The transcription factor LEF1 promotes the expansion and Th2-type polarization of invariant NKT cells in part by directly inducing the expression of the IL-7 receptor component CD127 and the transcription factors c-myc and Gata3.
Abstract
Invariant natural killer T cells (iNKT cells) are innate-like T cells that rapidly produce cytokines that impact antimicrobial immune responses, asthma, and autoimmunity. These cells acquire multiple effector fates during their thymic development that parallel those of CD4+ T helper cells. The number of Th2-type effector iNKT cells is variable in different strains of mice, and their number impacts CD8 T, dendritic, and B cell function. Here we demonstrate a unique function for the transcription factor lymphoid enhancer factor 1 (LEF1) in the postselection expansion of iNKT cells through a direct induction of the CD127 component of the receptor for interleukin-7 (IL-7) and the transcription factor c-myc. LEF1 also directly augments expression of the effector fate–specifying transcription factor GATA3, thus promoting the development of Th2-like effector iNKT cells that produce IL-4, including those that also produce interferon-γ. Our data reveal LEF1 as a central regulator of iNKT cell number and Th2-type effector differentiation.
Invariant NKT cells (iNKT cells) are a subset of TCRαβ+ T cells that recognize glycolipid antigens in the context of the unconventional major histocompatibility complex class I protein CD1D. In peripheral tissues, iNKT cells exist in a primed effector state capable of rapidly producing large quantities of diverse cytokines in response to antigenic stimulation, and these cytokines can modulate the ensuing adaptive immune response (Brennan et al., 2013). iNKT cells express a TCR of limited diversity and acquire their activated state and effector program during their thymic development. Although iNKT cells can contribute to the clearance of microbial pathogens, alterations in their activation or effector differentiation are associated with diseased states such as asthma and autoimmunity (Van Kaer et al., 2013).
iNKT cells diverge from the conventional αβ T cell pathway during their selection from CD4+CD8+ (double positive [DP]) thymocytes (Gapin et al., 2001; Benlagha et al., 2005; Egawa et al., 2005). Expression of the semi-invariant iNKT cell TCR, comprised of an invariant TCRα chain (Vα14Jα18) paired with a biased TCR Vβ repertoire (Vβ8, -7, and -2), allows for recognition of glycolipid–CD1D complexes on neighboring cortical DP thymocytes, which results in their selection into the iNKT cell lineage. This DP–DP interaction promotes engagement of the signaling lymphocytic activation molecule (SLAM) family receptors SLAMf1 (CD150) and SLAMf6 (Ly108), which are associated with the adaptor protein SAP that activates the src family kinase FynT, leading to the selection of DP thymocytes into the iNKT cell lineage (Griewank et al., 2007). iNKT cell selection also involves the EGR2-dependent induction of the transcription factor promyelocytic leukemia zinc finger (PLZF), which is required and sufficient for the primed effector state of iNKT cells and for their characteristic coproduction of Th1 and Th2 cytokines (Kovalovsky et al., 2008; Savage et al., 2008; Seiler et al., 2012).
The earliest postselection iNKT cell precursors, stage zero (ST0) iNKT cells, have a CD24hi cortical thymocyte phenotype, and are CD4+CD8loCD69+, similar to conventional TCRαβ CD4 transitional (CD4t) thymocytes (Benlagha et al., 2005). Unlike positive selection of conventional αβ T cells, positive selection of iNKT cells is followed by a phase of massive proliferation that is accompanied by decreased expression of CD24 and CD8 and transition to ST1. Proliferation and effector fate differentiation continue into ST2, where the cells acquire CD44 and they exhibit an activated memory-like phenotype. The iNKT program of cell division is dependent on the α chain of the receptor for IL-7 (CD127) and the transcription factor c-myc (Dose et al., 2009; Mycko et al., 2009; Tani-ichi et al., 2013). During the phase of intrathymic expansion, a subset of cells down-regulates CD4 to become CD4−CD8− (double negative [DN]) cells, whereas a majority of ST2 and 50% of ST3 cells retain CD4 (Benlagha et al., 2005). ST3 iNKT cells are characterized by a lower rate of cell division and expression of an NK cell–associated transcriptome, including expression of the surface markers NK1.1, DX5, and CD122 (Benlagha et al., 2002; Cohen et al., 2013).
The segregation of iNKT cells into stages based on expression of cell surface receptors has been a useful strategy for investigating their development and function (Benlagha et al., 2002; Pellicci et al., 2002). However, recent studies revealed that these stages do not exclusively define cells with a precursor progeny relationship, but rather, each stage contains terminally differentiated effector cells (Coquet et al., 2008; Michel et al., 2008; Watarai et al., 2012; Lee et al., 2013). At least three thymic iNKT cell effector subsets have been identified that parallel those of CD4 helper T cells and the recently identified innate lymphoid cells (ILCs; Verykokakis et al., 2014). Thymic iNKT cells can be identified as Th1-like PLZFloTBET+ iNKT1 cells, which are largely found among ST3 cells, Th2-like PLZFhiGATA3hi iNKT2 cells, which have an ST1/ST2 phenotype, and Th17-like PLZFintRORγt+ iNKT17 cells, which have an ST2 phenotype (Lee et al., 2013). Thus, ST2 cells are a heterogeneous population that consists of terminally differentiated iNKT2 and iNKT17 effector cells, as well as a few iNKT1 progenitors. These effectors also display differential expression of CD4, with iNKT1 cells being CD4+ and CD4− and iNKT2 cells being predominantly CD4+, whereas iNKT17 cells are mostly CD4−. Importantly, these effector subsets do not show interconversion after intrathymic injection (Lee et al., 2013).
A recent study revealed substantial heterogeneity in the number of thymic iNKT2 cells, with most inbred mouse strains having an iNKT2 bias compared with iNKT1 cells. iNKT2 cells were shown to contribute to basal levels of IL-4, and high numbers of iNKT2 cells promote a memory phenotype on CD8 T cells, increased serum IgE, and specific chemokine production from thymic dendritic cells, all of which can influence the immune response of these mice (Lee et al., 2013). It is unclear what drives the altered representation of iNKT cell effector fates in different mouse strains; however, TBET, GATA3, and RORγt regulate the development of iNKT1, iNKT2, and iNKT17 effector cells, respectively. Deletion of GATA3 favors the development of iNKT1 cells, whereas TBET deficiency leads to an outgrowth of iNKT2 and iNKT17 cells, suggesting that these transcription factors may interact in a common precursor cell (Kim et al., 2006; Wang et al., 2010; Lee et al., 2013).
Although the signature transcription factors for the iNKT lineage and the iNKT1, iNKT2, and iNKT17 cell fates have been defined, little is known about how these lineages are established and whether similar transcriptional networks control the CD4, ILC, and iNKT cell effector programs (Verykokakis et al., 2014). The transcription factor TCF1, a member of the TCF/lymphoid enhancer factor (LEF) family of high-mobility group (HMG) box proteins, plays multiple roles in T cell development and is critical for the development of CD4 Th2 cells, Th2-like ILCs (ILC2), and a subset of Th17-like ILCs (ILC3; Yu et al., 2009; Yang et al., 2013). In contrast, the TCF1-related transcription factor LEF1 is not required for conventional T cell development, although it plays a supportive role in the absence of TCF1 (Okamura et al., 1998; Yu et al., 2012). To date, no critical functions for LEF1 have been identified in T cells. Here we demonstrate essential, TCF1-independent functions for LEF1 in iNKT cell expansion and iNKT2 effector differentiation. We show that LEF1-deficient mice had reduced iNKT cell numbers in the thymus and peripheral tissues. LEF1 directly regulated the CD127 component of the receptor for IL-7 and the oncogenic transcription factor c-myc, both of which are critical for iNKT cell expansion. In addition, the remaining iNKT cells showed a specific deficiency in production of iNKT2 cells. We also demonstrated direct binding of LEF1 to regions proximal to the Gata3 gene, and we showed that GATA3 failed to be highly expressed in LEF1-deficient iNKT cells. Although TCF1 and LEF1 are coexpressed in a subset of GATA3hi iNKT cells, TCF1 expression was not altered by LEF1 deficiency. Therefore, LEF1 nonredundantly regulated Cd127, c-myc, and Gata3 to control iNKT cell numbers and iNKT2 cell effector differentiation.
RESULTS
Reduced iNKT cell development in the absence of LEF1
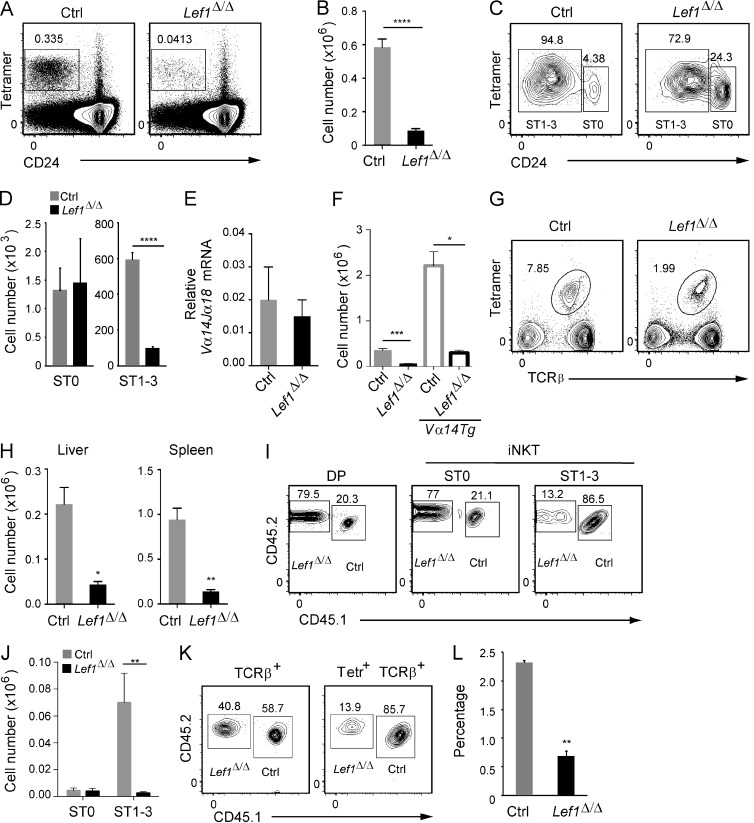
While investigating the requirements for LEF1 in T cell development, we discovered that VavCreLef1f/f (Lef1Δ/Δ) mice had a decreased number of iNKT cells in the thymus compared with their littermate controls (Fig. 1, A and B). In contrast, and consistent with a recent study (Steinke et al., 2014), conventional αβ T cell development was largely unaffected by the absence of LEF1 (not depicted). LEF1 has been implicated in the expression of Cd1d (Chen et al., 2010); however, we found no difference in the expression of CD1D or the SLAM family coreceptors LY108 or CD150 on Lef1Δ/Δ DP thymocytes, indicating that dysregulation of these essential iNKT cell selection molecules was not the cause of the reduced number of iNKT cells (not depicted). To directly determine whether LEF1 was required for the positive selection of iNKT cells, we determined the number of CD24hi (ST0) and CD24lo (ST1–3) cells among TCRβ+Tetramer+ iNKT cells after their enrichment from the thymus of Lef1Δ/Δ and control mice using CD1D tetramers loaded with the glycolipid PBS57 (Fig. 1 C). The number of ST0 iNKT cells was similar in the thymi from Lef1Δ/Δ and littermate control mice (Fig. 1 D). In contrast, there was a significant reduction in the number of Lef1Δ/Δ ST1–3 iNKT cells relative to control mice (Fig. 1, C and D). These data indicate that LEF1 was not required for the selection of iNKT cells, but rather it played a critical role in their subsequent expansion, survival, or differentiation. Consistent with these findings, Vα14Jα18 mRNA, which encodes the TCRα chain of the iNKT cell receptor, was expressed at similar levels in control and Lef1Δ/Δ DP thymocytes, and transgenic expression of a pre-rearranged Vα14Jα18 chain did not rescue the development of ST1–3 iNKT cells in Lef1Δ/Δ mice (Fig. 1, E and F). Reduced TCR signal strength can result in impaired iNKT cell development (Bedel et al., 2014); however, we found no difference in the expression of CD5 or CD69, surface proteins whose expression reflects TCR signal strength, and there was no difference in TCRβ usage or expression on ST1–3 iNKT cells from control and Lef1Δ/Δ mice (not depicted). Therefore, LEF1 was required for iNKT cell development after the emergence of ST0 iNKT cells. The number of iNKT cells was also reduced in the liver and spleen of Lef1Δ/Δ mice (Fig. 1, G and H), a finding which is consistent with the failure to generate CD24lo iNKT cells in the thymus.
Figure 1.
Lef1Δ/Δ mice lacked iNKT cells in the thymus and the periphery. (A) Thymi from control (Ctrl) and Lef1Δ/Δ mice were stained for CD24 and CD1d-PBS57 tetramers (Tetr) and analyzed by flow cytometry. The percentage of total thymocytes that were Tetramer+CD24lo iNKT cells is shown. (B) Mean number of thymic Tetr+CD24lo iNKT cells in control and Lef1Δ/Δ mice. Bars represent the mean from 10 independent experiments, with one pair of mice each. (C) The frequency of Tetr+CD24hi (ST0) and Tetr+CD24lo/− (ST1–3) among total thymic iNKT cells was determined by flow cytometry after MACS-based tetramer enrichment. (D) Absolute number of ST0 and ST1–3 iNKT cells from control and Lef1Δ/Δ mice. Data represent the mean from six independent experiments with one to two mice per genotype. (E) Expression of Vα14Jα18 mRNA transcripts relative to Hprt in sorted DP thymocytes from control and Lef1Δ/Δ mice was determined by qRT-PCR. Data represent the mean from four independent experiments. (F) Mean number of thymic Tetr+CD24lo iNKT cells in the indicated mouse strains from five independent experiments. (G) TCRβ and CD1d-PBS57 tetramer staining on liver cells from control and Lef1Δ/Δ mice was determined by flow cytometry. Numbers indicate the percentage of liver lymphocytes that are Tetr+TCRβ+. (H) Mean number of Tetr+TCRβ+ iNKT cells in the liver and spleen of control and Lef1Δ/Δ mice. Bars represent the mean from four independent experiments. (I) CD45.1+CD45.2+ control and CD45.1−CD45.2+ Lef1Δ/Δ FACS-sorted LK bone marrow cells were mixed at a 1:1 ratio, and 5 × 104 total cells were transferred into lethally irradiated CD45.1+ C57BL/6 mice. Mice were analyzed 6–8 wk after transplantation. Expression of CD45.1 and CD45.2 on DP thymocytes, ST0 iNKT cells, or ST1–3 iNKT cells from competitive bone marrow chimeras was determined by flow cytometry. The percentage of DP cells or CD24hi ST0 and CD24lo ST1–3 iNKT cells derived from CD45.1+CD45.2+ control or CD45.1−CD45.2+ Lef1Δ/Δ bone marrow cells is shown. Data are representative of three independent experiments with two to three chimeras each. (J) The mean number of ST0 CD24hi and ST1–3 CD24lo thymic iNKT cells derived from the CD45.1+CD45.2+ control or CD45.1−CD45.2+ Lef1Δ/Δ bone marrow cells is shown. (K) Expression of CD45.1 and CD45.2 on TCRβ+ (Tetr−) cells and Tetr+TCRβ+ iNKT cells in the liver of competitive mixed bone marrow chimeras was determined by flow cytometry. The gated regions show the percentage of cells that are derived from the control or Lef1Δ/Δ bone marrow. (L) Mean percentage of iNKT cells in the competitive chimeric mice that are derived from the control or Lef1Δ/Δ bone marrow. Numbers are the mean from three independent experiments with two to three chimeras each. All error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
LEF1 was intrinsically required for iNKT cell development
To determine whether the defect in iNKT cell development was intrinsic to the Lef1Δ/Δ cells, we generated mixed bone marrow chimeric mice in which Lef1Δ/Δ CD45.2+ bone marrow progenitors were mixed with control CD45.1+CD45.2+ competitor cells and transplanted into lethally irradiated CD45.1+ mice. Lef1Δ/Δ CD45.2+CD45.1− cells represented 79% of Tetramer− DP thymocytes and 77% of ST0 iNKT cells, confirming that LEF1 was not required for development of ST0 cells (Fig. 1 I). In contrast, only 13.2% of the ST1–3 iNKT cells were derived from the Lef1Δ/Δ CD45.2+CD45.1− bone marrow cells, demonstrating that the Lef1Δ/Δ cells were disadvantaged after ST0 (Fig. 1 I). The loss of Lef1Δ/Δ ST1–3 iNKT cells was particularly evident when the total number of ST0 and ST1–3 iNKT cells was calculated (Fig. 1 J). Lef1Δ/Δ iNKT cells were similarly disadvantaged in the liver, where only 13.9% of iNKT cells were derived from the CD45.2+ population, whereas 40% of TCRβ+ cells were CD45.2+CD45.1− (Fig. 1, K and L). Our data indicate that LEF1 was required for the normal generation of ST1–3 iNKT cells, as well as peripheral iNKT cells.
LEF1 controlled iNKT cell proliferation and survival
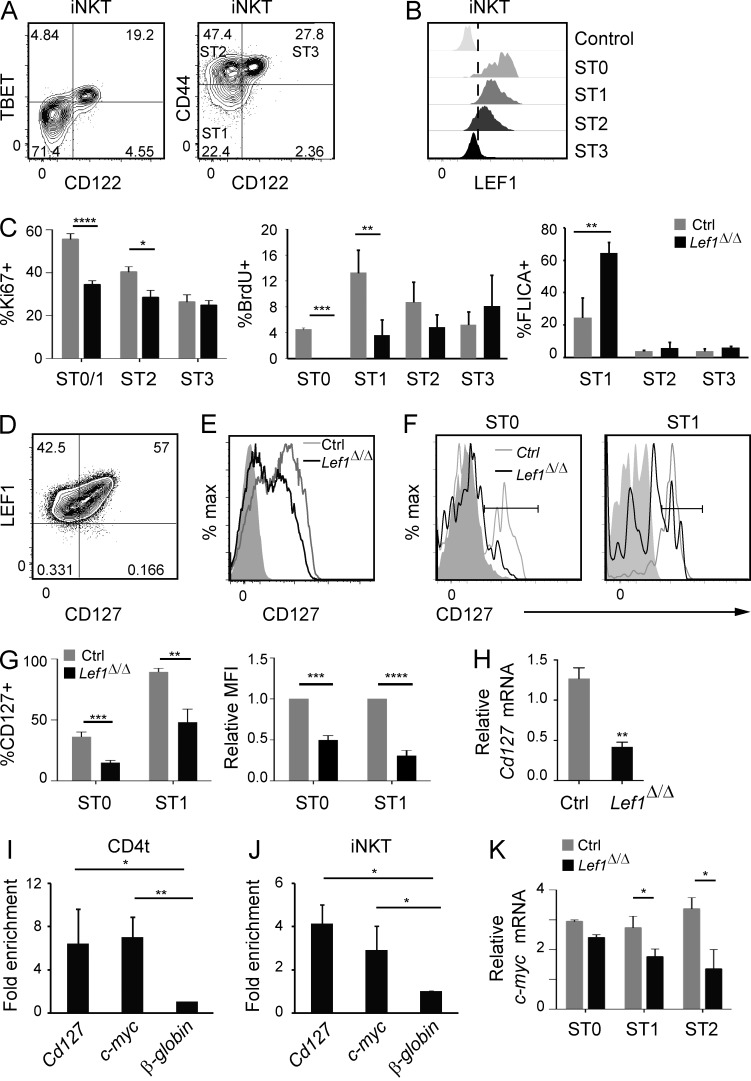
To better understand how LEF1 functioned, we examined LEF1 expression in cells at each stage of iNKT cell development. We used the β chain of the IL-2/15 receptor (CD122) to resolve ST3 iNKT cells because our mice were on the FVB/NJ background, which fails to express the classic NK cell marker NK1.1. CD122 expression was directly correlated with the ST3 transcription factor TBET, and CD44 and CD122 fractionated CD24lo iNKT cells into ST1 (CD44loCD122−), ST2 (CD44+CD122−), and ST3 (CD44+CD122+) cells (Fig. 2 A). We found that LEF1 was highly expressed in ST1 and ST2 cells and was absent from the majority of cells at ST3 (Fig. 2 B). LEF1 was also highly expressed in ST0 iNKT cells (Fig. 2 B). Therefore, LEF1 was expressed at the earliest stages of iNKT cell development.
Figure 2.
LEF1 promoted iNKT cell proliferation and regulated expression of the Cd127 and c-myc genes. (A) Expression of TBET (left) or CD44 (right) and CD122 on WT FVB/NJ Tetr+CD24lo thymic iNKT cells was determined by flow cytometry. The percentage of cells and iNKT cell stage in each quadrant is shown. (B) Expression of LEF1 was determined by flow cytometry in the indicated iNKT cell stages from WT FVB/NJ mice and is shown as a histogram. iNKT cell stages were gated as shown in A (ST1, ST2, and ST3) and in Fig. 1 C (ST0). The control sample was LEF1 staining in Lef1Δ/Δ iNKT cells. Plots are representative of at least 10 independent experiments with one to two mice per genotype. (C) Percentage of the indicated iNKT cell stage staining positive for Ki67, BrdU, or FLICA in the thymus of control and Lef1Δ/Δ mice. Graphs represent the mean of four independent experiments. (D) Expression of LEF1 and CD127 in CD4t (gated as CD4hiCD8loTCRβ+CD69+) WT FVB/NJ thymocytes was determined by flow cytometry. (E and F) CD127 expression in CD4t thymocytes (E) or ST0 and ST1 (F) iNKT cells from control and Lef1Δ/Δ mice was determined by flow cytometry. Data are representative of four to eight independent experiments with one to two mice per genotype. Solid gray histograms show staining with the isotype control antibody. (G) The mean percentage of CD127+ cells (left) or the MFI of CD127 (right) within ST0 and ST1 iNKT cells in control and Lef1Δ/Δ mice. Numbers were averaged from six independent experiments with one mouse per genotype. (H) Expression of Cd127 mRNA relative to Hprt from sorted ST0 iNKT cells (Tetr+TCRβ+CD24hi) was determined by qRT-PCR. The bar graphs represent the mean from three independent experiments. (I and J) LEF1 chromatin binding was determined by α-LEF1 ChIP on sorted CD4t thymocytes (I) or ST1/2 Vα14Tg thymic iNKT cells (J; Tetr+TCRβ+CD24loCD122−), followed by qPCR using primers spanning a TCF1/LEF1 binding site at the Cd127 and c-myc promoter region or an unrelated sequence at the β-globin gene locus. Numbers indicate mean fold enrichment of the immunoprecipitated DNA relative to the β-globin locus from three independent experiments. (K) Expression of c-myc mRNA relative to Hprt mRNA from sorted ST0, ST1, and ST2 iNKT cells was determined by qRT-PCR. Bar graphs represent the mean of three independent experiments. All error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Our data indicated that LEF1 was required for the expansion, survival, or differentiation of postselection iNKT cells. To determine whether Lef1Δ/Δ iNKT cells were signaled to enter the cell cycle appropriately, we examined their expression of Ki67, a marker of cells that have entered the cell cycle, and their ability to incorporate BrdU, a thymidine analogue incorporated into DNA during replication. Consistent with previous studies (D’Cruz et al., 2014), we found that nearly 60% of control ST0/ST1 iNKT cells expressed Ki67 and that proliferation, as marked by incorporation of BrdU, was greatest as cells transitioned to ST1 (Fig. 2 C; Gapin et al., 2001; Benlagha et al., 2002). In contrast to the control cells, only 35% of Lef1Δ/Δ ST0/ST1 iNKT cells were Ki67+, indicating that LEF1 influenced the number of cells that entered the cell cycle. Although ∼4% of control ST0 cells incorporated BrdU, there was essentially no BrdU incorporation into Lef1Δ/Δ ST0 iNKT cells, and Lef1Δ/Δ ST1 cells also showed significantly lower incorporation of BrdU (Fig. 2 C). ST2 and ST3 iNKT cells from control mice had lower Ki67 expression and incorporated less BrdU than ST1 cells, and LEF1 was not as essential for Ki67 or BrdU incorporation in these cells (Fig. 2 C). We also found that Lef1Δ/Δ ST1 iNKT cells had a higher frequency of cells relative to control mice that stained with fluorescent-labeled inhibitor of caspases (FLICA), a reagent which irreversibly binds to the activated caspases that are triggered during apoptosis (Fig. 2 C). Ki67 staining, BrdU incorporation, and FLICA reactivity were not changed in CD4 thymocytes from Lef1Δ/Δ as compared with control mice, indicating that LEF1 deficiency affected iNKT cells specifically (not depicted). Our data reveal that LEF1 was required for the intrathymic expansion of ST0/ST1 iNKT cells and that it protected these cells from apoptosis.
LEF1 directly regulated the Cd127 and c-myc genes in iNKT cells
iNKT cell expansion requires CD127, the α chain of the IL-7 receptor (IL-7Rα; Tani-ichi et al., 2013). We found that LEF1 expression was directly correlated with CD127 in CD4+ transitional (CD4t; CD4+CD8loCD69+TCRβ+) thymocytes, which include ST0 iNKT cells, leading us to question whether LEF1 might regulate CD127 expression (Fig. 2 D). Indeed, CD127 expression was decreased on Lef1Δ/Δ CD4t thymocytes compared with control cells (Fig. 2 E). Moreover, significantly fewer Lef1Δ/Δ ST0 and ST1 iNKT cells expressed CD127 when compared with control mice, and the mean fluorescence intensity (MFI) of CD127 was also reduced (Fig. 2, F and G). CD127 expression was decreased slightly in CD4 single-positive thymocytes and remained unaltered in CD8 thymocytes (not depicted). Consistent with these findings, Cd127 mRNA was reduced by >50% in ST0 iNKT cells from Lef1Δ/Δ as compared with control mice (Fig. 2 H).
The LEF1-related transcription factor TCF1 directly regulates Cd127 expression in Th2 cells and ILC2 (Yu et al., 2009; Yang et al., 2013). Therefore, we questioned whether LEF1 might regulate Cd127 directly. Indeed, we found that LEF1 was bound specifically near the Cd127 gene in CD4t cells and iNKT cells, as determined by anti-LEF1 chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR; Fig. 2, I and J), indicating that LEF1 directly regulated the expression of Cd127 in these cells.
While IL-7R signaling promotes intrathymic proliferation of iNKT cells, the oncogenic transcription factor c-myc is also critical for optimal expansion (Dose et al., 2009; Mycko et al., 2009). We found that c-myc mRNA was reduced in Lef1Δ/Δ ST1-ST2 iNKT cells (Fig. 2 K). c-myc is regulated downstream of IL-7R signaling in B and T cells, but it is also a target of LEF1 in epithelial cells (Morrow et al., 1992; Yip-Schneider et al., 1993; Lim and Hoffmann, 2006; Nakashima et al., 2013). We found that LEF1 bound to the c-myc locus in both CD4t cells and in iNKT cells, demonstrating that LEF1 could regulate both Cd127 and c-myc (Fig. 2, I and J). Collectively, these data demonstrate that LEF1 was required for proper expression of Cd127 and c-myc, which likely contributed to the reduced intrathymic expansion of Lef1Δ/Δ iNKT cells.
LEF1 promoted the development of iNKT2 cells
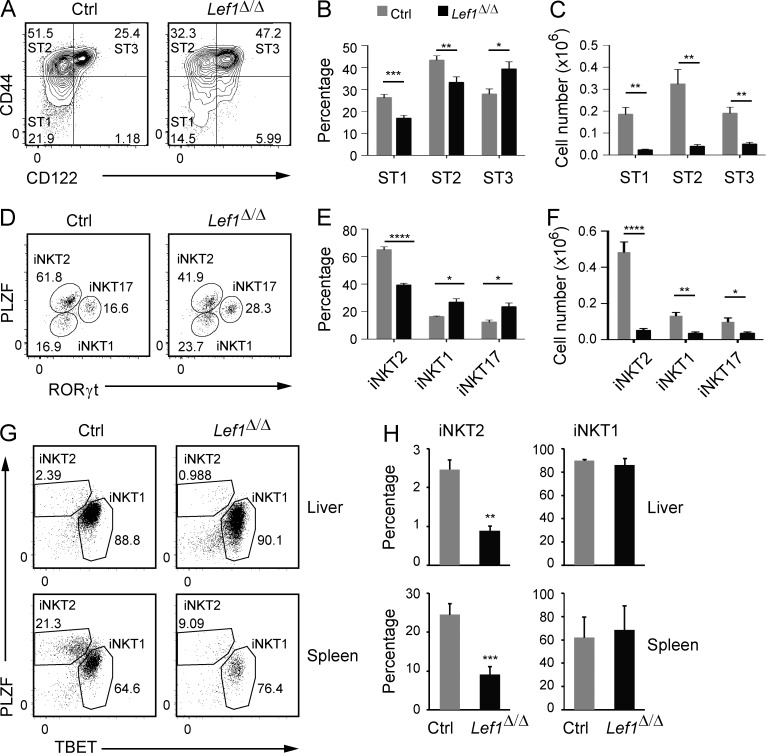
Our data demonstrated that LEF1 played an important role in the development of all iNKT cells. However, when compared with control thymic iNKT cells, there was a decreased frequency of ST1 and ST2 cells and an increased frequency of ST3 cells among the remaining Lef1Δ/Δ thymic iNKT cells (Fig. 3, A and B), despite the fact that the all iNKT cell populations were decreased in number (Fig. 3 C). Recent studies have shown that ST3 iNKT cells are largely synonymous with TBET+ iNKT1 cells, whereas ST2 cells are a mixture of RORγt+ iNKT17 cells and GATA3hi iNKT2 cells, which are also found among ST1 cells. The signature iNKT cell transcription factor PLZF is also differentially expressed among iNKT cell subsets, with the highest expression in iNKT2 cells (Lee et al., 2013). To determine how LEF1 deficiency affected iNKT cell effector subsets, we examined Lef1Δ/Δ iNKT cells for expression of PLZF and RORγt, which can be used to resolve iNKT1 (PLZFloRORγt−), iNKT2 (PLZFhiRORγt−), and iNKT17 (PLZFintRORγthi) cells (Fig. 3 D). By this definition, the frequency of iNKT1 and iNKT17 cells was increased slightly, whereas iNKT2 cells were decreased among Lef1Δ/Δ iNKT cells as compared with controls (Fig. 3, D and E), although, as expected, the absolute number of all iNKT effector subsets was reduced. (Fig. 3 F). Total iNKT cell numbers were also reduced in the liver and spleen of Lef1Δ/Δ mice as compared with control mice (Fig. 1 H); however, in these tissues the frequency of iNKT2 cells was reduced relative to that of iNKT1 cells (Fig. 3, G and H). Collectively, our data indicated that after the expansion of iNKT cells, LEF1 promoted the development of iNKT2 cells.
Figure 3.
LEF1 was required for iNKT2 development. (A) Expression of CD44 and CD122 on gated Tetr+CD24lo iNKT cells from the thymus of control and Lef1Δ/Δ mice was determined by flow cytometry. (B and C) The mean percentage (B) and number (C) of ST1, ST2, and ST3 cells among total iNKT cells in control and Lef1Δ/Δ mice. Data are the mean from eight independent experiments with one mouse per genotype. (D) PLZF versus RORγt expression in Tetr+CD24lo iNKT cells from control and Lef1Δ/Δ mice was used to resolve the iNKT1 (PLZFloRORγt−), iNKT2 (PLZFhiRORγt−), and iNKT17 (PLZFintRORγt+) effector subsets by flow cytometry. Numbers show the percentage of each population among total iNKT cells. (E and F) The mean percentage (E) and number (F) of iNKT1, iNKT2, and iNKT17 cells in the thymus of control and Lef1Δ/Δ mice. Data are the mean from five independent experiments. (G) Expression of PLZF and TBET was used to identify iNKT1 and iNKT2 cells in the liver and spleen of control and Lef1Δ/Δ mice by flow cytometry. (H) The mean percentage of iNKT2 and iNKT1 among total iNKT cells in the liver and spleen from control and Lef1Δ/Δ mice. Numbers were averaged from three independent experiments. All error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
LEF1 was required for the development of IL-4–producing iNKT2 cells
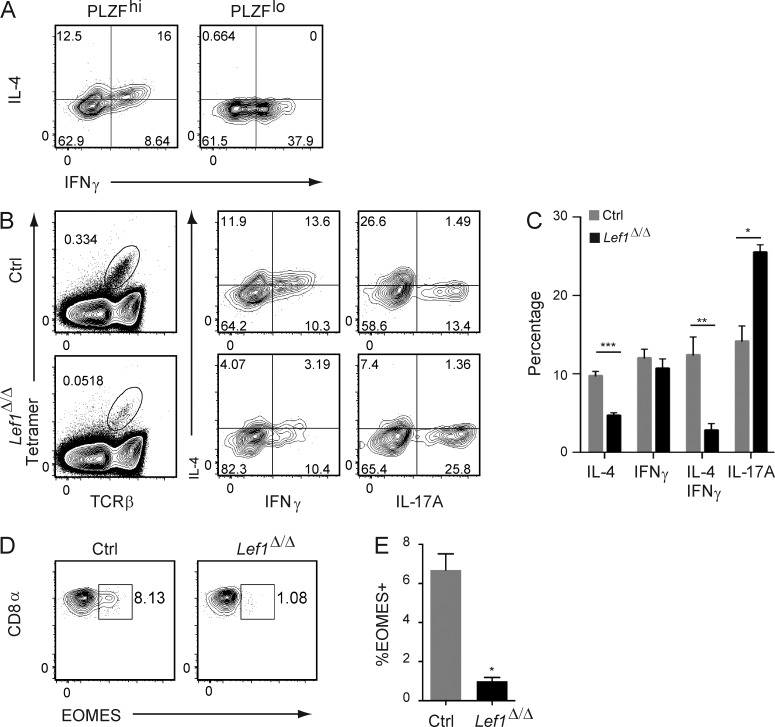
The cytokine production profiles for the iNKT cell effector fates are substantially more flexible than in their CD4+ Th cell counterparts. Dual production of IL-4 and IFNγ is a hallmark of iNKT cells that has been attributed to their high expression of PLZF (Savage et al., 2008). Indeed, some PLZFhi iNKT cells produce both IFNγ and IL-4 after in vitro stimulation with PMA and ionomycin, whereas PLZFlo iNKT cells produced IFNγ exclusively (Fig. 4 A). Given that the majority of PLZFhi cells are iNKT2 cells, we anticipated that LEF1 would be required for the development of the dual IL-4 plus IFNγ–producing population. Indeed, in vitro stimulation of total iNKT cells with PMA and ionomycin revealed that a lower frequency of Lef1Δ/Δ iNKT cells produced the Th2 cytokine IL-4 and the frequency of IL-4 plus IFNγ producers was also reduced (Fig. 4, B and C). In contrast, the frequency of Lef1Δ/Δ iNKT cells producing IFNγ only was not different from control mice, and IL-17A–producing cells were increased (Fig. 4, B and C). Therefore, LEF1 was also required for the development of functional iNKT2 cells that produced IL-4 and IFNγ.
Figure 4.
LEF1 promoted development of iNKT cells that produce IL-4 or IL-4 plus IFNγ. (A) Expression of IL-4 and IFNγ in PLZFhi and PLZFlo iNKT cells was determined by flow cytometry, after a 4-h in vitro stimulation of thymocytes with PMA and ionomycin. (B) Expression of IL-4, IFNγ, or IL-17A in thymic Tetr+TCRβ+ iNKT cells from control and Lef1Δ/Δ mice after a 4-h in vitro stimulation of thymocytes with PMA and ionomycin. Plots are representative of four to six independent experiments. (C) The mean percentage of control and Lef1Δ/Δ iNKT cells that expressed IL-4, IFNγ, IL-4 plus IFNγ, or IL-17A. Data were averaged from six independent experiments. (D) Expression of CD8α and EOMES in TCRβ+CD8α+ thymocytes was determined by flow cytometry. (E) The mean percentage of EOMES+ cells among TCRβ+CD8α+ thymocytes from four independent experiments. All error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In some strains of mice IL-4 production from iNKT2 cells reaches levels that induce EOMES expression in developing CD8 T cells, resulting in their conversion to an innate effector-like cell (Verykokakis et al., 2010; Weinreich et al., 2010; Lee et al., 2013). We found that ∼8% of CD8 T cells in FVB/NJ mice expressed EOMES, consistent with the high frequency of iNKT2 cells (Fig. 4, D and E). Moreover, this phenotype was dependent on LEF1 because these EOMES+ CD8 T cells were severely reduced in Lef1Δ/Δ mice (Fig. 4, D and E). These observations are consistent with the reduced number of functional IL-4–producing iNKT2 cells in Lef1Δ/Δ mice.
LEF1 expression correlated with GATA3, PLZF, and Th-POK
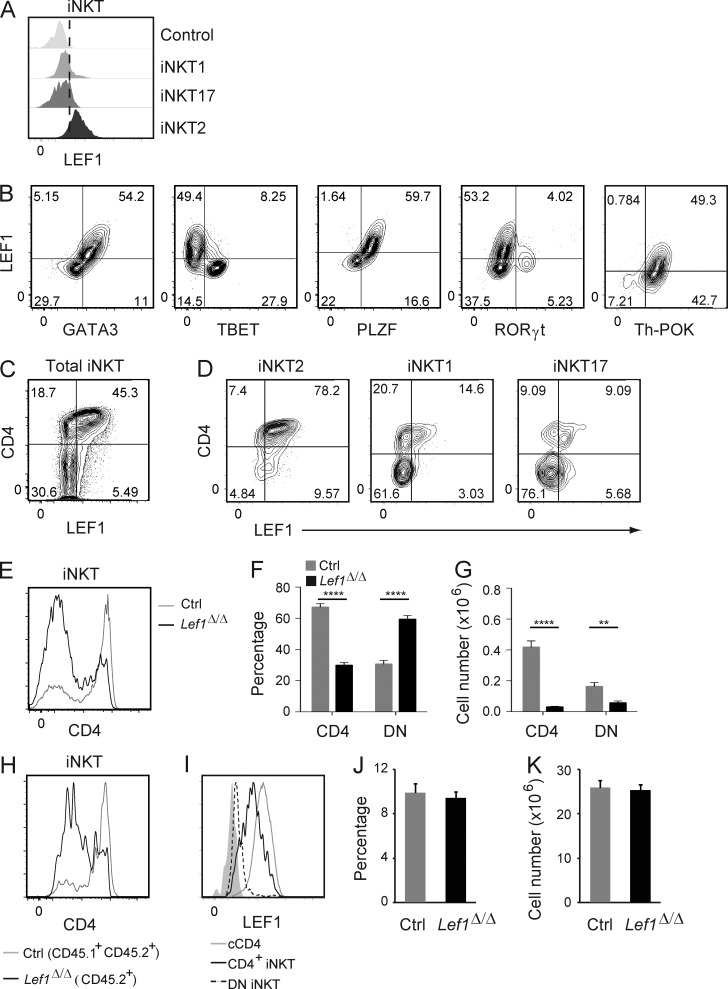
To determine how LEF1 regulated the development of iNKT2 cells, we further considered which iNKT cell subsets expressed LEF1. As shown in Fig. 2 B, LEF1 was highly expressed in ST0, ST1, and ST2 cells but was very low in ST3 cells. When we isolated iNKT cell effector subsets based on expression of PLZF and TBET, we found that LEF1 expression was enriched in iNKT2 (PLZFhiTBET−) cells and was low in iNKT1 (PLZFloTBET+) or iNKT17 (PLZFintRORγt+) cells (Fig. 5 A). When compared with other transcription factors, LEF1 expression was proportional to GATA3 and PLZF and was largely anticorrelated with TBET, consistent with the highest expression of LEF1 in iNKT2 cells (Fig. 5 B). LEF1 was present at very low levels in RORγt+ cells and was coexpressed with Th-POK, a transcription factor which inhibits development of iNKT17 cells and promotes development of CD4+ iNKT cells (Fig. 5 B; Enders et al., 2012; Engel et al., 2012). These data indicate that LEF1 was most highly expressed in iNKT2 cells and that its expression was directly correlated with GATA3, PLZF, and Th-POK, transcription factors which are associated with CD4 expression and the iNKT2 fate.
Figure 5.
LEF1 expression correlated with CD4, GATA3, PLZF, and Th-POK and supported development of CD4+ iNKT cells. (A) Expression of LEF1 in the indicated iNKT effector cells from WT FVB/NJ mice was determined by flow cytometry and is shown as a histogram. The control sample was LEF1 staining in Lef1Δ/Δ iNKT cells. Plots are representative of at least four independent experiments with one to two mice per genotype. (B) Expression of LEF1 and GATA3, TBET, PLZF, RORγt, or Th-POK in WT FVB/NJ thymic iNKT cells was determined by flow cytometry. The percentage of cells in each quadrant is indicated. Plots are representative of at least six independent experiments with one to two mice per genotype. (C and D) Expression of LEF1 and CD4 in total iNKT cells (C) or iNKT2 (PLZFhiTBET−), iNKT1 (PLZFloTBET+) and iNKT17 (RORγt+TBET−; D) cells from the thymus of WT FVB/NJ mice was determined by flow cytometry. Results are representative of at least four independent experiments. (E) CD4 expression on total thymic iNKT cells from control and Lef1Δ/Δ mice was determined by flow cytometry and is shown as histograms. (F and G) The mean percentage (F) and number (G) of CD4+ and CD4−CD8− (DN) thymic iNKT cells in the indicated mouse strains. Data were averaged from >10 independent experiments. (H) CD45.1+CD45.2+ control and CD45.1−CD45.2+ Lef1Δ/Δ FACS-sorted LK bone marrow cells were mixed at 1:1 ratio and transferred into lethally irradiated CD45.1+ C57BL/6 mice. 6–8 wk later, reconstituted mice were analyzed by flow cytometry for CD4 expression on control (CD45.1+CD45.2+) and Lef1Δ/Δ (CD45.2+) thymic iNKT cells. (I) LEF1 expression in cCD4+ thymocytes (cCD4), thymic CD4+ iNKT, and thymic DN iNKT cells was determined by flow cytometry. Solid gray histogram was LEF1 staining in LEF1-deficient iNKT cells. Histograms are representative of at least 10 independent experiments with one to two mice per genotype. (J and K) The percentage (J) and number (K) of cCD4 cells in the thymus of control or Lef1Δ/Δ mice. Graphs show the mean from 10 independent experiments. Error bars represent SEM. **, P < 0.01; ****, P < 0.0001.
LEF1 supported CD4+ iNKT cell development and prevented CD8 expression on peripheral iNKT cells
iNKT cells lack expression of the coreceptor protein CD8, but a subset of these cells expresses CD4, which has been correlated with the ability to produce IL-4 (Lee et al., 2013). In conventional T cells, and also in iNKT cells, CD4 expression and repression of CD8 are mediated by the Th2 transcription factor GATA3 and its downstream target Th-POK (Wang et al., 2010). Given that LEF1 expression was directly correlated with GATA3 and required for the development of functional iNKT2 cells, we questioned whether deletion of Lef1 would also phenocopy GATA3 deficiency with respect to CD4 and CD8 expression (Kim et al., 2006; Wang et al., 2010). As expected, we found that LEF1 was highest in a subset of CD4+ iNKT cells (Fig. 5 C). A majority of CD4+ iNKT cells are iNKT2 cells, but a small subset of iNKT1 and iNKT17 cells also express CD4 and LEF1 (Fig. 5 D). Importantly, the frequency of CD4+ cells was decreased among thymic Lef1Δ/Δ iNKT cells as compared with controls, although the total number of both CD4+ and DN iNKT cells was reduced (Fig. 5, E–G). The selective loss of CD4+ cells was intrinsic to the Lef1Δ/Δ iNKT cells because it was observed in mixed bone marrow chimeric animals (Fig. 5 H). Notably, although LEF1 was highly expressed in conventional CD4 (cCD4) T cells (Fig. 5 I), their frequency and total number was not altered in Lef1Δ/Δ mice as compared with controls (Fig. 5, J and K). Therefore, LEF1 was specifically required for the proper development of CD4+ iNKT cells in the thymus.
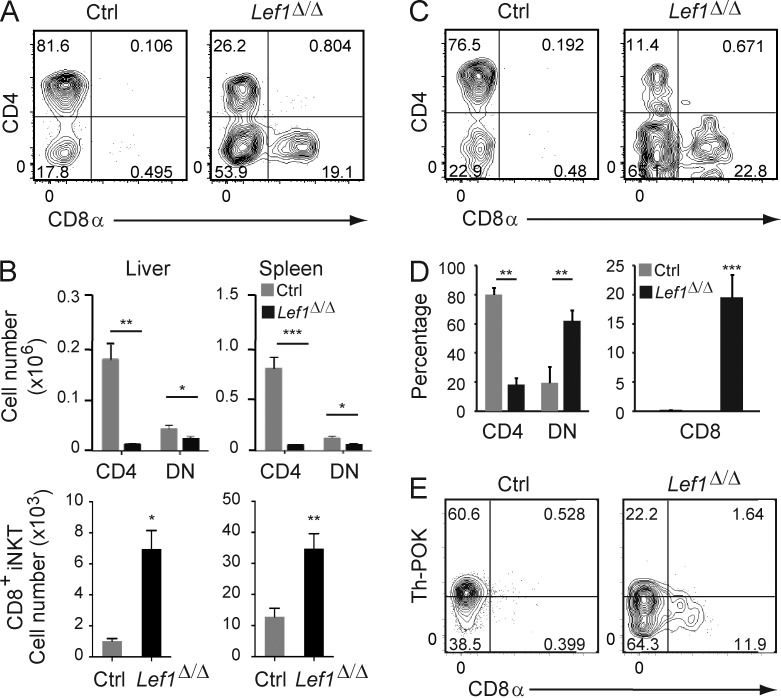
In the liver and spleen of Lef1Δ/Δ mice, the frequency of CD4+ iNKT cells was reduced and a novel population of CD8+ iNKT cells emerged (Fig. 6, A and B). The loss of CD4 and increased expression of CD8 on liver iNKT cells was intrinsic to the Lef1Δ/Δ cells as determined in mixed bone marrow chimeras (Fig. 6, C and D). We examined the expression of Th-POK in Lef1Δ/Δ splenic iNKT cells because Th-POK has been implicated as a GATA3 target that represses Cd8 expression in these cells (Wang et al., 2010), and LEF1 expression was correlated with both GATA3 and Th-POK (Fig. 5 B). We found that Th-POK expression was reduced in the absence of LEF1, particularly in the CD8-expressing iNKT cells (Fig. 6 E). Therefore, similar to GATA3, LEF1 was required for proper expression of Th-POK and promoted the development of CD4+ cells while preventing the expression of CD8 on peripheral iNKT cells.
Figure 6.
LEF1 prevented CD8α expression on peripheral iNKT cells. (A) CD4 and CD8 expression on Tetr+TCRβ+ iNKT cells from the liver of control or Lef1Δ/Δ mice was determined by flow cytometry. (B) The mean number of CD4+, DN, and CD8+ iNKT cells from the indicated tissues in control and Lef1Δ/Δ mice. Data were averaged from at least six independent experiments. (C) CD45.1+CD45.2+ control and CD45.1−CD45.2+ Lef1Δ/Δ FACS-sorted LK bone marrow cells were mixed at 1:1 ratio and transferred into lethally irradiated CD45.1+ C57BL/6 mice. 6–8 wk later, reconstituted mice were analyzed by flow cytometry for CD4 and CD8 expression on control or Lef1Δ/Δ iNKT cells from the liver. (D) The mean percentage of the CD4+, DN, and CD8+ liver iNKT cells from competitive BM chimeras. Data are representative of three independent experiments with two to three chimeras each. (E) Th-POK and CD8α expression in splenic iNKT cells from the indicated mice, as determined by flow cytometry. Data are representative of five independent experiments with one to two mice per genotype. All error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
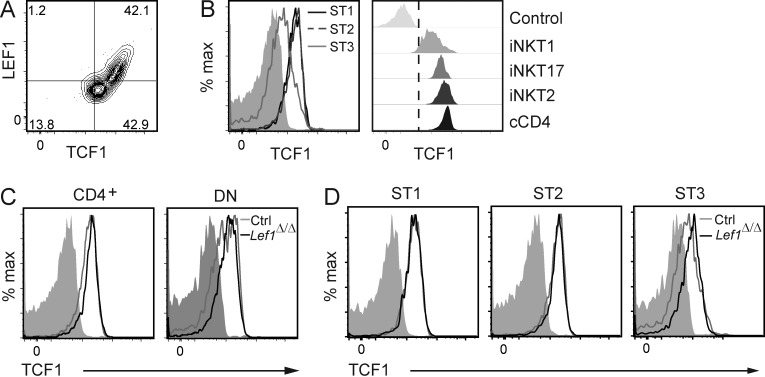
LEF1 functioned independently of TCF1 in iNKT cells
We questioned whether LEF1 deficiency was affecting the expression of TCF1 in iNKT cells because TCF1 is essential for CD4+ Th2 cell differentiation and ILC2 development and is generally redundant with or dominant to LEF1. TCF1 was expressed in all iNKT cells, and LEF1 expression was directly correlated with TCF1 in a subset of these cells (Fig. 7 A). Like LEF1, TCF1 was expressed most highly in ST1 and ST2 iNKT cells (Fig. 7 B); however, TCF1 was expressed in both CD4+ and DN iNKT cells and in ST3 iNKT cells (Fig. 7, B and C). Consistent with this expression pattern, TCF1 was expressed in iNKT2 and iNKT17 cells at levels comparable with cCD4+ T cells and at lower levels in iNKT1 cells (Fig. 7 B). Importantly, TCF1 was expressed equally well in control and Lef1Δ/Δ ST1 and ST2 iNKT cells, indicating that the alterations in iNKT cell development caused by LEF1 deficiency were not a consequence of a loss of TCF1 (Fig. 7 D). TCF1 expression was increased slightly in Lef1Δ/Δ ST3 iNKT cells, suggesting that there may be a selection for TCF1 in these cells or their progenitors when LEF1 is absent (Fig. 7 D). Therefore, our data revealed critical functions for LEF1 in iNKT cell expansion and the development of iNKT2 cells that were not compensated for by TCF1 despite TCF1 expression in all iNKT cells.
Figure 7.
TCF1 expression was not affected by deletion of Lef1 in iNKT cells. (A) Expression of LEF1 and TCF1 in total WT FVB/NJ iNKT cells, as determined by flow cytometry. (B) TCF1 expression in the indicated iNKT stages (left) or iNKT subsets (right) from WT FVB/NJ thymocytes was determined by flow cytometry and is shown as histograms. Solid gray or control represents anti-TCF1 staining in TCF1-deficient iNKT cells and serves as a negative control. (C and D) TCF1 expression in CD4+ and DN (C) or ST1, ST2, and ST3 (D) thymic iNKT cells from control and Lef1Δ/Δ mice was determined by flow cytometry. Data are representative of four independent experiments.
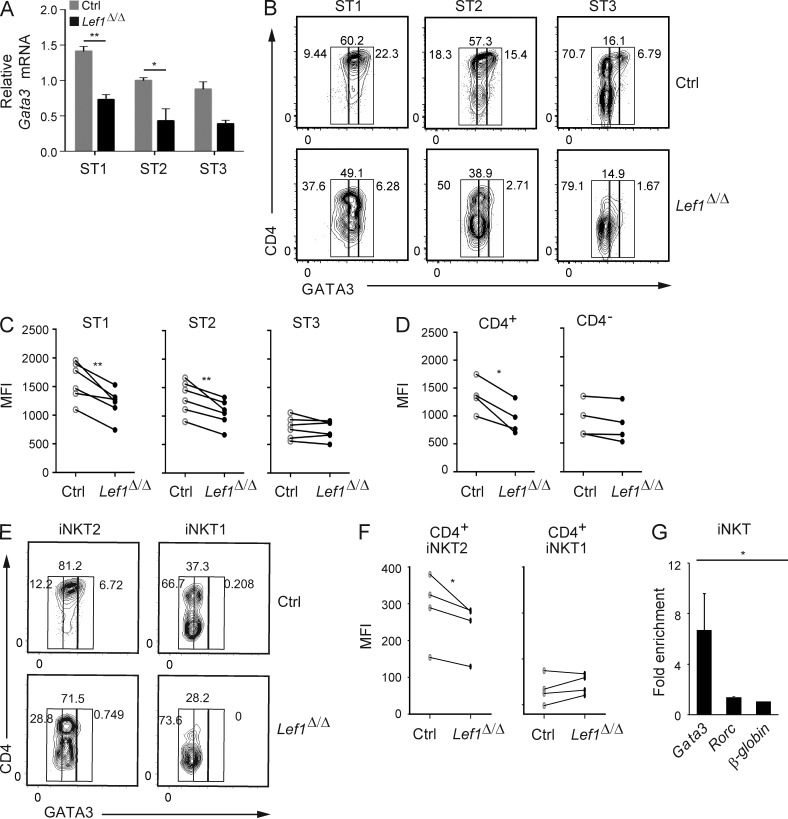
LEF1 directly regulated the expression of GATA3 in CD4+ iNKT cells
Our data demonstrate that LEF1 contributed to the maintenance of iNKT2 cells and was required for CD8 repression in peripheral iNKT cells. These findings are consistent with the hypothesis that LEF1 controls GATA3 expression. To address this hypothesis, we examined Gata3 mRNA in conventionally staged iNKT cells. Gata3 mRNA was decreased by >50% in ST1 and ST2 iNKT cells from Lef1Δ/Δ as compared with control mice (Fig. 8 A). We also analyzed GATA3 expression at the single cell level by flow cytometry in CD4+ and CD4− iNKT cells, which differ in their expression of LEF1. At each stage, the frequency of CD4+ iNKT cells was decreased in the absence of LEF1, and in the remaining CD4+ iNKT cells, the intensity of GATA3 staining was reduced relative to that in control cells (Fig. 8 B). This decline in GATA3 expression was evidenced as a decrease in the MFI at ST1 and ST2 iNKT cells, but not in ST3 cells, regardless of whether CD4 expression was considered (Fig. 8 C). However, total CD4+ iNKT cells, which correlate with iNKT2 cells, showed a more dramatic decline in GATA3 than CD4− iNKT cells (Fig. 8 D). Consistent with these findings, GATA3 expression was reduced in Lef1Δ/Δ CD4+ iNKT2 cells, whereas it remained low in CD4+ iNKT1 cells (Fig. 8, E and F). Importantly, GATA3 expression was not reduced in cCD4 T cells (not depicted). Using ChIP, we found that LEF1 bound specifically to the Gata3 gene, but not to a previously identified site at the Rorc locus (Malhotra et al., 2013), supporting the hypothesis that Gata3 was a direct target of LEF1 in iNKT cells (Fig. 8 G).
Figure 8.
LEF1 was required for proper expression of GATA3 in CD4+ iNKT cells. (A) Expression of Gata3 mRNA relative to Hprt in sorted ST1, ST2, and ST3 iNKT cells from control and Lef1Δ/Δ mice was determined by qRT-PCR. Data were combined from three independent experiments. (B) GATA3 versus CD4 expression in ST1, ST2, and ST3 thymic iNKT cells from control and Lef1Δ/Δ mice, as determined by flow cytometry. (C and D) MFI for GATA3 in ST1, ST2, and ST3 iNKT cells (C) or in CD4+ and CD4− iNKT cells (D) from the thymus of control and Lef1Δ/Δ mice. (E) GATA3 versus CD4 expression in iNKT1 (PLZFloTBET+) and iNKT2 (PLZFhiTBET−) cells from control and Lef1Δ/Δ mice, as determined by flow cytometry. Data are representative of four independent experiments. (F) MFI for GATA3 in thymic CD4+ iNKT2 and CD4+ iNKT1 cells from the indicated mouse strains. (C, D, and F) Each circle represents one mouse (paired Student’s t test: *, P < 0.05; **, P < 0.01). (G) LEF1 chromatin binding was determined by a-LEF1 ChIP on sorted thymic ST1/ST2 Vα14Tg iNKT cells, followed by qPCR using primers spanning a TCF1/LEF1-binding site upstream of the Gata3b promoter, the Rorc promoter, or an unrelated sequence at the β-globin gene locus. Numbers indicate mean fold enrichment of the immunoprecipitated DNA relative to the β-globin locus from three independent experiments is shown. Error bars represent SEM. *, P < 0.05; **, P < 0.01.
DISCUSSION
We have shown that LEF1 played critical and nonredundant roles in iNKT cell expansion and iNKT2 effector fate differentiation. We showed that in the absence of LEF1, iNKT cell expansion at ST0 and ST1 failed to occur and that LEF1 directly regulated expression of the CD127 component of the IL-7 receptor and the transcription factor c-myc, both of which are required for this phase of expansion (Dose et al., 2009; Mycko et al., 2009; Tani-ichi et al., 2013). LEF1 was also required for the development of iNKT2 cells, which include both IL-4 only and IL-4 plus IFNγ–producing iNKT cells, and LEF1 directly regulated the expression of the iNKT2 signature transcription factor GATA3. Our findings are particularly striking given that LEF1 deficiency has only subtle effects on conventional T cells during their differentiation and in the periphery during their activation and acquisition of the memory phenotype as the result of redundancy with the related transcription factor TCF1 (Okamura et al., 1998; Yu et al., 2012; Zhou and Xue, 2012; Steinke et al., 2014). However, TCF1 was unable to compensate for the loss of LEF1 during iNKT cell development, despite the fact that TCF1 was expressed in all LEF1-expressing iNKT cells.
We identified Cd127 as a LEF1 target gene in both iNKT cells and CD4t cells. In contrast, CD127 expression was only modestly decreased on cCD4 thymocytes and remained unchanged on CD8 T cells from Lef1Δ/Δ mice, and conventional αβ T cell development was normal in Lef1Δ/Δ mice. The slight reduction in CD127 expression on Lef1Δ/Δ cCD4 T cells had no effect on their proliferation or survival, and cCD4 T cell numbers were comparable with control mice. Our findings are consistent with a previous study that showed a significant reduction of iNKT cells but a marginal decrease in cCD4 T cell numbers, in mice with a Cd127 deletion at the DP stage (Tani-ichi et al., 2013). Similarly, deletion of c-myc in DP thymocytes resulted in a specific loss of iNKT cells, whereas conventional thymocyte development was unperturbed (Dose et al., 2009; Mycko et al., 2009). We also found that Lef1Δ/Δ ST1 iNKT cells were more apoptotic than their control counter parts. It is possible that the IL-7 receptor signaling pathway is required for the survival of these cells because this pathway regulates the expression of antiapoptotic members of the BCL2 family of proteins (Jiang et al., 2004). However, IL-7Rα and c-MYC are not essential for iNKT cell survival (Dose et al., 2009; Tani-ichi et al., 2013); therefore, LEF1 is likely to have additional targets in iNKT cells that control this process. Alternatively, the combined loss of CD127 and c-myc may be more catastrophic than loss of either protein alone. Further studies into the targets of LEF1 in iNKT cells and the role of IL-7R signaling will help to clarify the role of LEF1 in the survival of iNKT cells. CD127 expression was normal in the mature iNKT effector subsets from LEF1-deficient mice (unpublished data), indicating that LEF1 was not essential for CD127 expression in these cells or that there was a strong selection for CD127-expressing cells. However, we propose that LEF1 is required for Cd127 expression only during the early stages of iNKT cell differentiation and intrathymic expansion, rather than during iNKT cell effector differentiation, where it may be redundant with TCF1. We provide compelling evidence that LEF1 plays an essential role in the postselection proliferative burst that determines iNKT cell numbers, a unique developmental characteristic of this innate-like lymphocyte subset. LEF1 is also crucial for cycling and survival of pro-B cells, another IL-7R–dependent lymphocyte population (Reya et al., 2000), suggesting a broader role for this transcription factor in regulating proliferation and apoptosis during lymphocyte development.
LEF1 was required for the proper emergence of CD4+ iNKT cells, a majority of which are iNKT2 cells. GATA3 expression in iNKT cells was directly proportional to and regulated by LEF1. The effect of Lef1 deletion on iNKT effector cells was strikingly similar to that of Gata3 deletion, including the decreased frequency of CD4+ iNKT cells, decreased expression of Th-POK, and increased expression of CD8 on peripheral iNKT cells (Kim et al., 2006; Wang et al., 2010). Although TCF1 can bind to the Thpok locus and control its expression in cCD4 T cells (Steinke et al., 2014), it is currently unclear whether LEF1 directly or indirectly, through GATA3, influenced Th-POK expression in iNKT cells. The number of iNKT2 cells differs among strains of mice and can have a dramatic effect on immune responsiveness. In strains with a high number of iNKT2 cells, IL-4 can reach levels that promote a memory phenotype on thymic CD8 T cells, systemic IgE production, and Th2-biased chemokine production from thymic dendritic cells, and these effects correlate with Th2-biased immune responses (Verykokakis et al., 2010; Weinreich et al., 2010; Lee et al., 2013). We showed here that LEF1-deficient iNKT cells failed to produce IL-4 when stimulated and that in vivo EOMES+ “memory phenotype” CD8 T cells were diminished in the thymus. Therefore, our findings place LEF1 as a key regulator of immune system function through effects on the number and composition of effector fates among iNKT cells.
iNKT cells are agonist-selected cells and iNKT2 cells show the highest expression of Nur77GFP, a correlate of TCR signaling intensity (Moran et al., 2011), suggesting that iNKT2 cells are strongly stimulated by self-ligands. Therefore, decreased rearrangement, or decreased expression, of the iNKT cell receptor could lead to diminished overall iNKT cell numbers and/or iNKT2 cells. However, LEF1 deficiency did not influence expression or rearrangement of the iNKT cell receptor or other molecules involved in iNKT cell selection, and it did not affect the intensity of TCR signaling in ST0 iNKT cells. Indeed, ST0 iNKT cells developed normally in Lef1Δ/Δ mice, and the forced expression of a pre-rearranged Vα14Jα18 TCR did not restore iNKT cell numbers. In C57BL/6 mice the majority of iNKT cells express Vα14Jα18 in conjunction with Vβ8, but the few iNKT2 cells in these mice are enriched for Vβ7 and Vβ2. FVB/NJ mice do not express Vβ8; rather, they express Vβ7 and Vβ2 in association with Vα14Jα18 as the dominant iNKT cell receptors, indicating that the Vβ usage of FVB/NJ mice is skewed toward iNKT2 specificity (Lee et al., 2013). This altered receptor usage may contribute to the increased frequency of iNKT2 cells in FVB/NJ mice; however, no alterations in Vβ7 or Vβ2 usage were found between control and Lef1Δ/Δ iNKT cells (unpublished data). These findings indicate that LEF1 was not required for the proper expression of the iNKT cell receptor or selection into the iNKT cell lineage.
During positive selection, TCR signals promote β-catenin accumulation and may influence transcriptional activation by LEF1/TCF1 (Kovalovsky et al., 2009). Therefore, LEF1 and TCF1 function may be directly influenced by the strong TCR signals received by iNKT cells. Consistent with this hypothesis, constitutive activation of β-catenin in iNKT cells results in an increased number PLZFhi cells (likely iNKT2 cells) and promotes a memory phenotype on CD8 T cells through production of IL-4 (Sharma et al., 2012). However, in our hands, deletion of β-catenin using VavCre had a minimal effect on iNKT cell development (unpublished data). Therefore, in iNKT cells, LEF1 likely activated gene expression through mechanisms that did not require β-catenin even though TCR signaling and β-catenin may regulate these genes when they are activated. A recent study revealed that activating transcription factor 2 family transcription factors (ATF2) can promote transcription activation via interactions with LEF1/TCF1 in hematopoietic tumors, including a T cell leukemia line, suggesting that LEF1 can use means beyond β-catenin to activate gene expression (Grumolato et al., 2013). One possible mechanism could be through an interaction with ATF2, whose nuclear localization is mediated by the mitogen-activated protein kinase p38, which is activated via strong TCR signals and coreceptor signaling (Zhang et al., 1999). However, the effects of SLAM coreceptor ligation on ATF2 nuclear localization have not been investigated in iNKT cells. Nonetheless, our data are consistent with the conclusion that β-catenin is not essential for the functions of LEF1 in iNKT cells even though LEF1 transcriptional activity may be promoted by strong TCR signaling.
Although TCF1 expression was not decreased in Lef1Δ/Δ iNKT cells, we found that LEF1 was markedly reduced in ST1 iNKT cells from TCF1-deficient mice (unpublished data), indicating that LEF1 may be a target of TCF1 during iNKT cell development. In innate Vγ2+ γδTCR+ cells, LEF1 expression is dependent on TCF1, indicating that TCF1 may regulate Lef1 in multiple ILCs (Malhotra et al., 2013). Therefore, TCF1 deficiency should phenocopy LEF1 deficiency in iNKT cells. However, TCF1 may have LEF1-independent functions in iNKT cells given that TCF1, unlike LEF1, is expressed at high levels in CD4− iNKT cells. Interestingly, Cd127 and c-myc are also targets of GATA3 downstream of TCR signaling in CD8 T cells (Wang et al., 2013), indicating that there may be an intricate self-reinforcing loop that occurs between these factors, with LEF1 and/or TCF1 connecting this loop to TCR signaling events in iNKT cells. Our data also raise the intriguing possibility that differential expression of LEF1, or LEF1 function, may control the number of iNKT2 cells in different strains of mice. Indeed, because LEF1 may integrate TCR signals with the induction of high levels of GATA3, this is a possibility worth further consideration.
MATERIALS AND METHODS
Mice.
Littermate control and VavCre;Lef1f/f mice were on an FVB/NJ background. VavCre, Lef1f/f, or Lef1+/+ mice were used as controls. VavCre;Lef1f/f in the Vα14Tg background and their littermate controls were backcrossed five times onto the C57BL/6J background, and for mixed bone marrow chimera experiments, nontransgenic VavCre;Lef1f/f mice crossed five times onto C57BL/6J were used. Vα14Tg mice were described previously (Griewank et al., 2007). Mice were housed at The University of Chicago Animal Resource Center, and experiments were performed according to the guidelines of The University of Chicago Institutional Animal Care and Use Committee.
Flow cytometry, cell sorting, and antibodies.
Thymocyte suspensions from 3–6-wk-old mice were incubated with anti-FcγR before staining with fluorochrome-conjugated antibodies. Cells were acquired in an LSRII Fortessa (BD) or sorted in a FACSAria (BD) and analyzed with FlowJo. Enrichment of thymocytes for iNKT cells was performed by staining total thymocytes with APC- or PE-conjugated CD1dPBS57 tetramers (National Institutes of Health Tetramer Facility at Emory University), followed by anti-APC or anti-PE microbeads and subjected to MACS-based magnetic cell separation. Propidium iodide was included in all samples to exclude dead cells from the analysis. Pan-caspase staining was performed with the carboxyfluorescein (FAM)-FLICA in vitro Caspase Detection kit (ImmunoChemistry Technologies) according to the manufacturer’s instructions. Antibodies specific for the following antigens were purchased from BD, eBioscience, BioLegend, and Cell Signaling Technology: CD127 (A7R34), CD4 (GK1.5), CD8α (53–6.7), TCRβ (H57-597), CD122 (5H4), CD44 (IM7), CD24 (M1/69), IFNγ (XMG1.2), IL-4 (11B11), IL-17A (eBio17B7), CD69 (H1.2F3), NK1.1 (PK136), CD45.1 (A20), CD45.2 (104), PLZF (R17-809), RORγt (AFKJS-9), T-BET (4B10), GATA3 (L50-823), Th-POK (2POK), LEF1 (C12A5), and TCF-1 (C63D9). An anti-PLZF antibody from Santa Cruz Biotechnology, Inc. (clone D9) that was conjugated to Pacific Blue using the Pacific Blue Monoclonal Antibody Labeling kit (Invitrogen) was also used. Detection of TCF1 was performed with secondary anti–rabbit IgG directly conjugated with PE or FITC. Intracellular staining for PLZF, RORγt, GATA3, Th-POK, LEF1, TCF-1, and T-BET was performed using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience).
Adoptive transfers of bone marrow.
FACS-sorted Lin−cKit+ (LK) bone marrow cells from control C57BL/6 or Lef1Δ/Δ FVB/NJ background mice backcrossed onto C57BL/6 for five generations were prepared from femur and tibiae. CD45.1+CD45.2+ control and CD45.1−CD45.2+ Lef1Δ/Δ LK bone marrow cells were mixed in 1:1 ratio. Recipient CD45.1+ C57BL/6 mice were lethally irradiated (1,000 rad) 5 h before i.v. injection of total 5 × 104 cells. Reconstituted animals were analyzed by flow cytometry 6–8 wk after transplantation. For each independent experiment, one donor bone marrow was used to reconstitute two to three irradiated hosts.
Cell culture, cytokine production, and staining.
Thymocytes were treated with 50 ng PMA, 1 µg ionomycin, and Brefeldin A for 4 h, before harvesting and staining intracellularly for IFNγ, IL-4, and IL-17A using the BD Cytofix/Cytoperm kit.
In vivo BrdU incorporation.
In vivo BrdU incorporation assays were performed as previously described (Verykokakis et al., 2013). In brief, mice were injected i.p. with 1 mg/6 g BrdU 12–15 h before euthanasia. Subsequently, thymocytes were AUTOMACS-enriched for Tetr+ cells and stained for surface markers and then for BrdU according to the manufacturer’s instructions (BD).
RNA analysis and real-time qPCR.
RNA was extracted from sorted thymocytes using the RNeasy mini kit (QIAGEN), DNase-treated, and reverse transcribed using Superscript III (Invitrogen). qPCR was performed with gene-specific primers in an iCycler (Bio Rad Laboratories), using the iQ SYBR Green Supermix (Bio Rad Laboratories). Hprt was used to normalize expression. Relative expression was calculated using the ΔCT method.
ChIP.
Primary mouse CD4t thymocytes from control mice or Vα14Tg iNKT cells (2–5 × 106) were cross-linked with 1% formaldehyde and sheared in a water bath sonicator. Protein–DNA complexes were immunoprecipitated with 5 µg anti-LEF1 (C-19; Santa Cruz Biotechnology, Inc.). Protein G–coupled magnetic beads were used to isolate immune complexes. Cross-links were reversed by heating at 65°C, followed by proteinase K treatment. DNA was purified with DNA purification PCR spin columns (QIAGEN) and amplified by qPCR with primers specific for the c-Myc (Nakashima et al., 2013), Cd127 (Yang et al., 2013), Gata3, Rorc, or irrelevant (β-globin) genomic regions. For each independent ChIP experiment in iNKT cells, thymi from 8–10 mice were pooled together before staining and sorting.
Statistics.
A standard unpaired Student’s t test was used (unless otherwise stated) to determine the statistical probability of the differences observed between two populations of cells using Prism software (GraphPad Software): *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Acknowledgments
We thank members of the Kee, Gounari, and Bendelac laboratories for helpful discussions and reagents, Fotini Gounari for comments on the manuscript, Aditya Rao for help with mouse genotyping, the University of Chicago Flow Cytometry Core Facility for help with cell sorting, and Brian Green for animal husbandry.
This work was supported by grants from the National Institutes of Health to B.L. Kee (R01 CA99978/AI106352 and R56 AI104303). B.L. Kee was a Scholar of the Leukemia and Lymphoma Society. T. Carr was supported by the Interdisciplinary Training Program in Immunology (T32 AI007090). H.-H. Xue was supported by the American Cancer Society (RSG-11-161-01-MPC) and National Institutes of Health (R01 AI105351).
The authors declare no competing financial interests.
Author contributions: T. Carr, M. Verykokakis, and V. Krishnamoorthy performed experiments; S. Yu and H.-H. Xue provided the Lef1f/f mouse strain; B.L. Kee and M. Verykokakis supervised research; B.L. Kee obtained funding; and T. Carr, B.L. Kee, and M. Verykokakis designed and analyzed experiments, interpreted the data, and wrote the paper.
Footnotes
Abbreviations used:
- cCD4
- conventional CD4
- CD4t
- CD4 transitional
- ChIP
- chromatin immunoprecipitation
- DN
- double negative
- DP
- double positive
- FLICA
- fluorescent-labeled inhibitor of caspases
- ILC
- innate lymphoid cell
- iNKT cell
- invariant NKT cell
- LEF
- lymphoid enhancer factor
- LK
- Lin−cKit+
- MFI
- mean fluorescence intensity
- PLZF
- promyelocytic leukemia zinc finger
- SLAM
- signaling lymphocytic activation molecule
References
- Bedel R., Berry R., Mallevaey T., Matsuda J.L., Zhang J., Godfrey D.I., Rossjohn J., Kappler J.W., Marrack P., and Gapin L.. 2014. Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc. Natl. Acad. Sci. USA. 111:E119–E128 10.1073/pnas.1320777110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlagha K., Kyin T., Beavis A., Teyton L., and Bendelac A.. 2002. A thymic precursor to the NK T cell lineage. Science. 296:553–555 10.1126/science.1069017 [DOI] [PubMed] [Google Scholar]
- Benlagha K., Wei D.G., Veiga J., Teyton L., and Bendelac A.. 2005. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202:485–492 10.1084/jem.20050456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P.J., Brigl M., and Brenner M.B.. 2013. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 13:101–117 10.1038/nri3369 [DOI] [PubMed] [Google Scholar]
- Chen Q.Y., Zhang T., Pincus S.H., Wu S., Ricks D., Liu D., Sun Z., Maclaren N., and Lan M.S.. 2010. Human CD1D gene expression is regulated by LEF-1 through distal promoter regulatory elements. J. Immunol. 184:5047–5054 10.4049/jimmunol.0901912 [DOI] [PubMed] [Google Scholar]
- Cohen N.R., Brennan P.J., Shay T., Watts G.F., Brigl M., Kang J., and Brenner M.B.. ImmGen Project Consortium. 2013. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat. Immunol. 14:90–99 10.1038/ni.2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquet J.M., Chakravarti S., Kyparissoudis K., McNab F.W., Pitt L.A., McKenzie B.S., Berzins S.P., Smyth M.J., and Godfrey D.I.. 2008. Diverse cytokine production by NKT cell subsets and identification of an IL-17–producing CD4−NK1.1− NKT cell population. Proc. Natl. Acad. Sci. USA. 105:11287–11292 10.1073/pnas.0801631105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz L.M., Stradner M.H., Yang C.Y., and Goldrath A.W.. 2014. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J. Immunol. 192:2227–2236 10.4049/jimmunol.1302904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M., Sleckman B.P., Han J., Bredemeyer A.L., Bendelac A., and Gounari F.. 2009. Intrathymic proliferation wave essential for Vα14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA. 106:8641–8646 10.1073/pnas.0812255106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Eberl G., Taniuchi I., Benlagha K., Geissmann F., Hennighausen L., Bendelac A., and Littman D.R.. 2005. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 22:705–716 10.1016/j.immuni.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Enders A., Stankovic S., Teh C., Uldrich A.P., Yabas M., Juelich T., Altin J.A., Frankenreiter S., Bergmann H., Roots C.M., et al. . 2012. ZBTB7B (Th-POK) regulates the development of IL-17–-producing CD1d-restricted mouse NKT cells. J. Immunol. 189:5240–5249 10.4049/jimmunol.1201486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., Zhao M., Kappes D., Taniuchi I., and Kronenberg M.. 2012. The transcription factor Th-POK negatively regulates Th17 differentiation in Vα14i NKT cells. Blood. 120:4524–4532 10.1182/blood-2012-01-406280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L., Matsuda J.L., Surh C.D., and Kronenberg M.. 2001. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2:971–978 10.1038/ni710 [DOI] [PubMed] [Google Scholar]
- Griewank K., Borowski C., Rietdijk S., Wang N., Julien A., Wei D.G., Mamchak A.A., Terhorst C., and Bendelac A.. 2007. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 27:751–762 10.1016/j.immuni.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumolato L., Liu G., Haremaki T., Mungamuri S.K., Mong P., Akiri G., Lopez-Bergami P., Arita A., Anouar Y., Mlodzik M., et al. . 2013. β-Catenin-independent activation of TCF1/LEF1 in human hematopoietic tumor cells through interaction with ATF2 transcription factors. PLoS Genet. 9:e1003603 10.1371/journal.pgen.1003603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Li W.Q., Hofmeister R.R., Young H.A., Hodge D.R., Keller J.R., Khaled A.R., and Durum S.K.. 2004. Distinct regions of the interleukin-7 receptor regulate different Bcl2 family members. Mol. Cell. Biol. 24:6501–6513 10.1128/MCB.24.14.6501-6513.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.J., Pai S.Y., Brigl M., Besra G.S., Gumperz J., and Ho I.C.. 2006. GATA-3 regulates the development and function of invariant NKT cells. J. Immunol. 177:6650–6659 10.4049/jimmunol.177.10.6650 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. . 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D., Yu Y., Dose M., Emmanouilidou A., Konstantinou T., Germar K., Aghajani K., Guo Z., Mandal M., and Gounari F.. 2009. β-Catenin/Tcf determines the outcome of thymic selection in response to αβTCR signaling. J. Immunol. 183:3873–3884 10.4049/jimmunol.0901369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.K., and Hoffmann F.M.. 2006. Smad4 cooperates with lymphoid enhancer-binding factor 1/T cell-specific factor to increase c-myc expression in the absence of TGF-β signaling. Proc. Natl. Acad. Sci. USA. 103:18580–18585 10.1073/pnas.0604773103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N., Narayan K., Cho O.H., Sylvia K.E., Yin C., Melichar H., Rashighi M., Lefebvre V., Harris J.E., Berg L.J., and Kang J.. Immunological Genome Project Consortium. 2013. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 38:681–693 10.1016/j.immuni.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M.L., Mendes-da-Cruz D., Keller A.C., Lochner M., Schneider E., Dy M., Eberl G., and Leite-de-Moraes M.C.. 2008. Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc. Natl. Acad. Sci. USA. 105:19845–19850 10.1073/pnas.0806472105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow M.A., Lee G., Gillis S., Yancopoulos G.D., and Alt F.W.. 1992. Interleukin-7 induces N-myc and c-myc expression in normal precursor B lymphocytes. Genes Dev. 6:61–70 10.1101/gad.6.1.61 [DOI] [PubMed] [Google Scholar]
- Mycko M.P., Ferrero I., Wilson A., Jiang W., Bianchi T., Trumpp A., and MacDonald H.R.. 2009. Selective requirement for c-Myc at an early stage of Vα14i NKT cell development. J. Immunol. 182:4641–4648 10.4049/jimmunol.0803394 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Arai S., Suzuki A., Nariai Y., Urano T., Nakayama M., Ohara O., Yamamura K., Yamamoto K., and Miyazaki T.. 2013. PAD4 regulates proliferation of multipotent haematopoietic cells by controlling c-myc expression. Nat. Commun. 4:1836 10.1038/ncomms2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura R.M., Sigvardsson M., Galceran J., Verbeek S., Clevers H., and Grosschedl R.. 1998. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 8:11–20 10.1016/S1074-7613(00)80454-9 [DOI] [PubMed] [Google Scholar]
- Pellicci D.G., Hammond K.J., Uldrich A.P., Baxter A.G., Smyth M.J., and Godfrey D.I.. 2002. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1−CD4+ CD1d-dependent precursor stage. J. Exp. Med. 195:835–844 10.1084/jem.20011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., O’Riordan M., Okamura R., Devaney E., Willert K., Nusse R., and Grosschedl R.. 2000. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 13:15–24 10.1016/S1074-7613(00)00004-2 [DOI] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M.P., Mathew R., Liszewski M.K., Spooner C.J., Barr K., Meng F., Singh H., and Bendelac A.. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13:264–271 (published erratum appears in Nat. Immunol. 2013. 14:413) 10.1038/ni.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Chen Q., Nguyen T., Yu Q., and Sen J.M.. 2012. T cell factor-1 and β-catenin control the development of memory-like CD8 thymocytes. J. Immunol. 188:3859–3868 10.4049/jimmunol.1103729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke F.C., Yu S., Zhou X., He B., Yang W., Zhou B., Kawamoto H., Zhu J., Tan K., and Xue H.H.. 2014. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4+ T cell fate and interact with Runx3 to silence Cd4 in CD8+ T cells. Nat. Immunol. 15:646–656 10.1038/ni.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani-ichi S., Shimba A., Wagatsuma K., Miyachi H., Kitano S., Imai K., Hara T., and Ikuta K.. 2013. Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc. Natl. Acad. Sci. USA. 110:612–617 10.1073/pnas.1219242110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kaer L., Parekh V.V., and Wu L.. 2013. Invariant natural killer T cells as sensors and managers of inflammation. Trends Immunol. 34:50–58 10.1016/j.it.2012.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M., Boos M.D., Bendelac A., and Kee B.L.. 2010. SAP protein-dependent natural killer T-like cells regulate the development of CD8+ T cells with innate lymphocyte characteristics. Immunity. 33:203–215 10.1016/j.immuni.2010.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M., Krishnamoorthy V., Iavarone A., Lasorella A., Sigvardsson M., and Kee B.L.. 2013. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J. Immunol. 191:5973–5983 10.4049/jimmunol.1301521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M., Zook E.C., and Kee B.L.. 2014. ID’ing innate and innate-like lymphoid cells. Immunol. Rev. 261:177–197 10.1111/imr.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Carr T., Xiong Y., Wildt K.F., Zhu J., Feigenbaum L., Bendelac A., and Bosselut R.. 2010. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur. J. Immunol. 40:2385–2390 10.1002/eji.201040534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Misumi I., Gu A.D., Curtis T.A., Su L., Whitmire J.K., and Wan Y.Y.. 2013. GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat. Immunol. 14:714–722 10.1038/ni.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai H., Sekine-Kondo E., Shigeura T., Motomura Y., Yasuda T., Satoh R., Yoshida H., Kubo M., Kawamoto H., Koseki H., and Taniguchi M.. 2012. Development and function of invariant natural killer T cells producing TH2- and TH17-cytokines. PLoS Biol. 10:e1001255 10.1371/journal.pbio.1001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M.A., Odumade O.A., Jameson S.C., and Hogquist K.A.. 2010. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11:709–716 10.1038/ni.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Monticelli L.A., Saenz S.A., Chi A.W., Sonnenberg G.F., Tang J., De Obaldia M.E., Bailis W., Bryson J.L., Toscano K., et al. . 2013. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 38:694–704 10.1016/j.immuni.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip-Schneider M.T., Horie M., and Broxmeyer H.E.. 1993. Characterization of interleukin-7-induced changes in tyrosine phosphorylation and c-myc gene expression in normal human T cells. Exp. Hematol. 21:1648–1656. [PubMed] [Google Scholar]
- Yu Q., Sharma A., Oh S.Y., Moon H.G., Hossain M.Z., Salay T.M., Leeds K.E., Du H., Wu B., Waterman M.L., et al. . 2009. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-γ. Nat. Immunol. 10:992–999 10.1038/ni.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Zhou X., Steinke F.C., Liu C., Chen S.C., Zagorodna O., Jing X., Yokota Y., Meyerholz D.K., Mullighan C.G., et al. . 2012. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 37:813–826 10.1016/j.immuni.2012.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Salojin K.V., Gao J.X., Cameron M.J., Bergerot I., and Delovitch T.L.. 1999. p38 mitogen-activated protein kinase mediates signal integration of TCR/CD28 costimulation in primary murine T cells. J. Immunol. 162:3819–3829. [PubMed] [Google Scholar]
- Zhou X., and Xue H.H.. 2012. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J. Immunol. 189:2722–2726 10.4049/jimmunol.1201150 [DOI] [PMC free article] [PubMed] [Google Scholar]