Abstract
The circadian timing system in plants synchronizes their physiological functions with the environment. This is achieved by a global control of gene expression programs with a considerable part of the transcriptome undergoing 24-h oscillations in steady-state abundance. These circadian oscillations are driven by a set of core clock proteins that generate their own 24-h rhythm through periodic feedback on their own transcription. Additionally, post-transcriptional events are instrumental for oscillations of core clock genes and genes in clock output. Here we provide an update on molecular events at the RNA level that contribute to the 24-h rhythm of the core clock proteins and shape the circadian transcriptome. We focus on the circadian system of the model plant Arabidopsis thaliana but also discuss selected regulatory principles in other organisms.
Keywords: circadian oscillation, RNA-binding protein, post-transcriptional regulation
Introduction
Regulation of the Flow of Genetic Information at the RNA Level
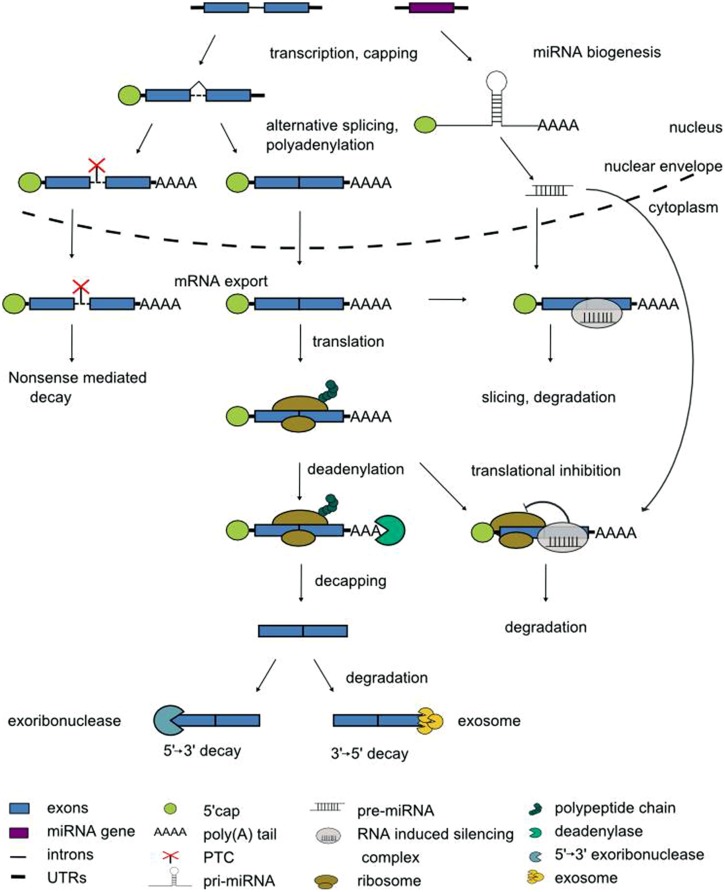
The expression of eukaryotic genes is regulated at multiple levels. In the nucleus, transcription factors recruit RNA polymerase II to the gene’s promoter. The access of transcriptional activators or repressors to the DNA in turn is licensed by chromatin remodeling factors. Once transcription has been initiated, pre-mRNAs enter a series of processing steps to mature mRNAs (Darnell, 2013). When the nascent pre-mRNA is ca. 20 nucleotides in length, its 5′ end receives the 7-methylguanosine cap to protect the mRNA against degradation. Pre-mRNAs are spliced in the nucleus to remove introns, and differential usage of splice sites can give rise to multiple alternatively spliced transcript isoforms of one and the same pre-mRNA (Syed et al., 2012; Kornblihtt et al., 2013; Reddy et al., 2013). Polyadenylation signals determine the processing at the 3′ end, i.e., the cleavage at a specific position before addition of the poly(A) tail that protects against degradation from the 3′ end (Proudfoot, 2011). Subsequently, the mature mRNA is exported from the nucleus for translation (Figure 1).
FIGURE 1.
Steps in pre-mRNA processing. See text for details.
During this journey from the site of transcription in the nucleus to the cytoplasm the fate of the mRNAs is influenced by two main types of trans-acting regulators, RNA-binding proteins and non-coding RNAs. RNA-binding proteins, through dedicated domains, specifically recognize cis-active motifs in mRNAs to regulate their processing. The most frequent type of RNA-binding domain is the RNA recognition motif (RRM) of approximately 80 amino acids with a well conserved structure of four antiparallel β strands packed against two α helices (Maris et al., 2005). Non-coding RNAs (ncRNAs) do not have a protein-coding potential. Numerous ncRNAs have emerged as important regulators of gene expression through the identification of cognate mRNA targets via base-pairing to sites with a variable degree of complementarity (Cech and Steitz, 2014). Non-coding RNAs are generally classified according to their size. Long non-coding RNAs (lncRNAs) vary in length between ca. 200 nucleotides and dozens of kilobases (Kim and Sung, 2012). Small non-coding RNAs are between 20 and 25 nucleotides in length and are further classified according to their mode of biogenesis. Small interfering RNAs (siRNAs) are derived from double-stranded precursors whereas microRNAs (miRNAs) are derived from transcripts with partially double-stranded, hairpin-like structures (Figure 1; Rogers and Chen, 2013). Collectively, the pre-mRNA processing steps equip the cell with obvious checkpoints to rapidly modulate the transcriptome.
A prominent example of gene expression programs that continuously require fine tuning are periodic fluctuations in mRNA steady-state abundance across the day. Such daily rhythms in gene expression are controlled by an endogenous timing mechanism, the “circadian” clock. The circadian clock acts predominantly by directing promoter activity to defined time intervals of the day. However, off-switching of promoter activity is in many cases not sufficient to account for a steep decline from peak transcript levels to trough levels within a few hours to maintain correct phase, period and amplitude of transcript oscillations.
The Circadian Timing System
Plant physiology, biochemistry and behavior are orchestrated by the circadian system which serves to optimally align metabolic functions of the plant with the periodic changes in ambient light and dark phases (Barak et al., 2000; Eriksson and Millar, 2003; McClung, 2006; de Montaigu et al., 2010; Yerushalmi et al., 2011). Conceptually, the circadian timing system is divided into three functional units. The core clockwork is responsible for self-sustained 24-h rhythms of clock proteins. Input pathways ensure synchrony of the core clockwork with the day/night cycles through perception of periodic changes in light and temperature. Output pathways drive the expression of a large part of the circadian transcriptome with around a third of the protein-coding genes regulated by the circadian clock (Covington et al., 2008; Hazen et al., 2009). These gene expression rhythms translate into physiological and biochemical output rhythms.
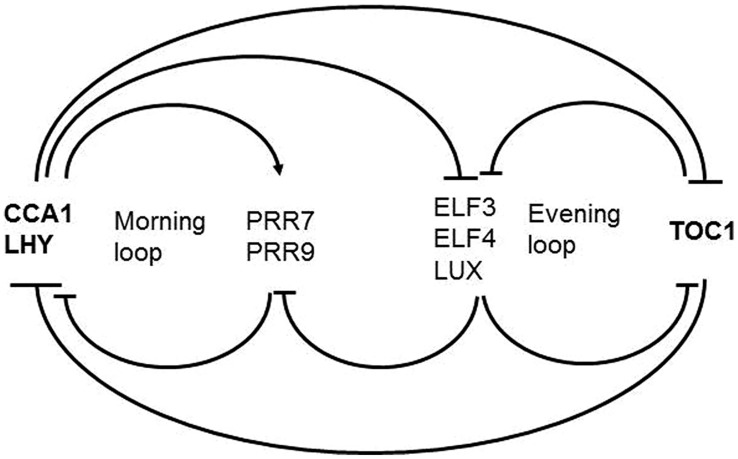
The framework of the plant circadian timekeeping system has been established in Arabidopsis thaliana. Below we describe the components that are relevant for the topic we cover here. For a complete picture readers are referred to dedicated reviews (Yanovsky and Kay, 2001; Staiger, 2002; Harmer, 2009; McClung, 2011; Herrero and Davis, 2012; Nagel and Kay, 2012; Staiger et al., 2013; Hsu and Harmer, 2014). The core clockwork is made up by a series of autoregulatory circuits of clock proteins (Figure 2). The central loop consists of two Myb transcription factors LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) peaking at dawn, and the pseudoresponse regulator TIMING OF CAB EXPRESSION1 (TOC1) peaking at dusk, that reciprocally repress their own expression (Alabadi et al., 2001; Gendron et al., 2012; Huang et al., 2012). This core loop is interconnected with a loop preferentially active in the morning and another loop preferentially active in the evening. Through the morning loop, LHY and CCA1 activate the expression of the PSEUDORESPONSE REGULATORS PRR9 and PRR7 which in turn repress CCA1 and LHY. PRR7 and PRR9 expression is switched off during the night through the evening complex (EC) consisting of the Myb-type transcription factor LUX ARRHYTHMO (LUX), EARLY FLOWERING 3 (EFL3) and ELF4 proteins (Dixon et al., 2011; Helfer et al., 2011; Herrero et al., 2012). As a result, CCA1 and LHY transcription resumes. In the evening loop, the EC and TOC1 reciprocally regulate their expression. The interconnection of these feedback loops is thought to contribute to robustness of the rhythmic expression patterns.
FIGURE 2.
Scheme of the Arabidopsis core clockwork. In the central loop LHY and CCA1 on the one hand and TOC1 on the other hand reciprocally repress their own expression. In the morning loop, LHY and CCA1 activate PRR9 and PRR7 which in turn repress CCA1 and LHY. PRR7 and PRR9 expression is switched off during the night through the EC consisting of LUX, EFL3, and ELF4. In the evening loop, the EC and TOC1 reciprocally regulate their expression.
Modification of clock proteins, time-of-day dependent proteolysis and regulated nuclear uptake contribute to maintaining a period of 24 h (Mas et al., 2003; Fujiwara et al., 2008; Wang et al., 2010). These post-translational processes operating at the level of clock proteins are beyond the scope of this manuscript and have been reviewed elsewhere (Schöning and Staiger, 2005; Mehra et al., 2009; Seo and Mas, 2014).
Rhythms in eukaryotes generally arise in feedback loops of clock gene transcription/translation. The molecular players, however, are different in plants, arguing for an independent origin of plant and animal clocks (Roden and Carré, 2001). Below, we briefly touch upon the core components of the mammalian and fungal timing systems.
In mammals, the core clock loop comprises two transcription factors, CLOCK and BMAL1 that activate the Period (Per) 1, 2, and 3 and Cryptochrome (Cry) 1 and 2 genes (Young and Kay, 2001; Dibner et al., 2010; Partch et al., 2013). PER and CRY proteins undergo heterodimerization and are taken up into the nucleus to inhibit activation of their own genes by CLOCK and BMAL1. Upon proteoloytic degradation of CLOCK and BMAL1 a new cycle can then be initiated.
In Neurospora crassa, the transcription factors White collar 1 (WC-1) and WC-2 form the white collar complex (WCC), which drives the rhythmic expression of the frequency (frq) clock gene (Liu et al., 1999; Bell-Pedersen et al., 2005). FRQ interacts with frequency interacting RNA helicase (FRH). The resulting FRQ/FRH complex in turn inhibits WCC activity.
Layers of Post-Transcriptional Control in the Circadian Timing System
Transcription is considered the prime mechanism driving rhythmic gene expression both in the core clockwork and in clock output. However, mRNA steady-state abundance is determined by the rates of both transcription and degradation and theoretical considerations have led to the conclusion that transcriptional rhythms are manifest in high amplitude mRNA cycling only when the mRNA has a sufficiently short half-life (Wuarin et al., 1992; Lück et al., 2014).
Enhancer trapping using a promoter-less luciferase reporter in Arabidopsis unveiled that one third of the genome is under transcriptional control by the clock (Michael and McClung, 2003). However, the LHCB1∗3 (LIGHT HARVESTING CHLOROPHYLL BINDING PROTEIN) promoter is clock-regulated but transcript levels are constitutive, suggesting that changes in mRNA stability obscure rhythmic transcription (Millar and Kay, 1991). CATALASE3 mRNA oscillations damp to a high level in constant darkness while CATALASE3 promoter-driven luciferase activity still oscillates with an evening peak (Zhong et al., 1997; Michael and McClung, 2002). On the contrary, NITRATE REDUCTASE mRNA oscillates despite a time-of-day independent transcription rate (Pilgrim et al., 1993).
In mammals, about 10% of the transcripts in the liver undergo circadian oscillations (Akhtar et al., 2002). A comprehensive RNA-seq analysis uncovered that rhythms of only 22% of them are driven by de novo transcription (Koike et al., 2012). Furthermore, a recent transcriptome analysis with 2-h resolution around the clock found that 10% of the N. crassa transcriptome is reproducibly rhythmic at the mRNA level under normal growth conditions, and that the circadian clock may influence as much as 40% of the genome under other conditions (Hurley et al., 2014). A parallel high-throughput assay for timing of promoter activity using the luciferase reporter unveiled significant discordance between promoter activity and transcript oscillations. These discrepancies between clock-controlled transcription and oscillations in mRNA steady-state abundance pointed to additional levels of control impinging on clock-regulated transcripts and thus post-transcriptional regulation moved centre-stage in chronobiology.
Here, we discuss our current view on RNA-based regulation of gene expression in the Arabidopsis circadian timing system. Additionally, selected examples of RNA processing steps that have been shown to shape the daily pattern of transcripts in other model organisms including mammals, the fly Drosophila melanogaster and the bread mold N. crassa are presented for an integrated view. For a general survey of post-transcriptional regulation in these circadian clock systems readers are referred to comprehensive reviews (Crosthwaite, 2004; Harms et al., 2004; Keene, 2007; Kojima et al., 2011; Staiger and Green, 2011; Staiger and Köster, 2011; Zhang et al., 2011; Wang et al., 2013; Kojima and Green, 2014).
Alternative Splicing in the Circadian System
In plants, our understanding of post-transcriptional regulation of circadian timekeeping is most advanced for alternative splicing. We begin by briefly describing key points of the mechanism, the players and the outcome of alternative splicing before turning to its relevance for circadian timekeeping in Arabidopsis and conclude by selected examples of alternative splicing in other model organisms of chronobiology.
Regulation of Alternative Splicing
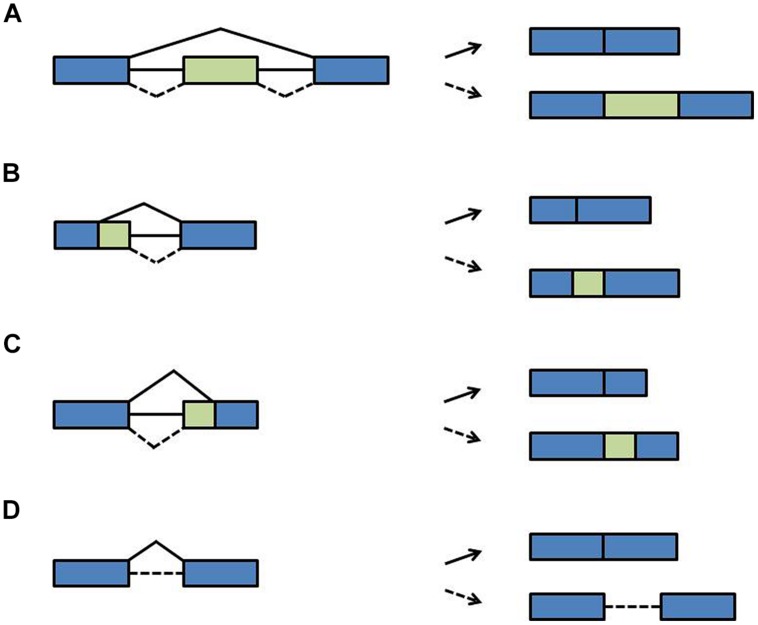
During pre-mRNA splicing, introns are excised and the flanking exons are joined. However, not every splice site is used each time a pre-mRNA is processed. Rather, through the variable use of splice sites exonic sequences can be lost or intronic sequences can remain in the mRNA, designated as alternative splicing. During exon skipping, exons are removed together with their flanking introns (Figure 3A). The use of alternative 5′ splice sites or alternative 3′ splice sites causes variable portions of introns to be removed and variable portions of exons to remain in the mRNA (Figures 3B,C). During intron retention, introns can stay in the pre-mRNA (Figure 3D). Due to this variation in splicing patterns the corresponding proteins can be composed of distinct domains and thus have different functions (Nilsen and Graveley, 2010; Carvalho et al., 2012; Syed et al., 2012; Reddy et al., 2013). This tremendously increases the coding capacity of the genome. At the RNA level, alternative splice isoforms can have a different inventory of cis-regulatory sequence motifs and thus be differentially recognized by RNA-binding proteins or miRNAs. Alternative splice isoforms can also be identified as being “aberrant” and targeted for degradation. For example, if intronic sequences are retained, the open reading frame (ORF) may terminate at a premature termination codon (PTC). Such PTCs are recognized by the nonsense-mediated decay (NMD) pathway, a surveillance mechanism that eliminates aberrant transcripts (Arciga-Reyes et al., 2006; Isken and Maquat, 2008). Through linkage with NMD, alternative splicing can lead to quantitative changes in overall transcript levels (McGlincy and Smith, 2008; Nicholson and Mühlemann, 2010). Of note is that the NMD pathway has recently been shown to contribute to innate immunity in plant-pathogen-interaction and thus may have more widespread physiological roles (Gloggnitzer et al., 2014).
FIGURE 3.
Types of alternative splicing events. (A) Exon skipping (B) alternative 5′ splice site (C) alternative 3′ splice site (D) intron retention. Blue boxes: exons; green boxes: alternative exons; lines: introns; solid diagonal lines: constitutive splicing events; broken diagonal lines: alternative splicing events.
Pre-mRNA splicing is executed by a high molecular weight machine in the cell, the spliceosome. The main components of the spliceosome are five ribonucleoprotein (RNP) complexes with specific RNAs designated the U-rich small nuclear RNAs (U snRNAs). U1, U2, U4, and U5 assemble with the Sm proteins B/B’, D1, D2, D3, E, F, and G to form U snRNPs. The U6 snRNP contains the related LSM2 (Like-Sm2) to LSM8 proteins (Tharun, 2009; Golisz et al., 2013).
The decision whether a particular splice site is used is influenced by additional RNA-binding proteins that interact with distinct motifs in the pre-mRNAs to favor or inhibit the recruitment of the spliceosome to neighboring alternative splice sites (Reddy et al., 2012, 2013; Kornblihtt et al., 2013). These regulators are mainly serine/arginine-rich (SR) proteins or heterogeneous nuclear ribonucleoparticle proteins (hnRNPs). The SR proteins contain one or two RRMs as well as a domain with a high proportion of arginine/serine repeats (Reddy, 2004; Barta et al., 2010). The hnRNPs are a diverse class of RNA-binding proteins with one or multiple RRMs or RNA binding domains of the K homology (KH) motif type, originally found in human hnRNP K (Wachter et al., 2012). Numerous components involved in pre-mRNA splicing have been identified in A. thaliana, based on homology to yeast and mammalian sequences (Koncz et al., 2012; Reddy et al., 2013).
Alternative Splicing in the Core Clockwork
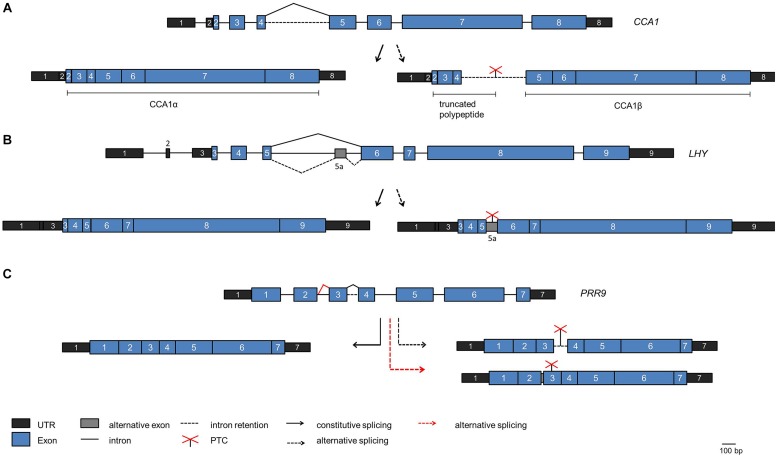
Whole transcriptome sequencing has uncovered a prominent role of alternative pre-mRNA splicing in the plant circadian system (Sanchez et al., 2011; Syed et al., 2012; Henriques and Mas, 2013; Staiger and Brown, 2013; Cui et al., 2014). For the core clock gene CCA1 an alternative splice isoform was found to increase upon exposure of the plants to high light and decrease upon exposure to cold (Filichkin et al., 2010). This splice isoform retains intron 4, the long intron following the Myb-domain encoding exons, and thus can give rise to a truncated protein due to a PTC (Figure 4A). The splicing factor SR45 is able to bind to the intron in vitro, suggesting that it may function in alternative splicing of intron 4 (Filichkin et al., 2015).
FIGURE 4.
Alternative splicing events in core clock genes. The scheme of the genes are shown on the top of each panel, the fully spliced isoforms encoding the full length proteins are indicated on the left side and alternative splice isoforms are indicated on the right side. (A) CCA1. The fully spliced isoform encoding full length CCA1 [designated CCA1α by Seo et al. (2012)] is indicated on the left. The splice isoform with intron 4 retained is shown on the right. It is predicted to encode a polypeptide terminating at the PTC within intron 4, thus comprising only the DNA-binding Myb domain, or the CCA1β polypeptide comprising only the dimerization domain (Seo et al., 2012). (B) LHY. The splice isoform with alternative splicing at intron 5 leading to inclusion of exon 5a including a PTC is shown on the right. (C) PRR9. The splice isoform retaining intron 3 including a PTC is shown on the right (black broken arrow). The spliced isoform with additional eight nucleotides at exon 2 due to the use of an alternative 5′ splice site entailing a frame shift is shown on the right at the bottom (red broken line). This splice isoform is elevated in prmt5.
A targeted search for alternative splice isoforms of the Arabidopsis clock genes using a high resolution Reverse transcription-PCR based alternative splicing panel uncovered that alternative splicing is widespread in the core clock (James et al., 2012). For LHY, an alternative splice isoform containing an alternative exon was found due to alternative splicing at the long intron 5 (that corresponds to intron 4 in the related CCA1 transcript; Figure 4B). This splice isoform accumulates when plants are shifted to low temperature. Because it contains a PTC and is a substrate for the NMD pathway the production of full length LHY protein is precluded and thus LHY protein levels decline at low temperature (James et al., 2012). In contrast, CCA1 intron 4 retention decreases at low temperature, and the CCA1 transcript transiently shows a higher and broader peak upon shift to low temperature. These data suggest that alternative splicing may adjust the core oscillator to low temperatures, and the differential behavior of CCA1 and LHY in this response may contribute to the overlapping but not redundant action of these two proteins in the clock mechanism.
Subsequently, autoregulation of CCA1 has been proposed to underlie this low temperature response. The CCA1 alternative splice isoform retaining intron 4 including a PTC accumulates at low temperature, as described above, and can produce a polypeptide comprising only the N-terminal MYB domain (Figure 4A). This transcript has been predicted to produce a protein that consists of the C-terminal dimerization domain without the DNA-binding MYB domain, designated CCA1β (Figure 4A; Seo et al., 2012). Upon targeted over-expression in transgenic plants CCA1β interferes with complex formation of the full length CCA1α protein both with itself and with LHY and thus with their function as transcriptional repressors in the core clock. Indeed, over-expression of CCA1β from a constitutive promoter leads to a short period phenotype, as observed in cca1 lhy mutants, consistent with CCA1β acting as a dominant negative inhibitor. It remains to be demonstrated whether such a reinitiation of translation downstream of an ORF occurs in planta to produce the CCA1β protein.
Similar to CCA1 and LHY, PRR7, and PRR9 are thought to act partially redundantly in the clock. For PRR7 non-functional alternative splice isoforms transiently accumulate to substantial levels upon exposure to low temperatures and thus would lead to reduced PRR7 levels. In contrast, PRR9 transiently increases at low temperatures, but the PRR9 alternative splicing events that lead to non-functional transcripts due to intron 3 retention or inclusion of eight nucleotides at the end of exon 2 are not influenced by temperature (Figure 4C; James et al., 2012). This points to differences in the regulation of this pair of clock proteins. TOC1/PRR1 and PRR5 are also alternatively spliced to PTC-containing isoforms at low temperatures.
Alternative splicing of clock genes has also been observed upon exposure to high temperatures, e.g., for the LUX, LOV KELCH PROTEIN 2 and TIME FOR COFFEE (TIC) transcripts but the physiological consequences for clock function have not been addressed (Filichkin and Mockler, 2012).
For PRR7 and LHY rapid changes in alternative splicing patterns in response to red light activation of the phytochrome photoreceptor have been uncovered (Shikata et al., 2014). Given the role of phytochrome in mediating light input (Fankhauser and Staiger, 2002; Millar, 2004), it is conceivable that such changes in alternative splicing may impact light entrainment of the clock.
Alternative Splicing in Clock Output
Currently it is estimated that alternative splicing affects more than 60% of all intron-containing genes in Arabidopsis (Marquez et al., 2012). The functional relevance of most of the alternative splice isoforms remains to be demonstrated (Carvalho et al., 2012). A differential function was found for two RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE ACTIVASE (RCA) protein variants encoded by splice isoforms. The RCA transcript undergoes circadian oscillations in steady-state abundance as well as circadianly regulated alternative splicing (Sanchez et al., 2010). A short alternative splice isoform encodes a protein that acts independent of light, and a long alternative splice isoform encodes a protein isoform regulated by light (Zhang et al., 2002). Alternative splicing of the mRNA isoform that encodes the light-regulated protein increases during the day (Sanchez et al., 2010).
The use of whole-genome tiling arrays for transcript profiling around the clock unveiled circadian rhythms in the steady-state level of numerous introns (Hazen et al., 2009). In cases where these retained introns are embedded in rhythmically expressed genes and oscillate in phase with their surrounding exons, the retained introns should lead to truncated protein variants.
A Circadian Feedback Loop based on Alternative Splicing and NMD
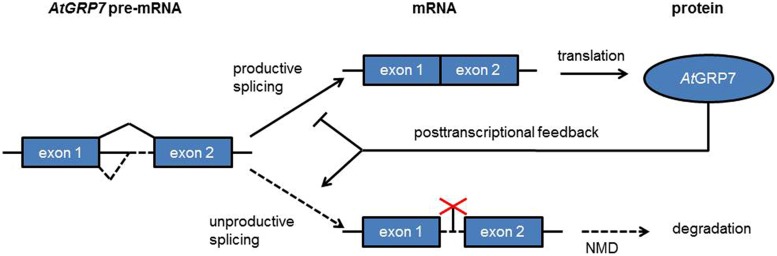
AtGRP7 (A. thaliana glycine-rich RNA binding protein 7) and AtGRP8 are clock-regulated RNA binding proteins which peak at the end of the day. Both AtGRP7 and AtGRP8 negatively autoregulate through alternative splicing (Figure 5). A transcript isoform retaining part of the intron including a PTC is generated that rapidly decays via NMD (Staiger and Heintzen, 1999; Schöning et al., 2008). Upon mutation of a conserved arginine residue in the RRM the in vitro and in vivo RNA binding activities as well as the negative autoregulation of AtGRP7 are lost (Schöning et al., 2007; Köster et al., 2014a; Leder et al., 2014). AtGRP7 and AtGRP8 thus represent two clock-regulated feedback circuits that additionally cross-regulate via alternative splicing and NMD. These were the first examples of feedback loops based on post-transcriptional regulation in the circadian system. AtGRP7 and AtGRP8 in turn regulate steady-state abundance or alternative splicing of several transcripts which undergo circadian oscillations themselves, suggesting that the AtGRP7/AtGRP8 feedback loops pass timing information from the core oscillator to clock output (Rudolf et al., 2004; Streitner et al., 2010, 2012; Schmal et al., 2013).
FIGURE 5.
A clock-regulated negative autoregulatory circuit based on alternative splicing and NMD. In response to increasing AtGRP7 levels the use of the cryptic intronic 5′ splice site is favored, leading to unproductive splicing with the splice isoform being degraded via NMD.
Apart from circadian rhythms, AtGRP7 has been shown to be involved in the low temperature response, response to oxidative stress, flowering time control and pathogen defense (Kim et al., 2007b, 2008; Schmidt et al., 2010; Nicaise et al., 2013; Hackmann et al., 2014; Löhr et al., 2014). This suggests that it may exert widespread post-transcriptional control in the cell and integrate responses to external stimuli with endogenous timing cues.
A Defective Clock in Mutants of Candidate Splicing Components
A long period circadian phenotype has been observed in mutants deficient in PROTEIN METHYLTRANSFERASE 5 (PRMT5; Sanchez et al., 2010, 2011). PRMT5 is a type II protein arginine methyltransferase that symmetrically dimethylates arginine side chains, i.e., deposits a methyl group on either one of the two terminal guanidino nitrogens. PRMT5 acts upon a broad spectrum of substrates that can be detected by antibodies against symmetrically dimethylated arginine in wild type plants, but not in the prmt5 mutant. Among those are numerous proteins involved in RNA processing including AtSmD1, AtSmD3, and LSM4 (Deng et al., 2010). In Arabidopsis, a complex of LSM2 to LSM8 complex is part of U snRNPs and thus functions in pre-mRNA splicing (Perea-Resa et al., 2012). In prmt5, PRR9 levels are greatly elevated compared to wild type plants. As PRR9 over-expression leads to a short period of the clock, altered PRR9 steady-state abundance seemed unlikely to cause the prmt5 long period phenotype (Matsushika et al., 2002). However, the PRR9 transcript was aberrantly spliced in prmt5. An alternative splice isoform with a PTC due to the use of an alternative 5′ splice site at intron 2 accumulate at the expense of the functional isoform (Figure 4B). This suggests that the long period in prmt5 can be partly attributed to aberrant PRR9 splicing. When the prmt5 mutant was analyzed on tiling arrays, a global impact of PRMT5 on alternative splicing was found. The use of the high resolution alternative splicing panel then unveiled that PRMT5 function is required in particular to activate weak 5′ splice sites (Sanchez et al., 2010).
Subsequently, mutants defective in the PRMT5 substrate LSM4 were also linked to circadian regulation. The lsm4 mutant shows long period oscillations of gene expression as well as aberrant splicing of several clock genes (Perez-Santángelo et al., 2014). The lsm5 mutant, previously identified because of its hypersensitivity to abscisic acid and thus named sad1 (supersensitive to abscisic acid 1) also led to long period leaf movement rhythms (Perez-Santángelo et al., 2014). The expression of several clock genes is altered in lsm5. Furthermore, there is an increase in TOC1 intron 4 retention, similar to wild-type plants exposed to low temperature conditions, and intron 2 is retained in CCA1. A genome-wide analysis of both mutants uncovered a more widespread role of LSM4 in the control of alternative splicing. Notably, the LSM5 transcript undergoes circadian oscillations itself. Such a clock regulation of splicing regulators like LSM5 or AtGRP7 may serve as a means to coordinate time-of-day dependent changes in splicing of a cohort of target genes (Staiger, 2001; Perez-Santángelo et al., 2013).
Mutations in two other splicing factors, Ski-interacting protein (SKIP) and SPLICEOSOMAL TIMEKEEPER LOCUS 1 (STIPL1), led also to a long period of the clock (Jones et al., 2012; Wang et al., 2012). The yeast and human SKIP counterparts have a demonstrated role as splicing factors (Gahura et al., 2009). AtSKIP associates with the splicing factor SR45 and in the skip-1 mutant, alternative splicing of PRR7 and PRR9 is defective, contributing to the long period phenotype (Wang et al., 2012). STIPL1 encodes a homolog of TUFTELIN-INTERACTING PROTEIN 11 (TFIP11) in humans and Ntr1p in yeast involved in spliceosome disassembly (Tannukit et al., 2009; Jones et al., 2012). The altered expression of CCA1, LHY, PRR9, GI, and TOC1 caused by the aberrant splicing is likely to contribute to the circadian defects in the stipl1 mutant. The mechanisms of how the Arabidopsis proteins impact splicing of their targets remain to be described.
In mice, a very interesting rhythmic and light-induced alternative splicing event was described for the splicing factor U2AF26, a homolog of the small U2 auxiliary factor (U2AF) subunit U2AF35 involved in recognition of the 3′ splice site (Preußner et al., 2014). Through this alternative splicing events, translation of U2AF26 extends into the 3′ untranslated region (3′ UTR), generating a C-terminal extension of the ORF. This additional domain shows homology to the Drosophila clock protein Timeless, an interaction partner of Period. U2AF26-deficient mice show nearly arrhythmic Period1 protein levels and aberrant mRNA cycling in peripheral clocks. Moreover, lack in U2AF26 leads to increased phase advance in response to alterations in the environmental light–dark cycles. These data suggest that light induced U2AF26 alternative splicing serves to limit Period1 induction in response to changes in ambient light and thus is involved in entrainment (Preußner et al., 2014).
RNA Stability
Eukaryotic mRNAs vary widely in their stabilities and mRNA turnover is exquisitely regulated (Houseley and Tollervey, 2009). The first step in mRNA degradation is the removal of the poly(A) tail, followed by exoribonuclease digestion from the 5′ end after removal of the cap structure or digestion from the 3′ end by the exosome, a multi-subunit machinery for RNA degradation in eukaryotes.
It has long been predicted that changes in mRNA half-life across the circadian cycle contribute to circadian transcript oscillations (So and Rosbash, 1997). In Arabidopsis, a suite of clock-controlled transcripts were identified in an approach to globally identify short-lived transcripts using DNA microarrays (Gutierrez et al., 2002). For two of them, CCR-LIKE (CCL) and SENESCENCE ASSOCIATED GENE 1, mRNA stability changes across the day. The changes in CCL mRNA stability continue under free-running conditions, indicating that they are controlled by the circadian clock (Lidder et al., 2005). Furthermore, degradation of these mRNAs is mediated by the downstream (DST) element first identified in the 3′ UTR of the auxin inducible SMALL AUXIN-UP RNAs and shown to destabilize mRNA (Newman et al., 1993). Of note, disruption of the DST-mediated RNA decay pathway leads to circadian defects (Lidder et al., 2005). Components of the DST pathway have not yet been reported.
For the core clock gene CCA1, a dependence of transcript stability on light quality has been found (Yakir et al., 2007). The CCA1 mRNA is relatively stable in the dark but has a short half-life in the light. The light-dependent CCA1 mRNA degradation in combination with light-regulated CCA1 transcription has been implicated in entrainment of the clock. Again, little is known about mechanisms such as, for example, RNA-binding proteins that regulate the access of nucleases, e.g., through steric hindrance or conformational changes in the mRNA depending on the light quality.
In mammals, changes in RNA stability contribute to oscillations of the core clock genes Cry1, Per1, Per2, and Per3, with a higher stability during the upswing and a lower stability during the downswing. Several cellular RNA-binding proteins including hnRNP D and hnRNP I, also known as the polypyrimidine tract binding protein PTB, have been shown to bind to the 3′ UTRs of clock transcripts in a circadian phase dependent manner, entailing their degradation (Kwak et al., 2006; Woo et al., 2009, 2010; Lee et al., 2014; Kim et al., 2015).
In N. crassa, the exosome is involved in regulation of several rhythmic transcripts (Guo et al., 2009). For example, downregulation of the Neurospora ortholog of RRP44, the 3′→5′ exonuclease subunit of the exosome, leads to enhanced frq mRNA stability, higher frq levels and a longer period of frq mRNA oscillations. Because FRH interacts with the RRP44 ortholog, FRQ, FRH and the exosome are part of a post-transcriptional negative feedback loop interlocked with the clock transcriptional feedback loop that regulates WCC activity. The rrp44 transcript itself is clock-controlled suggesting that time-of-day-dependent exosome activity may play a wider role in circadian regulation.
In the green algae Chlamydomonas reinhardtii knockdown of XRN1, a 5′→3′ exoribonuclease leads to low amplitude and rapid dampening of the bioluminescence rhythm (Matsuo et al., 2008). XRN1 has been shown to interact with the C3 subunit of the RNA-binding protein CHLAMY1 that is required for correct period and phase of circadian rhythms (Dathe et al., 2012).
Alternative Polyadenylation and Poly(A) Tail Length
Processing at the mRNA 3′ end comprises pre-mRNA cleavage at the poly(A) site followed by the addition of tracts of adenosines (Xing and Li, 2011). This poly(A) tail influences both mRNA stability and translation. A dedicated group of RNA-binding proteins, the poly(A) binding proteins, bind to the poly(A) tail and additionally interact with the translation initiation factor eIF4G that in turn interacts with the Cap binding protein eIF4E, thus bending the mRNA into a circle. This enables translational control by the poly(A) tail.
New insights into polyadenylation in Arabidopsis came from implementation of direct RNA sequencing, a single molecule technique where native mRNA is used as the template (Sherstnev et al., 2012). This allowed the determination of the site of RNA cleavage and polyadenylation without errors cause by aberrant reverse transcription or PCR amplification during library generation and unveiled a widespread heterogeneity in 3′ ends through alternative polyadenylation (Sherstnev et al., 2012). Such variation in the length of the 3′ UTRs can have functional significance for the resulting transcript isoforms, e.g., due to the presence of different regulatory motifs or miRNA binding sites in 3′ UTRs of different length. The functional consequences of the usage of the alternative polyadenylation sites for core clock genes and genes of clock output remain to be resolved.
In the cytoplasm, poly(A) tail shortening from the 3′ end is catalyzed by deadenylases. This variation in poly(A) tail length can affect mRNA stability. A connection of variation in poly(A) tail length with circadian rhythms has not been addressed in plants.
In mammals, daily fluctuations in poly(A) tail size have initially been observed for the mRNA encoding the neuroactive peptide vasopressin that is synthesized within the suprachiasmatic nuclei and displays rhythmic concentration changes in the cerebrospinal fluid (Robinson et al., 1988). More recently, 2.5% of transcripts in mouse liver turned out to undergo circadian oscillations in poly(A) tail length in a genome-wide poly(A)denylome analysis using microarrays (Kojima et al., 2012). Interestingly, this variation in poly(A) tail length rather than variation in mRNA steady-state abundance correlated with rhythms in protein abundance, suggesting that the rhythmic variation in the length of the poly(A) tail leads to rhythmic translation.
One of the deadenylases in mammals, Nocturnin, is rhythmically expressed (Wang et al., 2001). Mice deficient for Nocturnin have defects in lipid homeostasis and response to glucose, suggesting that Nocturnin mediates post-transcriptional regulation of metabolic events by the circadian clock.
Translational Control
Changes in translation efficiency allow a rapid adjustment in the proteome without prior regulation of transcription or RNA processing. Regulation at the level of translation initiation is a topic intensely studied in plants (Roy and von Arnim, 2013). In particular, upstream ORF (uORFs) located 5′ to a reading frame can alter the usage of its start codons. When such uORFs are longer than 25 codons they inhibit translation of the downstream ORF (Nyiko et al., 2009; Roy et al., 2010). About a third of all Arabidopsis genes harbor uORFs but few of them have been functionally studied (Kim et al., 2007a). Alternative splicing of introns in the 5′ UTR can affect the inventory of uORFs. Moreover, miRNAs can affect translation of their target mRNAs (Brodersen et al., 2008; Li et al., 2013).
Several studies in Arabidopsis indicate that protein levels often do not follow rhythmic mRNA levels (Piques et al., 2009; Baerenfaller et al., 2012). In rice seedlings, a number of proteins display oscillations at the protein level that do not correlate with mRNA rhythms (Hwang et al., 2011). On the one hand, such observations point to rhythms in translation. On the other hand, cycling of proteins made from rhythmic mRNAs can also be blunted through long half-lives. In tomato, in vivo labeling with 35S methionine uncovered circadian oscillations of the translation rate of the photosystem II light harvesting complex polypeptide that closely correlate with the transcript oscillations (Riesselmann and Piechulla, 1992). In contrast, no oscillation was detected at the protein level, perhaps due to the low turnover of this membrane protein.
In Arabidopsis, translation of the core clock gene LHY has been shown to be influenced by light (Kim et al., 2003). Thus, when LHY transcript levels fall after the dawn peak, light promotes LHY translation. This simultaneous translational induction and transcriptional repression has been suggested to sharpen the LHY protein peak.
Clearly, a more widespread inventory of changes in translation would provide insights into clock-control over translation. Comparing the pool of mRNAs associated with polysomes to the mRNAs not associated with polysomes serves as an indication for active translation. Such a global translatome profiling across the circadian cycle would allow conclusions about changes in the translation status of each transcript in the course of the day (Missra and von Arnim, 2014).
In mammals, dedicated RNA-binding proteins have been identified that control translation of clock genes. The mLARK protein binds to the 3′ UTR of Per1 and boosts PER1 protein levels, most likely through stimulation of translation (Kojima et al., 2007). Additionally, hnRNPQ rhythmically binds to the Per1 5′ UTR to stimulate its translation in a time-of-day dependent manner (Lee et al., 2012).
Notably, as many as 50% of the proteins that cycle in liver are translated from constitutively expressed mRNAs (Reddy et al., 2006). More recently, it has been observed that the clock exerts also a widespread control of the translation apparatus through coordinated transcription of translation initiation factors, ribosomal proteins, and rRNAs (Jouffe et al., 2013). Additionally, distinct signaling pathways impinging on translation initiation factors are rhythmically activated.
In C. reinhardtii, the RNA-binding protein CHLAMY1 regulates translation of output genes. CHLAMY1 binds, in a circadian manner, to transcripts with UG repeats in their 3′ UTR (Zhao et al., 2004). Among these transcripts is NITRITE REDUCTASE (Waltenberger et al., 2001). Consistent with a role for CHLAMY1 as a translational repressor, the activity of NITRITE REDUCTASE in reciprocal to the levels of CHLAMY1 C1 and 3 subunits.
Non-Coding RNAs
microRNAs
Plant miRNAs regulate a wide range of mRNAs predominantly by mRNA cleavage and subsequent degradation but also via inhibition of translation (Figure 1; Rogers and Chen, 2013). In Arabidopsis, a suite of miRNAs were interrogated for rhythmic expression. miR171, miR398, miR168, and miR167 oscillate diurnally but are not under clock-control (Sire et al., 2009). In contrast, the precursors of miR157A, miR158A, miR160B, and miR167D are clock-controlled (Hazen et al., 2009). Whether these daily fluctuations in expression have functional consequences for their targets has not been addressed. Notably, clock-regulated RNA-binding protein AtGRP7 impacts processing of several miRNA precursors (Köster et al., 2014b).
In mammals, rhythmic miRNA expression has been widely observed. Several clock genes are targets of miRNAs, and miRNAs have been implicated in the regulation of period length and light resetting of the clock (Nagel et al., 2009). Recently, miR122 was shown to control the expression of the deadenylase nocturnin, another post-transcriptional regulator in the circadian system (Kojima et al., 2010).
Natural Antisense Transcripts
Arabidopsis contains a large number of convergently overlapping gene pairs that can give rise to natural antisense transcripts (NATs) which might act as regulators of the sense gene (Zubko et al., 2011). Using tiling arrays, rhythmic NATs have been detected for 7% of the protein coding genes (Hazen et al., 2009). Among these are the oscillator genes LHY, CCA1, TOC1, PRR3, PRR5, PRR7, and PRR9. The functional significance for the oscillator mechanism has not been addressed.
A prime example of antisense RNA regulation in circadian timekeeping is the N. crassa frq locus that gives rise to a long non-coding antisense RNA qrf oscillating in antiphase to frq (Kramer et al., 2003). Light-dependent qrf expression is involved in resetting of the clock (Xue et al., 2014). Moreover, frq transcription and qrf transcription are mutually inhibitory, resulting in the antiphasic frq and qrf oscillations. Antisense Per1 transcripts that cycle in antiphase to Per1 have been detected in mouse liver and in silkmoth, suggesting that such pairs of rhythmic antisense RNAs may also play a role in circadian clocks (Sauman and Reppert, 1996; Koike et al., 2012; Menet et al., 2012).
As the number of identified ncRNAs including ncRNAs and siRNAs is increasing, it is conceivable that more ncRNAs will be found to fulfill a role in regulating circadian gene expression.
The Epitranscriptome
Modification of cellular components by methylation is mostly known for DNA and proteins. Methylation generally depends on the availability of S-adenosylmethionine (SAM) that donates the methyl group and the concentration of S-adenosylhomocysteine (SAH), a by-product that is a competitive inhibitor of the methylation. The drug 3-deazaadenosine inhibits SAH hydrolysis and thus indirectly inhibits methylation though accumulation of the competitive inhibitor (Chiang, 1998).
The role of histone methylation in clock gene transcription has been established in Arabidopsis (Malapeira et al., 2012; Song and Noh, 2012). The role of non-histone protein methylation in the clock is obvious from the pervasive effect of the prmt5 mutant on the pace of Arabidopsis clock (Hong et al., 2010; Sanchez et al., 2010).
In contrast to DNA and protein methylation, the physiological role of mRNA modification by methylation of nucleobases is less well understood. Nevertheless, the importance of RNA methylation including methylation of adenine (m6A) has recently been recognized in plants (Bodi et al., 2012). Impaired m6A methylation affects embryonic development and leads to aberrant growth phenotypes in adult plants. Within transcripts, m6A was found predominantly about 150 nucleotides upstream of the polyadenylation site.
RNA methylation has recently been shown to affect the mammalian clock (Fustin et al., 2013). Treatment of human cells with 3-deazaadenosin led to global changes in gene expression. The gene ontology category “rhythmic processes” ranked fourth among significantly affected processes, suggesting an exquisite sensitivity of the circadian clock to imbalanced methylation. Among the upregulated genes were RNA processing factors including RNA m7G cap methylases, m6A demethylases, RNA methylases, and splicing factors. RNA immunoprecipitation using an antibody against m6A then unveiled the presence of m6A in many clock gene transcripts. Inhibition of m6A RNA methylation by knockdown of the methyltransferase Mettl3 led to a long period of Per2 driven luciferase activity and locomoter activity. The processing of clock genes including Per2 was delayed which may slow down the speed of the clock.
Conclusion
Ample evidence has accumulated for discordances between rhythms in promoter activity and rhythms in mRNA levels on the one hand and mRNA and protein rhythms on the other hand, respectively. In particular, the importance of correct alternative pre-mRNA splicing in the Arabidopsis clock has been recognized either through aberrant clock function in splicing factor mutants or the appearance of specific alternative splice isoforms of clock genes. In the future, the recent establishment of STABLE ISOTOPE LABELING by amino acids in cell culture (SILAC) for Arabidopsis seedlings will allow quantitative proteomics and identification of predicted polypeptides corresponding to alternative splice isoforms (Lewandowska et al., 2013). A next logical step is to prove the relevance of alternative splice isoforms by testing their association with polysomes and through complementation of mutants with forced isoform expression. Furthermore, the development of robust RNA immunoprecipitation protocols provides an entré to identify direct targets of candidate splicing factors (Terzi and Simpson, 2009; Köster et al., 2014a).
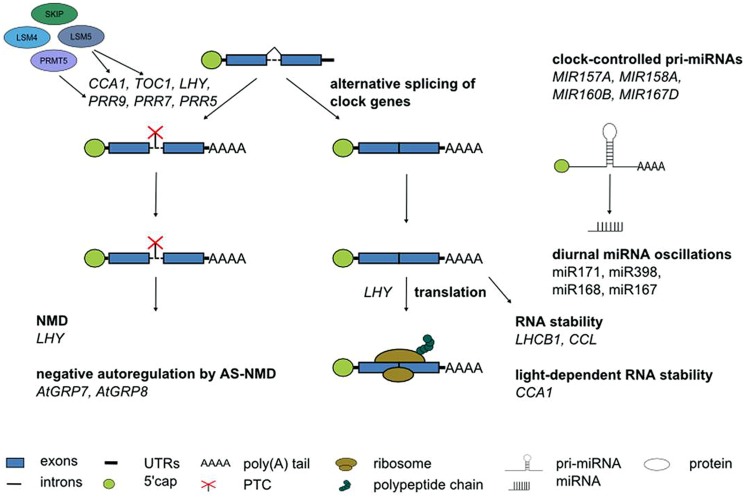
Although alternative splicing undoubtedly is the layer of post-transcriptional regulation currently understood in most detail in Arabidopsis, it would be premature to assume that it is the most important one. Other RNA processing steps clearly contribute to shaping the circadian transcriptome (Figure 6). This is evident from the anecdotal reports on regulated stability of a few circadian genes, oscillations of a handful of miRNAs or translational regulation of clock genes summarized here. In particular, in the light of fundamental differences in the mechanism of pri-miRNA processing and in target mRNA regulation by miRNAs between animals and plants it will be interesting to see the impact of miRNAs on rhythmic gene expression programs. Novel developments in high throughput techniques combined with more powerful bioinformatics pipelines will help to further shift the focus from individual gene expression patterns to genome-wide impact of these regulatory events. This will advance our knowledge on the importance of other RNA processing steps for the circadian system also in plants, as it has been the case in animals.
FIGURE 6.
Post-transcriptional processes in the Arabidopsis circadian system. Numerous core clock genes undergo alternative splicing, in particular in response to temperature changes. The spliceosomal Lsm4 and Lsm5 proteins control alternatives splicing. PRMT5 affects alternative splicing of core clock genes likely through modification of snRNP proteins and splicing factors. Alternative splice isoforms of clock genes containing a PTC can undergo NMD. The RNA-binding proteins AtGRP7 and AtGRP8 are part of a clock-controlled post-transcriptional feedback loop based on alternative splicing and NMD. Translation of the core clock component LHY is regulated. The stability of LHCB1 and CCL is time-of-day dependent. CCA1 stability depends on the light quality. Some miRNA show diurnal oscillations. Several pri-miRNAs undergo clock-controlled oscillations. See text for further details.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CN is supported by the German National Academic Foundation. The work in our laboratory is supported by the DFG (STA653 and priority program 1530).
References
- Akhtar R. A., Reddy A. B., Maywood E. S., Clayton J. D., King V. M., Smith A. G., et al. (2002). Circardian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12 540–550 10.1016/S0960-9822(02)00759-5 [DOI] [PubMed] [Google Scholar]
- Alabadi D., Oyama T., Yanovsky M. J., Harmon F. G., Mas P., Kay S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 10.1126/science.1061320 [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes L., Wootton L., Kieffer M., Davies B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47 480–489 10.1111/j.1365-313X.2006.02802.x [DOI] [PubMed] [Google Scholar]
- Baerenfaller K., Massonnet C., Walsh S., Baginsky S., Buhlmann P., Hennig L., et al. (2012). Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol. Syst. Biol. 8:606 10.1038/msb.2012.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak S., Tobin E. M., Green R. M., Andronis C., Sugano S. (2000). All in good time: the Arabidopsis circadian clock. Trends Plant Sci. 5 517–522 10.1016/S1360-1385(00)01785-4 [DOI] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Reddy A. S. (2010). Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR Proteins) in plants. Plant Cell 22 2926–2929 10.1105/tpc.110.078352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., et al. (2005). Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6 544–556 10.1038/nrg1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z., Zhong S., Mehra S., Song J., Li H., Graham N., et al. (2012). Adenosine methylation in Arabidopsis mRNA is associated with the 3’ end and reduced levels cause developmental defects. Front. Plant Sci. 3:48 10.3389/fpls.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190 10.1126/science.1159151 [DOI] [PubMed] [Google Scholar]
- Carvalho R., Feijao C., Duque P. (2012). On the physiological significance of alternative splicing events in higher plants. Protoplasma 250 639–650 10.1007/s00709-012-0448-9 [DOI] [PubMed] [Google Scholar]
- Cech T. R., Steitz J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157 77–94 10.1016/j.cell.2014 [DOI] [PubMed] [Google Scholar]
- Chiang P. K. (1998). Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol. Ther. 77 115–134 10.1016/S0163-7258(97)00089-2 [DOI] [PubMed] [Google Scholar]
- Covington M. F., Maloof J. N., Straume M., Kay S. A., Harmer S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130 10.1186/gb-2008-9-8-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite S. K. (2004). Circadian clocks and natural antisense RNA. FEBS Lett. 567 49–54 10.1016/j.febslet.2004.04.073 [DOI] [PubMed] [Google Scholar]
- Cui Z., Xu Q., Wang X. (2014). Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis. J. Exp. Bot. 65 1973–1980 10.1093/jxb/eru085 [DOI] [PubMed] [Google Scholar]
- Darnell J. E. (2013). Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA 19 443–460 10.1261/rna.038596.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe H., Prager K., Mittag M. (2012). Novel interaction of two clock-relevant RNA-binding proteins C3 and XRN1 in Chlamydomonas reinhardtii. FEBS Lett. 586 3969–3973 10.1016/j.febslet.2012.09.046 [DOI] [PubMed] [Google Scholar]
- de Montaigu A., Toth R., Coupland G. (2010). Plant development goes like clockwork. Trends Genet. 26 296–306 10.1016/j.tig.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., et al. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 107 19114–19119 10.1073/pnas.1009669107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C., Schibler U., Albrecht U. (2010). The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72 517–549 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- Dixon L. E., Knox K., Kozma-Bognar L., Southern M. M., Pokhilko A., Millar A. J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 25 120–125 10.1016/j.cub.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M. E., Millar A. J. (2003). The circadian Clock. A plant’s best friend in a spinning world. Plant Physiol. 132 732–738 10.1104/pp.103.022343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Staiger D. (2002). Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216 1–16 10.1007/s00425-002-0831-4 [DOI] [PubMed] [Google Scholar]
- Filichkin S. A., Cumbie J. S., Dharmawadhana J. P., Jaiswal P., Chang J. H., Palusa S. G., et al. (2015). Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant. 10.1093/mp/ssu130 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Filichkin S., Mockler T. (2012). Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol. Direct. 7:20 10.1186/1745-6150-7-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Priest H. D., Givan S. A., Shen R., Bryant D. W., Fox S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20 45–58 10.1101/gr.093302.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S. S., Salome P. A., McClung C. R., et al. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283 23073–23083 10.1074/jbc.M803471200 [DOI] [PubMed] [Google Scholar]
- Fustin J.-M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., et al. (2013). RNA-Methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155 793–806 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Gahura O., Abrhámová K., Skružný M., Valentová A., Munzarová V., Folk P., et al. (2009). Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J. Cell. Biochem. 106 139–151 10.1002/jcb.21989 [DOI] [PubMed] [Google Scholar]
- Gendron J. M., Pruneda-Paz J. L., Doherty C. J., Gross A. M., Kang S. E., Kay S. A. (2012). Arabidopsis circadian clock protein, TOC1 is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109 3167–3172 10.1073/pnas.1200355109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer J., Akimcheva S., Srinivasan A., Kusenda B., Riehs N., Stampfl H., et al. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16 376–390 10.1016/j.chom.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Golisz A., Sikorski P. J., Kruszka K., Kufel J. (2013). Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 41 6232–6249 10.1093/nar/gkt296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Cheng P., Yuan H., Liu Y. (2009). The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138 1236–1246 10.1016/j.cell.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R. A., Ewing R. M., Cherry J. M., Green P. J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. U.S.A. 99 11513–11518 10.1073/pnas.152204099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackmann C., Korneli C., Kutyniok M., Köster T., Wiedenlübbert M., Müller C., et al. (2014). Salicylic acid-dependent and –independent impact of an RNA-binding protein on plant immunity. Plant Cell Environ. 37 696–706 10.1111/pce.12188 [DOI] [PubMed] [Google Scholar]
- Harmer S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60 357–377 10.1146/annurev.arplant.043008.092054 [DOI] [PubMed] [Google Scholar]
- Harms E., Kivimae S., Young M. W., Saez L. (2004). Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 19 361–373 10.1177/0748730404268111 [DOI] [PubMed] [Google Scholar]
- Hazen S. P., Naef F., Quisel T., Gendron J. M., Chen H., Ecker J. R., et al. (2009). Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 10:R17 10.1186/gb-2009-10-2-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D. A., Chow B. Y., Gehrke A. R., Bulyk M. L., Kay S. A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21 126–133 10.1016/j.cub.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Mas P. (2013). Chromatin remodeling and alternative splicing: pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semi. Cell Dev. Biol. 24 399–406 10.1016/j.semcdb.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Herrero E., Davis S. J. (2012). Time for a nuclear meeting: protein trafficking and chromatin dynamics intersect in the plant circadian system. Mol. Plant 5 554–565 10.1093/mp/sss010 [DOI] [PubMed] [Google Scholar]
- Herrero E., Kolmos E., Bujdoso N., Yuan Y., Wang M., Berns M. C., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24 428–443 10.1105/tpc.111.093807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Song H. R., Lutz K., Kerstetter R. A., Michael T. P., McClung C. R. (2010). Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107 21211–21216 10.1073/pnas.1011987107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. (2009). The many pathways of RNA degradation. Cell 136 763–776 10.1016/j.cell.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Hsu P. Y., Harmer S. L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19 240–249 10.1016/j.tplants.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Perez-Garcia P., Pokhilko A., Millar A. J., Antoshechkin I., Riechmann J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336 75–79 10.1126/science.1219075 [DOI] [PubMed] [Google Scholar]
- Hurley J. M., Dasgupta A., Emerson J. M., Zhou X., Ringelberg C. S., Knabe N., et al. (2014). Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc. Natl. Acad. Sci. U.S.A. 111 16995–17002 10.1073/pnas.1418963111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H. J., Cho M.-H., Hahn B.-S., Lim H., Kwon Y.-K., Hahn T.-R., et al. (2011). Proteomic identification of rhythmic proteins in rice seedlings. Biochim. Biophys. Acta 1840 470–479 10.1016/j.bbapap.2011.01.010 [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L. E. (2008). The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9 699–712 10.1038/nrg2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. B., Syed N. H., Bordage S., Marshall J., Nimmo G. A., Jenkins G. I., et al. (2012). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24 961–981 10.1105/tpc.111.093948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. A., Williams B. A., Mcnicol J., Simpson C. G., Brown J. W. S., Harmer S. L. (2012). Mutation of Arabidopsis SPLICEOSOMAL TIMEKEEPER LOCUS1 causes circadian clock defects. Plant Cell 24 4907–4916 10.1105/tpc.112.104828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C., Cretenet G., Symul L., Martin E., Atger F., Naef F., et al. (2013). The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11:e1001455 10.1371/journal.pbio.1001455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D. (2007). Biological clocks and the coordination theory of RNA operons and regulons. Cold. Spring Harb. Symp. Quant. Biol. 72 157–165 10.1101/sqb.2007.72.013 [DOI] [PubMed] [Google Scholar]
- Kim B.-H., Cai X., Vaughn J., von Arnim A. (2007a). On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 8:R60 10.1186/gb-2007-8-4-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Park S. J., Kwak K. J., Kim Y. O., Kim J. Y., Song J., et al. (2007b). Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 35 506–516 10.1093/nar/gkl1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.-D., Sung S. (2012). Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 17 16–21 10.1016/j.tplants.2011.10.00 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Jung H. J., Lee H. J., Kim K. A., Goh C. H., Woo Y., et al. (2008). Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 55 455–466 10.1111/j.1365-313X.2008.03518.x [DOI] [PubMed] [Google Scholar]
- Kim J.-Y., Song H.-R., Taylor B. L., Carré I. A. (2003). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22 935–944 10.1093/emboj/cdg075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Lee K.-H., Kim D.-Y., Kwak E., Kim S., Kim K.-T. (2015). Rhythmic control of mRNA stability modulates circadian amplitude of mouse period3 mRNA. J. Neurochem. 132 642–656 10.1111/jnc.13027 [DOI] [PubMed] [Google Scholar]
- Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K., et al. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338 349–354 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Gatfield D., Esau C. C., Green C. B. (2010). MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE 5:e11264 10.1371/journal.pone.0011264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Green C. B. (2014). Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry 54 124–133 10.1021/bi500707c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Matsumoto K., Hirose M., Shimada M., Nagano M., Shigeyoshi Y., et al. (2007). LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. U.S.A. 104 1859–1864 10.1073/pnas.0607567104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Sher-Chen E. L., Green C. B. (2012). Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26 2724–2736 10.1101/gad.208306.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Shingle D. L., Green C. B. (2011). Post-transcriptional control of circadian rhythms. J. Cell Sci. 124 311–320 10.1242/jcs.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z. (2012). The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3:9 10.3389/fpls.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A. R., Schor I. E., Allo M., Dujardin G., Petrillo E., Munoz M. J. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14 153–165 10.1038/nrm3525 [DOI] [PubMed] [Google Scholar]
- Köster T., Haas M., Staiger D. (2014a). The RIPper Case: identification of RNA-binding protein targets by RNA immunoprecipitation. Methods Mol. Biol. 1158 107–121 10.1007/978-1-4939-0700-7_7 [DOI] [PubMed] [Google Scholar]
- Köster T., Meyer K., Weinholdt C., Smith L. M., Lummer M., Speth C., et al. (2014b). Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 42 9925–9936 10.1093/nar/gku716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C., Loros J. J., Dunlap J. C., Crosthwaite S. K. (2003). Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421 948–952 10.1038/nature01427 [DOI] [PubMed] [Google Scholar]
- Kwak E., Kim T. D., Kim K. T. (2006). Essential role of 3’-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 281 19100–19106 10.1074/jbc.M511927200 [DOI] [PubMed] [Google Scholar]
- Leder V., Lummer M., Tegeler K., Humpert F., Lewinski M., Schüttpelz M., et al. (2014). Mutational definition of binding requirements of an hnRNP-like protein in Arabidopsis using fluorescence correlation spectroscopy. Biochem. Biophys. Res. Commun. 453 69–74 10.1016/j.bbrc.2014.09.056 [DOI] [PubMed] [Google Scholar]
- Lee K.-H., Kim S.-H., Kim H.-J., Kim W., Lee H.-R., Jung Y., et al. (2014). AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 42 3590–3606 10.1093/nar/gkt1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Woo K. C., Kim D. Y., Kim T. D., Shin J., Park S. M., et al. (2012). Rhythmic interaction between Period1 mRNA and HnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol. 32 717–728 10.1128/MCB.06177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska D., Ten Have S., Hodge K., Tillemans V., Lamond A. I., Brown J. W. S. (2013). Plant SILAC: stable-isotope labelling with amino acids of Arabidopsis seedlings for quantitative proteomics. PLoS ONE 8:e72207 10.1371/journal.pone.0072207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu L., Zhuang X., Yu Y., Liu X., Cui X., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the Endoplasmic Reticulum in Arabidopsis. Cell 153 562–574 10.1016/j.cell.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidder P., Gutierrez R. A., Salome P. A., McClung C. R., Green P. J. (2005). Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 138 2374–2385 10.1104/pp.105.060368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Heintzen C., Loros J., Dunlap J. C. (1999). Regulation of clock genes. Cell Mol. Life Sci. 55 1195–1205 10.1007/s000180050366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhr B., Streitner C., Steffen A., Lange T., Staiger D. (2014). A glycine-rich RNA-binding protein affects gibberellin biosynthesis in Arabidopsis. Mol. Biol. Rep. 41 439–445 10.1007/s11033-013-2878-7 [DOI] [PubMed] [Google Scholar]
- Lück S., Thurley K., Thaben P. F., Westernmark P. O. (2014). Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep 9 741–751 10.1016/j.celrep.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Malapeira J., Khaitova L. C., Mas P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109 21540–21545 10.1073/pnas.1217022110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F. H. (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272 2118–2131 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W. S., Simpson C. G., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22 1184–1195 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P., Kim W. Y., Somers D. E., Kay S. A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570 10.1038/nature02163 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Okamoto K., Onai K., Niwa Y., Shimogawara K., Ishiura M. (2008). A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 22 918–930 10.1101/gad.1650408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A., Imamura A., Yamashino T., Mizuno T. (2002). Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol. 43 833–843 10.1093/pcp/pcf118 [DOI] [PubMed] [Google Scholar]
- McClung C. R. (2006). Plant circadian rhythms. Plant Cell 18 792–803 10.1105/tpc.106.040980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C. R. (2011). The genetics of plant clocks. Adv. Genet. 74 105–139 10.1016/B978-0-12-387690-4.00004-0 [DOI] [PubMed] [Google Scholar]
- McGlincy N. J., Smith C. W. (2008). Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 33 385–393 10.1016/j.tibs.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Mehra A., Baker C. L., Loros J. J., Dunlap J. C. (2009). Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 34 483–490 10.1016/j.tibs.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J. S., Rodriguez J., Abruzzi K. C., Rosbash M. (2012). Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife Sci. 1:e00011 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T. P., McClung C. R. (2002). Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 130 627–638 10.1104/pp.004929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T. P., McClung C. R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in arabidopsis. Plant Physiol. 132 629–639 10.1104/pp.021006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. J. (2004). Input signals to the plant circadian clock. J. Exp. Bot. 55 277–283 10.1093/jxb/erh034 [DOI] [PubMed] [Google Scholar]
- Millar A. J., Kay S. (1991). Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3 541–550 10.1105/tpc.3.5.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A., von Arnim A. (2014). “Analysis of mRNA translation states in Arabidopsis over the diurnal cycle by polysome microarray,” in Plant Circadian Networks, ed. Staiger D. (New York, NY: Springer; ), 157–174 10.1007/978-1-4939-0700-7_10 [DOI] [PubMed] [Google Scholar]
- Nagel D. H., Kay S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22 R648–R657 10.1016/j.cub.2012.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R., Clijsters L., Agami R. (2009). The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 276 5447–5455 10.1111/j.1742-4658.2009.07229.x [DOI] [PubMed] [Google Scholar]
- Newman T. C., Ohme-Takagi M., Taylor C. B., Green P. J. (1993). DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5 701–714 10.1105/tpc.5.6.701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V., Joe A., Jeong B., Korneli C., Boutrot F., Wested I., et al. (2013). Pseudomonas HopU1 affects interaction of plant immune receptor mRNAs to the RNA-binding protein GRP7. EMBO J. 32 701–712 10.1038/emboj.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P., Mühlemann O. (2010). Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 38 1615–1620 10.1042/BST0381615 [DOI] [PubMed] [Google Scholar]
- Nilsen T. W., Graveley B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463 457–463 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyiko T., Sonkoly B., Merai Z., Benkovics A. H., Silhavy D. (2009). Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 71 367–378 10.1007/s11103-009-9528-4 [DOI] [PubMed] [Google Scholar]
- Partch C. L., Green C. B., Takahashi J. S. (2013). Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24 90–99 10.1016/j.tcb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C., Hernandez-Verdeja T., Lopez-Cobollo R., Castellano M. D. M., Salinas J. (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24 4930–4947 10.1105/tpc.112.103697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Santángelo S., Mancini E., Francey L. J., Schlaen R. G., Chernomoretz A., Hogenesch J. B., et al. (2014). Role for LSM genes in the regulation of circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 111 15166–15171 10.1073/pnas.1409791111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Santángelo S., Schlaen R. G., Yanovsky M. J. (2013). Genomic analysis reveals novel connections between alternative splicing and circadian regulatory networks. Brief. Funct. Genomics 12 13–24 10.1093/bfgp/els052 [DOI] [PubMed] [Google Scholar]
- Pilgrim M. L., Caspar T., Quail P. H., McClung C. R. (1993). Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Mol. Biol. 23 349–364 10.1007/BF00029010 [DOI] [PubMed] [Google Scholar]
- Piques M., Schulze W. X., Hohne M., Usadel B., Gibon Y., Rohwer J., et al. (2009). Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5:314 10.1038/msb.2009.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preußner M., Wilhelmi I., Schultz A.-S., Finkernagel F., Michel M., Möröy T., et al. (2014). Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice. Mol. Cell 54 651–662 10.1016/j.molcel.2014.04.015 [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. (2011). Ending the message: poly(A) signals then and now. Genes Dev. 25 1770–1782 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O’neill J. S., et al. (2006). Circadian orchestration of the hepatic proteome. Curr. Biol. 16 1107–1115 10.1016/j.cub.2006.04.026 [DOI] [PubMed] [Google Scholar]
- Reddy A. S. (2004). Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 9 541–547 10.1016/j.tplants.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Reddy A. S. N., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25 3657–3683 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S. N., Rogers M. F., Richardson D. N., Hamilton M., Ben-Hur A. (2012). Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 3:18 10.3389/fpls.2012.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesselmann S., Piechulla B. (1992). Diurnal and circadian light-harvesting complex and quinone B-binding protein synthesis in leaves of tomato (Lycopersicon esculentum). Plant Physiol. 100 1840–1845 10.1104/pp.100.4.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. G., Frim D. M., Schwartz W. J., Majzoub J. A. (1988). Vasopressin mRNA in the suprachiasmatic nuclei: daily regulation of polyadenylate tail length. Science 241 342–344 10.1126/science.3388044 [DOI] [PubMed] [Google Scholar]
- Roden L. C., Carré I. A. (2001). The molecular genetics of circadian rhythms in Arabidopsis. Semin. Cell Dev. Biol. 12 305–315 10.1006/scdb.2001.0258 [DOI] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, Turnover, and mode of action of plant microRNAs. Plant Cell 25 2383–2399 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., Vaughn J. N., Kim B.-H., Zhou F., Gilchrist M. A., von Arnim A. G. (2010). The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA 16 748–761 10.1261/rna.2056010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., von Arnim A. G. (2013). Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 11:e0165 10.1199/tab.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf F., Wehrle F., Staiger D. (2004). Slave to the rhythm. Biochemistry 26 11–13. [Google Scholar]
- Sanchez S. E., Petrillo E., Beckwith E. J., Zhang X., Rugnone M. L., Hernando C. E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468 112–116 10.1038/nature09470 [DOI] [PubMed] [Google Scholar]
- Sanchez S. E., Petrillo E., Kornblihtt A. R., Yanovsky M. J. (2011). Alternative splicing at the right time. RNA Biol. 8 954–959 10.4161/rna.8.6.17336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauman I., Reppert S. M. (1996). Circadian clock neurons in the silkmoth Antheraea pernyi: novel mechanisms of period protein regulation. Neuron 17 889–900 10.1016/S0896-6273(00)80220-2 [DOI] [PubMed] [Google Scholar]
- Schmal C., Reimann P., Staiger D. (2013). A circadian clock-regulated toggle switch explains AtGRP7 and AtGRP8 oscillations in Arabidopsis thaliana. PLoS Comput. Biol. 9:e1002986 10.1371/journal.pcbi.1002986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Marnef A., Cheung M. K., Wilson I., Hancock J., Staiger D., et al. (2010). A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol. Biol. Rep. 37 839–845 10.1007/s11033-009-9636-x [DOI] [PubMed] [Google Scholar]
- Schöning J. C., Staiger D. (2005). At the pulse of time: protein interactions determine the pace of circadian clocks. FEBS Lett. 579 3246–3252 10.1016/j.febslet.2005.03.028 [DOI] [PubMed] [Google Scholar]
- Schöning J. C., Streitner C., Meyer I. M., Gao Y., Staiger D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36 6977–6987 10.1093/nar/gkn847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning J. C., Streitner C., Page D. R., Hennig S., Uchida K., Wolf E., et al. (2007). Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52 1119–1130 10.1111/j.1365-313X.2007.03302.x [DOI] [PubMed] [Google Scholar]
- Seo P. J., Mas P. (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26 79–87 10.1105/tpc.113.119842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Park M.-J., Lim M.-H., Kim S.-G., Lee M., Baldwin I. T., et al. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24 2427–2442 10.1105/tpc.112.098723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A., Duc C., Cole C., Zacharaki V., Hornyik C., Ozsolak F., et al. (2012). Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol. Biol. 19 845–852 10.1038/nsmb.2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H., Hanada K., Ushijima T., Nakashima M., Suzuki Y., Matsushita T. (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 18781–18786 10.1073/pnas.1407147112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire C., Moreno A. B., Garcia-Chapa M., Lopez-Moya J. J., San Segundo B. (2009). Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167 miR168 miR171 and miR398. FEBS Lett. 583 1039–1044 10.1016/j.febslet.2009.02.024 [DOI] [PubMed] [Google Scholar]
- So W. V., Rosbash M. (1997). Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16 7146–7155 10.1093/emboj/16.23.7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.-R., Noh Y.-S. (2012). Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells 34 279–287 10.1007/s10059-012-0103-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D. (2001). RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philo. Trans. R. Soc. Lond. B Biol. Sci. 356 1755–1759 10.1098/rstb.2001.0964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D. (2002). Circadian rhythms in Arabidopsis: time for nuclear proteins. Planta 214 334–344 10.1007/s004250100662 [DOI] [PubMed] [Google Scholar]
- Staiger D., Brown J. W. S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25 3640–3656 10.1105/tcp.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Green R. (2011). RNA-based regulation in the plant circadian clock. Trends Plant Sci. 16 517–523 10.1016/j.tplants.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Staiger D., Heintzen C. (1999). The circadian system of Arabidopsis thaliana: forward and reverse genetic approaches. Chronobiol. Int. 16 1–16 10.3109/07420529908998708 [DOI] [PubMed] [Google Scholar]
- Staiger D., Köster T. (2011). Spotlight on post-transcriptional control in the circadian system. Cell Mol. Life. Sci. 68 71–83 10.1007/s00018-010-0513-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Shin J., Johansson M., Davis S. J. (2013). The circadian clock goes genomic. Genome Biol. 14:208 10.1186/gb-2013-14-6-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C., Hennig L., Korneli C., Staiger D. (2010). Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 10:221 10.1186/1471-2229-10-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C., Köster T., Simpson C. G., Shaw P., Danisman S., Brown J. W. S., et al. (2012). An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with target transcripts in Arabidopsis thaliana. Nucleic Acids Res. 40 11240–11255 10.1093/nar/gks873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed N. H., Kalyna M., Marquez Y., Barta A., Brown J. W. S. (2012). Alternative splicing in plants - coming of age. Trends Plant Sci. 17 616–623 10.1016/j.tplants.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannukit S., Crabb T. L., Hertel K. J., Wen X., Jans D. A., Paine M. L. (2009). Identification of a novel nuclear localization signal and speckle-targeting sequence of tuftelin-interacting protein 11 a splicing factor involved in spliceosome disassembly. Biochem. Biophys. Res. Commun. 390 1044–1050 10.1016/j.bbrc.2009.10.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi L. C., Simpson G. G. (2009). Arabidopsis RNA immunoprecipitation. Plant J. 59 163–168 10.1111/j.1365-313X.2009.03859.x [DOI] [PubMed] [Google Scholar]
- Tharun S. (2009). Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell. Mol. Biol. 272 149–189 10.1016/S1937-6448(08)01604-3 [DOI] [PubMed] [Google Scholar]
- Wachter A., Rühl C., Stauffer E. (2012). The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 3:81 10.3389/fpls.2012.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltenberger H., Schneid C., Grosch J. O., Bareiss A., Mittag M. (2001). Identification of target mRNAs for the clock-controlled RNA-binding protein chlamy 1 from Chlamydomonas reinhardtii. Mol. Genet. Genomics 265 180–188 10.1007/s004380000406 [DOI] [PubMed] [Google Scholar]
- Wang D., Liang X., Chen X., Guo J. (2013). Ribonucleoprotein complexes that control circadian clocks. Int. J. Mol. Sci. 14 9018–9036 10.3390/ijms14059018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fujiwara S., Somers D. E. (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29 1903–1915 10.1038/emboj.2010.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wu F., Xie Q., Wang H., Wang Y., Yue Y., et al. (2012). SKIP Is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24 3278–3295 10.1105/tpc.112.100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Osterbur D. L., Megaw P. L., Tosini G., Fukuhara C., Green C. B., et al. (2001). Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev. Biol. 1:9 10.1186/1471-213X-1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C., Ha D. C., Lee K. H., Kim D. Y., Kim T. D., Kim K. T. (2010). Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol. Cell. Biol. 30 197–205 10.1128/MCB.01154-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C., Kim T. D., Lee K. H., Kim D. Y., Kim W., Lee K. Y., et al. (2009). Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 37 26–37 10.1093/nar/gkn893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Falvey E., Lavery D., Talbot D., Schmidt E., Ossipow V., et al. (1992). The role of the transcriptional activator protein DBP in circadian liver gene expression. J. Cell Sci. 16 123–127 10.1242/jcs.1992.Supplement_16.15 [DOI] [PubMed] [Google Scholar]
- Xing D., Li Q. Q. (2011). Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip. Rev. RNA 2 445–458 10.1002/wrna.59 [DOI] [PubMed] [Google Scholar]
- Xue Z., Ye Q., Anson S. R., Yang J., Xiao G., Kowbel D., et al. (2014). Transcriptional interference by antisense RNA is required for circadian clock function. Nature 514 650–653 10.1038/nature13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Hassidim M., Green R. M. (2007). CCA1 Transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol. 145 925–932 10.1104/pp.107.103812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M. J., Kay S. A. (2001). Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 4 429–435 10.1016/S1369-5266(00)00196-5 [DOI] [PubMed] [Google Scholar]
- Yerushalmi S., Yakir E., Green R. M. (2011). Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 20 1155–1165 10.1111/j.1365-294X.2010.04962.x [DOI] [PubMed] [Google Scholar]
- Young M. W., Kay S. A. (2001). Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2 702–715 10.1038/35088576 [DOI] [PubMed] [Google Scholar]
- Zhang L., Weng W., Guo J. (2011). Posttranscriptional mechanisms in controlling eukaryotic circadian rhythms. FEBS Lett. 585 1400–1405 10.1016/j.febslet.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Zhang N., Kallis R. P., Ewy R. G., Portis A. R. (2002). Light modulation of Rubisco in Arabidopsis requires a capacity for redox regulation of the larger Rubisco activase isoform. Proc. Natl. Acad. Sci. U.S.A. 99 3330–3334 10.1073/pnas.042529999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Schneid C., Iliev D., Schmidt E. M., Wagner V., Wollnik F., et al. (2004). The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot. Cell 3 815–825 10.1128/EC.3.3.815-825.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H. H., Resnick A. S., Straume M., McClung C. R. (1997). Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell 9 947–955 10.1105/tpc.9.6.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E., Kunova A., Meyer P. (2011). Sense and antisense transcripts of convergent gene pairs in Arabidopsis thaliana can share a common polyadenylation region. PLoS ONE 6:e16769 10.1371/journal.pone.0016769 [DOI] [PMC free article] [PubMed] [Google Scholar]