 10.1016/j.molcel.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., and Galande S.. 2007. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 9:45–56 10.1038/ncb1516 [DOI] [PubMed] [Google Scholar]
- Kuo T.C., and Schlissel M.S.. 2009. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr. Opin. Immunol. 21:173–178 10.1016/j.coi.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X., et al. . 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 498:516–520 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund R., Ahlfors H., Kainonen E., Lahesmaa A.M., Dixon C., and Lahesmaa R.. 2005. Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur. J. Immunol. 35:3307–3319 10.1002/eji.200526079 [DOI] [PubMed] [Google Scholar]
- McMurry M.T., Hernandez-Munain C., Lauzurica P., and Krangel M.S.. 1997. Enhancer control of local accessibility to V(D)J recombinase. Mol. Cell. Biol. 17:4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., and Odom D.T.. 2013. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 152:1285–1297 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Monroe R.J., Chen F., Ferrini R., Davidson L., and Alt F.W.. 1999. RAG2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc. Natl. Acad. Sci. USA. 96:12713–12718 10.1073/pnas.96.22.12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G., and Andrau J.C.. 2012. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 46:1–19 10.1146/annurev-genet-110711-155459 [DOI] [PubMed] [Google Scholar]
- Notani D., Gottimukkala K.P., Jayani R.S., Limaye A.S., Damle M.V., Mehta S., Purbey P.K., Joseph J., and Galande S.. 2010. Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 8:e1000296 10.1371/journal.pbio.1000296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G., et al. . 2013. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41(D1):D56–D63 10.1093/nar/gks1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y., Yokota T., Sudo T., Kondo M., Lai A., Kincade P.W., Kouro T., Iida R., Kokame K., Miyata T., et al. . 2013. The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity. 38:1105–1115 10.1016/j.immuni.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F., Davila A., Nechanitzky R., De-La Rosa-Velazquez I., Pereira C.F., Engelke R., Takahashi K., Jenuwein T., Kohwi-Shigematsu T., Fisher A.G., et al. . 2009. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 23:2625–2638 10.1101/gad.1815709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., and Swanson P.C.. 2011. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202 10.1146/annurev-genet-110410-132552 [DOI] [PubMed] [Google Scholar]
- Seitan V.C., Hao B., Tachibana-Konwalski K., Lavagnolli T., Mira-Bontenbal H., Brown K.E., Teng G., Carroll T., Terry A., Horan K., et al. . 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 476:467–471 10.1038/nature10312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H.Y., and Krangel M.S.. 2013. Chromatin architecture, CCCTC-binding factor, and V(D)J recombination: managing long-distance relationships at antigen receptor loci. J. Immunol. 190:4915–4921 10.4049/jimmunol.1300218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D., Ma Q., Schwartz M., Scully K., Li W., Liu Z., Taylor H., Tollkuhn J., Ohgi K.A., Notani D., et al. . 2014. Required enhancer-matrin-3 network interactions for a homeodomain transcription program. Nature. 514:257–261 10.1038/nature13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R., Canfield T., et al. . Mouse ENCODE Consortium. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 13:418 10.1186/gb-2012-13-8-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B., Vogler T.O., Bartholdy B., Garrett-Bakelman F., Mayer J., Barreyro L., Pandolfi A., Todorova T.I., Okoye-Okafor U.C., Stanley R.F., et al. . 2013. Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nat. Immunol. 14:437–445 10.1038/ni.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoutsos N., Wilson P., Yu W., Chen H.T., Nussenzweig A., Petrie H., and Nussenzweig M.C.. 2001. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J. Exp. Med. 194:471–480 10.1084/jem.194.4.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoutsos N., Barreto V., Misulovin Z., Gazumyan A., Yu W., Rajewsky N., Peixoto B.R., Eisenreich T., and Nussenzweig M.C.. 2004. A cis element in the recombination activating gene locus regulates gene expression by counteracting a distant silencer. Nat. Immunol. 5:443–450 10.1038/ni1053 [DOI] [PubMed] [Google Scholar]
- Yasui D., Miyano M., Cai S., Varga-Weisz P., and Kohwi-Shigematsu T.. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 419:641–645 10.1038/nature01084 [DOI] [PubMed] [Google Scholar]
- Yu W., Misulovin Z., Suh H., Hardy R.R., Jankovic M., Yannoutsos N., and Nussenzweig M.C.. 1999. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 285:1080–1084 10.1126/science.285.5430.1080 [DOI] [PubMed] [Google Scholar]
- Zhang W., Sommers C.L., Burshtyn D.N., Stebbins C.C., DeJarnette J.B., Trible R.P., Grinberg A., Tsay H.C., Jacobs H.M., Kessler C.M., et al. . 1999. Essential role of LAT in T cell development. Immunity. 10:323–332 10.1016/S1074-7613(00)80032-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.